Significance
To date, therapies are lacking that efficiently target the Kirsten rat sarcoma (KRAS) oncogene, which is responsible for approximately a quarter of all lung cancer cases in the United States. This study provides genetic evidence that the insulin/insulin-like growth factor 1 (IGF1) signaling, which modulates cellular survival, growth, and metabolism, is required for KRAS-driven lung cancer initiation. It further identifies a metabolic vulnerability in tumors with loss of such signaling, that is, the dependence on autophagy (a self-eating process), and protein degradation, to compensate for decreased amino acid levels. Such vulnerability could be exploited therapeutically using available autophagy and protein degradation inhibitors, in combination with insulin receptor/IGF1 receptor inhibitors, in patients with KRAS-mutant lung cancer.
Keywords: non–small-cell lung cancer, Kras, insulin receptor substrates, autophagy, amino acids
Abstract
Non–small-cell lung cancer (NSCLC) is a leading cause of cancer death worldwide, with 25% of cases harboring oncogenic Kirsten rat sarcoma (KRAS). Although KRAS direct binding to and activation of PI3K is required for KRAS-driven lung tumorigenesis, the contribution of insulin receptor (IR) and insulin-like growth factor 1 receptor (IGF1R) in the context of mutant KRAS remains controversial. Here, we provide genetic evidence that lung-specific dual ablation of insulin receptor substrates 1/2 (Irs1/Irs2), which mediate insulin and IGF1 signaling, strongly suppresses tumor initiation and dramatically extends the survival of a mouse model of lung cancer with Kras activation and p53 loss. Mice with Irs1/Irs2 loss eventually succumb to tumor burden, with tumor cells displaying suppressed Akt activation and strikingly diminished intracellular levels of essential amino acids. Acute loss of IRS1/IRS2 or inhibition of IR/IGF1R in KRAS-mutant human NSCLC cells decreases the uptake and lowers the intracellular levels of amino acids, while enhancing basal autophagy and sensitivity to autophagy and proteasome inhibitors. These findings demonstrate that insulin/IGF1 signaling is required for KRAS-mutant lung cancer initiation, and identify decreased amino acid levels as a metabolic vulnerability in tumor cells with IR/IGF1R inhibition. Consequently, combinatorial targeting of IR/IGF1R with autophagy or proteasome inhibitors may represent an effective therapeutic strategy in KRAS-mutant NSCLC.
Non–small-cell lung cancer (NSCLC) accounts for the majority of lung cancer, which to date remains the leading cause of cancer death in the United States and worldwide (1, 2). About a quarter of NSCLC cases harbor Kirsten rat sarcoma (KRAS) activating mutations (3, 4). However, effective therapies targeting KRAS are still lacking and alternative approaches are urgently needed (5). Previous reports showed that KRAS can directly bind to and activate the p110α catalytic subunit of PI3K and that this interaction is required for in vivo Kras-driven tumor initiation and maintenance in mouse models of lung cancer (6, 7). However, the sufficiency of KRAS–PI3K interaction in driving lung cancer development remains largely controversial, with evidence from cell culture studies implicating additional input from insulin receptor (IR) and insulin-like growth factor 1 receptor (IGF1R) (8, 9). Most insulin/IGF1 signaling in the lungs converges intracellularly onto the adaptor proteins insulin receptor substrates IRS1 and IRS2 (10) before diverging to a complex network of downstream signaling effectors, including PI3K/AKT (11). Here, using a robust genetic approach, we provide evidence that concomitant ablation of Irs1 and Irs2 in the lungs of a well-established genetically engineered mouse model of lung cancer with conditional Kras activation and p53 loss strongly suppresses tumor initiation, doubling tumor latency and significantly extending survival. Lung cells with Irs1/Irs2 ablation eventually overcome this suppression, although at a stochastic rate, forming advanced lung adenocarcinomas. Interestingly, cells derived from these tumors not only display suppressed Akt activation in response to insulin/IGF1 stimulation but also significantly decreased intracellular levels of essential amino acids. We find that acute loss or knockdown of IRS1/IRS2 in KRAS-mutant human NSCLC cells, or pharmacological inhibition of IR/IGF1R in KRAS-mutant NSCLC or murine lung cancer cells results in the decreased uptake and intracellular levels of essential amino acids, accompanied by enhanced basal autophagy. NSCLC cells with acute loss of IRS1/IRS2 also display increased sensitivity to lysosomal and proteasomal inhibitors. Our findings provide evidence that IRS1 and IRS2 are required for KRAS-mutant lung cancer formation. They further shed light on the metabolic vulnerabilities that arise in tumors treated with IR/IGF1R inhibitors, pointing to potential combinatorial approaches for treating KRAS-driven NSCLC.
Results
Lung-Specific Genetic Ablation of Irs1 and Irs2 Significantly Delays Tumor Formation in a Kras-Driven Mouse Model of Lung Cancer.
The conditional genetically engineered mouse model of lung cancer Lox-STOP-Lox (LSL)-KrasG12D/+;p53fl/fl (12–14), herein referred to as “KP,” was bred to mice harboring floxed alleles for both Irs1 and Irs2 genes (15), leading to the generation of KrasG12D/+;p53fl/fl;Irs1fl/fl;Irs2fl/fl or “KPI” mice. Intranasal administration of adenoviral Cre into KP or KPI mice at 5–6 wk of age led to the concomitant expression of activated Kras and loss of p53 alone (KP mice) or the triple loss of p53, Irs1, and Irs2 (KPI mice) in mouse lung cells (Fig. 1A). As previously reported (14, 16), 10 wk post-Cre administration, KP mice developed extensive lung adenomas and adenocarcinomas (Fig. 1 B and C) and succumbed to tumor burden by 140 d, with median survival of 107 or 103 d for males and females, respectively (Fig. 1D). In contrast, lungs of KPI mice were devoid of any tumors or hyperplasia at 8–10 wk postinfection and retained normal histology (Fig. 1 B, C, E, and F). Surprisingly, however, KPI mice eventually overcame the loss of Irs1/Irs2 and developed lung adenocarcinomas at a significantly extended and highly variable tumor latency (16–30 wk for KPI compared with 8 wk for KP; Fig. 1 E and F). Moreover, both the median survival (191 d for males and 209 d for females) and maximal survival (∼300 d) of KPI mice were twice as long as those of KP mice (Fig. 1D). Interestingly, when lung tumors from moribund KP and KPI mice were analyzed and compared histopathologically, KPI tumors displayed a significantly increased proportion of higher-grade adenocarcinomas and carcinomas, characterized by nuclear pleomorphism and invasion of basement membrane (Fig. 1 G and H). These results indicate that Irs1 and Irs2 are required for Kras-driven lung tumor initiation and that Kras-transformed cells can eventually bypass Irs1/Irs2 loss, developing more aggressive tumors at a highly variable and extended latency.
Fig. 1.
Loss of Irs1 and Irs2 significantly delays Kras-driven lung tumorigenesis. (A) Breeding schematic for KP and KPI mice. (B) H&E staining of KP and KPI lungs 10 wk postadenoviral Cre infection (Left) with 10-fold magnification of framed area (Right). [Scale bars: 400 μm (Left) and 40 μm (Right).] (C) Tumor burden in KP and KPI lungs 10 wk postadenoviral Cre infection. Males, n = 21 (KP) and n = 9 (KPI); females, n = 6 (KP) and n = 9 (KPI); ***P < 0.001; ****P < 0.0001. (D) Kaplan–Meier survival curves. Males, n = 53 (KP) and n = 20 (KPI); females, n = 46 (KP) and n = 17 (KPI); ****P < 0.0001 by log-rank test. (E) MRI showing axial planes of representative KP lungs at weeks 0–16, and KPI lungs at weeks 0–35 postadenoviral Cre infection. Tumor areas are delineated in red. (Scale bars: 5 mm.) (F) Volumes of KP and KPI tumors quantified from MRI images represented in E. Each data point represents one animal. *P < 0.05; **P < 0.01. (G) H&E staining of KP and KPI lungs from moribund mice (Left) with 10-fold magnification of framed area (Right). [Scale bars: 400 μm (Left) and 40 μm (Right).] (H) Percentage of KP and KPI mice with medium- or high-grade tumors at moribund stage (Left) with Fisher’s exact test (Right). In C and F, data represent the mean ± SD. H&E, hematoxylin and eosin.
Loss of Irs1 and Irs2 Suppresses Akt Signaling and Leads to Decreased Amino Acid Levels in Murine Kras-Driven Lung Tumor Cells.
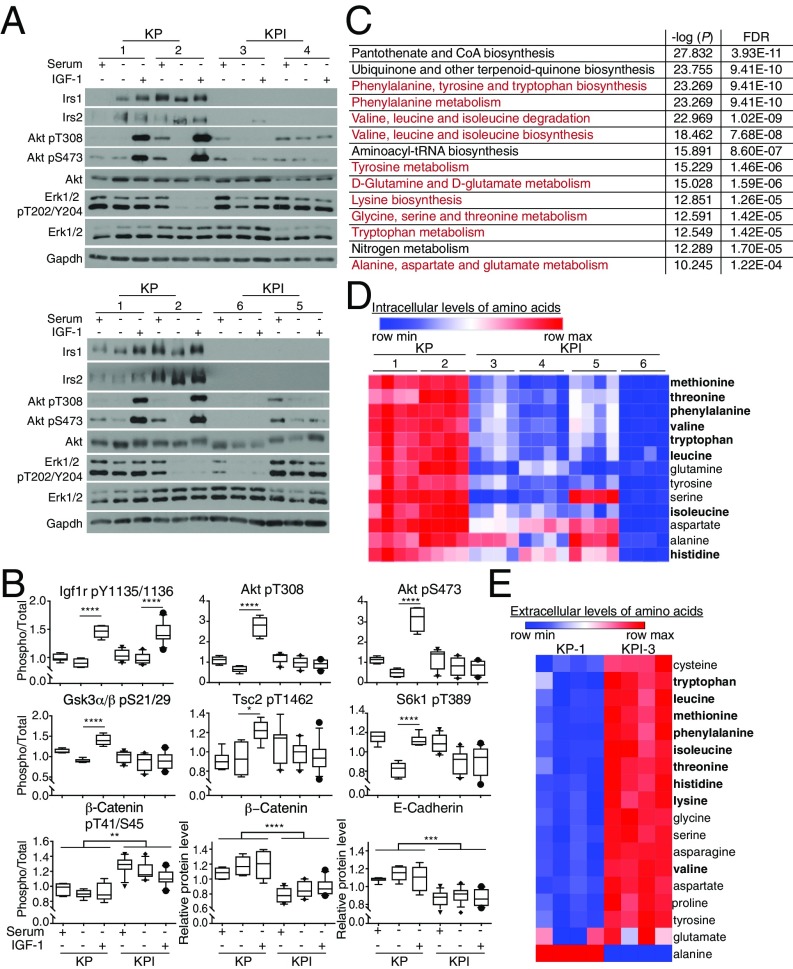
To characterize the signaling and metabolism of cells with Irs1/Irs2 loss, KP and KPI lung tumors were isolated and used to establish several cell lines in culture. Genotyping confirmed the concomitant excision of the STOP codon upstream of KrasG12D and loss of p53 in both KP and KPI cells, with the additional deletion of Irs1 and Irs2 in KPI but not KP cells, thus validating that the KPI tumors did not escape Irs1/Irs2 loss (Fig. S1A). Although one of the KPI cell lines (KPI-6) seemed to harbor incomplete Cre-mediated recombination, the faint genotyping bands for LSL-KrasG12D and floxed Irs1 and Irs2 were due to the rare residual presence of nontransformed cells at this early cell line passage. Indeed, in future passages, Irs1 and Irs2 proteins were completely lost in all KPI cell lines tested (Fig. 2A and Fig. S1B).
Fig. 2.
Murine Irs1/Irs2-null Kras-driven lung tumor cells have impaired Akt signaling and decreased intracellular amino acid levels. (A) Levels of Irs1, Irs2, total or phosphorylated Akt and Erk1/2 in KP and KPI cells grown in 10% serum or serum-starved for 1 h with or without IGF1 (50 ng/mL) stimulation for 10 min. Gapdh was used as a loading control. (B) Box plots representing total or phosphorylated levels of selected effectors of Akt signaling quantified by reverse-phase protein array (RPPA) in KP and KPI cells treated as in A. Levels of phosphorylated proteins were normalized to total levels of the respective proteins. Data represent the median ±10th to 90th percentile for each protein/phosphoprotein; n = 3 biological replicates per condition per cell line. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (C) List of top 14 metabolic pathways that are significantly different between KP (-1, -2) and KPI (-3 to -6) cells grown in 10% serum for 24 h. In red are pathways involved in amino acid metabolism. Data were processed by Metaboanalyst 3.0, and pathways were ranked by −log of the P value. FDR indicates false-discovery rate. (D and E) Heat maps listing in descending order of statistical significance (P < 0.05 by t test) amino acids whose intracellular (A) or extracellular media (B) levels are different between KP and KPI cells described in C. In bold are essential amino acids. Red indicates higher expression, and blue indicates lower expression relative to the mean expression level within each group; n = 4 biological replicates per cell line.
To confirm the relevance and engagement of Irs1 and Irs2 in insulin/IGF1 signaling in lung tumor cells, we first performed a Luminex bead-based immunoassay (17) in KP and KPI cell lines that is optimized to assess the interaction between Irs1 and Pi3k catalytic subunit p110α (Fig. S1C). Acute stimulation of serum-starved KP but not KPI cells with either insulin or IGF1 dramatically enhanced Irs1-p110α interaction, confirming signal transduction from Igf1r to Pi3k in the lung cancer cells (Fig. S1C). We then assessed alterations in Akt phosphorylation (pT308 and pS473) upon ligand stimulation in KP and KPI cells with single or double knockdown of Irs1 and Irs2. Loss of either Irs1 or Irs2 alone did not significantly affect Akt activation in the murine KP cells (Fig. S2) or in human KRAS-mutant NSCLC cells (Fig. S3). In contrast, concomitant silencing of both Irs1 and Irs2 strongly suppressed Akt activation in response to insulin or IGF1 stimulation (Fig. 2A). These results provide evidence of a functional redundancy in Irs1 and Irs2 signal transduction in Kras-driven lung tumor cells, and are consistent with a recent report demonstrating increased rather than decreased tumor formation in a Kras-driven mouse model of lung cancer with loss of Irs1 alone (18). Intriguingly, however, Erk1/2 activation (pT202/Y204) levels varied significantly among the different cell lines upon ligand treatment, and did not correlate with the loss of Irs1/Irs2, reflecting heterogeneity in the tumor cell populations (Fig. 2A and Figs. S2 and S3). This observation is consistent with the previously reported intratumoral stage-heterogeneity in Kras-driven lung tumors, where MAPK signal amplification, a driver of malignant progression, was found to only mark a fraction of the tumor cell populations (19).
To acquire a broader assessment of the effects of Irs1 and Irs2 loss on signaling networks, we performed reverse-phase protein array (RPPA) analysis (20) of 300 proteins and phosphoproteins representing many major signaling pathways (Fig. 2B and Dataset S1). We used protein lysates from KP and KPI cells that were either nonstarved (10% serum), 1-h serum-starved or serum-starved and acutely stimulated with IGF1. The most striking differences were detected under acute IGF1-stimulated conditions, representing significant suppression of Akt activation and its downstream signaling in KPI compared with KP cells. As expected, IGF1 stimulation resulted in the phosphorylation of its receptor at Y1135/1136 in both KP and KPI cells (Fig. 2B). However, Akt activation (pT308 and S473) upon IGF1 stimulation was blunted in KPI, compared with KP cells. This was mirrored by the blunted inactivating phosphorylation of the Akt targets, Gsk-3α/β (pS21/29) and Tsc2 (T1462). Indeed, the increased activity in KPI cells, of Tsc2, a repressor of mTORC1 signaling, led to decreased phosphorylation of the mTORC1 target S6k1 (T389) upon IGF1 stimulation, compared with KP cells (Fig. 2B). However, because mTORC1 can be activated by growth factor signaling independent of insulin/IGF1 (21), S6k1 phosphorylation was maintained in KPI cells cultured in 10% serum, independent of Irs1/Irs2 loss. Consistent with suppressed Akt signaling in KPI cells, an increase in Gsk-3 α/β activity resulted in constitutive phosphorylation of β-catenin (T41/S45), which leads to its proteasomal degradation (Fig. 2B). Indeed, lower levels of β-catenin were detected in KPI cells, accompanied by a decrease in the levels of its binding partner E-cadherin (Fig. 2B). Interestingly, this phenotype is known to correlate with tumor progression (22) and a transition to a more mesenchymal, invasive phenotype (23), consistent with the more aggressive nature of KPI tumors at a moribund stage (Fig. 1 G and H).
We then sought to explore the metabolic differences between KP and KPI cells, with the goal of identifying metabolic dependencies upon Irs1/Irs2 loss. To that end, we profiled over 150 polar metabolites in the tumor cells grown for 24 h under either non–serum-starved or serum-starved conditions. KP and KPI cells displayed no significant metabolic changes under serum-starved conditions, indicating that loss of Irs1/Irs2 closely mimics conditions of growth factor deprivation. In contrast, striking differences were observed under nonstarved conditions, with pathway enrichment analysis identifying amino acid synthesis or degradation as top impacted metabolic pathways between KP and KPI cells (Fig. 2C). In particular, KPI cells exhibited significantly lower intracellular levels of essential amino acids, including leucine, isoleucine, valine, methionine, phenylalanine, threonine, tryptophan, in addition to glutamine and tyrosine (Fig. 2D), suggesting a role for Irs1/Irs2 in amino acid uptake and/or metabolism. Indeed, in contrast to their intracellular levels, extracellular media levels of most amino acids were strikingly higher in KPI compared with KP cells (Fig. 2E), implying a decrease in amino acid uptake upon Irs1/Irs2 loss. These results are consistent with previous reports describing regulation of amino acid transport in diverse cell types by different growth factors, including insulin/IGF1 (24–27).
Loss of IRS1 and IRS2 in Human KRAS-Mutant NSCLC Cells Leads to Impaired AKT Signaling and Reduced Intracellular Amino Acid Levels.
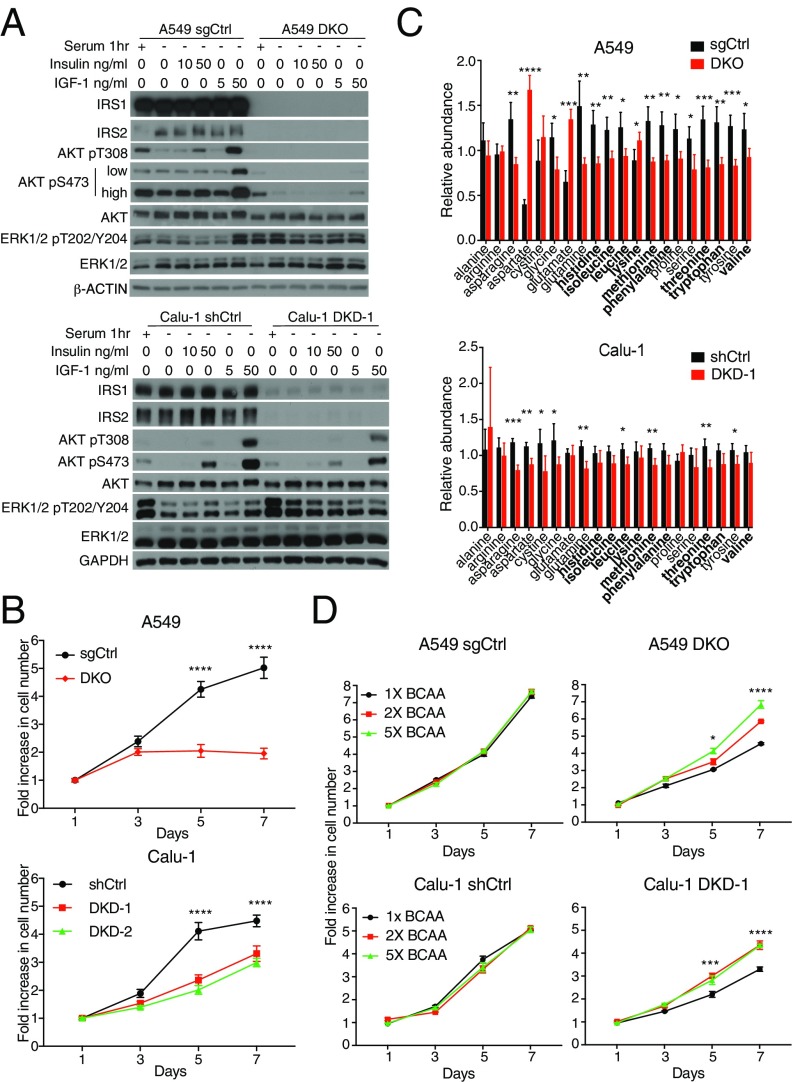
To investigate the relevance of IRS1 and IRS2 in human lung cancer, we extended our studies to established NSCLC cell lines that harbor activating KRAS mutations. Dual loss of IRS1 and IRS2 was engineered via CRISPR/Cas9 double knockout (DKO) in A549 cells (Fig. 3A), whereas IRS1/IRS2 double knockdown (DKD) was achieved via stable small hairpin expression in both A549 and Calu-1 cells (Fig. 3A and Fig. S4 A–C). In A549 DKO, A549 DKD, and Calu-1 DKD cells, AKT activation was severely mitigated in response to insulin or IGF1 stimulation (Fig. 3A and Fig. S4 A and B), consistent with the results obtained from murine KP and KPI cells (Fig. 2A). Furthermore, in vitro cellular proliferation was impaired by loss of IRS1/IRS2 (Fig. 3B and Fig. S4D).
Fig. 3.
Loss of IRS1 and IRS2 in human KRAS-mutant NSCLC leads to impaired AKT signaling and reduced intracellular amino acid levels. (A) Levels of IRS1, IRS2, total or phosphorylated AKT and ERK1/2 in NSCLC A549 cells with IRS1/IRS2 (A549 DKO) or control (A549 sgCtrl) DKO as well as Calu-1 cells with IRS1/IRS2 (Calu-1 DKD-1) or control (Calu-1 shGFP/shScramble, termed shCtrl) DKD cells. Cells were serum-starved for 1 h and then stimulated with insulin or IGF1 for 10 min. β-ACTIN and GAPDH were used as loading controls. (B) Proliferation curves of cells described in A that were grown under low serum conditions (0.1% serum for A549 and 2% serum for Calu-1) over 7 d; n = 6. (C) Levels of amino acids in NSCLC cells described in A that were first normalized to protein levels, and then normalized to the median of all samples for each amino acid; n = 4 biological replicates per cell line; data are representative of two independent experiments. (D) Proliferation curves of cells described in A that were grown under low serum conditions (0.5% serum for A549 and 2% serum for Calu-1) in RPMI media containing either fivefold (5×), twofold (2×), or the standard (onefold or 1×) concentrations of BCAAs: leucine (0.05 g/L), isoleucine (0.05 g/L), and valine (0.02 g/L) over 7 d; n = 4. In B–D, data represent the mean ± SD (B and C) or ±SEM (D). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. In B, significance is between shCtrl and DKO or DKD conditions. In D, significance is between 1× BCAA and both 2× and 5× conditions.
To assess the effects of loss of IRS1 and IRS2 on cellular metabolism, metabolite profiling was performed on A549 and Calu-1 cells cultured under non–serum-starved conditions. Strikingly, and consistent with our findings in the mouse KPI cells, the intracellular levels of all essential amino acids, except for lysine, were significantly decreased or trended toward a decrease in A549 DKO and Calu-1 DKD cells compared with the control cells (Fig. 3C). This was accompanied by a moderate yet consistent decrease in the uptake of extracellular amino acids from the media (Fig. S5A). Furthermore, supplementation of their media with essential branched-chain amino acids (BCAAs) (i.e., leucine, isoleucine and valine) partially rescued the growth of A549 DKO and Calu-1 DKD cells (Fig. 3D). These results led us to investigate a potential regulation of amino acid transporter expression levels by loss of IRS1/IRS2. Although a variety of such transport systems exist that display overlapping substrate specificity, two of the mostly extensively studied are the SLC7A5 (LAT-1)/SLC3A2 heterodimer, which mediates the uptake of large neutral amino acids (including BCAAs), and SLC7A1 (CAT-1), a cationic amino acid (arginine, histidine, lysine) transporter. Loss of IRS1/IRS2 in A549 DKO cells resulted in a significant decrease in the protein levels of SLC7A5 and SLC3A2, and a concomitant increase in SLC7A1 levels (Fig. S5B). However, despite similarly affecting amino acid uptake (Fig. 3C and Fig. S5A), or cellular proliferation (Fig. 3B and Fig. S4D), silencing of IRS1/IRS2 in A549 DKD or Calu-1 DKD cells did not lead to similar or consistent changes, indicating a potential effect on other transporters and/or regulation at the activity level. This is consistent with previous reports indicating growth factor regulation of amino acid transport systems in variety of cell types, at both the expression and activity levels (24–27).
Acute Loss of IRS1 and IRS2 Promotes Autophagy in Mouse and Human KRAS-Mutant Lung Cancer Cells.
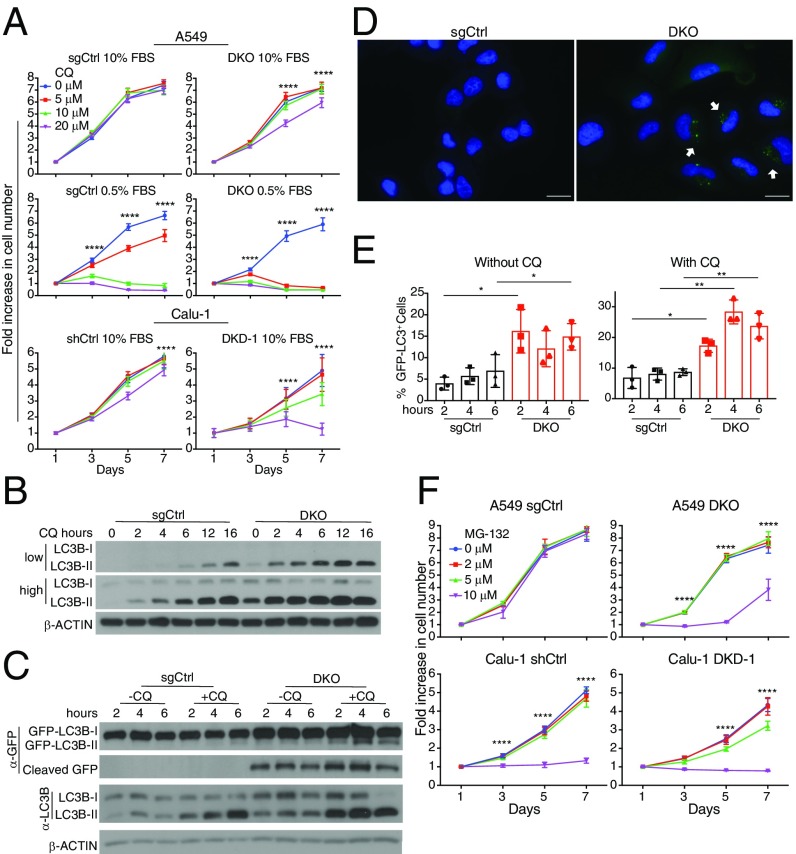
Because amino acid levels were significantly decreased upon IRS1/IRS2 loss, we investigated a potential effect on autophagy, a self-catabolic process induced by nutrient starvation and decreased mTORC1 activity (28). We found that, compared with control cells, both A549 DKO and Calu-1 DKD cells are more sensitive to chloroquine (CQ) treatment, indicating that loss of IRS1/IRS2 sensitizes NSCLC cells to autophagy inhibition (Fig. 4A). Indeed, A549 DKO cells demonstrated higher basal autophagic flux than control cells, as evidenced by increased levels and enhanced accumulation, in the presence of full media, of the autophagosome marker LC3-II upon CQ treatment (Fig. 4B). Moreover, upon GFP-LC3 overexpression, these cells demonstrated significantly higher levels of GFP cleavage independent of CQ treatment (Fig. 4C). Consistently, compared with control cells, a higher percentage of A549 DKO cells displayed GFP-LC3 punctae, resulting from enhanced autophagosome formation (Fig. 4 D and E). These results imply that NSCLC cells promote intracellular protein catabolic pathways to compensate for the sharp decrease in amino acid levels upon IRS1/IRS2 loss. Indeed, compared with their respective control cells expressing IRS1 and IRS2, both A549 DKO and Calu-1 DKD cells were more sensitive to the proteasome inhibitor MG-132 (Fig. 4F).
Fig. 4.
Acute loss of IRS1 and IRS2 induces autophagy in human KRAS-mutant NSCLC cells. (A) Proliferation curves of NSCLC A549 DKO or sgCtrl cells as well as Calu-1 DKD or shCtrl cells cultured in 10% serum and treated with chloroquine (CQ); n = 6; ****P < 0.0001 between 0 and 20 μM conditions. (B) Levels of LC3B-I and LC3B-II in A549 cells described in A treated with 10 μM CQ for 0–16 h. Compared with sgCtrl cells, A549 DKO cells have enhanced LC3B-II accumulation, indicating enhanced autophagic flux. (C) Increased accumulation with or without CQ treatment, of endogenous LC3B-II as well as GFP cleavage from exogenously expressed GFP-LC3B in A549 DKO compared with A549 sgCtrl cells. Cells were cultured in the presence of 10% serum with or without 10 μM CQ for 2, 4, or 6 h. In B and C, β-ACTIN was used as a loading control. (D) Fluorescence microscopy images demonstrating increased GFP-LC3 punctae (white arrows) upon loss of IRS1/IRS2 in A549 DKO compared with sgCtrl cells described in C that were treated with 10 μM CQ for 6 h. (Scale bars, 25 μM.) (E) Quantification of GFP-LC3 punctae in cells from D; n = 15–20 images per condition; *P < 0.05; **P < 0.01. Data are pooled from three independent experiments. (F) Proliferation curves of A549 and Calu-1 cells described in A, which were grown in 10% serum and treated with MG-132 for 7 d; n = 6; ****P < 0.0001 between 0 and 10 μM conditions for A549 and between 0 and 5 μM as well as 0 and 10 μM for Calu-1. In A, E, and F, data represent the mean ± SD.
Inhibition of Insulin and IGF1 Signaling Hinders in Vivo NSCLC Growth and Sensitizes Tumors to Autophagy Inhibition.
To assess whether acute inhibition of insulin/IGF1 signaling can result in a similar metabolic phenotype, murine KP cells (Fig. 5 A and B) and KRAS-mutant human NSCLC cells (Fig. 5 C and D) were treated with the pharmacological IR/IGF1R inhibitor NVP-AEW541. The latter led to enhanced basal autophagy detected by rapid accumulation of LC3B-II (Fig. 5 A and C). It also led to a significant decrease in intracellular amino acid levels (Fig. 5 B and D). These data indicate that acute suppression of insulin/IGF1 signaling decreases amino acid availability, generating an increased dependency on protein catabolic pathways to compensate for lower nutrient levels.
Fig. 5.
Acute inhibition of insulin/IGF1 signaling in KRAS-mutant lung cancer cells leads to in vivo growth suppression and dependence on autophagy. (A and C) Levels of total and phosphorylated IGF1R and IR, total and phosphorylated AKT, as well as LC3B-I and LC3B-II in murine Kras-mutant, p53-null lung cancer (KP) cells (A) or NSCLC A549 and Calu-1 cells (C) grown in 10% serum and treated with 2 μM NVP-AEW541 for 0, 4, or 24 h; β-ACTIN was used as a loading control. (B and D) Levels of amino acids in KP cells (B) or Calu-1 cells (D) cultured in the presence of 10% serum with or without 2 μM NVP-AEW541 for 24 h. Metabolite levels were first normalized to protein levels, and then to the median of all samples for each amino acid; n = 4 biological replicates per condition. (E) Proliferation curves of Calu-1 shCtrl and DKD cells, which were grown in 10% serum and treated with either vehicle control (DMSO), CQ, NVP-AEW541, or both drugs for 7 d; n = 4. Data represent the mean ± SEM. ****P < 0.0001 between single compound treatment and combinatorial treatment conditions; ns, nonsignificant. (F) Growth of individual s.c. xenograft tumors derived from A549 cells (Top) over 67 d (one mouse had to be killed at 51 d) and Calu-1 (Bottom) over 84 d (Left) with endpoint average tumor volume (Right); n = 12 (A549 sgCtrl); n = 11 (A549 DKO); n = 10 (Calu-1 shCtrl); n = 12 (Calu-1 DKD-1). (G) Percent growth of s.c. xenograft Calu-1 tumors with control or IRS1/IRS2 knockdown in mice treated with vehicle control (PBS) or chloroquine (CQ) (60 mg/kg), 5 d per week for 32 d after median tumor volume reached ∼150 mm3; n = 8 (shCtrl PBS); n = 10–12 (shCtrl CQ); n = 10 (DKD-1 PBS); n = 10 (DKD-1 CQ). In B, D, F, Right, and G, data represent the mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
To validate the effect of combinatorial inhibition of IR/IGF1R and autophagy, we treated NSCLC cells with either NVP-AEW541, chloroquine, or a combination of both. Although neither drug affected the proliferation of Calu-1 control cells when administered alone at the indicated doses, simultaneous addition of both compounds significantly suppressed their cellular growth (Fig. 5E, Left). As expected, silencing of IRS1/IRS2 also sensitized Calu-1 DKD cells to chloroquine treatment. Importantly, however, treatment with NVP-AEW541 either alone or in combination with chloroquine did not result in any additional, nonspecific effects upon IRS1/IRS2 knockdown (Fig. 5E, Right). Similar results were observed in A549 cells (Fig. S4E), although the combinatorial treatment of A549 DKD cells using the higher dose (10 μM) but not lower dose (5 μM) of chloroquine resulted in nonspecific toxic effects, indicating that the combinatorial treatment needs to be carefully titrated for different NSCLC cells.
Interestingly, the murine KPI cells, which overcame chronic rather than acute Irs1/Irs2 loss in vivo following prolonged tumor latency (Fig. 1), did not display enhanced autophagic flux nor did they exhibit enhanced sensitivity to CQ treatment (Fig. S6 A and B). This implies that the KPI cells have adapted alternative mechanisms to overcome the metabolic and growth impairment resulting from chronic loss of Irs1 and Irs2. Indeed, RPPA results revealed that three of four KPI cells had enhanced activation of growth factor receptors other than IR/IGF1R (Fig. S6C). In particular, increased phosphorylation of epidermal growth factor (Egfr) on Y1173 and Y1068 was found in both KPI-3 and KPI-5 cells. Moreover, KPI-6 cell line displayed significantly increased phospho-Y1289 Her3. On the other hand, KPI-5 cells exhibited increased levels of platelet-derived growth factor-β (Pdgf-β). Up-regulation or activation of these alternative receptor tyrosine kinases may have enabled KPI cells to overcome the loss of Irs1 and Irs2 and the resulting suppression of lung tumor growth.
Recently, Kras-driven lung cancer was shown to be dependent on BCAA metabolism for nucleotide synthesis and in vivo tumor growth (29). Consistently, we found that loss of IRS1/IRS2, which results in decreased intracellular levels of essential amino acids, significantly hinders the ability of NSCLC cells to form tumors in vivo. Indeed, whereas all A549 control cells formed xenograft tumors over a period of 10 wk, only 3 of 11 A549 DKO cell injections yielded tumors that, however, grew to a significantly lesser extent than controls (Fig. 5F). Similarly, the growth of all Calu-1 xenograft tumors was mitigated upon IRS1/IRS2 knockdown (Fig. 5F). Importantly, IRS1/IRS2 silencing sensitized NSCLC preformed tumors to autophagy inhibition, as demonstrated by significantly decreased growth of Calu-1 DKD tumors following chloroquine treatment, compared with vehicle control. In contrast, no significant changes were observed in tumor growth upon chloroquine treatment of mice bearing control knockdown Calu-1 tumors (Fig. 5G). Altogether, these results indicate that loss or silencing of IRS1/IRS2 suppresses in vivo growth of KRAS-mutant NSCLC and leads to enhanced sensitivity to autophagy inhibition.
To evaluate the respective roles of IR and IGF1R in KRAS-driven NSCLC cells, we knocked down each receptor individually in A549 and Calu-1 cells and assessed the knockdown effect on downstream insulin/IGF1 signaling and cellular proliferation. We found that, compared with IR, IGF1R is indeed a dominant receptor in these cancer cells, as its silencing resulted in a more profound suppression of AKT activation (Fig. S7 A and B). Consistently, whereas IR knockdown did not affect the in vitro proliferation of A549 or Calu-1 cells, that of IGF1R resulted in significant suppression of cellular growth (Fig. S7 C and D). These data support the translational relevance and therapeutic potential of using specific IGF1R inhibitors in combination with autophagy/proteasome inhibitors to target NSCLC.
Discussion
Previous work demonstrated that KRAS can bind to and activate PI3K p110α, and that this interaction is required for KRAS-driven transformation. Disrupting the Kras–p110α interaction in genetically engineered mouse models of lung cancer suppresses tumor initiation and causes partial regression of established tumors (6, 7). However, despite its requirement, the sufficiency of KRAS–p110α interaction in driving KRAS-driven lung tumor formation, and the need for additional input from growth factor receptors upstream of KRAS and PI3K has remained largely controversial (9). Here, using a conditional genetically engineered mouse model of lung cancer with Irs1/Irs2 loss, we provide robust evidence that insulin/IGF1 signaling is required for Kras-driven lung tumor initiation. We show that concomitant expression of Kras oncogene and the triple loss of p53, Irs1, and Irs2 specifically in lung cells, strongly suppresses Kras-driven lung tumor formation. Moreover, we show that Kras-transformed lung cells can eventually overcome this suppression, albeit at a stochastic rate and with extended latency.
It is noteworthy that genetic loss of Irs1 alone in a similar Kras-driven mouse model of lung cancer, which however expresses wild-type p53, resulted in increased rather than decreased tumor burden and reduced survival (18). This seemingly contradictory result is however not surprising, given that the tumor cells had retained wild-type Irs2 expression. Although the report did not mention or discuss Irs2 expression, we find that Irs2 protein levels are significant in both mouse and human Kras-mutant lung cancer cells, and that knockdown of Irs1 either does not affect or rather causes a compensatory increase in the expression of Irs2. As a result, Akt activation remains intact in Irs1 knockdown cells, in response to insulin/IGF1 stimulation (Figs. S2 and S3). Thus, dual genetic ablation of Irs1 and Irs2, but not Irs1 alone, is required for suppression of Akt activation upon ligand stimulation and suppression of tumor growth.
We find that the late-arising, higher-grade, Kras-driven tumors that overcome Irs1/Irs2 loss have decreased E-cadherin levels, a feature of enhanced invasiveness, and a recognized marker of an epithelial-to-mesenchymal transition (EMT) (23). These cells also display significantly decreased intracellular levels of essential amino acids, likely due to decreased amino acid uptake resulting from suppressed growth factor signaling (24–27). Interestingly, in a recent report, EMT was shown to induce autophagy in breast cancer stem cell populations, endowing them with an enhanced ability to escape immune surveillance (30). Thus, a link between EMT, tumor invasiveness, and induction of autophagy deserves further investigation in the KRAS-driven NSCLC tumors with suppressed insulin/IGF1 signaling.
Whereas preclinical studies indicated a role for insulin/IGF1 signaling in lung tumor growth, clinical trials showed adverse effects of systemic IGF1R inhibition in unselected patients (8, 31), leaving questionable the relevance of therapeutic targeting of IR/IGF1R in NSCLC patients. Our study highlights the translational relevance of blocking IR/IGF1R signaling specifically in the lungs, leading to strong suppression of tumor formation. Furthermore, it provides a metabolic link between IR/IGF1R signaling and amino acid utilization, as inhibition of such signaling results in decreased intracellular amino acid levels, generating a metabolic dependency of KRAS-driven lung tumors on protein catabolic pathways. Consequently, combinatorial targeting of IGF1R and either autophagy or the proteasome may represent a valuable therapeutic strategy in treating KRAS-mutant NSCLC.
Materials and Methods
Mouse Studies.
Animal studies were approved by the Institutional Animal Care and Use Committee at Boston Children’s Hospital. Breeding and adenoviral-Cre infection of KP and KPI mice, xenograft studies, as well as tumor burden quantification and grading are described in detail in SI Materials and Methods.
Cell Culture Studies.
Tumor dissociation, cell culture, genotyping, imaging of GFP-LC3, immunoblotting and Luminex assay, RPPA protocol, and metabolite quantification are described in detail in SI Materials and Methods.
Statistical Analyses.
Detailed statistical analyses for both in vitro and in vivo data are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the N.Y.K. and M.F.W. laboratory members for thoughtful comments and discussions. This research was supported by Boston Children’s Hospital (H.X., M.-S.L., P.-Y.T., A.S.A., N.L.C., S.C., and N.Y.K.) and NIH/National Cancer Institute Grant R01 CA211944 (to N.Y.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718414115/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization 2017. Cancer (WHO, Geneva), Fact Sheet No. 297.
- 3.Project CG. 2017. Catalogue of Somatic Mutations in Cancer (COSMIC) (Wellcome Trust Sanger Institute, Hinxton, UK), Version 80.
- 4.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Castellano E, et al. Requirement for interaction of PI3-kinase p110α with RAS in lung tumor maintenance. Cancer Cell. 2013;24:617–630. doi: 10.1016/j.ccr.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 8.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: An update. Nat Rev Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 9.Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov. 2013;3:548–563. doi: 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White MF, Maron R, Kahn CR. Insulin rapidly stimulates tyrosine phosphorylation of a Mr-185,000 protein in intact cells. Nature. 1985;318:183–186. doi: 10.1038/318183a0. [DOI] [PubMed] [Google Scholar]
- 11.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 12.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 14.Oliver TG, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong X, et al. Irs1 and Irs2 signaling is essential for hepatic glucose homeostasis and systemic growth. J Clin Invest. 2006;116:101–114. doi: 10.1172/JCI25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curry NL, et al. Pten-null tumors cohabiting the same lung display differential AKT activation and sensitivity to dietary restriction. Cancer Discov. 2013;3:908–921. doi: 10.1158/2159-8290.CD-12-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Copps KD, Hançer NJ, Qiu W, White MF. Serine 302 phosphorylation of mouse insulin receptor substrate 1 (IRS1) is dispensable for normal insulin signaling and feedback regulation by hepatic S6 kinase. J Biol Chem. 2016;291:8602–8617. doi: 10.1074/jbc.M116.714915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metz HE, et al. Insulin receptor substrate-1 deficiency drives a proinflammatory phenotype in KRAS mutant lung adenocarcinoma. Proc Natl Acad Sci USA. 2016;113:8795–8800. doi: 10.1073/pnas.1601989113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldser DM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468:572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennessy BT, et al. A technical assessment of the utility of reverse phase protein arrays for the study of the functional proteome in non-microdissected human breast cancers. Clin Proteomics. 2010;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: How does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 24.Boerner P, Resnick RJ, Racker E. Stimulation of glycolysis and amino acid uptake in NRK-49F cells by transforming growth factor beta and epidermal growth factor. Proc Natl Acad Sci USA. 1985;82:1350–1353. doi: 10.1073/pnas.82.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duclos MJ, Chevalier B, Goddard C, Simon J. Regulation of amino acid transport and protein metabolism in myotubes derived from chicken muscle satellite cells by insulin-like growth factor-I. J Cell Physiol. 1993;157:650–657. doi: 10.1002/jcp.1041570327. [DOI] [PubMed] [Google Scholar]
- 26.Durante W, Liao L, Iftikhar I, Cheng K, Schafer AI. Platelet-derived growth factor regulates vascular smooth muscle cell proliferation by inducing cationic amino acid transporter gene expression. J Biol Chem. 1996;271:11838–11843. doi: 10.1074/jbc.271.20.11838. [DOI] [PubMed] [Google Scholar]
- 27.Obata T, et al. Insulin signaling and its regulation of system A amino acid uptake in cultured rat vascular smooth muscle cells. Circ Res. 1996;79:1167–1176. doi: 10.1161/01.res.79.6.1167. [DOI] [PubMed] [Google Scholar]
- 28.Perera RM, Zoncu R. The lysosome as a regulatory hub. Annu Rev Cell Dev Biol. 2016;32:223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayers JR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akalay I, et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013;73:2418–2427. doi: 10.1158/0008-5472.CAN-12-2432. [DOI] [PubMed] [Google Scholar]
- 31.Fidler MJ, Shersher DD, Borgia JA, Bonomi P. Targeting the insulin-like growth factor receptor pathway in lung cancer: Problems and pitfalls. Ther Adv Med Oncol. 2012;4:51–60. doi: 10.1177/1758834011427576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.