Significance
Despite the importance of liverworts as the earliest diverging land plant lineage to support fungal symbiosis, it is unknown whether filamentous pathogens can establish intracellular interactions within living cells of these nonvascular plants. Here, we demonstrate that an oomycete pathogen invades Marchantia polymorpha and related liverworts to form intracellular infection structures inside cells of the photosynthetic layer. Plants lacking this tissue layer display enhanced resistance to infection, revealing an architectural susceptibility factor in complex thalloid liverworts. Moreover, we show that dedicated host cellular trafficking proteins are recruited to pathogen interfaces within liverwort cells, supporting the idea that intracellular responses to microbial invasion originated in nonvascular plants.
Keywords: bryophyte, liverworts, oomycetes, Phytophthora, haustoria
Abstract
The expansion of plants onto land was a formative event that brought forth profound changes to the earth’s geochemistry and biota. Filamentous eukaryotic microbes developed the ability to colonize plant tissues early during the evolution of land plants, as demonstrated by intimate, symbiosis-like associations in >400 million-year-old fossils. However, the degree to which filamentous microbes establish pathogenic interactions with early divergent land plants is unclear. Here, we demonstrate that the broad host-range oomycete pathogen Phytophthora palmivora colonizes liverworts, the earliest divergent land plant lineage. We show that P. palmivora establishes a complex tissue-specific interaction with Marchantia polymorpha, where it completes a full infection cycle within air chambers of the dorsal photosynthetic layer. Remarkably, P. palmivora invaginates M. polymorpha cells with haustoria-like structures that accumulate host cellular trafficking machinery and the membrane syntaxin MpSYP13B, but not the related MpSYP13A. Our results indicate that the intracellular accommodation of filamentous microbes is an ancient plant trait that is successfully exploited by pathogens like P. palmivora.
Plant–microbe associations are ubiquitous throughout the plant kingdom, which suggests that the ability to support microbial colonization occurred early during the evolution of land plants. Extensive evidence has revealed that ancient land plants were colonized by filamentous eukaryotic microbes, as several fossils from the Rhynie chert (Early Devonian, 400 to 480 Mya) display symbiosis-like microbial structures within plant cells (1–4). Indeed, symbiotic interactions with arbuscular mycorrhizal fungi are widespread among extant early divergent land plant lineages (5–8). Moreover, recent studies suggest that early divergent land plants and their algal predecessors were preadapted for symbiosis, as these organisms encode functionally equivalent homologs of core symbiosis signaling components (9, 10). In comparison, our understanding of how early divergent land plants interact with pathogenic microbes remains extremely limited.
Interactions between plants and filamentous eukaryotic microbes are often associated with the development of specialized microbial structures that protrude into plant cells. Such structures include the finely branched arbuscules of symbiotic arbuscular mycorrhizal fungi and the digit-, knob-, or peg-like haustoria of oomycete and fungal pathogens (11, 12). Arbuscules and haustoria are both involved in the manipulation of host cell function to improve microbial colonization; however, symbiotic structures participate in the mutually beneficial exchange of resources, while pathogenic structures do not (13, 14). Numerous host tissues and cells are capable of supporting filamentous microbes (13, 15, 16), which suggests that partially overlapping mechanisms are employed to accommodate symbiotic and pathogenic microbes. This has led to the idea that pathogens establish intracellular interfaces by exploiting host machinery designed to accommodate endosymbiotic structures. Whether this dynamic was established early during the coevolution of plants and microbes is unknown. Extensive evidence has demonstrated that early divergent land plants can accommodate arbuscules within their cells (7, 17), whereas specialized pathogenic structures like haustoria have not been observed in these plants.
Bryophytes (liverworts, hornworts, and mosses) are a paraphyletic group of nonvascular, gametophyte (haploid)-dominant plants that quickly diverged from other embryophyte plant lineages when ancient plants colonized land. Phylogenetic analyses often place liverworts basal to mosses and hornworts (18, 19). This suggests that liverworts represent the earliest divergent land plant lineage, although this has yet to yield a consensus among the field (20, 21). Many bryophytes are colonized by symbiotic microbes; however, intracellular endosymbiotic structures (arbuscules) have been observed only in liverworts and hornworts (7, 17, 19). Unfortunately, our understanding of plant–pathogen interactions in these plants is extremely limited. In comparison, pathogenic interactions in the model moss Physcomitrella patens have been described by several groups (22–24). The colonization of moss tissues is associated with intracellular hyphal growth; however, specialized infection structures similar to haustoria are not observed within moss cells (23). Therefore, an alternative bryophyte model system is required to study intracellular interactions with pathogenic microbes. Liverworts are the most suitable candidate to accomplish this, given their ability to accommodate endosymbiotic structures and the recent establishment of molecular genetic tools in these plants (25).
In this study, we investigated plant–pathogen interactions in the model liverwort Marchantia polymorpha, which propagates clonally via small propagules (gemmae) or sexually when the sperm of male plants fertilizes eggs housed in the archegonia of female plants. Both male and female plants have a thalloid body plan comprising a ventrally located epidermis with rhizoids and scales, a central nonphotosynthetic storage region, and a dorsal photosynthetic layer. The photosynthetic layer of complex liverworts like M. polymorpha is made up of air chambers, which are walled enclosures that contain plastid-rich photosynthetic filaments and have open pores to facilitate gas exchange (26).
To determine whether liverworts support intracellular colonization by a filamentous eukaryotic pathogen, we challenged M. polymorpha with Phytophthora palmivora, a broad host-range oomycete pathogen with demonstrated virulence in root and leaf tissues of monocot and dicot (angiosperm) hosts (27–29). P. palmivora is a hemibiotroph—that is, initially it is parasitic in living tissues and then continues to grow and sporulate in dead tissue. Infection begins when motile zoospores contact plant surfaces, causing the spores to encyst, germinate, and form appressoria to penetrate the surface (30). Upon accessing plant tissues, the pathogen develops digit-like haustoria that protrude into living plant cells for the release of virulence effector proteins that manipulate the host (11). Precise mechanisms for oomycete effector delivery/uptake remain to be clarified; however, secreted effector proteins containing the RXLR motif translocate into host cells and interfere with immunity (31–33). The pathogen eventually transitions to a necrotrophic phase, where it actively destroys plant tissues and completes its asexual lifestyle by releasing motile zoospores contained within sporangia (34).
Here, we demonstrate that P. palmivora preferentially colonizes the photosynthetic layer of liverworts and causes extensive disease. Molecular and microscopic analyses revealed that the phase when P. palmivora colonizes living tissues is associated with the up-regulation of virulence effector molecules and the deployment of haustoria-like intracellular infection structures. Several endogenous host proteins accumulated at intracellular infection structures, including Rab GTPases and the homolog of a membrane-localized syntaxin associated with mutually beneficial symbioses in angiosperms. Together, these markers clearly defined atypically branched infection structures and intracellular hyphae inside living Marchantia cells, revealing a prevalent intracellular phase of P. palmivora pathogenesis that is not commonly observed during interactions with vascular plants. Furthermore, we demonstrate that P. palmivora requires liverwort air chambers to fully exploit M. polymorpha, as pathogen fitness is greatly reduced in air-chamberless nop1 mutants.
Results
P. palmivora Colonizes the Photosynthetic Layer of M. polymorpha.
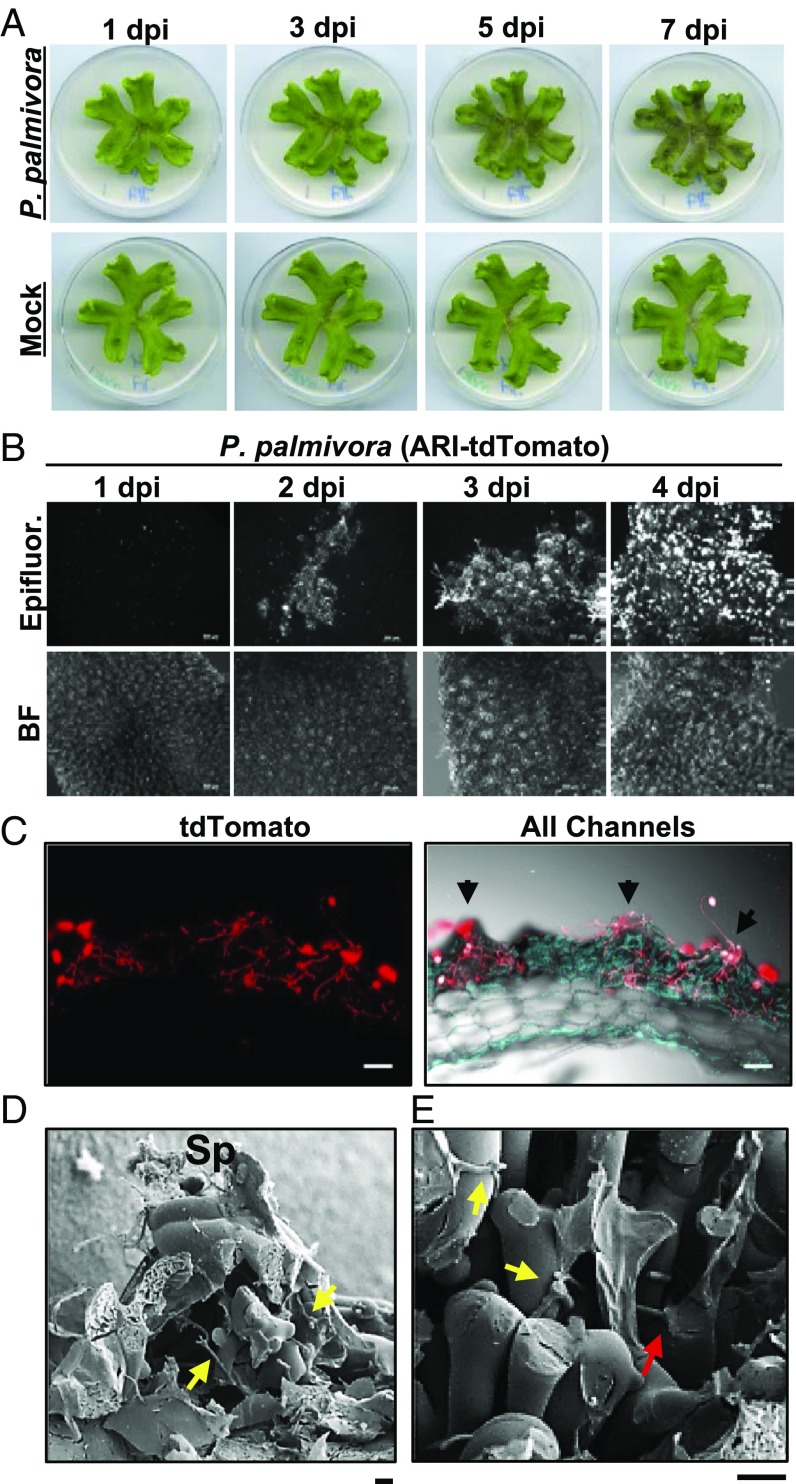
To determine whether liverworts support colonization by a broad host-range filamentous pathogen, we challenged 3-wk-old M. polymorpha TAK1 (male) plants with zoospores of a tdTomato-expressing isolate of P. palmivora [accession no. P3914, derived from Arizona (ARI-td)] and tracked pathogen growth and disease progression. Since P. palmivora colonizes multiple tissue types in angiosperm model plants (28, 29, 35), initial experiments were performed to determine whether colonization occurs in the rhizoids or thalli of Marchantia. Rhizoid inoculations led to variable disease phenotypes, such that plants occasionally exhibited disease symptoms by 7 d postinoculation (dpi) (SI Appendix, Fig. S1A). Confocal fluorescence microscopy indicated that rhizoids were mostly devoid of intracellular P. palmivora (ARI-td) growth, with rare instances of intracellular hyphae in damaged rhizoids (SI Appendix, Fig. S1B). In contrast, Marchantia thalli inoculated with ARI-td zoospores exhibited disease symptoms that increased in severity from 3 to 7 dpi (Fig. 1A). Epifluorescence and confocal microscopy supported these observations, demonstrating the gradual spread of ARI-td hyphae across TAK1 thalli from 1 to 4 dpi (Fig. 1B and SI Appendix, Fig. S2). Plants were completely colonized by 7 dpi, demonstrating that M. polymorpha thalli are highly susceptible to P. palmivora.
Fig. 1.
P. palmivora colonizes the photosynthetic layer of M. polymorpha. (A) Disease symptoms of 3-wk-old M. polymorpha TAK1 (male) thalli inoculated with P. palmivora ARI-td zoospores or water (Mock) over a 7-d time course. (B) Epifluorescence microscopy demonstrating the spread of P. palmivora growth across TAK1 thalli from 1 to 4 dpi. Epifluorescence (Epifluor.) from the pathogen is displayed alongside bright-field (BF) images. (Scale bars, 500 μm.) (C) Confocal fluorescence microscopy of sectioned TAK1 thalli infected with P. palmivora at 7 dpi. Z-stack projections of red fluorescence from the pathogen are displayed alone (Left, tdTomato) or merged with all channels (Right, bright-field and plastid autofluorescence in turquoise). Arrowheads indicate air pores. (Scale bars, 100 μm.) (D) Cryo-SEM image of TAK1 thalli colonized by P. palmivora at 7 dpi. Mechanically fractured air chamber demonstrating hyphal growth within the chamber (yellow arrows) and sporangia (Sp) at the air pore. (E) Cryo-SEM image showing intercellular (yellow arrows) and intracellular (red arrow) associations between P. palmivora hyphae and photosynthetic filaments within M. polymorpha air chambers at 7 dpi. (Scale bars, 20 μm.) All experiments were performed at least three times, with similar results.
Confocal fluorescence microscopy demonstrated that P. palmivora colonized the surface of M. polymorpha thalli in a discrete manner that overlapped with air chamber morphology (SI Appendix, Fig. S2). We therefore analyzed cross-sections of infected M. polymorpha thalli, which revealed high levels of colonization within air chambers at 7 dpi (Fig. 1C). Pathogen growth was largely limited to the photosynthetic layer, although intercellular hyphae were observed in the nonphotosynthetic storage tissue of plants with ongoing necrosis (SI Appendix, Fig. S3). In support of these findings, experiments performed using additional liverwort species (Lunularia cruciata and Marchantia paleacea) similarly exhibited colonization within air chambers (SI Appendix, Fig. S4A). Cryogenic scanning electron microscopy (cryo-SEM) of colonized Marchantia thalli demonstrated P. palmivora sporangia and hyphae traversing through the central pores of air chambers (Fig. 1D), and hyphal growth was observed within air chambers (Fig. 1 D and E). P. palmivora hyphae often associated with photosynthetic filament cells and were sometimes observed to penetrate them (Fig. 1E). Together, our results indicate that P. palmivora preferentially colonizes the air chambers of M. polymorpha.
Natural diversity of pathogen isolates has classically been used to probe host–microbe interactions to identify loci or mechanisms important for plant resistance to disease. To explore this paradigm in Marchantia, we assessed the ability of several P. palmivora isolates to colonize TAK1 thalli (SI Appendix, Fig. S5). Disease progression was similar to ARI-td for the majority of strains tested, with the exception of the MAZI (P6375) and TAZI (P6802) isolates that displayed an accelerated disease progression (SI Appendix, Fig. S5). We also tested compatibility between Marchantia and Phytophthora infestans, the causal agent of potato and tomato late blight. TAK1 thalli infected with P. infestans (Pi-88069-td) were asymptomatic over a 7-d infection time course, similar to mock-treated plants (SI Appendix, Fig. S6). In contrast, TAK1 thalli inoculated with P. palmivora (ARI-td) zoospores were highly susceptible to pathogen ingress. These results demonstrate that P. infestans is unable to overcome preexisting or induced barriers to colonization in Marchantia, unlike several P. palmivora isolates that cause extensive disease.
M. polymorpha Responds to P. palmivora Colonization.
Studies in angiosperms have revealed a common set of host responses to microbial invasion that includes the accumulation of reactive oxygen species (ROS) and the induction of defense-associated gene expression (36). These responses are also observed during successful plant–pathogen interactions in the moss P. patens (24), which suggests that they arose early during the evolution of land plants. To further support this idea, we characterized host responses activated during the colonization of Marchantia thalli with P. palmivora ARI-td. We first performed DAB (diaminobenzidine) staining to assess hydrogen peroxide (ROS) accumulation in mock-treated and P. palmivora-colonized TAK1 thalli. Brown precipitates indicative of hydrogen peroxide accumulation were observed in colonized thalli at 3 and 5 dpi, while mock-treated thalli displayed background levels of staining (SI Appendix, Fig. S7A). Next, we identified candidate plant response genes based on studies demonstrating the up-regulation of PRX (peroxidase) and DIR (dirigent-like) transcripts in pathogen-treated moss (37, 38), and then quantified their expression levels in colonized and mock-treated TAK1 plants using qRT-PCR. The expression levels of MpPRX (Mapoly0106s0049) and MpDIR (Mapoly0117s0014) were significantly higher in P. palmivora ARI-td-colonized TAK1 plants relative to mock-treated controls by 3 dpi (SI Appendix, Fig. S7B). In addition, MpPRX:GUS reporter lines showed a strong induction of GUS activity in the photosynthetic layer of ARI-td–infected plants at 4 dpi compared with mock-treated controls and wild-type plants (SI Appendix, Fig. S7C). Collectively, our results demonstrate that the colonization of M. polymorpha thalli activates a response to microbial invasion that is insufficient to block P. palmivora growth.
P. palmivora Forms Intracellular Hyphae in Living Host Cells.
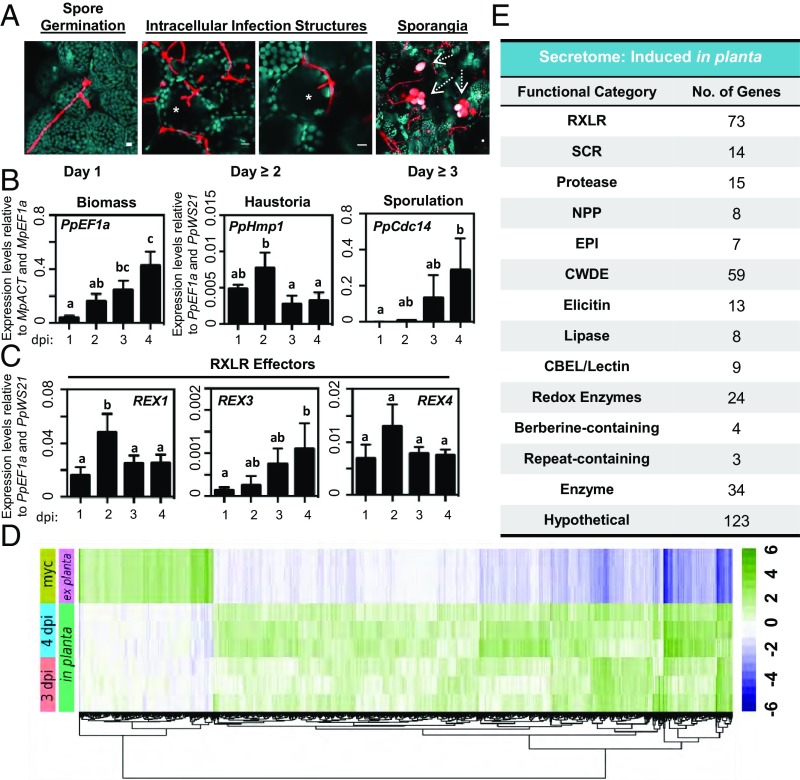
The colonization of angiosperm tissues by P. palmivora is associated with distinct transitions in pathogen lifestyle. These transitions are typically characterized by the presence of specialized penetration/infection structures and clear shifts in the P. palmivora transcriptome (29). We first performed confocal fluorescence microscopy to document the pathogen lifestyle transitions that occur during the colonization of TAK1 thalli (Fig. 2A). At 1 dpi, hyphae of germinated zoospores appeared to wander along the surface of the thallus without developing appressorial penetration structures (Figs. 1B and 2A and SI Appendix, Fig. S2). The colonization of plant tissues occurred by 2 dpi and was associated with the development of intracellular infection structures. We observed typical digit-like haustoria; however, branched intracellular structures and intracellular hyphae were observed in greater abundance (Fig. 2A). For every digit-like haustorium, there were approximately four times as many branched intracellular structures and twice as many instances of intracellular hyphal growth. Intracellular structures were also observed during interactions with L. cruciata and M. paleacea; however, digit- or knob-like haustoria appeared to be more prevalent than branched infection structures (SI Appendix, Fig. S4B). Sporangia were observed by 3 to 4 dpi (Fig. 2A), indicating that P. palmivora completes a full infection cycle in Marchantia.
Fig. 2.
P. palmivora completes a full infection cycle that includes the intracellular colonization of living Marchantia cells. (A) Confocal fluorescence microscopy demonstrating key morphological transitions in P. palmivora lifestyle during the colonization of TAK1 plants from 1 to 3 dpi. Z-stack projections of pathogen fluorescence merged with plastid autofluorescence (turquoise). Intracellular infection structures are denoted by an asterisk. Sporangia are indicated by dashed arrows. (Scale bars, 10 μm.) (B) Quantification of P. palmivora lifestyle marker genes during the colonization of TAK1 thalli from 1 to 4 dpi via qRT-PCR analysis. Pathogen biomass (PpEF1a) was quantified relative to M. polymorpha biomass markers (MpACT and MpEF1a). Haustoria (PpHmp1) and sporulation (PpCdc14) marker genes were quantified relative to pathogen biomass controls (PpEF1a and PpWS21). Different letters signify statistically significant differences in transcript abundance [ANOVA, Tukey’s honest significant difference (HSD), P < 0.05]. (C) Quantification of P. palmivora RXLR effector gene transcripts during the colonization of TAK1 thalli from 1 to 4 dpi via qRT-PCR analysis. RXLR effector (REX1, REX3, and REX4) gene expression was quantified relative to P. palmivora biomass (PpEF1a and PpWS21). Different letters signify statistically significant differences in transcript abundance (ANOVA, Tukey’s HSD, P < 0.05). (D) P. palmivora transcriptome. Hierarchical clustering of differentially expressed genes between in planta (3 and 4 dpi) and axenically grown mycelium transcriptomes (LFC of ≥2, P value of ≤10−3). rLog-transformed counts, median-centered by gene, are shown. (E) P. palmivora secretome. Summary of functional categories of 394 genes encoding putative secreted proteins up-regulated during P. palmivora infection of M. polymorpha. Experiments in A–C were performed at least three times, with similar results.
To further clarify the timing of pathogen lifestyle transitions during the Marchantia–Phytophthora interaction, we monitored the expression of lifestyle-associated P. palmivora marker genes over a 4-d infection time course by qRT-PCR analysis (Fig. 2B). Levels of the P. palmivora biomass marker PpEF1a significantly increased over time, which is consistent with our microscopic analyses (Fig. 1B and SI Appendix, Fig. S2). The haustoria-associated marker gene PpHmp1 [haustoria membrane protein 1 (39)] peaked in expression at 2 dpi, while the sporulation-specific cell-cycle marker gene PpCdc14 (40) was induced by 3 to 4 dpi. These results are consistent with our microscopy data that demonstrate intracellular infection structures at 2 dpi and sporangia at 3 to 4 dpi (Fig. 2A). Collectively, our results indicate that the P. palmivora infection cycle in Marchantia begins with spore germination and hyphal growth at 1 dpi; is followed by the colonization of living tissues and cells (including intracellular infection structures) at 2 to 3 dpi; and ends with the completion of the pathogen’s asexual life cycle after 3 dpi.
The transcriptional induction of pathogen genes encoding secreted effector proteins is a hallmark of Phytophthora–angiosperm interactions (29). Among these secreted proteins, those of the RXLR class of effectors are believed to act within host cells to suppress immunity and enhance pathogen growth (31–33). To determine if P. palmivora up-regulates RXLR effectors during the colonization of Marchantia, we analyzed the expression profiles of the P. palmivora RXLR effectors REX1, REX3, and REX4 (29). Significant up-regulation of REX1 transcripts occurred at 2 dpi (during the stage when P. palmivora forms intracellular structures within living cells), and REX3 levels increased throughout infection to a maximum observed at 4 dpi (Fig. 2C). REX4 expression peaked at 2 dpi, although this was not statistically significant in all experiments. REX expression profiles were similar to those observed in colonized Nicotiana benthamiana roots (29), demonstrating that P. palmivora up-regulates RXLR effector transcripts during the colonization of Marchantia thalli.
To assess broad transcriptional changes in P. palmivora during liverwort colonization, we performed RNA sequencing (RNA-seq) of infected thalli (at 3 and 4 dpi) and of axenically grown P. palmivora mycelia, which served as an ex planta control. Differential expression analysis revealed 3601 up-regulated and 932 down-regulated P. palmivora genes expressed in planta, relative to the ex planta control [absolute log fold change (LFC) of ≥2, adjusted P value of <10−3] (Fig. 2D and Dataset S1). qRT-PCR analysis validated a subset of colonization-induced P. palmivora genes, which were either up-regulated throughout colonization or were induced late during infection (SI Appendix, Fig. S8). Among all up-regulated genes, 394 (11%) were predicted to encode putative secreted proteins, with RXLR effectors and cell wall-degrading enzymes (CWDE) being the most abundant functional categories (Fig. 2E). Together, these results suggest that P. palmivora infection of M. polymorpha involves large-scale transcriptional induction of genes typical of successful Phytophthora–angiosperm interactions (29, 41–43), including effectors and hydrolytic enzymes.
To further support these data, we analyzed the expression of Phytophthora lifestyle marker genes during an additional compatible interaction with the P. palmivora MAZI isolate in thalli, and we contrasted this with incompatible interactions between P. infestans and thalli or between P. palmivora ARI-td and rhizoids. The hypervirulent MAZI isolate displayed increased levels of transcripts associated with pathogen biomass (PpEF1a and PpWS21) and sporulation (PpCdc14) during the colonization of Marchantia thalli (1 to 4 dpi) compared with ARI-td (SI Appendix, Fig. S9A). Moreover, haustoria-associated PpHmp1, previously characterized RXLR effectors [REX1, REX3, and REX4 (29)], and two additional RXLR effectors identified in our RNA-seq analysis (PpRXLR-06960 and PpRXLR-06188) were all significantly up-regulated in MAZI during thalli infections (SI Appendix, Fig. S9B). In contrast, the attempted colonization of Marchantia thalli by P. infestans showed no significant differences in oomycete biomass (PiWS21), sporulation (PiCdc14), haustoria-associated PiHmp1, or the RXLR effectors PiAVR3a and PiAVRblb2 (SI Appendix, Fig. S9C). Importantly, we also demonstrate that P. palmivora ARI-td fails to up-regulate haustoria-associated transcripts (PpHmp1), RXLR effectors (PpREX1 and PpREX4), or sporulation-specific PpCdc14 during unsuccessful interactions with Marchantia rhizoids (SI Appendix, Fig. S10). Collectively, our expression data further support the idea that RXLR effectors are expressed concomitantly with the development of intracellular infection structures in living plant cells during the biotrophic phase of plant colonization. This validates the use of commonly used Phytophthora lifestyle marker genes during pathogenic interactions with early divergent land plants and demonstrates the intricacies associated with the successful and unsuccessful colonization of different liverwort tissues by filamentous pathogens.
Host Cellular Responses to Intracellular Infection Structures.
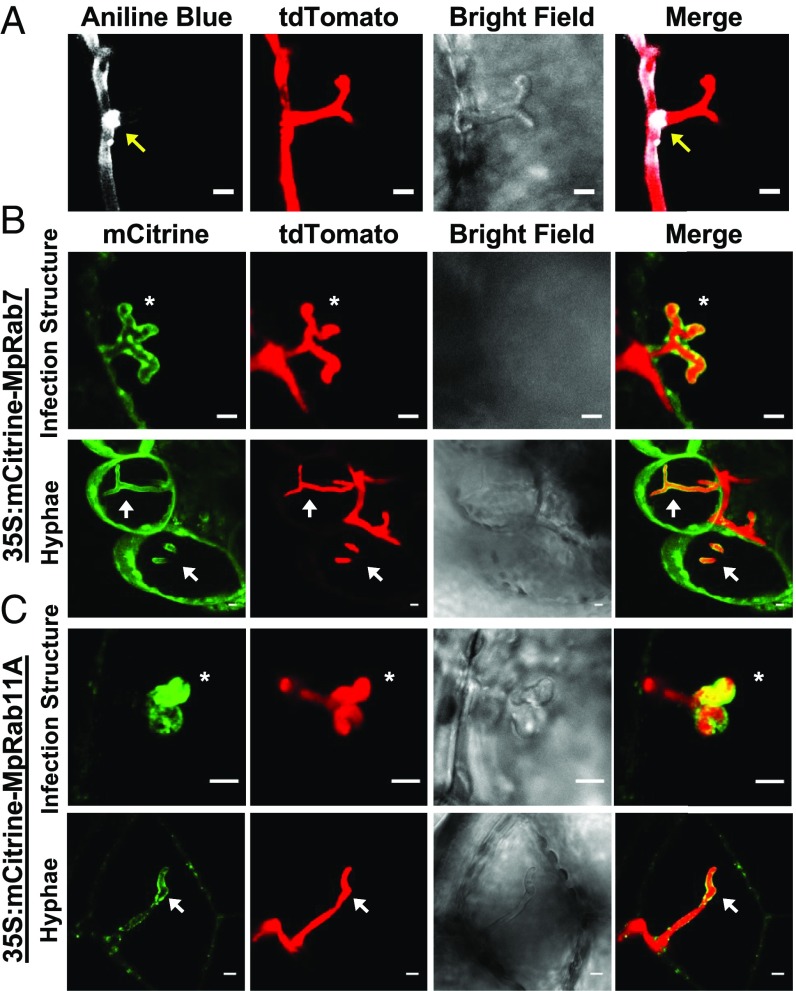
The accommodation of intracellular hyphal structures requires reorganization of the host cell and the biogenesis of novel membranes to separate the host cell from the pathogen. Moreover, plants responding to invading microbes often accumulate callose, a β-1,3-glucan associated with cell wall strengthening. We therefore applied callose staining and live-cell imaging of fluorescently tagged proteins labeling membrane compartments to investigate the subcellular changes associated with accommodating intracellular P. palmivora structures in living Marchantia cells. An extended callosic envelope is characteristic of dysfunctional intracellular structures, while functional interfaces display callosic collars that are limited to the neck region or no callosic depositions at all (44, 45). Aniline blue staining of ARI-td–infected TAK1 thalli revealed that callose deposition was limited to the peripherally located neck region of intracellular infection structures (Fig. 3A and SI Appendix, Fig. S11), suggestive of functional P. palmivora–Marchantia interfaces. Next, we localized homologs of host Rab GTPases that label oomycete haustoria in angiosperms and are believed to direct vesicle delivery to host–pathogen interfaces (46). We generated transgenic plants that constitutively overexpress mCitrine-MpRab7 or mCitrine-MpRab11A, since homologs of these proteins strongly localize to P. infestans haustoria in N. benthamiana leaves (i.e., NbRabG3c or NbRabA1e, respectively) (46, 47). In the absence of P. palmivora colonization, mCitrine-MpRab7 and mCitrine-MpRab11A localized to the tonoplast and to endosomes, respectively (SI Appendix, Fig. S12), which is consistent with RabG3c/Rab7 and RabA1e/Rab11 localization in vascular plants (47, 48). During infection, we observed strong labeling of intracellular infection structures and intracellular hyphae by mCitrine-MpRab7 (Fig. 3B) and mCitrine-MpRab11A (Fig. 3C) within living Marchantia cells at 3 dpi (SI Appendix, Fig. S13). Phylogenetic analyses of both Rab7- and Rab11-related proteins in selected algae, bryophytes, lycophytes, and flowering plants demonstrate that MpRab7 (SI Appendix, Fig. S14) and MpRab11A (49) were present in algal predecessors and subsequently expanded in land plants. Together, our results reveal a conserved host cellular response to invasive haustoria-like intracellular infection structures and hyphae during the colonization of Marchantia cells by P. palmivora.
Fig. 3.
Host cellular responses to invading oomycete structures. (A) Detection of callose deposition at P. palmivora ARI-td intracellular infection structures at 3 dpi in M. polymorpha TAK1. Arrows indicate callose deposition at the peripheral neck region of the invading intracellular infection structure. (Scale bars, 5 μm.) (B) MpRab7 colocalization with invading P. palmivora ARI-td structures in 35S:mCitrine-MpRab7 plants at 3 dpi and (C) MpRab11A colocalization with invading P. palmivora ARI-td structures in 35S:mCitrine-MpRab11A plants at 3 dpi. Infection structures are indicated with asterisks, while intracellular hyphae are denoted by arrows. Z-stack projections are displayed. (Scale bars, 5 μm.) Experiments were performed three times, with similar results.
A Colonization-Induced Host Syntaxin Is Targeted to Intracellular Infection Structures.
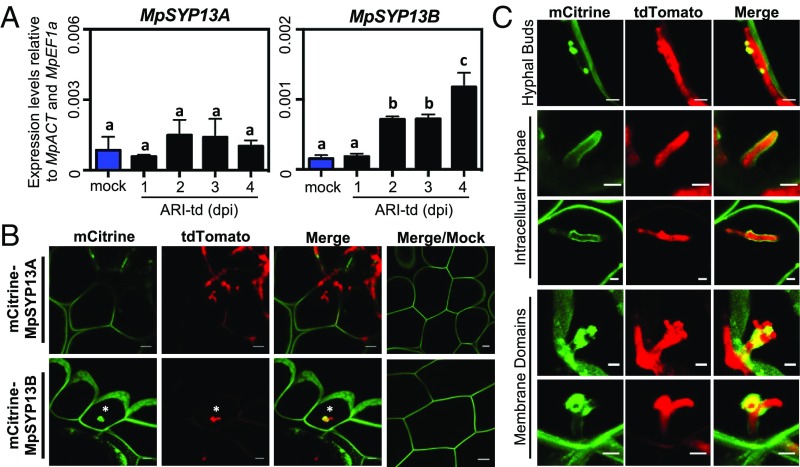
Plasma membrane-resident syntaxins mediate exocytosis and have been associated with penetration structures of symbiotic and pathogenic microbes in several angiosperm systems (50, 51). To determine if symbiosis-associated membrane syntaxins function at intracellular infection structures, we monitored the expression levels of Marchantia SYP13 family homologs during infection and assessed MpSYP13 localization at pathogen interfaces using mCitrine-tagged reporter lines (52). Expression levels of MpSYP13A were not affected during colonization with P. palmivora, whereas MpSYP13B displayed significant up-regulation from 2 to 4 dpi (Fig. 4A). In addition, mCitrine-MpSYP13B strongly labeled intracellular infection structures while maintaining localization at the cell periphery. Conversely, mCitrine-MpSYP13A localization was unaffected by the presence of intracellular infection structures (Fig. 4B). We observed a number of distinct MpSYP13B localization patterns, including focal accumulation proximal to hyphal buds of early developing intracellular infection structures, complete labeling around intracellular hyphae, and localization to membrane domains of branched intracellular infection structures (Fig. 4C). These results demonstrate that the colonization of Marchantia by P. palmivora includes a complex intracellular phase that recruits the MpSYP13B syntaxin to specific extrainvasive hyphal domains.
Fig. 4.
A colonization-induced host syntaxin accumulates at intracellular infection structures. (A) qRT-PCR analysis of MpSYP13A and MpSYP13B transcripts in mock-treated or P. palmivora-colonized (ARI-td) TAK1 plants from 1 to 4 dpi. Expression values are shown relative to internal MpACT and MpEF1a controls. Different letters signify statistically significant differences in transcript abundance (ANOVA, Tukey’s HSD, P < 0.05). (B) Confocal fluorescence microscopy demonstrating mCitrine-MpSYP13A/B localization in cells containing P. palmivora (ARI-td) intracellular infection structures at 3 dpi. Asterisks denote intracellular infection structures. (Scale bars, 10 μm.) (C) Patterns of mCitrine-MpSYP13B localization in P. palmivora-colonized (ARI-td) plants, including close-up images of the structure displayed in B. (Scale bars, 5 μm.) Experiments were performed three times, with similar results.
Co-Option of Marchantia Air Chambers for Pathogen Colonization.
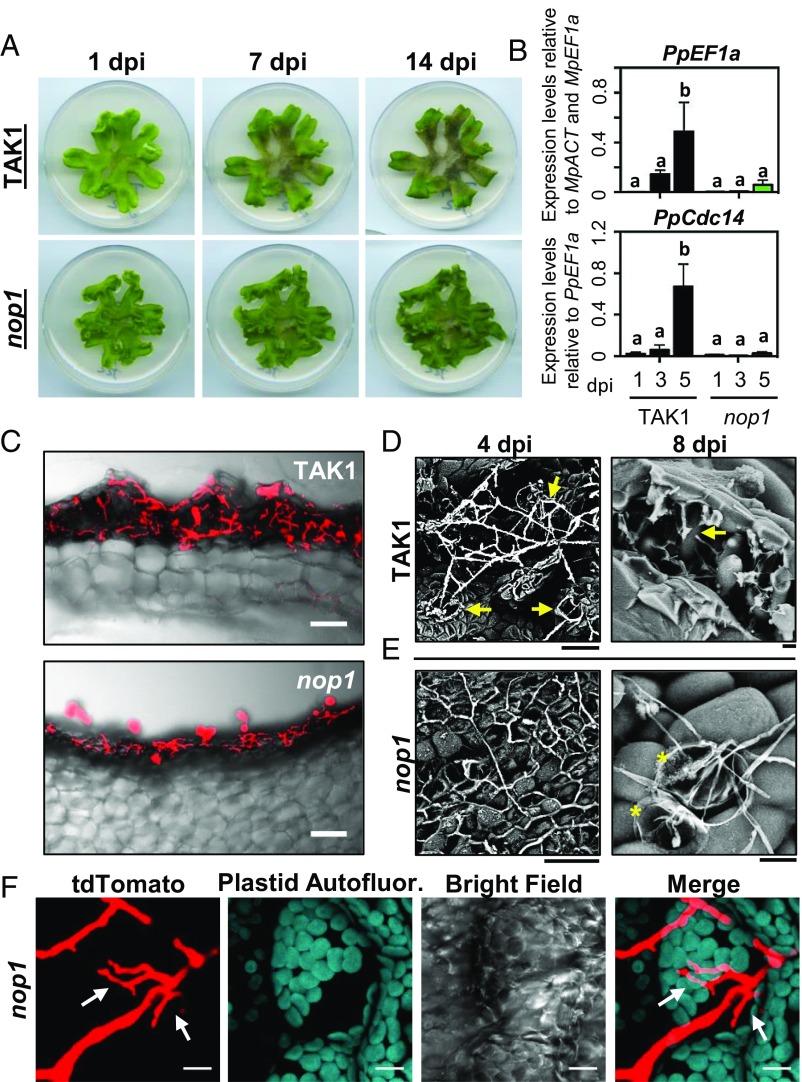
The colonization dynamics described thus far have occurred in epidermal cells and within air chambers of the photosynthetic layer of M. polymorpha. In Marchantia, loss-of-function mutations in the E3 ubiquitin ligase NOPPERABO (NOP1) result in the development of a photosynthetic layer that lacks air chambers entirely (53). To determine if air chambers facilitate P. palmivora colonization, we compared the colonization phenotypes of wild-type TAK1 and air-chamberless nop1 mutants (Fig. 5). As expected, P. palmivora ARI-td zoospores inoculated onto TAK1 thalli caused extensive disease symptoms by 7 dpi and continued to 14 dpi, when plants were essentially dead. In comparison, air-chamberless nop1 mutants displayed reduced disease symptoms, with plants remaining relatively healthy throughout the experiment (Fig. 5A). To support these observations, we quantified the expression of pathogen-specific marker genes indicative of biomass (PpEF1a) and sporulation (PpCdc14) by qRT-PCR. Compared with wild-type TAK1, pathogen biomass and sporulation were both significantly reduced in nop1 mutants (Fig. 5B), which indicates that P. palmivora fitness is largely dependent on the presence of air chambers. This was further supported by microscopic analyses of P. palmivora growth in TAK1 compared with nop1 plants. Confocal fluorescence microscopy of sectioned thalli revealed extensive hyphal growth within TAK1 air chambers at 7 dpi, whereas nop1 plants displayed only a thin layer of P. palmivora hyphae on the dorsal epidermis (Fig. 5C). Moreover, cryo-SEM images confirmed that hyphae travel from chamber to chamber during the colonization of TAK1 thalli, with widespread hyphal growth observed within air chambers (Fig. 5D). In comparison, a network of surface hyphae was observed on nop1 thalli, and collapsed epidermal cells containing P. palmivora hyphae were occasionally detected late during infection (Fig. 5E). In support of this, confocal fluorescence microscopy identified invasive P. palmivora hyphal growth within nop1 epidermal cells by 3 dpi (Fig. 5F). In addition, sporangia were detected on nop1 thalli late during infection (Fig. 5C), which indicates that P. palmivora can complete a full infection cycle on nop1 hosts despite the limited amount of colonizable tissue.
Fig. 5.
P. palmivora requires air chambers for the successful colonization of M. polymorpha thalli. (A) Disease symptoms of 3-wk-old M. polymorpha TAK1 (wild-type) and nop1 mutant plants inoculated with P. palmivora ARI-td zoospores at 1, 7, and 14 dpi. (B) Quantification of P. palmivora biomass (PpEF1a) and sporulation (PpCdc14) marker genes during the colonization of wild-type TAK1 and nop1 plants at 1, 3, and 5 dpi. PpEF1a expression was quantified relative to M. polymorpha biomass markers (MpACT and MpEF1a). PpCdc14 was quantified relative to pathogen biomass (PpEF1a). Different letters signify statistically significant differences in transcript abundances (ANOVA, Tukey’s HSD, P < 0.05). (C) Confocal fluorescence microscopy of sectioned TAK1 and nop1 thalli infected with P. palmivora ARI-td at 7 dpi. Micrographs display merged z-stack projections of red pathogen fluorescence and bright-field images. (Scale bars, 100 μm.) (D) Cryo-SEM images of TAK1 plants demonstrate ARI-td hyphae traveling between air pores (Left, arrows) at 4 dpi and hyphal growth (Right, arrow) within air chambers at 8 dpi. (Scale bars, 20 μm.) (E) Cryo-SEM images of nop1 plants demonstrate a network of ARI-td surface hyphae (Left) at 4 dpi, and collapsed epidermal cells (Right, asterisks) that are sometimes observed at 8 dpi. (Scale bars, 20 μm.) (F) Confocal fluorescence microscopy demonstrating invasive ARI-td hyphal growth (arrows) in nop1 epidermal cells at 3 dpi. Micrographs display z-stack projections of red pathogen fluorescence (tdTomato), plastid autofluorescence, bright-field images, and plastid and tdTomato fluorescence merged together. (Scale bars, 10 μm.) Experiments were performed three times (A–C and F) or twice (D and E), with similar results.
Collectively, our data suggests that liverwort air chambers support P. palmivora colonization. To address a potential role for NOP1 in immunity, we quantified the levels of pathogen-inducible MpPRX and MpDIR transcripts in TAK1 and nop1 mutants infected with P. palmivora ARI-td. Both MpPRX and MpDIR were more highly expressed in wild-type TAK1 compared with nop1 plants at 3 and 5 d after P. palmivora infection, which demonstrates that resistance in nop1 is not associated with the heightened expression of these markers (SI Appendix, Fig. S15A). Importantly, transcript levels of both MpPRX and MpDIR are similar in nop1 and wild-type plants when quantified relative to pathogen biomass markers rather than endogenous controls (SI Appendix, Fig. S15B). Together, our results demonstrate that nop1-dependent changes in thallus architecture impair P. palmivora proliferation, suggesting a key role for air chambers in supporting pathogen colonization.
Discussion
In this study, we demonstrate that the broad host-range oomycete P. palmivora colonizes the photosynthetic layer of nonvascular liverworts and forms intracellular hyphal structures within living host cells. Several lines of evidence lead us to conclude that the interaction between P. palmivora and Marchantia includes an infection stage with features of biotrophy. First, we noted profuse hyphal colonization without significant perturbations of host cell viability at early infection stages (Figs. 3 and 4 and SI Appendix, Fig. S13). Second, P. palmivora formed haustoria-like intracellular structures in living Marchantia cells, which is a well-established marker of biotrophic plant-pathogen interactions in vascular plants (39, 54). Third, the development of these structures in compatible Marchantia–Phytophthora interactions was associated with the up-regulation of Phytophthora effector genes, which are widely acknowledged markers for the establishment of a biotrophic (nondestructive) phase in vascular plants (41–43). Fourth, Phytophthora haustoria-like structures displayed a permissive callose distribution pattern, and, remarkably, a host gene associated with a biotrophic symbiotic interaction in Medicago [SYP13B, the Medicago SYP132 homolog (55)] was similarly up-regulated and localized to intracellular microbial structures in Marchantia. Considered alongside observations of arbuscules in liverworts and hornworts (7, 17, 19, 56), the data indicate that descendants of the earliest diverging land plants have the capacity to accommodate pathogenic filamentous eukaryotes within their cells. P. palmivora eventually completes its asexual life cycle on Marchantia thalli, as demonstrated by the formation of sporangia and up-regulation of the sporulation-specific PpCdc14 transcript (40). Similar to interactions with vascular plants, the completion of the P. palmivora life cycle is associated with the switch to a necrotrophic lifestyle (28, 29), as plant tissues are necrotized to further sustain pathogen growth at later stages of infection.
The colonization of Marchantia thalli by P. palmivora was largely limited to the photosynthetic layer, with prolific hyphal growth within and between air chambers. Colonization assays with the air-chamberless nop1 mutant revealed that these structures largely contribute to disease susceptibility in Marchantia. This is likely due to the fact that air chambers offer photosynthetic filament cells rich in carbohydrates and provide a microenvironment with stable humidity, which is ideal for water molds such as Phytophthora. Our results underscore the need for plants to protect intercellular spaces important for gas exchange. Such adaptations are present in higher plants, where analogous intercellular spaces (spongy mesophyll) are protected by stomata. In many cases, the early detection of pathogens causes stomatal closure to prevent the colonization of the intercellular space (57). However, several pathogens have developed mechanisms to circumvent this strategy, either by accessing the tissue by other means or by the direct regulation of stomatal guard cells (58). Most filamentous pathogens bypass stomata by penetrating host surfaces using appressoria (penetration structures) that do not appear to play a major role during the P. palmivora–Marchantia interaction. Rather, air chambers were accessed via intra- or intercellular hyphal growth, or through constitutively open pores. P. palmivora sporangia often emanated from air pores, similar to observations of sporangia traversing stomata in angiosperms. The sporophytes of mosses and hornworts contain stomata, but liverworts do not. How these early forms of stomata function in response to biotic cues is unknown; however, elevated levels of CO2 do not alter bryophyte stomata as they do in angiosperms (59). It is therefore possible that interactions with pathogenic microbes contributed to the development of stomata in early divergent land plants. Future comparative studies are required to explore the relationship between microbial colonization and the development and regulation of pores involved in gas exchange.
The cell walls and surfaces of extant early diverging land plants are less complex than those of vascular plants (60). How this impacts interactions with pathogenic or symbiotic microbes remains to be explored. It is tempting to speculate that simple cell walls are susceptible to invading microbes; however, this may not be the case. For example, previous work in Marchantia demonstrated a differential capacity for powdery mildew spore establishment, such that Erysiphe trifoliorum spores germinated but were destroyed on M. polymorpha surfaces, while Oidium neolycopersici survived and developed appressoria (61). The authors did not explore whether O. neolycopersici penetrates M. polymorpha cells and establishes infection. A recent study showed that P. infestans and Phytophthora capsici hyphae penetrate protonemal cells during nonhost interactions in moss, whereas colonization by P. palmivora and Phytophthora sojae is rarely detected (23). Both P. infestans and P. capsici fail to establish haustoria-like structures in moss (23). In contrast, we demonstrated the colonization of Marchantia thalli by P. palmivora (rather than by P. infestans), which was associated with the development of haustoria-like infection structures. Susceptibility to P. palmivora was greatly enhanced when thalli, rather than rhizoids, were inoculated with zoospores. In addition, intercellular hyphal growth within the central storage parenchyma was less prevalent than growth within air chambers, although plants eventually succumbed to disease and were likely colonized throughout. Based on these observations, we hypothesize that the central storage region is resistant to pathogen colonization and requires prolific biotrophic growth within air chambers and a subsequent switch to necrotrophy for colonization. In contrast, the colonization of liverworts by symbiotic fungi is abundant within the central storage region but does not extend to the photosynthetic layer (5, 17). Collectively, these observations suggest that the characteristics of bryophyte tissues and surfaces act as barriers to microbial growth. Future efforts to identify the structural, genetic, or chemical components responsible for these phenotypes may reveal interesting similarities and differences compared to vascular plants.
We identified several aspects of P. palmivora colonization that differ in Marchantia compared to interactions with angiosperms. Appressorial penetration structures were not prevalent in Marchantia, as they are in angiosperms (27, 28). The development of oomycete and fungal appressoria requires cues such as hydrophobicity and the perception of cutin monomers characteristic of host cuticles (62, 63). The lack of P. palmivora appressoria on Marchantia surfaces suggests that key differences in cuticle composition impact pathogen development on liverwort surfaces. Indeed, bryophytes are poikilohydric plants that contain simple cuticle-like layers which afford desiccation tolerance and allow water absorption through plant bodies (64). Interestingly, the fungus O. neolycopersici develops appressoria on Marchantia (61), which may suggest differences in the detection of host surfaces/chemicals compared with P. palmivora. The P. palmivora–Marchantia interaction was also associated with highly branched intracellular infection structures and intracellular hyphae, whereas interactions in angiosperms are associated with digit-like haustoria and intercellular hyphal growth. Phytophthora haustoria have been associated with the manipulation of host cells through the secretion of RXLR effectors and other secreted proteins (29, 31–33). In support of this, genetic markers associated with haustoria development and function (PpHmp1 and RXLR effectors) were expressed during the infection stages when branched and digit-type structures are formed within Marchantia cells. A comparison of up-regulated candidate secreted P. palmivora proteins in Marchantia and N. benthamiana (SI Appendix, Table S1) points to a shared set (>50%), which may be even larger in a more comprehensive comparison utilizing the same pathogen isolate infecting comparable (i.e., photosynthetic) tissues. Importantly, branched intracellular infection structures were labeled in living cells by host proteins that are typically associated with the extrahaustorial matrix/membrane (46, 47), which suggests a function similar to digit-like haustoria. Interestingly, a range of intracellular infection structure morphologies were observed in different liverworts, with L. cruciata and M. paleacea also demonstrating the presence of digit- and knob-like haustoria, respectively. This suggests that characteristics of liverwort cells influence the development of intracellular infection structures. Whether this applies to other bryophyte species remains to be determined, as intracellular infection structures have not been described in mosses or hornworts. To date, there is no evidence to support a role for Phytophthora haustoria in nutrient acquisition during interactions with vascular or nonvascular plants. Importantly, biotrophic pathogens do not always rely on intracellular hyphae for nutrient acquisition. For example, growth of the biotrophic fungus Cladosporium fulvum is limited to the apoplast in tomato leaves, where the pathogen thrives on apoplastic nutrients despite the lack of dedicated infection structures (65, 66).
The membrane-localized SYP132 syntaxin family is associated with symbiosis in higher plants. In Medicago truncatula, the presence of symbiotic microbes induces alternative splicing of the SYP132 locus to produce SYP132a, which in turn labels infection threads and symbiosomes of nitrogen-fixing rhizobacteria (55, 67). Moreover, the silencing of SYP132a suppressed mycorrhizal colonization in Medicago roots, demonstrating the importance of SYP132a in promoting multiple symbiotic associations (55). We similarly observed colonization-induced up-regulation and interface localization of a specific membrane syntaxin variant in Marchantia (MpSYP13B); however, this was afforded by homologous gene copies rather than by alternative splicing. These data suggest that the evolution of membrane-localized plant syntaxins was influenced by interactions with symbiotic and pathogenic microbes, perhaps diverging for roles in general secretory processes or for the accommodation of intracellular microbial structures. Indeed, phylogenetic analysis of membrane-localized syntaxins in the green-plant lineage indicates their presence before the diversification of land plants (49). We hypothesize that MpSYP13B represents a specialized syntaxin that evolved to label intracellular structures in liverworts (this work), similar to SYP132a function in Medicago (55).
Early divergent descendants of the first land plants can accommodate filamentous eukaryotic microbes as diverse as oomycetes (this study) and mycorrhizal fungi (5, 17). Our data demonstrate that liverworts encode proteins that respond and localize to intracellular infection structures and pathogen hyphae, which suggests that the molecular machinery required to support intracellular colonization by eukaryotic microbes emerged early during land plant evolution. Previous work has established conserved genetic components required for endosymbiosis, with experimental evidence demonstrating the presence of functionally conserved homologs of key symbiosis genes in certain bryophyte and algal species (9, 10). Together, these studies suggest an ancient origin for the evolution of microbial accommodation in plants, which is further supported by observations of highly branched microbial structures and hyphae inside fossilized plant cells (1–4). In liverworts, we observed highly branched haustoria-like structures and prolific intracellular hyphal growth that could appear symbiosis-like in morphology. We therefore suggest caution in attributing the presence of branched intracellular structures to mutually beneficial symbiotic interactions based solely on the interpretation of fossilized microbial structures or microscopy-based analyses. Our work further underscores the need to consider pathogenic microbes alongside symbiotic counterparts when discussing the evolution of microbial accommodation processes in land plants.
Materials and Methods
Plant Growth.
The plants used in this study are described in SI Appendix, Table S2. All plants were cultivated from gemmae under axenic conditions. M. polymorpha and M. paleacea were grown on one-half–strength MS (Murashige and Skoog) media (pH 6.7) supplemented with B5 vitamins under continuous light (70 μE⋅m−2⋅s−1) at 22 °C. L. cruciata were grown in short-day photoperiod conditions (9 h light) at 20 °C on M (mycorrhization) media (10).
Pathogen Growth and Infection Assays.
Pathogen strains used in this study are described in SI Appendix, Table S3. P. palmivora ARI-td mycelia were maintained by routine passaging on rye sucrose agar (RSA) plates, and zoospores were collected from 7- to 10-d-old cultures on V8 juice agar plates as previously described (28). P. infestans zoospores were collected similar to P. palmivora, except the pathogen was grown solely on RSA plates incubated at 18 °C. Colonization experiments were performed by applying 10-μL droplets of a zoospore suspension inoculum (105 zoospores per milliliter) along the thallus or directly onto rhizoids of 3-wk-old M. polymorpha and M. paleacea plants. Slower-growing L. cruciata liverworts grown from gemmae were infected 6 to 8 wk postplating.
Microscopy.
Epifluorescence microscopy was conducted using a Zeiss Axio Imager stereo fluorescence microscope with a DsRED filter. All images were processed with AxioVision software (release 4.8). Confocal fluorescence microscopy was performed using a Leica TCS SP8 equipped with HyD detectors. Settings for pathogen detection (tdTomato) are described in ref. 28, mCitrine in ref. 52, and aniline blue in ref. 68. All experiments were performed at least three times, with similar results. Images were collected from at least three independent plants in at least two separate infection sites per plant. Cross-sections of 200- to 300-μm thickness were prepared from 3% agarose-embedded samples using a vibratome. All images were processed using ImageJ. Cryo-SEM was performed essentially as described in ref. 69, except that ARI-td–infected M. polymorpha tissue was used. All samples were sublimated for 30 to 60 s. Where indicated, samples were analyzed in backscatter mode to enhance contrast between oomycete and liverwort tissue.
Histochemical Staining.
Callose staining was performed by gently rinsing whole plants in 0.07 M phosphate buffer (pH 9), and then submerging whole plants in freshly prepared 0.05% aniline blue (wt/vol) solution (dissolved in 0.07 M phosphate buffer, pH 9). Plants were incubated in staining solution for 60 min and imaged immediately. A total of six infection sites (two per plant; three independent plants) were assessed.
RNA Isolation, cDNA Synthesis, and qRT-PCR Analysis.
Total RNA was extracted from flash-frozen M. polymorpha (TAK1) plants that were mock-inoculated (water) or infected with P. palmivora (ARI-td) zoospores at 1, 2, 3, or 4 dpi using the Concert Plant RNA Reagent (catalog no. 12322-012; Invitrogen) following the manufacturer’s instructions. Total RNA was extracted from axenically cultivated P. palmivora ARI-td using the Qiagen Plant RNeasy kit, followed by on-column DNase treatment (for RNA-seq), or by using the Concert Plant RNA Reagent (for qRT-PCR analysis). All samples extracted using the Concert Plant RNA Reagent were treated with Turbo-DNase reagent (Ambion) to degrade residual DNA contamination before further use. cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen) using 2 μg of total RNA following the manufacturer’s instructions. All cDNA samples were diluted 10× with nuclease-free water and stored at −20 °C until further use. qRT-PCR analyses were carried out with 2.5 μL of diluted cDNA and LightCycler 480 SYBR Green I Master Mix (Roche) in a 10-μL volume, according to the manufacturer’s instructions. Primers for qRT-PCR analyses were designed using Primer3 (70, 71) and are listed in SI Appendix, Table S4. Specificity was validated by analyzing melt curves after each run. Three technical replicates were analyzed for each of three independent sample replicates at any given time point/treatment. Calculations of expression levels normalized to internal controls and statistical analyses were performed using R software. Graphs were generated in GraphPad Prism6.
Cloning and Marchantia Transformation.
The Marchantia MpRab7 and MpRab11A homologs were first identified by BlastP analysis using homologous sequences from N. benthamiana (NbRabG3c and NbRabA1e, respectively). Full-length MpRab7 (Mapoly0946s0001) and MpRab11A (MapolyY_A0041.1) were cloned from cDNA generated from untreated, 3-wk-old M. polymorpha (TAK1) plants using Phusion DNA Polymerase (New England Biolabs) and Gateway-compatible primers described in SI Appendix, Table S4. Full-length attB sequences were generated using universal attB primers that allow for N-terminal fusions. Amplicons were recombined into pDONR221 using BP Clonase II (Invitrogen) following manufacturer instructions. Sequence-verified entry clones were then recombined into pMpGWB105 (72) using LR Clonase II Enzyme Mix (Invitrogen) following manufacturer instructions. To generate the MpPRX:GUS reporter line, a 1.8-kb fragment of the MpPRX promoter region (SI Appendix, Materials and Methods) flanked by attL sites was synthesized by Genewiz and recombined into pMpGWB104 (72) using LR Clonase II Enzyme Mix (Invitrogen) following manufacturer instructions. Completed destination constructs were transformed into Agrobacterium tumefaciens GV3101 (pMP90) by electroporation. M. polymorpha transformation was carried out using the Agrobacterium-mediated thallus regeneration method using TAK1 plants (73). Transformants were selected on solidified one-half–strength MS media (pH 5.6) supplemented with hygromycin B (15 to 25 μg/mL) and cefotaxime (125 μg/mL). Stable, nonchimeric transgenic plants were obtained by propagating gemmae from T1 thalli.
Library Preparation and Sequencing.
mRNAs from M. polymorpha plants infected with P. palmivora at 3 and 4 dpi and P. palmivora mRNAs from the mycelium sample were purified using Poly(A) selection from total RNA sample, and then fragmented. cDNA library preparation was performed with the TruSeq RNA Sample Preparation Kit (Illumina) according to the manufacturer’s protocol. cDNA sequencing of the nine samples (at 3 dpi, at 4 dpi, and the mycelium sample; all in triplicate) was performed with Illumina HiSeq 2500 in 100 paired-end mode. Samples were demultiplexed and analyzed further. The raw fastq data are accessible at https://www.ncbi.nlm.nih.gov/sra/ (accession no. SRP115544).
Expression Analysis.
Raw reads after quality control with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) were aligned back to the P. palmivora reference genome (74) using STAR (version 2.5.2b) aligner. Raw counts were obtained with FeatureCounts (75), and only uniquely mapped and properly paired reads were considered further. Differentially expressed genes were identified with DESeq2 Bioconductor package (76) following pair-wise comparisons between in planta and mycelium samples. Differentially expressed genes (absolute LFC of ≥2 and adjusted P value of ≤10−3) were used to perform hierarchical clustering of samples. Heatmaps for the differentially expressed genes were generated using the pheatmap R package (77). For the final heatmaps, rlog-transformed counts, median-centered by gene, were used. Scripts used to analyze the P. palmivora RNA-seq dataset and to visualize differentially expressed genes are available at https://github.com/gogleva/public-palmivora.
Secretome Prediction.
Putative secreted P. palmivora proteins were predicted using the SecretSanta R package (78) based on the gene models from ref. 74.
Supplementary Material
Acknowledgments
We thank Dr. Takashi Ueda (University of Tokyo), Dr. Jim Haseloff (University of Cambridge), and Dr. Pierre-Marc Delaux (Paul Sabatier University) for providing materials; our colleagues at the Joint Genome Institute (JGI) for access to M. polymorpha genomic information (https://phytozome.jgi.doe.gov); Dr. Raymond Wightman (Sainsbury Laboratory, University of Cambridge) for assistance with cryo-SEM analysis; Dr. Sophien Kamoun (The Sainsbury Laboratory–Norwich) for critically assessing an early draft of this manuscript; and Anthony Bridgen, Dr. Edouard Evangelisti, Dr. Ruth Le Fevre, Dr. Temur Yunusov, Lara Busby, and all members of the Schornack group for additional critical and technical support. This research was funded by grants from the Gatsby Charitable Foundation (RG62472), the Royal Society (RG69135), the Biotechnology and Biological Sciences Research Council OpenPlant initiative (BB/L014130/1), and the Natural Environment Research Council (NE/N00941X/1). The work conducted by the US Department of Energy (DOE) JGI, a DOE Office of Science User Facility, is supported by the Office of Science of the DOE under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw fastq data have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra/ (accession no. SRP115544).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717900115/-/DCSupplemental.
References
- 1.Remy W, Taylor TN, Hass H, Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor TN, Remy W, Hass H, Kerp H. Fossil arbuscular mycorrhizae from the Early Devonian. Mycologia. 1995;87:560–573. [Google Scholar]
- 3.Krings M, et al. Fungal endophytes in a 400-million-yr-old land plant: infection pathways, spatial distribution, and host responses. New Phytol. 2007;174:648–657. doi: 10.1111/j.1469-8137.2007.02008.x. [DOI] [PubMed] [Google Scholar]
- 4.Strullu-Derrien C, et al. Fungal associations in Horneophyton ligneri from the Rhynie Chert (c. 407 million year old) closely resemble those in extant lower land plants: novel insights into ancestral plant-fungus symbioses. New Phytol. 2014;203:964–979. doi: 10.1111/nph.12805. [DOI] [PubMed] [Google Scholar]
- 5.Ligrone R, et al. Glomeromycotean associations in liverworts: a molecular, cellular, and taxonomic analysis. Am J Bot. 2007;94:1756–1777. doi: 10.3732/ajb.94.11.1756. [DOI] [PubMed] [Google Scholar]
- 6.Adams DG, Duggan PS. Cyanobacteria-bryophyte symbioses. J Exp Bot. 2008;59:1047–1058. doi: 10.1093/jxb/ern005. [DOI] [PubMed] [Google Scholar]
- 7.Pressel S, Bidartondo MI, Ligrone R, Duckett JG. Fungal symbioses in bryophytes: New insights in the twenty first century. Phytotaxa. 2010;9:238–253. [Google Scholar]
- 8.Desiro A, Duckett JG, Pressel S, Villarreal JC, Bidartondo MI. Fungal symbioses in hornworts: A chequered history. Proc Biol Sci. 2013;280:20130207. doi: 10.1098/rspb.2013.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang B, et al. Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 2010;186:514–525. doi: 10.1111/j.1469-8137.2009.03137.x. [DOI] [PubMed] [Google Scholar]
- 10.Delaux P-M, et al. Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci USA. 2015;112:13390–13395. doi: 10.1073/pnas.1515426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petre B, Kamoun S. How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 2014;12:e1001801. doi: 10.1371/journal.pbio.1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth R, Paszkowski U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr Opin Plant Biol. 2017;39:50–56. doi: 10.1016/j.pbi.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Parniske M. Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr Opin Plant Biol. 2000;3:320–328. doi: 10.1016/s1369-5266(00)00088-1. [DOI] [PubMed] [Google Scholar]
- 14.Rey T, Schornack S. Interactions of beneficial and detrimental root-colonizing filamentous microbes with plant hosts. Genome Biol. 2013;14:121. doi: 10.1186/gb-2013-14-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov S, Fedorova E, Bisseling T. Intracellular plant microbe associations: secretory pathways and the formation of perimicrobial compartments. Curr Opin Plant Biol. 2010;13:372–377. doi: 10.1016/j.pbi.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Zeilinger S, et al. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol Rev. 2016;40:182–207. doi: 10.1093/femsre/fuv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carella P, Schornack S. Manipulation of bryophyte hosts by pathogenic and symbiotic microbes. Plant Cell Physiol. 2017 doi: 10.1093/pcp/pcx182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu YL, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renzaglia KS, et al. Bryophyte phylogeny: Advancing the molecular and morphological frontiers. Bryologist. 2007;110:179–213. [Google Scholar]
- 20.Cox CJ, Li B, Foster PG, Embley TM, Civán P. Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Syst Biol. 2014;63:272–279. doi: 10.1093/sysbio/syt109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 2014;14:23. doi: 10.1186/1471-2148-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponce de León I. The moss Physcomitrella patens as a model system to study interactions between plants and phytopathogenic fungi and oomycetes. J Pathogens. 2011;2011 doi: 10.4061/2011/719873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overdijk EJ, et al. Interaction between the moss Physcomitrella patens and Phytophthora: a novel pathosystem for live-cell imaging of subcellular defence. J Microsc. 2016;263:171–180. doi: 10.1111/jmi.12395. [DOI] [PubMed] [Google Scholar]
- 24.Ponce de León I, Montesano M. Adaptation mechanisms in the evolution of moss defenses to microbes. Front Plant Sci. 2017;8:366. doi: 10.3389/fpls.2017.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizaki K, Nishihama R, Yamato KT, Kohchi T. Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 2016;57:262–270. doi: 10.1093/pcp/pcv097. [DOI] [PubMed] [Google Scholar]
- 26.Shimamura M. Marchantia polymorpha: Taxonomy, phylogeny, and morphology of a model system. Plant Cell Physiol. 2016;57:230–256. doi: 10.1093/pcp/pcv192. [DOI] [PubMed] [Google Scholar]
- 27.Rey T, Chatterjee A, Buttay M, Toulotte J, Schornack S. Medicago truncatula symbiosis mutants affected in the interaction with a biotrophic root pathogen. New Phytol. 2015;206:497–500. doi: 10.1111/nph.13233. [DOI] [PubMed] [Google Scholar]
- 28.Le Fevre R, O’Boyle B, Moscou MJ, Schornack S. Colonization of barley by the broad-host hemibiotrophic pathogen Phytophthora palmivora uncovers a leaf development-dependent involvement of Mlo. Mol Plant Microbe Interact. 2016;29:385–395. doi: 10.1094/MPMI-12-15-0276-R. [DOI] [PubMed] [Google Scholar]
- 29.Evangelisti E, et al. Time-resolved dual root-microbe transcriptomics reveals early induced Nicotiana benthamiana genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biol. 2017;15:39. doi: 10.1186/s12915-017-0379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Judelson HS, Blanco FA. The spores of Phytophthora: weapons of the plant destroyer. Nat Rev Microbiol. 2005;3:47–58. doi: 10.1038/nrmicro1064. [DOI] [PubMed] [Google Scholar]
- 31.Morgan W, Kamoun S. RXLR effectors of plant pathogenic oomycetes. Curr Opin Microbiol. 2007;10:332–338. doi: 10.1016/j.mib.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Anderson RG, Deb D, Fedkenheuer K, McDowell JM. Recent progress in RXLR effector research. Mol Plant Microbe Interact. 2015;28:1063–1072. doi: 10.1094/MPMI-01-15-0022-CR. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, et al. Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol. 2017;216:205–215. doi: 10.1111/nph.14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardham AR. Cell biology of plant-oomycete interactions. Cell Microbiol. 2007;9:31–39. doi: 10.1111/j.1462-5822.2006.00833.x. [DOI] [PubMed] [Google Scholar]
- 35.Torres GA, et al. Bud rot caused by Phytophthora palmivora: A destructive emerging disease of oil palm. Phytopathology. 2016;106:320–329. doi: 10.1094/PHYTO-09-15-0243-RVW. [DOI] [PubMed] [Google Scholar]
- 36.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 37.Lehtonen MT, et al. Quickly-released peroxidase of moss in defense against fungal invaders. New Phytol. 2009;183:432–443. doi: 10.1111/j.1469-8137.2009.02864.x. [DOI] [PubMed] [Google Scholar]
- 38.Ponce De León I, et al. Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol Plant Pathol. 2012;13:960–974. doi: 10.1111/j.1364-3703.2012.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avrova AO, et al. A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol. 2008;10:2271–2284. doi: 10.1111/j.1462-5822.2008.01206.x. [DOI] [PubMed] [Google Scholar]
- 40.Ah Fong AMV, Judelson HS. Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus-like oomycete Phytophthora infestans. Mol Microbiol. 2003;50:487–494. doi: 10.1046/j.1365-2958.2003.03735.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, et al. Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell. 2011;23:2064–2086. doi: 10.1105/tpc.111.086082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jupe J, et al. Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 2013;14:R63. doi: 10.1186/gb-2013-14-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 44.Bellincampi D, Cervone F, Lionetti V. Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front Plant Sci. 2014;5:228. doi: 10.3389/fpls.2014.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faulkner C. A cellular backline: specialization of host membranes for defence. J Exp Bot. 2015;66:1565–1571. doi: 10.1093/jxb/erv021. [DOI] [PubMed] [Google Scholar]
- 46.Bozkurt TO, et al. Rerouting of plant late endocytic trafficking toward a pathogen interface. Traffic. 2015;16:204–226. doi: 10.1111/tra.12245. [DOI] [PubMed] [Google Scholar]
- 47.Lu YJ, et al. Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell Microbiol. 2012;14:682–697. doi: 10.1111/j.1462-5822.2012.01751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C, Hicks GR, Raikhel NV. Plant vacuole morphology and vacuolar trafficking. Front Plant Sci. 2014;5:476. doi: 10.3389/fpls.2014.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowman JL, et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304.e15. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 50.Gu Y, Zavaliev R, Dong X. Membrane trafficking in plant immunity. Mol Plant. 2017;10:1026–1034. doi: 10.1016/j.molp.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanazawa T, Ueda T. Exocytic trafficking pathways in plants: why and how they are redirected. New Phytol. 2017;215:952–957. doi: 10.1111/nph.14613. [DOI] [PubMed] [Google Scholar]
- 52.Kanazawa T, et al. SNARE molecules in Marchantia polymorpha: Unique and conserved features of the membrane fusion machinery. Plant Cell Physiol. 2016;57:307–324. doi: 10.1093/pcp/pcv076. [DOI] [PubMed] [Google Scholar]
- 53.Ishizaki K, et al. Essential role of the E3 ubiquitin ligase nopperabo1 in schizogenous intercellular space formation in the liverwort Marchantia polymorpha. Plant Cell. 2013;25:4075–4084. doi: 10.1105/tpc.113.117051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bozkurt TO, et al. The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol. 2014;165:1005–1018. doi: 10.1104/pp.114.235804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan H, et al. A symbiotic SNARE protein generated by alternative termination of transcription. Nat Plants. 2016;2:15197. doi: 10.1038/nplants.2015.197. [DOI] [PubMed] [Google Scholar]
- 56.Schussler A. Glomus claroideum forms an arbuscular mycorrhiza-like symbiosis with the hornwort Anthoceros punctatus. Mycorrhiza. 2000;10:15–21. [Google Scholar]
- 57.McLachlan DH, Kopischke M, Robatzek S. Gate control: guard cell regulation by microbial stress. New Phytol. 2014;203:1049–1063. doi: 10.1111/nph.12916. [DOI] [PubMed] [Google Scholar]
- 58.Melotto M, Zhang L, Oblessuc PR, He S-Y. Stomatal defense a decade later. Plant Physiol. 2017;174:561–571. doi: 10.1104/pp.16.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Field KJ, Duckett JG, Cameron DD, Pressel S. Stomatal density and aperture in non-vascular land plants are non-responsive to above-ambient atmospheric CO2 concentrations. Ann Bot. 2015;115:915–922. doi: 10.1093/aob/mcv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarkar P, Bosneaga E, Auer M. Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J Exp Bot. 2009;60:3615–3635. doi: 10.1093/jxb/erp245. [DOI] [PubMed] [Google Scholar]
- 61.Takikawa Y, et al. Targeted destruction of fungal structures of Erysiphe trifoliorum on flat leaf surfaces of Marchantia polymorpha. Plant Biol (Stuttg) 2014;16:291–295. doi: 10.1111/plb.12089. [DOI] [PubMed] [Google Scholar]
- 62.Latijnhouwers M, de Wit PJGM, Govers F. Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 2003;11:462–469. doi: 10.1016/j.tim.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 63.Wang E, et al. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol. 2012;22:2242–2246. doi: 10.1016/j.cub.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 64.Ligrone R, Duckett JG, Renzaglia KS. Major transitions in the evolution of early land plants: a bryological perspective. Ann Bot. 2012;109:851–871. doi: 10.1093/aob/mcs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Solomon PS, Oliver RP. Evidence that gamma-aminobutyric acid is a major nitrogen source during Cladosporium fulvum infection of tomato. Planta. 2002;214:414–420. doi: 10.1007/s004250100632. [DOI] [PubMed] [Google Scholar]
- 66.de Wit PJ. Cladosporium fulvum effectors: Weapons in the arms race with tomato. Annu Rev Phytopathol. 2016;54:1–23. doi: 10.1146/annurev-phyto-011516-040249. [DOI] [PubMed] [Google Scholar]
- 67.Catalano CM, Czymmek KJ, Gann JG, Sherrier DJ. Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta. 2007;225:541–550. doi: 10.1007/s00425-006-0369-y. [DOI] [PubMed] [Google Scholar]
- 68.Caillaud M-C, et al. The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 2014;10:e1004496. doi: 10.1371/journal.ppat.1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wightman R, Wallis S, Aston P. Hydathode pit development in the alpine plant Saxifraga cochlearis. Flora. 2017;233:99–108. [Google Scholar]
- 70.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 71.Untergasser A, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishizaki K, et al. Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One. 2015;10:e0138876. doi: 10.1371/journal.pone.0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kubota A, Ishizaki K, Hosaka M, Kohchi T. Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci Biotechnol Biochem. 2013;77:167–172. doi: 10.1271/bbb.120700. [DOI] [PubMed] [Google Scholar]
- 74.Ali SS, et al. Phytophthora megakarya and P. palmivora, closely related causal agents of cacao black pod rot, underwent increases in genome sizes and gene numbers by different mechanisms. Genome Biol Evol. 2017;9:536–557. doi: 10.1093/gbe/evx021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 76.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kolde R. 2015 Package “pheatmap”: Pretty heatmaps. R package, version 1.0.8. Available at https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf. Accessed April 1, 2017.
- 78.Gogleva A, Drost H-G, Schornack S. SecretSanta: flexible pipelines for functional secretome prediction. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.