Significance
Osteoporosis and sarcopenia are common and costly comorbid diseases of aging, and there is an urgent need to prevent and treat both to reduce morbidity and mortality. We found that, in humans, genetic variation near FAM210A, a gene of previously unknown function, was strongly associated with both appendicular and whole body lean mass, as well as bone mineral density. In mice, Fam210a was expressed in muscle mitochondria and cytoplasm but not in bone. Nevertheless, in genetically modified mouse models, Fam210a strongly influenced the structure and strength of both muscle and bone. These studies therefore disclose a novel pathway whose further study may lead to the development of new treatments for both osteoporosis and sarcopenia.
Keywords: FAM210A, bone, muscle, osteoporosis, sarcopenia
Abstract
Osteoporosis and sarcopenia are common comorbid diseases, yet their shared mechanisms are largely unknown. We found that genetic variation near FAM210A was associated, through large genome-wide association studies, with fracture, bone mineral density (BMD), and appendicular and whole body lean mass, in humans. In mice, Fam210a was expressed in muscle mitochondria and cytoplasm, as well as in heart and brain, but not in bone. Grip strength and limb lean mass were reduced in tamoxifen-inducible Fam210a homozygous global knockout mice (TFam210a−/−), and in tamoxifen-inducible Fam210 skeletal muscle cell-specific knockout mice (TFam210aMus−/−). Decreased BMD, bone biomechanical strength, and bone formation, and elevated osteoclast activity with microarchitectural deterioration of trabecular and cortical bones, were observed in TFam210a−/− mice. BMD of male TFam210aMus−/− mice was also reduced, and osteoclast numbers and surface in TFam210aMus−/− mice increased. Microarray analysis of muscle cells from TFam210aMus−/− mice identified candidate musculoskeletal modulators. FAM210A, a novel gene, therefore has a crucial role in regulating bone structure and function, and may impact osteoporosis through a biological pathway involving muscle as well as through other mechanisms.
Osteoporosis is a systemic skeletal disease characterized by increased bone fragility due to a decrease in bone quantity and/or impaired bone quality, which leads to an increase in fracture risk (1). Osteoporotic fractures in the elderly have been associated with reduced quality of life and increased mortality (2). The proportion of the population at risk for osteoporotic fractures has increased with the aging of the population. Sarcopenia is also an age-related disorder characterized by a progressive and generalized decrease in muscle mass and muscle strength leading to physical disability, reduced quality of life, and death (3). In the elderly over age 80, more than 30% may manifest osteoporosis and/or sarcopenia (4). Sarcopenia also leads to frailty, which in turn is a major cause of institutionalization of the elderly (5). Consequently, to reduce morbidity and mortality, there is an urgent need to prevent and treat both osteoporosis and sarcopenia.
It has been proposed that bones adapt their strength to maintain the strain caused by physiological loads close to a set point (6–8). Because the largest physiological loads are caused by skeletal muscle contractions, there should therefore be a close relationship between bone mineral density (BMD) and skeletal muscle mass and strength. Previous studies have shown that muscle tissue secretes various hormones called myokines, which affect bone tissues (9, 10). Conversely, osteokines, hormones secreted from bone, can affect muscle tissues (9, 10). Consequently, there is clearly biochemical cross-talk between muscle and bone (11). Furthermore, genes from multiple known pathways have been suggested as candidates for pleiotropic regulation of bone and muscle (12). Recently, variants with pleiotropic effects in seven established BMD loci were reported in children; variants near the TOM1L2/SREBF1 locus were found to exert opposing effects on total body lean mass and total body less head BMD (13). SREBP-1, and the product of the SREBF1 gene, is known to exert opposing effects on osteoblast and myoblast differentiation (14, 15). However, more commonly, bone loss coincides with a decrease in muscle mass and function, suggesting that there are shared biological mechanisms leading to both osteoporosis and sarcopenia.
In the largest genome-wide association studies (GWAS) that have identified genetic variants associated with a reduction of BMD, or increased osteoporotic fracture risk or both (16–18), six novel single-nucleotide polymorphisms (SNPs) involving rs4796995 were discovered to be strongly associated with the risk of fracture, but less so with BMD reduction. These SNPs mapped near the gene, family with sequence similarity 210, member A (FAM210A) [also called chromosome 18 ORF 19 (C18orf19)]. Since most fracture-associated genes predominantly affect BMD (17), this suggested that FAM210A might influence fracture through mechanisms partly independent of BMD. While GWAS have identified many novel loci for traits such as BMD, with few exceptions, the causal genes at these loci have rarely been elucidated (17, 19, 20). FAM210A is predicted to encode as many as four different-sized transcripts, ranging from an intracellular protein of 67 amino acids to mitochondrial and membrane proteins up to 272 amino acids, with the latter containing a common single-pass transmembrane region (ref. 21; www.proteinatlas.org). However, the protein is not predicted to contain a signal peptide to facilitate secretion. Although high mRNA expression in muscle, as well as other tissues, has been noted, the function of FAM210A remains unknown.
We therefore hypothesized that FAM210A, may play an important role in modulating both muscle and bone biology. Consequently, we examined whether this novel gene is associated with parameters of bone and skeletal muscle structure and function in humans and in genetically modified mice.
Results
Association Between SNPs Mapping to FAM210A and Lean Mass as Well as BMD in Humans.
We first examined whether SNPs mapping to FAM210A were associated with bone and muscle traits in adults and children using the largest GWAS to date. We observed that, for the genomic region of FAM210A and the surrounding 500 kilobases upstream and downstream (chr18: 13,163,346–14,226,591, build hg19), rs1941749 was significantly associated with BMD estimated from the heel calcaneous (eBMD) in 142,487 adults (P = 3.5 × 10−43; Table 1) and rs4796995 was significantly associated with fracture assessed in 31,016 cases and 102,444 controls (P = 8.8 × 10−13; Table 1). Interestingly, nearby SNPs, rs9955264 and rs1284201, were associated with appendicular lean mass (P = 5.2 × 10−3, n = 28,117) and whole body lean mass (P = 1.4 × 10−2, n = 38,318), respectively, in adults, after accounting for multiple testing in the region (Table 1). Further, rs9303793 was associated in a bivariate analysis of BMD and lean mass in 10,414 children (P = 1.1 × 10−3; Table 1) again surpassing multiple testing correction. Taken together, these findings indicate that genetic variation near FAM210A in humans is associated with BMD, fracture, and measures of sarcopenia. We therefore studied the expression of Fam210a and its effect on bone and muscle biology in genetically modified mouse models.
Table 1.
Summary of human genetic association data with bone and muscle traits at the FAM210A genomic region
| Trait | Lead SNP | EA | NEA | EAF | Effect estimate | SE | P | Alpha | N |
| Fracture | rs4796995 | G | A | 0.390 | 1.08* | 0.030 | 8.8 × 10−13 | N/A | 31,016 cases, 102,444 controls |
| Estimated BMD | rs1941749 | G | A | 0.359 | −0.048 | 0.004 | 3.5 × 10−43 | 2.5 × 10−2 | 142,487 |
| Appendicular lean mass | rs9955264 | A | G | 0.232 | 0.063 | 0.023 | 5.2 × 10−3 | 2.5 × 10−2 | 28,117 |
| Whole body lean mass | rs1284201 | C | T | 0.143 | 0.111 | 0.045 | 1.4 × 10−2 | 2.5 × 10−2 | 38,318 |
| Pediatric BMD and lean mass† | rs9303793 | T | G | 0.114 | 0.020 | 0.021 | 1.1 × 10−3 | 2.5 × 10−2 | 10,414 |
| 0.069 | 0.021 |
Alpha was determined by dividing P = 0.05 by the number of independent SNPs tested across the locus for each trait (see Materials and Methods). EA, effect allele; EAF, effect allele frequency; NEA, noneffect allele.
Effect on fracture reported as an odds ratio.
Effect estimates and SEs for (Top) pediatric BMD and (Bottom) pediatric lean mass reported.
Expression of Fam210a in Mouse Organs, Tissues, and Cells.
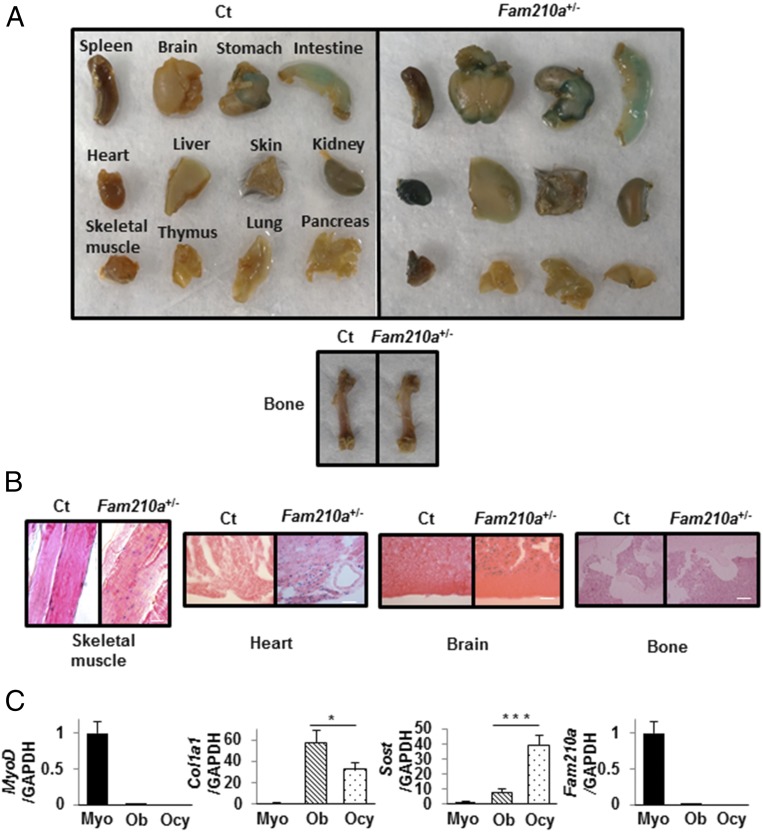
To clarify the expression of Fam210a, we performed histochemical X-Gal staining to detect β-galactosidase activity in organs and tissues of Fam210a heterozygous global knockout mice (Fam210a+/−) expressing a lacZ reporter under the control of the Fam210a promoter. Dark blue X-Gal staining reflecting Fam210a expression was observed in a variety of organs, including skeletal muscle, heart, and brain (Fig. 1A). In contrast, there was no X-Gal staining in bone (Fig. 1A). By microscopy, skeletal muscle cells, cardiac muscle cells, and nerve cells in white matter all stained strongly in Fam210a+/− mice, whereas staining was absent in control (Ct) mice (Fam210a WT mice with CMV-Cre) (Fig. 1B). There was again no X-Gal staining observed in bone (Fig. 1B). Both Fam210a and MyoD were expressed by quantitative RT-PCR in primary myoblasts isolated from WT mice (Fig. 1C); nevertheless, Fam210a expression was not detectable in primary osteoblasts or in primary osteocytes (Fig. 1C), although the osteoblasts were strongly positive for the marker Col1a1, and the osteocytes were strongly positive for the marker Sost (Fig. 1C).
Fig. 1.
X-Gal staining in mouse organs and tissues in Ct mice and in Fam210a+/− mice expressing the lacZ reporter under the control of the Fam210a promoter, and quantitative RT-PCR of RNA from primary myoblasts, osteoblasts, and osteocytes of WT mice. (A) X-Gal staining (blue) showing expression of Fam210a in diverse organs in Ct mice (Fam210a WT mice with CMV-Cre) (Left) and in Fam210a+/− mice expressing the lacZ reporter (Right). (B) Microscopy of X-Gal staining in tissues of Ct mice (Left) and Fam210a+/− mice expressing the lacZ reporter (Right). Scale bars represent 50 μm for skeletal muscle, heart, and brain, and 200 μm for bone. No blue staining reaction product was observed in Ct tissues. The highest levels of staining in Fam210a+/− mice were seen in skeletal muscle, heart, and brain; no reaction product was seen in bone. (C) Quantitative RT-PCR to assess the expression of MyoD, Col1a1 (encoding collagen type 1 alpha 1 chain), Sost (encoding sclerostin), and Fam210a in primary myoblasts (Myo), osteoblasts (Ob), and osteocytes (Ocy) from WT mice. Data are expressed relative to the GAPDH mRNA value. Data represent mean ± SEM of 5–6 determinations. *P < 0.05; ***P < 0.001.
Phenotypic Characterization of Fam210a Heterozygous Knockout Mice.
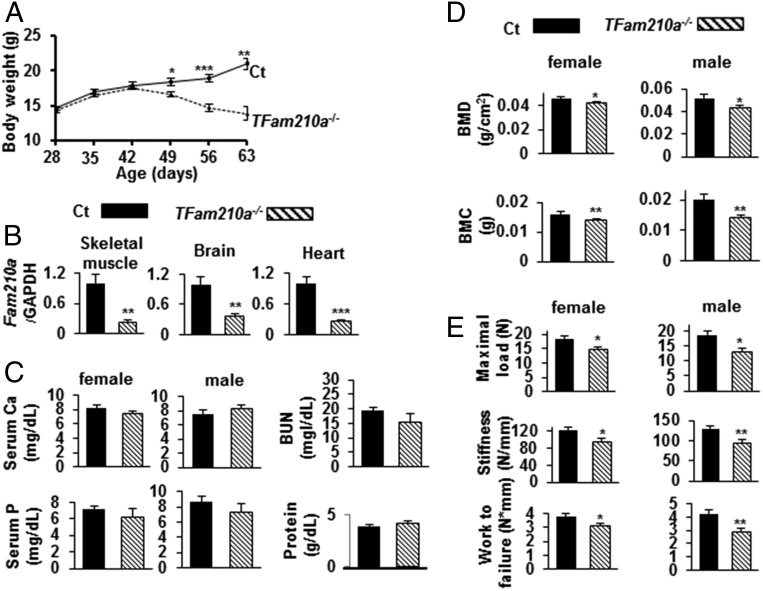
As shown in Fig. 2A, there was no difference in levels of serum calcium as well as phosphate between Ct and Fam210a+/− mice at 56 d of age, and, using ELISA, the level of Fam210a protein in mouse serum of both Fam210a+/− and Ct littermates was undetectable. In contrast, Fam210a protein was clearly measurable in Fam210a+/− mouse muscle extract and the level was ∼54% of that in Ct mouse muscle extract (Fig. 2A). Both BMD and bone mineral content (BMC) values of Fam210a+/− mice tended to be lower than those of Ct mice (Fig. 2B), but these differences were not significant (female: P = 0.060, and P = 0.102, respectively; male: P = 0.096, and P = 0.076). However, three-point bending tests, to assess biomechanical bone strength, showed that the maximal load and stiffness of Fam210a+/− mouse bones were significantly lower than those of Ct mouse bones in both females and males (Fig. 2C). Moreover, work-to-failure of male Fam210a+/− mouse bones was significantly lower than those of Ct mouse bones (Fig. 2C). We then investigated parameters of trabecular and cortical bone microarchitecture using microcomputed tomography (μCT). As shown in Fig. 2D and Table 2, cortical bone volume/tissue volume (BV/TV) and cortical thickness (Co.Th.) of Fam210a+/− mouse bones were significantly lower than those of Ct mouse bones, and cortical porosity (Po.V) of Fam210a+/− mouse bones was significantly greater than that of Ct mouse bones. In contrast, there was no difference in parameters of trabecular microarchitecture between Ct and Fam210a+/− mouse bones (Fig. 2D and Table 2). We next determined whether bone formation and/or resorption were altered. As shown in Fig. 2E, the mineral apposition rate (MAR) and bone formation rate (BFR) of Fam210a+/− mouse cortical bones were significantly lower than those of Ct mouse cortical bones. In addition, the osteoclast number relative to bone surface (Oc.N/BS) of Fam210a+/− mouse bones was significantly higher than that of Ct mouse trabecular and cortical bones, and the osteoclast surface relative to bone surface (Oc.S/BS) of Fam210a+/− mouse cortical bones was significantly higher than that of Ct mouse cortical bones (Fig. 2F). Finally, we assessed parameters of muscle function and structure. Grip strength and lean mass of all limbs of Fam210a+/− mice were significantly lower than those of Ct mice (Fig. 2G).
Fig. 2.
Phenotypic characterization of Fam210a+/− mice. (A) Serum calcium (Ca) and phosphorus (P) in Ct (female, n = 9; male, n = 7) and Fam210a+/− (female, n = 8; male; n = 8) mice, and the level of Fam210a protein in serum and muscle extracts of Ct (n = 5) and Fam210a+/− (n = 5) mice using ELISA. Undetectable levels were less than 0.3 ng/mL. (B) BMD and BMC of Ct (female, n = 9; male, n = 9) and Fam210a+/− (female, n = 10; male, n = 9) mouse bones. (C) Parameters of biomechanical bone strength, i.e., the maximal load, stiffness, and work-to-failure, measured by three-point bending tests, in Ct (female, n = 14; male, n = 11) and Fam210a+/− (female, n = 11; male, n = 12) mouse femurs. (D) Representative 3D reconstructions of μCT images of trabecular and cortical bone in Ct (n = 3) and Fam210a+/− (n = 3) mouse femurs. (E) Representative micrographs of double calcein-labeled sections of trabecular and cortical bone of Ct and Fam210a+/− mice; MAR and BFR in trabecular and cortical bone of Ct (n = 4) and Fam210a+/− (n = 4) mice calculated using calcein double labeling. (F) Oc.N/BS and Oc.S/BS in trabecular and cortical bone of Ct (trabecular bone, n = 8; cortical bone, n = 10) and Fam210a+/− (trabecular bone, n = 8; cortical bone, n = 9) mice using TRAP staining. (G) Grip strength in Ct (female, n = 5; male, n = 5) and Fam210a+/− (female, n = 5; male, n = 5) mice; lean mass of all limbs in Ct (female; n = 4, male; n = 4) and Fam210a+/− (female, n = 5; male, n = 4). Solid bars indicate values in Ct, and hatched bars indicate values in Fam210a+/− mice. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 using a two-sided Student’s t test. Analyses were performed at 56 d of age.
Table 2.
Parameters of trabecular and cortical bone calculated from micro-CT
| Bone parameter | Ct (CMV-Cre) ♀ | Fam210a+/− ♀ | Ct (Fam210aflox/flox) ♂ | TFam210a−/− ♂ | Ct (Fam210aflox/flox) ♀ | TFam210aMus−/− ♀ |
| Trabecular bone | ||||||
| BV/TV, % | 6.251 ± 0.640 | 5.198 ± 1.152 | 24.335 ± 0.732 | 15.975 ± 2.162* | 12.924 ± 2.979 | 7.897 ± 1.222 |
| Tb.Th, mm | 0.061 ± 0.001 | 0.059 ± 0.001 | 0.057 ± 0.002 | 0.058 ± 0.001 | 0.063 ± 0.002 | 0.059 ± 0.002 |
| Tb.N, 1/mm | 1.024 ± 0.099 | 0.935 ± 0.204 | 4.280 ± 0.168 | 2.804 ± 0.304** | 2.026 ± 0.416 | 1.360 ± 0.248 |
| Tb.Sp, mm | 0.302 ± 0.018 | 0.383 ± 0.089 | 0.257 ± 0.005 | 0.182 ± 0.006* | 0.231 ± 0.019 | 0.264 ± 0.027 |
| SM1 | 2.488 ± 0.036 | 2.476 ± 0.059 | 1.361 ± 0.072 | 1.717 ± 0.104* | 2.041 ± 0.194 | 2.376 ± 0.122 |
| Cortical bone | ||||||
| BV/TV, % | 42.320 ± 0.486 | 39.847 ± 0.285* | 48.492 ± 0.469 | 42.093 ± 0.659*** | 52.281 ± 2.722 | 47.709 ± 1.606 |
| Co.Th., mm | 0.166 ± 0.001 | 0.150 ± 0.004* | 0.211 ± 0.011 | 0.179 ± 0.005* | 0.204 ± 0.006 | 0.192 ± 0.003 |
| Po.V, % | 1.67 × 10−6 ± 3.33 × 10−6 | 3.33 × 10−5 ± 3.33 × 10−6* | 1.02 × 10−5 ± 1.38 × 10−6 | 3.00 × 10−5 ± 1.29 × 10−6** | 1.78 × 10−5 ± 2.87 × 10−6 | 2.48 × 10−5 ± 7.51 × 10−6 |
Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 according to a two-sided Student’s t test. Ct mice used were all Fam210a+/+, but, depending on the genotype of the experimental model, a different control was used. Thus, CMV-Cre mice were used as the Ct for the Fam210a+/−, and all were female aged 56 d; Fam210aflox/flox littermates (mice expressing allele Tm1c) were used as Ct for TFam210a−/−, and all were male aged 56 d; Fam210aflox/flox littermates (mice expressing allele Tm1c) were used as Ct for TFam210aMus−/−, and all were female aged 56 d. ♀ represents females; ♂ represents males.
Phenotypic Characterization of Tamoxifen-Inducible Fam210a Homozygous Knockout Mice.
In view of the fact that Fam210a homozygous global knockout (Fam210a−/−) mice died in utero shortly after embryonic day 9.5, we examined tamoxifen-inducible Fam210a homozygous global knockout (TFam210a−/−) mice, obtained by injection of tamoxifen at 28 d after birth for 5 d. Most TFam210a−/− mice died between 63 d and 70 d after birth. We therefore examined the phenotype of TFam210a−/− mice at 56 d after birth. As shown in Fig. 3A, body weight of TFam210a−/− mice was significantly lower than that of Ct mice at each time point from 49 d after birth. The expression level of Fam210a was significantly reduced (23 to 36% that of Ct mice) in muscle, brain, and heart of TFam210a−/− mice (Fig. 3B). There were no differences in the levels of serum calcium and phosphate between TFam210a−/− and Ct mice, or in the levels of urea nitrogen (BUN) and total protein (Fig. 3C). BMD and BMC of TFam210a−/− mice were significantly lower than those of Ct mice (Fig. 3D). By three-point bending test, the maximal load, stiffness, and work-to-failure of TFam210a−/− mouse bones were significantly lower than those of Ct mouse bones (Fig. 3E). As shown in Fig. 4A and Table 2, trabecular BV/TV and trabecular number (Tb.N) of TFam210a−/− mice were significantly lower than those of Ct mice, and trabecular separation (Tb.Sp) and structure model index (SMI) (trabecular rods relative to plates) of Fam210a−/− mice were significantly higher than those of Ct mice by μCT. Moreover, cortical BV/TV and Co.Th. of Fam210a−/− mice were significantly lower than those of Ct mice, and Po.V of TFam210a−/− mice was significantly greater (Fig. 4A and Table 2). By calcein double labeling, MAR and BFR of TFam210a−/− mouse trabecular and cortical bone were significantly lower than those of Ct mouse bone (Fig. 4B). In contrast, Oc.N/BS and Oc.S/BS of TFam210a−/− mouse trabecular and cortical bone were significantly increased (Fig. 4C). Finally, as shown in Fig. 4D, grip strength and lean mass of all limbs of TFam210a−/− mice were significantly lower than those of Ct mice.
Fig. 3.
Phenotypic characterization of TFam210a−/− mice. (A) Body weight of TFam210a−/− mice and of CT mice (Fam210aflox/flox littermates) (28 d, n = 6; 35 d, n = 6; 42 d, n = 6; 49 d, n = 6; 56 d, n = 6; 63 d, n = 3) and TFam210a−/− (28 d, n = 8; 35 d, n = 8; 42 d, n = 6; 49 d, n = 6; 56 d, n = 6; 63 d, n = 3) mice, after tamoxifen injection at 28 d after birth for 5 d. (B) Quantitative RT-PCR in mouse muscle, brain, and heart extracts to assess the expression of Fam210a in Ct (n = 5) and TFam210a−/− (n = 5) mice. Data are expressed relative to the GAPDH mRNA value. (C) Serum Ca and P, in Ct (calcium: female, n = 4; male, n = 5; phosphate: female, n = 5; male, n = 5) and TFam210a−/− (calcium: female, n = 5; male, n = 5; phosphate: female, n = 5; male, n = 5) mice. BUN and total protein in Ct and TFam210a−/− (mixture of one female and two male samples, i.e., n = 3 for each parameter in Ct and in TFam210a−/−). (D) BMD and BMC of Ct (female, n = 7; male, n = 8) and TFam210a−/− (female, n = 6; male, n = 9) mouse bones. (E) Biomechanical parameters of bone strength, by three-point bending tests in femurs, including the maximal load, stiffness, and work-to-failure, in Ct (maximal load: female, n = 4; male, n = 8; stiffness: female, n = 4; male, n = 6; work-to-failure: female, n = 5; male; n = 8) and TFam210a−/− (maximal load: female, n = 4; male; n = 8; stiffness: female, n = 4; male, n = 6; work-to-failure: female, n = 8; male; n = 8). Solid bars indicate values in Ct, and hatched bars indicate values in TFam210a−/− mice. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 using a two-sided Student’s t test. Analyses were done at 56 d of age.
Fig. 4.
Additional phenotypic characterization of TFam210a−/− mice. (A) Representative 3D reconstructions of μCT images of trabecular and cortical bone of Ct (n = 4) and TFam210a−/− (n = 4) mouse femurs. (B) Representative micrographs of double calcein-labeled sections of trabecular and cortical bone of Ct and TFam210a−/− mice; MAR and BFR in trabecular and cortical bone Ct (n = 4) and TFam210a−/− (n = 4) mice calculated using calcein double labeling. (C) Oc.N/BS and Oc.S/BS in trabecular and cortical bone of Ct (Oc.N/BS: trabecular bone, n = 5; cortical bone, n = 5; Oc.S/BS: trabecular bone, n = 5; cortical bone, n = 5) and TFam210a−/− (Oc.N/BS: trabecular bone, n = 4; cortical bone, n = 5; Oc.S/BS: trabecular bone, n = 5; cortical bone; n = 5) mice using TRAP staining. (D) Grip strength in Ct (female; n = 5, male; n = 5) and TFam210a−/−, (female; n = 4, male; n = 4) mice; lean mass of all limbs in Ct (female; n = 5, male; n = 3) and TFam210a−/− (female; n = 4, male; n = 4). Solid bars indicate values in Ct, and hatched bars indicate values in TFam210a−/− mice. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 using a two-sided Student’s t test.
Phenotypic Characterization of Tamoxifen-Inducible Fam210a Skeletal Muscle Cell-Specific Knockout Mice.
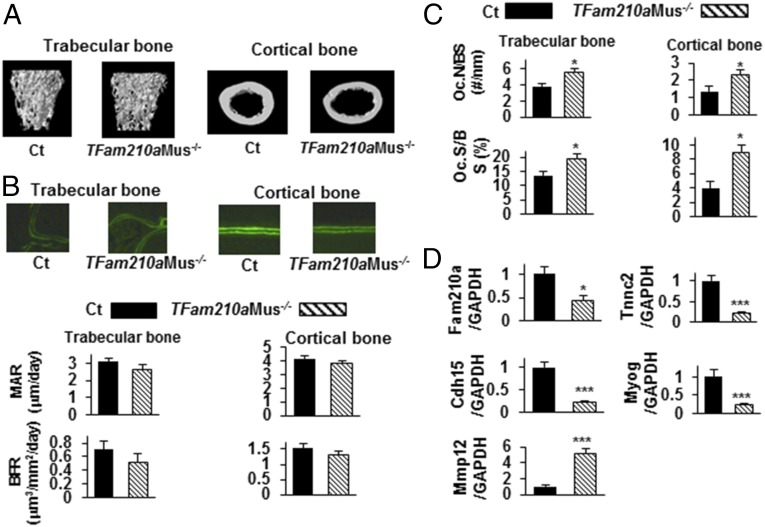
We first determined the localization of Fam210a in skeletal muscle cells. As shown in Fig. 5A, Fam210a protein was expressed in cytoplasm and mitochondria of primary myoblasts and myotubes from Ct mice (Fig. 5A). Unlike global TFam210a−/− mice, we found no difference in the body weight between Ct and tamoxifen-inducible Fam210a skeletal muscle cell-specific knockout (TFam210aMus−/−) mice, generated by tamoxifen injection at 28 d after birth for 5 d (Fig. 5B). We then examined the phenotype of TFam210aMus−/− mice at 56 d after birth as we had for TFam210a−/− mice. As shown in Fig. 5C, by RT-PCR, the expression of Fam210a mRNA in TFam210aMus−/− mouse muscles was 18% that of Ct mouse muscles. Grip strength and lean mass of all limbs of TFam210aMus−/− mice were significantly lower than those of Ct mice (Fig. 5D). BMD of male TFam210aMus−/− mice was significantly lower than that of Ct mice, and BMC of male TFam210aMus−/− mice tended to be lower than that of Ct mice (Fig. 5E). Although BMD and BMC of female TFam210aMus−/− mice tended to be lower than those of Ct mice, these differences were not significant (P = 0.059 and P = 0.054, respectively). Three-point bending tests showed no difference in the maximal load, stiffness, and work-to-failure between TFam210aMus−/− and Ct mouse bones (Fig. 5F). As shown in Fig. 6 and Table 2, in TFam210aMus−/− mouse bones, trabecular BV/TV, trabecular thickness (Tb.Th) and Tb.N tended to be lower, SMI tended to be higher, cortical BV/TV and Co.Th. tended to be lower, and MAR and BFR of trabecular and cortical bone tended to be lower than those of Ct mouse bone, but these parameters were not significantly different. However, Oc.N/BS and Oc.S/BS of TFam210aMus−/− mouse trabecular and cortical bone were significantly higher than those of Ct mouse bone (Fig. 6C).
Fig. 5.
Phenotypic characterization of TFam210aMus−/− mice. (A) Immunofluorescence was performed in primary myoblasts (Left) and myotubes (Right) from CT (Fam210a flox/flox) mice. Representative confocal microscopy images show localization of nuclei (DAPI; blue fluorescence), Fam210a (green fluorescence), and mitochondria (red fluorescence) or both Fam210a and mitochondria (yellow in merge panels). (B) Body weight of CT (Fam210a flox/flox littermates) (28 d; n = 9, 35 d; n = 9, 42 d; n = 9, 49 d; n = 9, 56 d; n = 9, 63 d; n = 9) and TFam210aMus−/− (28 d; n = 8, 35 d; n = 8, 42 d; n = 8, 49 d; n = 8, 56 d; n = 7, 63 d; n = 7) mice, following tamoxifen injection at 28 d after birth for 5 d. (C) Quantitative RT-PCR in mouse muscle to assess the expression of Fam210a in Ct (n = 5) and TFam210aMus−/− (n = 5) mice. Data are expressed relative to the GAPDH mRNA value. (D) Grip strength in Ct (female, n = 6; male, n = 9) and TFam210aMus−/− (female, n = 6; male, n = 5) mice; lean mass of all limbs in Ct (female, n = 5; male, n = 7), and TFam210aMus−/− (female, n = 4; male, n = 4) mice. (E) BMD and BMC of Ct (female, n = 12; male, n = 13) and TFam210aMus−/− (female, n = 13; male, n = 11) mouse bones. (F) Biomechanical parameters of bone strength, by three-point bending tests in femurs including the maximal load, stiffness, and work-to-failure, in Ct (maximal load: female, n = 6; male, n = 6; stiffness: female, n = 5; male; n = 6; work-to-failure: female, n = 6; male, n = 6) and TFam210aMus−/− (maximal load: female, n = 6; male, n = 5; stiffness: female, n = 5; male, n = 5; work-to-failure: female, n = 6; male; n = 5) mice. Data are expressed relative to the GAPDH mRNA value. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 using a two-sided Student’s t test.
Fig. 6.
Additional phenotypic characterization of TFam210aMus−/− mice. (A) Representative 3D reconstructions of μCT images of trabecular and cortical bone in Ct (n = 4) and TFam210aMus−/− (n = 4) mouse femurs. (B) Representative micrographs of double calcein-labeled sections of trabecular and cortical bone of Ct and TFam210aMus−/− mice; MAR and BFR in trabecular and cortical bone Ct (n = 4) and TFam210aMus−/− (n = 4) mice calculated using double calcein labeling. (C) Oc.N/BS and Oc.S/BS in trabecular and cortical bone of Ct (Oc.N/BS: trabecular bone, n = 5; cortical bone, n = 5; Oc.S/BS: trabecular bone, n = 5; cortical bone, n = 5) and TFam210aMus−/− (Oc.N/BS: trabecular bone, n = 5; cortical bone, n = 5; Oc.S/BS: trabecular bone, n = 5; cortical bone; n = 5) mice using TRAP staining. Solid bars indicate values in Ct, and hatched bars indicate values in TFam210aMus−/− mice. (D) RT-PCR to assess the expression of troponin c2, Tnnc2, Cdh15, Myog, and Mmp12 in primary muscle cells from Ct (n = 4) and TFam210aMus−/− (n = 4) mice. Data are expressed relative to the GAPDH mRNA value. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 using a two-sided Student’s t test.
Finally, we performed microarray analysis to evaluate differences between TFam210aMus−/− mice and Ct mice in the expression of genes which Fam210a modulates in differentiated primary muscle cells (Table S1). The levels of several muscle differentiation-related factors, such as myogenin (Myog) and cadherin 15 (Cdh15), were lower in primary muscle cells from TFam210aMus−/− mice compared with those from Ct mice. In addition, the levels of several muscle contraction-related factors, involving troponin c2, fast (Tnnc2), calsequestrin 2 (Casq2), and sarcalumenin (Srl) were decreased in primary muscle cells from TFam210aMus−/− mice compared with those from Ct mice. In contrast, the level of a bone resorption-related factor, matrix metalloproteinase 12 (Mmp12), was increased in primary muscle cells from TFam210aMus−/− mice compared with that from Ct mice (Table S1). As shown in Fig. 6D, expression of Fam210a was significantly reduced, relative to levels in Ct primary muscle cells, in the TFam210aMus−/− primary muscle cells, and the alterations of Tnnc2, Cdh15, Myog, and Mmp12 mRNA between primary muscle cells from Ct and TFam210aMus−/− mice were confirmed by quantitative real-time PCR.
Discussion
Understanding the shared biological determinants of osteoporosis and sarcopenia may help to provide targets for the prevention of osteoporotic fractures and frailty in our aging societies. Here, using human genetics to identify a novel locus influencing both traits, we provide evidence, through murine models, that Fam210a influences the mass and strength of both bone and muscle.
SNPs near FAM210A have previously been strongly associated with fracture and BMD in humans (16–18). However, while most SNPs identified to date have a predominant effect on BMD and a more attenuated effect on fracture, the FAM210A locus has an unusually strong effect on fracture, given its effect on BMD (16). This suggested that FAM210A may influence fracture through additional pathways that are independent of BMD. Since we observed that SNPs near FAM210A were associated with lean mass in adults, and, from a bivariate analysis, with lean mass and BMD in children, these data support that an additional pathway by which this locus influences fracture could be through muscle mass and strength.
We therefore investigated the tissue expression of Fam210a and the concomitant effects on bone and muscle after targeted deletion of Fam210a using genetically modified mice. By X-Gal staining, Fam210a was mainly expressed in muscle and especially skeletal muscle. In addition, Fam210a protein was localized to mitochondria and cytoplasm of muscle cells and myotubes by immunofluorescence. However, we detected no Fam210a protein in serum, which indicates that Fam210a may not be a secreted protein, although it conceivably may be released into the circulation from severely damaged muscle. Interestingly, parameters of muscle function and structure, that is, grip strength and lean mass of all limbs, were decreased in Fam210a homozygous and heterozygous knockout mice as well as in mice with muscle-specific knockout of Fam210a. By microarray analysis, the levels of several muscle differentiation-related factors, such as Myog, encoding myogenin, and Cdh15, encoding cadherin 15, were decreased in primary muscle cells from TFam210aMus−/− mice. Myogenin is a muscle-specific basic-helix–loop–helix transcription factor, whose expression is related to the initiation of myoblast differentiation (22), and which is essential for the development of functional skeletal muscle. Cadherin 15 is known to be up-regulated in myotube-forming cells and plays a crucial role in terminal muscle differentiation (23). Thus, Fam210a may affect muscle differentiation through its effect on myogenin and cadherin 15, an observation which appears consistent with our finding of the association of FAM 210A with lean body mass in children and of reduced lean mass in mouse limbs of 56-d-old TFam210aMus−/− mice. In addition, Tnnc2 and Srl were decreased in primary muscle cells from TFam210aMus−/− mice. Tnnc2 encodes troponin C2, fast, which is a component of the troponin complex and has two additional binding sites for Ca2+ in muscle contraction (24). Srl encodes sarcalumenin, a major luminal Ca2+-binding protein in the muscle sarcoplasmic reticulum, which has been associated with aging-related loss of muscle mass in rats (25). These findings suggest that Fam210a might affect muscle contraction by its effect on troponin C2, fast, and sarcalumenin.
Although Fam210a was not expressed in mouse bone, bone phenotypes were also observed in Fam210a+/− and especially TFam210a−/− mice. Thus, in TFam210a−/− mice, there were decreases in BMD, BMC, and biomechanical strength as well as increased microarchitectural deterioration of trabecular and cortical bones, decreases in bone formation, and increases in osteoclast activity. Similar but less pronounced changes were observed in Fam210a+/− mice, compatible with haploinsufficiency. Although significant weight loss occurred in TFam210a−/− mice, likely driven at least in part by reductions in lean body mass, and such weight loss may have contributed to an osteoporotic phenotype, serum calcium, phosphate, total protein, and BUN were not altered, suggesting that there was no evidence of gross malabsorption or renal impairment. Because Fam210a was not expressed in bone, Fam210a effects in other tissues seemed likely to be causal of the bone phenotypes. Interestingly, BMD of male TFam210aMus−/− mouse bone was low, and microarchitectural indices of trabecular and cortical bones of TFam210aMus−/− mice tended to be altered. Furthermore, osteoclast numbers and surface were increased in TFam210aMus−/− mice. These findings indicate that Fam210a in muscle may play a role in modifying both bone quantity and bone quality. Although changes appeared more severe in male mice, sex-stratified analyses for association of loci with BMD in human studies to date appear to have been limited by the lower number of men than women in the discovery and replication datasets used for analyses (e.g., ref. 16).
By microarray analysis, we found that the level of Mmp12 was increased in primary muscle cells from TFam210aMus−/− mice. MMPs are associated with multiple osteoclast functions, including recognition, migration, spreading, and attachment of osteoclasts to bone surface (26, 27), and MMP-12 has been reported to activate the putative functional domains of two bone matrix proteins, osteopontin and bone sialoprotein, that strongly enhance osteoclast activities. Osteoclasts were in fact increased, and Po.V was increased in the bone of TFam210aMus−/− mice compared with Ct mice. These findings suggest that Fam210a may induce the expression of MMP-12 in muscle, and that muscle-derived MMP-12 may in turn enhance osteoclast function in bone.
Because the bone phenotype of TFam210aMus−/− mice was less severe compared with that of TFam210a−/− mice, it seems possible that Fam210a expression in tissues other than skeletal muscle, in which Fam210a is clearly expressed, contribute to bone homeostasis. Thus, Fam210a was also highly expressed in heart and brain. Previous studies demonstrated that low BMD and hip fracture were associated with patients in heart failure (28, 29), and it has been reported that expression of the potent osteoclast-activating cytokine, receptor activator of nuclear factor kappa-B ligand is enhanced in heart failure, and circulating serum levels are increased (30). In addition, a variety of neuropeptides, including brain-derived serotonin, substance P, calcitonin-gene-related peptide, and the neurotransmitters norepinephrine and glutamine, have been reported to alter bone formation and resorption (31). Leptin, a hormone associated with appetite control, may also regulate bone metabolism through the sympathetic nervous system by binding to its receptors in hypothalamus (32). Thus, further studies are required to assess the role of Fam210a in tissues such as cardiac muscle and brain in regulating bone structure and function.
In conclusion, the present study shows that genetic variation near FAM210A is associated with reductions in both BMD and lean mass in humans and that Fam210a is associated with parameters of bone and muscle structure and function in genetically modified mice. We have therefore identified a molecule which mediates bone structure and strength as well as muscle structure and strength. Inasmuch as we could not identify release of Fam210a into the circulation, it seems unlikely that Fam210a per se functions as a myokine, although it is conceivable that it does have a direct effect on bone via a paracrine mechanism. Nevertheless, we have identified candidate musculoskeletal modulators such as muscle-derived MMP-12 which may enhance osteoclast function in bone. Further study should therefore lead to the development of novel treatments for osteoporosis and sarcopenia.
Materials and Methods
Assessment of FAM210A Genetic Variation.
We collected data from the largest GWAS to date for appendicular and whole body lean mass (33), pediatric BMD and lean mass (18), and eBMD from the heel calcaneus (34), and fracture (16). Our analyses were restricted to the genomic region of FAM210A and the surrounding 500 kb upstream and downstream, to only study genetic variation likely to be in linkage disequilibrium with those directly influencing FAM210A function. We tested whether SNPs mapping near FAM210A were associated with appendicular and whole body lean mass in adults, BMD, and lean mass in children, by correcting for multiple testing by dividing alpha = 0.05 by the number of independent SNPs assessed within each study. We did not correct the fracture association data for multiple testing, because FAM210A was already reported as significantly associated with fracture by Estrada et al. (16) as the C18orf19 gene. We tested for independence of SNPs using European genotype data from the 1000 Genomes Project (n = 503) and applying an r2 of 0.05. For appendicular and whole body lean mass in adults, pediatric BMD and lean mass in children, and eBMD in adults, we observed two independent sets of SNPs at the region (r2 < 0.05) and used a threshold of alpha = 0.025 (0.05/2) to determine an association.
Derivation of Mouse Strains.
All animal experiments were reviewed and approved by the McGill University Animal Care Committee. Mice were generated by the Toronto Center for Phenogenomics, as part of the North American Conditional Mouse Mutagenesis project, in which exon 2 of Fam210a was floxed with a targeted trap lacZ reporter cassette (Fig. S1) (35).
These mice were bred with CMV-Cre mice from The Jackson Laboratory, to obtain Fam210a+/− mice expressing the lacZ reporter or Fam210a−/−. To generate mice lacking Fam210a in muscle cells, mice in which the lacZ reporter cassette is flanked by flippase (FLP) recombinase target sequences were first bred to B6.Cg-Tg (ACTFLPe) mice from Thomas Ludwig at Columbia University to generate mice with a Fam210a conditional floxed allele. Then, Fam210aflox/flox mice were bred to CAG-Cre/Esr1 or ACTA1-cre/Esr1 mice, from The Jackson Laboratory, which contain tamoxifen-inducible sites, to generate TFam210a−/− or TFam210aMus−/− mice, respectively. Tamoxifen (Sidma Aldrich) was dissolved in corn oil with 2% ethanol. At 4 wk of age, 1 mg of tamoxifen was injected two times per day for five consecutive days. Fam210a WT mice with CMV-Cre were used as Ct for Fam210a+/− experiments. Fam210a flox/flox littermates were used as Ct for TFam210a−/− and TFam210aMus−/− experiments.
Genotyping of Mice.
Genomic DNA was isolated from tail fragments by Extra DNA Prep for PCR-Tissue (Quanta BioSciences) and analyzed by PCR. PCR was performed by a one-step method using a KAPA Mouse Genotyping Kit (Kapa Biosystems). To identify Fam210a+/− knockouts, the WT Fam210a allele was detected using forward primer (5′-GTTATCTGTGAGAACCCGGTAG-3′) and reverse primer (5′-GGAGCACAAATCCTGAGTACAG-3′), and the Fam210a knockout allele was detected using forward primer (5′-CCATTACCAGTTGGTCTGGTGTC-3′) and reverse primer (5′-AACCACTGCCACGTTCATAAGCAGG-3′). To detect the floxed-Fam210a, the WT Fam210a allele was detected using forward primer (5′-GTTATCTGTGAGAACCCGGTAG-3′) and reverse primer (5′-GGAGCACAAATCCTGAGTACAG-3′), and the null allele was detected using forward primer (5′-GCGCAACGCAATTAATGATAAC-3′) and reverse primer (5′-CTTACTTCTGCGTCCTCCTAAAC-3′). Forward primer (5′-CCTGGAAAATGCTTCTGTCCG-3′) and reverse primer (5′-CAGGGTGTTATAAGCAATCCC-3′) were used to detect the Cre transgene.
Quantitative RT-PCR.
RNA was prepared from mouse muscle, brain, and heart using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized using M-MLV Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol. For quantitative RT-PCR, the StepOnePlus Real-Time PCR System (Applied Biosystems) was used. The conditions were 10 μL of EvaGreen 2× qPCR MasterMix (Applied Biological Materials), 0.25 μM primers, and 5 μL of cDNA sample and/or H2O to a final volume of 20 μL. Samples were amplified for 40 cycles with a temperature transition rate of 20° per second for all three steps, and were denatured at 95° for 10 s, annealed at 95° for 15 s, and extended at 56° for 60 s. Eva green fluorescence was measured to determine the amount of double-stranded DNA. The mRNA value for each gene was normalized relative to the mouse GAPDH mRNA levels in RNA samples. Primer sequences (forward and reverse) were as follows: GAPDH, 5′-TCACCATCTTCCAGGAGCG-3′ and 5′-CTGCTTCACCACCTTCTTGA-3′; Fam210a, 5′-CCAGGCTCTTTGATCACCAT-3′ and 5′-GAGTTTCCTGGGATGTGGAA-3′; troponin c2, Tnnc2, 5′-ATCATCGAGGAGGTGGACGA-3′ and 5′-GCCAGTTCCTCTTCGCTCTT-3′; Cdh15, 5′-TGGGCAGTCCCTGAGCCCAAAC-3′ and 5′-TCCAGCGTGGCATTGAGGTACA-3′; Myog, 5′-GCTGCCTAAAGTGGAGATCCT-3′ and 5′-GCGCTGTGGGAGTTGCAT -3′; and Mmp12, 5′-GAAACCCCCATCCTTGACAA-3′ and 5′-TTCCACCAGAAGAACCAGTCTTTAA-3′. For quantitative RT-PCR of RNA from primary muscle cells, osteoblasts, and osteocytes, primer sequences (forward and reverse) were as follows: MyoD, 5′-GACGGCTCTCTCTGCTCCTT-3′ and 5′-AGTAGAGAAGTGTGCGTGCT-3′; Col1a1, 5′- AACCCGAGGTATGCTTGATCT-3′ and 5′-CCAGTTCTTCATTGCATTGC-3′; and Sost, 5′-TCCTCCTGAGAACAACCAGAC-3′ and 5′-TGTCAGGAAGCGGGTGTAGTG-3′
X-Gal Staining.
Histochemical X-Gal staining to detect β-galactosidase activity of the lacZ gene that had been inserted into the Fam210a gene under the control of regulatory elements was performed as described previously (35). Briefly, entire tissues from Ct and Fam210a+/− mice expressing the lacZ reporter under the control of the Fam210a promoter were fixed for 30 min on ice in fresh 4% paraformaldehyde and washed in PBS four times at room temperature. Tissues were stained with X-Gal solution [5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6], 1 mg/mL X-Gal] for 4 h at 37 °C in the dark. Entire tissues and 10-μm sections of the tissues from WT and Fam210a+/− mice were photographed using a Sony alpha 6000 Mirrorless Digital Camera (Sony Canada). Expression was marked by a dark blue stain and could be detected at the single-cell level.
Biochemistry.
Serum was obtained by centrifugation at 1,500 × g for 20 min and stored at −80°. Serum concentrations of calcium were measured by using a QuantiChrom Calcium Assay Kit (DICA-500) (BioAssays) according to the manufacturer’s instructions. Serum concentrations of phosphate were measured by a Phosphate Assay Kit (Abcam) according to the manufacturer’s instructions. BUN and total protein were measured by the Diagnostic Laboratory of the Animal Resource Center of McGill University, using colorimetric assays. Fam210a protein in serum or extract from muscle was determined using an ELISA Kit (EIAab) according to the instructions of the manufacturer. Briefly, 96 wells of a microtiter plate were precoated with antibody specific to Fam210a. Fifty microliters of serum, extract from muscle, or standards were placed in each of the 96 wells with a biotin-conjugated antibody specific to Fam210a. Next, avidin conjugated to Horseradish Peroxidase was added to each microplate well and incubated. After addition of 3,3′,5,5′-tetramethylbenzidine substrate solution, only those wells that contain Fam210a, biotin-conjugated antibody, and enzyme-conjugated avidin exhibited a change in color. The enzyme−substrate reaction was terminated by the addition of sulphuric acid solution, and the color change was measured spectrophotometrically at a wavelength of 450 nm. The concentration of Fam210a in the samples was then determined by comparing the optical density of the samples to the standard curve. The minimum detectable dose of this assay was 0.3 ng/mL.
Primary Cell Isolation and Culture.
Tibialis anterior muscles (TA) were isolated from 2-mo-old Ct or TFam210aMus−/− mice. Briefly, the isolated TA was digested with collagenase at 37 °C degrees in a 5% CO2 incubator for 1.5 h, and washed in PBS. After separating the fibers by pipetting, the fibers were transferred to matrigel-coated flasks containing DMEM (Invitrogen) with 1% penicillin−streptomycin (Invitrogen), 0.2% fungizone (GIBCO), 10% horse serum (HS), and 0.5% Chicken Embryo Extract (CEE) (GEMINI). The fibers were incubated at 37 °C in a 5% CO2 incubator for 4 d. Putative satellite cells appeared after 12 h to 24 h, and the media was changed to DMEM with 1% penicillin−streptomycin and 0.2% fungizone containing 20% FBS, 10% HS, and 1% CEE 4 d after the isolation. When cells were 70% confluent, the media was changed to DMEM with 1% penicillin−streptomycin and 0.2% fungizone containing 2% FBS, 10% HS, and 0.5% CEE for 1 wk. The cells were then used for experiments.
Primary osteoblasts and osteocytes were obtained from 2-mo-old WT mice by serial enzymatic digestion of femurs as previously described (36). In brief, femur fragments were digested with 300 active units per milliliter of collagenase type IA (Sigma-Aldrich) in α-MEM for 25 min at 37 °C; bones were then rinsed three times with Hanks’ balanced salt solution (HBSS), and the rinsate (digest 1) was discarded. The digestions were repeated twice (digests 2 and 3). The bones were then incubated in 5 mM EDTA for 25 min at 37 °C, and rinsed with HBSS, and the rinsate (digest 4) was discarded. Next, the bones were again incubated in 300 active units per milliliter collagenase for 25 min at 37 °C, and rinsed with HBSS (digest 5). The digestion was repeated four times (digests 6 to 9). Digests 6 and 9 contain mainly osteoblasts and osteocytes, respectively (36). Digests 6 and 9 were centrifuged at 1,000 × g, resuspended in culture media, and transferred to collagen-coated six-well plates. Confluent cells were then used for experiments.
Immunofluorescence.
Primary myoblasts and myotubes were fixed in 4% paraformaldehyde for 10 min. The cells were incubated with blocking buffer containing 1% BSA for 1 h. Then, the cells were incubated with Fam210a antibody (1:200 dilution, rabbit anti-Fam210a antibody; Abcam) and an antibody to adenosine triphosphate synthase subunit beta (ATPB) as a mitochondrial marker (1:200 dilution, mouse anti-ATPB antibody; Abcam) at 4 °C overnight. After incubating with DAPI (1:100 dilution, blue staining of DNA; Invitrogen) for 10 min, the cells were incubated with Alexa Fluor 488 (1:200 dilution, anti-rabbit; Invitrogen) and 568 (1:200 dilution, anti-mouse; Invitrogen) secondary antibodies at room temperature without light for 1 h. After washout of secondary antibodies, fluorescent images were captured by confocal laser scanning microscopy (Zeiss LSM 780 NLO; Zeiss). Fam210a and mitochondria were detected as green and red fluorescent images, respectively.
μCT.
μCT scans were performed, as previously described (37), on femurs using a high-resolution SkyScan-1072 MicroCT scanner (Bruker-microCT). The samples were scanned at an X-ray source power of 45 kV/222 mA, with spatial resolution of 5.63 mm/pixel. The rotation was set at 0.9°/step for 180°. The CT-Analyzer (version 1.10.0.2) was used for analyses of 3D architectural parameters including BV/TV, Tb.N, Tb.Th, Tb.Sp, and SMI. The volume of interest was built by drawing polygons including only trabecular bone over the range of 0.26 mm below the growth plate in distal femur. The cortical BV/TV, Co.Th., and Po.V were measured over the range of 0.57 mm starting from the section 2.80 mm away from growth plate.
Dual-Energy X-Ray Absorptiometry.
Using a Lunar PIXImus II Mouse Densitometer (GE Medical Systems), live mice were scanned to determine lean mass of forelimbs, and bone samples were scanned to determine BMD (grams per square centimeter) and BMC (grams). Before being scanned, the mice were anesthetized and then ventrally positioned on the tray so that the limbs were moderately stretched within the scanning area. To evaluate BMD and BMC bone samples, mouse tibias were placed in the center area of the tray and then scanned, using up to eight replicates. After scanning, lean mass of individual limbs and BMD and BMC of individual bone samples were analyzed using custom-defined region of interest functions.
Biomechanical Testing.
Three-point bending tests were performed at the right femur midshaft with a displacement rate of 10 mm/min (span length, 10 mm) using a Mach-1TM Micromechanical testing system (Biosyntech). Bone mechanical properties, including maximum load, stiffness, and work-to-failure, were determined.
Grip Strength.
Grip strength of mouse forelimbs was measured by a grip strength meter (Columbus Instruments and Bioseb), as described previously (38, 39), by allowing mice to hold a metal grid with their forelimbs, lifting mice by the tail so that the hind limbs were not in contact with the grid, and gently pulling backward until they could no longer hold the grid. Genotype was blind to observer, and grip strength was measured from 7 to 10 times in consecutive trials. The mean of at least five successful trials was taken for analysis.
Histochemical Staining for Tartrate-Resistant Acid Phosphatase.
Histochemical staining for tartrate-resistant acid phosphatase (TRAP) was performed after decalcification as previously described (39). Tibias were dehydrated and embedded in paraffin. Briefly, the sections were incubated in TRAP Staining Solution Mix, containing sodium acetate anhydrous (Sigma), l-(+) tartaric acid (Sigma), and glacial acetic acid at 37° for 30 min. The sections were counterstained with 0.02% Fast Green (Sigma) for 30 s. The number of osteoclasts was then determined from TRAP-stained sections. Cells were identified visually, traced manually, counted automatically, and reported as cell numbers per millimeter of bone perimeter in trabecular or cortical bone. To evaluate bone resorption, Oc.N/BS and Oc.S/BS were calculated as the number or the percent of bone surface occupied by osteoclasts.
Double Calcein Labeling.
Ten micrograms of calcein per gram of body weight (C-0875; Sigma) was injected intraperitoneally at the 10th and third day before killing as previously described (40). Images were taken using a fluorescence microscope (Zeiss LSM 780 NLO). The double calcein-labeled widths of cortical and trabecular bones were measured using Image J 1.38 software from the National Institutes of Health (https://imagej.nih.gov/), and the MAR was calculated as the interlabel width/labeling period (expressed in micrometers per day). BFR was calculated as mineralizing surface per bone surface multiplied by the MAR.
Gene Expression Microarray Analyses.
RNA was isolated from primary muscle cells differentiated by HS from Ct and TFam210Mus−/− mice by RNeasy Plus Mini Kit (QIAGEN) by following the manufacturer’s instructions. RNA quality testing, quantification, and hybridization to the Mouse ClariomTM S Assay (ThermoFisher Scientific) were performed at the Genome Quebec Innovation Centre. Background-corrected, normalized and summarized expression values at the transcript cluster level were calculated using the robust multichip average (rma) method available through the oligo Bioconductor R package (41). An estimate for background expression level was obtained by fitting a normal−exponential mixture on all expression values and used to nonspecifically filter features for which expression exceeded this threshold in at least three samples. Differential expression between Ct and TFam210Mus−/− sample groups was performed using the limma Bioconductor package (42) and to estimate false-discovery rates (FDR). Fold change > 1.7 and FDR < 0.25 were taken to indicate significance.
Statistical Analysis.
All analyses in mice were carried out using StatView (Abacus Concepts). Data from image analysis are presented as means ± SE. Statistical analysis was performed using t tests. A P value < 0.05 was taken to indicate a significant difference.
Supplementary Material
Acknowledgments
Grip strength of Fam210a+/− and Fam210a−/− mice was kindly measured by Vladimir V. Rymar and Abbas F. Sadikot (Cone Laboratory, Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montreal, QC, Canada). This study was supported by grants from the Canadian Institutes of Health Research, Merck Sharp and Dohme (MSD) Life Science Foundation, Public Interest Incorporated Foundation, The Uehara Memorial Foundation, and Mochida Memorial Foundation, and by a Richard and Edith Strauss Postdoctoral Fellowship in Medicine from the Research Institute of the McGill University Health Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719089115/-/DCSupplemental.
References
- 1.Cosman F, et al. National Osteoporosis Foundation Clinician’s guide to prevention and treatment of osteoporosis. Osteoporos Int. 2014;25:2359–2381. doi: 10.1007/s00198-014-2794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cauley JA, Thompson DE, Ensrud KC, Scott JC, Black D. Risk of mortality following clinical fractures. Osteoporos Int. 2000;11:556–561. doi: 10.1007/s001980070075. [DOI] [PubMed] [Google Scholar]
- 3.Visser M, Schaap LA. Consequences of sarcopenia. Clin Geriatr Med. 2011;27:387–399. doi: 10.1016/j.cger.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Cooper C, et al. Frailty and sarcopenia: Definitions and outcome parameters. Osteoporos Int. 2012;23:1839–1848. doi: 10.1007/s00198-012-1913-1. [DOI] [PubMed] [Google Scholar]
- 5.Rockwood K, Stolee P, McDowell I. Factors associated with institutionalization of older people in Canada: Testing a multifactorial definition of frailty. J Am Geriatr Soc. 1996;44:578–582. doi: 10.1111/j.1532-5415.1996.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 6.Frost HM. On our age-related bone loss: Insights from a new paradigm. J Bone Miner Res. 1997;12:1539–1546. doi: 10.1359/jbmr.1997.12.10.1539. [DOI] [PubMed] [Google Scholar]
- 7.Burr DB. Muscle strength, bone mass, and age-related bone loss. J Bone Miner Res. 1997;12:1547–1551. doi: 10.1359/jbmr.1997.12.10.1547. [DOI] [PubMed] [Google Scholar]
- 8.Schoenau E. From mechanostat theory to development of the “Functional Muscle-Bone-Unit.”. J Musculoskelet Neuronal Interact. 2005;5:232–238. [PubMed] [Google Scholar]
- 9.Tagliaferri C, Wittrant Y, Davicco MJ, Walrand S, Coxam V. Muscle and bone, two interconnected tissues. Ageing Res Rev. 2015;21:55–70. doi: 10.1016/j.arr.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Kawao N, Kaji H. Interactions between muscle tissues and bone metabolism. J Cell Biochem. 2015;116:687–695. doi: 10.1002/jcb.25040. [DOI] [PubMed] [Google Scholar]
- 11.Brotto M, Bonewald L. Bone and muscle: Interactions beyond mechanical. Bone. 2015;80:109–114. doi: 10.1016/j.bone.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karasik D, Kiel DP. Genetics of the musculoskeletal system: A pleiotropic approach. J Bone Miner Res. 2008;23:788–802. doi: 10.1359/jbmr.080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina-Gomez C, et al. Bivariate genome-wide association meta-analysis of pediatric musculoskeletal traits reveals pleiotropic effects at the SREBF1/TOM1L2 locus. Nat Commun. 2017;8:121. doi: 10.1038/s41467-017-00108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorski JP, et al. Inhibition of proprotein convertase SKI-1 blocks transcription of key extracellular matrix genes regulating osteoblastic mineralization. J Biol Chem. 2011;286:1836–1849. doi: 10.1074/jbc.M110.151647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecomte V, et al. A new role for sterol regulatory element binding protein 1 transcription factors in the regulation of muscle mass and muscle cell differentiation. Mol Cell Biol. 2010;30:1182–1198. doi: 10.1128/MCB.00690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrada K, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng HF, et al. AOGC Consortium UK10K Consortium Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp JP, et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet. 2017;49:1468–1475. doi: 10.1038/ng.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Movérare-Skrtic S, et al. Osteoblast-derived WNT16 represses osteoclastogenesis and prevents cortical bone fragility fractures. Nat Med. 2014;20:1279–1288. doi: 10.1038/nm.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo J, et al. LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat Med. 2016;22:539–546. doi: 10.1038/nm.4076. [DOI] [PubMed] [Google Scholar]
- 21.Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018;27:233–244. doi: 10.1002/pro.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright WE, Sassoon DA, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 23.Donalies M, Cramer M, Ringwald M, Starzinski-Powitz A. Expression of M-cadherin, a member of the cadherin multigene family, correlates with differentiation of skeletal muscle cells. Proc Natl Acad Sci USA. 1991;88:8024–8028. doi: 10.1073/pnas.88.18.8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gahlmann R, Kedes L. Cloning, structural analysis, and expression of the human fast twitch skeletal muscle troponin C gene. J Biol Chem. 1990;265:12520–12528. [PubMed] [Google Scholar]
- 25.O’Connell K, Gannon J, Doran P, Ohlendieck K. Reduced expression of sarcalumenin and related Ca2+ -regulatory proteins in aged rat skeletal muscle. Exp Gerontol. 2008;43:958–961. doi: 10.1016/j.exger.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Everts V, Delaissé JM, Korper W, Beertsen W. Cysteine proteinases and matrix metalloproteinases play distinct roles in the subosteoclastic resorption zone. J Bone Miner Res. 1998;13:1420–1430. doi: 10.1359/jbmr.1998.13.9.1420. [DOI] [PubMed] [Google Scholar]
- 27.Holliday LS, et al. Initiation of osteoclast bone resorption by interstitial collagenase. J Biol Chem. 1997;272:22053–22058. doi: 10.1074/jbc.272.35.22053. [DOI] [PubMed] [Google Scholar]
- 28.van Diepen S, Majumdar SR, Bakal JA, McAlister FA, Ezekowitz JA. Heart failure is a risk factor for orthopedic fracture: A population-based analysis of 16,294 patients. Circulation. 2008;118:1946–1952. doi: 10.1161/CIRCULATIONAHA.108.784009. [DOI] [PubMed] [Google Scholar]
- 29.Fohtung RB, et al. Bone mineral density and risk of heart failure in older adults: The cardiovascular health study. J Am Heart Assoc. 2017;6:e004344. doi: 10.1161/JAHA.116.004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueland T, et al. Dysregulated osteoprotegerin/RANK ligand/RANK axis in clinical and experimental heart failure. Circulation. 2005;111:2461–2468. doi: 10.1161/01.CIR.0000165119.62099.14. [DOI] [PubMed] [Google Scholar]
- 31.Masi L. Crosstalk between the brain and bone. Clin Cases Miner Bone Metab. 2012;9:13–16. [PMC free article] [PubMed] [Google Scholar]
- 32.Kajimura D, et al. Genetic determination of the cellular basis of the sympathetic regulation of bone mass accrual. J Exp Med. 2011;208:841–851. doi: 10.1084/jem.20102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zillikens MC, et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat Commun. 2017;8:80. doi: 10.1038/s41467-017-00031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moayyeri A, et al. Genetic determinants of heel bone properties: Genome-wide association meta-analysis and replication in the GEFOS/GENOMOS consortium. Hum Mol Genet. 2014;23:3054–3068. doi: 10.1093/hmg/ddt675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah KMA, et al. Osteocyte isolation and culture methods. Bonekey Rep. 2016;5:838. doi: 10.1038/bonekey.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue Y, et al. The calcium-sensing receptor complements parathyroid hormone-induced bone turnover in discrete skeletal compartments in mice. Am J Physiol Endocrinol Metab. 2012;302:E841–E851. doi: 10.1152/ajpendo.00599.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hockly E, et al. Environmental enrichment slows disease progression in R6/2 Huntington’s disease mice. Ann Neurol. 2002;51:235–242. doi: 10.1002/ana.10094. [DOI] [PubMed] [Google Scholar]
- 39.Davies JE, et al. Doxycycline attenuates and delays toxicity of the oculopharyngeal muscular dystrophy mutation in transgenic mice. Nat Med. 2005;11:672–677. doi: 10.1038/nm1242. [DOI] [PubMed] [Google Scholar]
- 40.Miao D, et al. Skeletal abnormalities in Pth-null mice are influenced by dietary calcium. Endocrinology. 2004;145:2046–2053. doi: 10.1210/en.2003-1097. [DOI] [PubMed] [Google Scholar]
- 41.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.