Significance
Cardiolipins are a unique class of phospholipids in mitochondrial membranes that are crucial for cellular bioenergetics as they stabilize respiratory chain complexes. In contrast to most other phospholipids, cardiolipins are substituted with four, rather than only two fatty acyl side chains. Consequently, this opens up a vast number of different theoretically possible molecular lipid species. Experimentally assessing the molecular diversity of cardiolipin species is analytically challenging. In this study we successfully combine tandem mass spectrometry with a mathematical structural modeling approach, to achieve the comprehensive characterization of complex biological cardiolipin compositions.
Keywords: cardiolipin, mitochondria, lipids, mass spectrometry, mathematical modeling
Abstract
Current strategies used to quantitatively describe the biological diversity of lipids by mass spectrometry are often limited in assessing the exact structural variability of individual molecular species in detail. A major challenge is represented by the extensive isobaric overlap present among lipids, hampering their accurate identification. This is especially true for cardiolipins, a mitochondria-specific class of phospholipids, which are functionally involved in many cellular functions, including energy metabolism, cristae structure, and apoptosis. Substituted with four fatty acyl side chains, cardiolipins offer a particularly high potential to achieve complex mixtures of molecular species. Here, we demonstrate how systematically generated high-performance liquid chromatography-mass spectral data can be utilized in a mathematical structural modeling approach, to comprehensively analyze and characterize the molecular diversity of mitochondrial cardiolipin compositions in cell culture and disease models, cardiolipin modulation experiments, and a broad variety of frequently studied model organisms.
Biological membranes contain highly diverse phospholipid compositions (1). Beside a general categorization on basis of their polar headgroups, it is especially the substitution with different fatty acyl (FA) side chains that generates a vast variety of molecular lipid species, a combinatorial multiplicity that can be of regulated and stochastic origin. This is particularly true for cardiolipins (CL), a lipid class with four possible acyl side chains, rather than the two present in most other phospholipids (2).
CLs are glycerol-bridged, dimeric phospholipids and represent an essential component of mitochondrial membranes. Their unique shape and charge state is responsible for a broad functional spectrum. CLs directly interact with and stabilize respiratory chain complexes (3, 4) and ATP synthase (5), thereby impacting on ATP production (6). They further support mitochondrial cristae formation (7), protect mtDNA (8), and their close association with cytochrome c functionally involves them into mitophagy and apoptosis (9, 10). Postbiosynthetic maturation and homeostasis of the CL side-chain substitution—and therewith their structural diversity—is ensured by an enzymatic remodeling process initiated by phospholipase catalyzed side-chain hydrolysis to form monolyso-cardiolipins (MLCL) (11). An acyltransferase activity, predominantly mediated by the transacylase Tafazzin, is then required for reestablishing functional CL molecules (12). Indicative for the importance of this process is the X-linked genetic disorder Barth Syndrome (BTHS, OMIM: #302060), in which Tafazzin activity is impaired, leading to abnormal CL patterns (13) and CL instability (14). This causes severe symptoms, such as cardiomyopathy, neutropenia, muscle weakness, and 3-methylglutaconic aciduria (15).
Importantly, when assessing CL patterns in biological samples, a detailed description of their structural diversity is often missing. A major problem is caused by the numerous possible isobaric CL molecules exhibiting largely similar chromatographic behaviors. Features measured in lipidomic experiments are often attributed to their most plausible molecular species or (more accurately) described in a collective term for multiple isobaric possibilities (16–18). Furthermore, the problem is often circumvented by focusing only on some selected species (19, 20). Even when using high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS), where large amounts of structural data can be recorded, these details are typically lost during data extraction and analysis, or have to be manually curated in a time-consuming and laborious process (18, 19, 21).
In this study, we present a highly comprehensive analysis of the molecular diversity of CLs in BTHS patient-derived cells, mammalian cell culture models, CL composition modulation experiments, as well as a broad variety of frequently studied model organisms. Generating this detailed structural data was made possible due to a mathematical structural modeling strategy that, in combination with HPLC-MS/MS experiments, solves the problem of structural data loss when analyzing lipidomic datasets and allows to comprehensively characterize a broad spectrum of CLs on the basis of fragment spectra recorded by data-dependent acquisition.
Results
Quantification of CLs by HPLC-MS/MS.
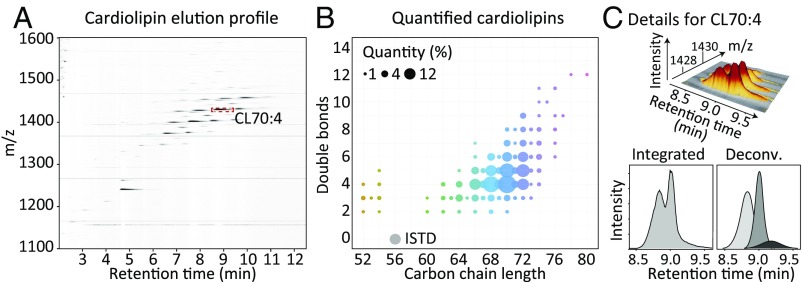
Cellular lipids were extracted from pellets of the murine RAW 264.7 cell line and used for method development and validation. The optimized HPLC-MS/MS method allowed clear chromatographic separation of up to 135 different CL mass-species within 12 min and their MS1-based identification and quantitation as single charged ions in the mass range of 1,000–1,600 m/z (Fig. 1A). Each of these species can be characterized by its total carbon chain length and double-bond count (Fig. 1B). The elution profiles of individual peaks indicated the presence of several isobaric CL subspecies, with only slightly differing chromatographic behavior (Fig. 1C). These differences are caused by variable FA substitutions, generating different molecular CL species with the same total carbon chain length and number of double bonds (For nomenclature, see SI Appendix, Text S1). A detailed description of analytic method development and validation can be found in SI Appendix, Text S2, including linearity and detection limits (SI Appendix, Fig. S1), as well as repeatability, precision, accuracy, and recovery information (SI Appendix, Fig. S2).
Fig. 1.
Quantifying CLs by HPLC-MS/MS. (A) A typical mass-chromatogram obtained for a RAW 264.7 cell lipid extract is shown. (B) Integrated CL peak areas projected back onto their respective carbon chain length and double-bond components. Circle sizes indicate the CL mass-species peak areas, fill colors correspond to the respective total number of side-chain carbons. The ISTD CL(14:0)4 is shown in gray. (C) Representation of the monoisotopic mass peak and the first three isotope peaks corresponding to CL70:4 (Upper) and as sum of the mass range 1,428.1 ± 0.3 (Lower Left), which elutes at least as double peak. (Lower Right) Hypothetical deconvolution of CL70:4 into contributions of individual molecular CL species.
Structural Characterization of CL Compositions by Mathematical Modeling of MS/MS Data.
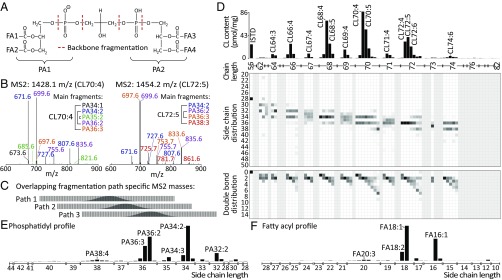
In light of the broad structural CL diversity, even within individual isobaric CL peaks (Fig. 1C), we established a data-dependent MS/MS fragmentation strategy that allows recording fragment spectra for all relevant CL species within a chromatographic run (SI Appendix, Fig. S3). These spectra can be interpreted on basis of the backbone fragmentation behavior of CLs (Fig. 2A). Fig. 2B illustrates two exemplary fragment spectra and their side-chain distribution annotation according to the three major fragmentation paths and their respective annotation (details in SI Appendix, Fig. S4).
Fig. 2.
Mathematical modeling of fragment spectra allows detailed structural characterization of individual CL species. (A) Initial fragmentation of CLs occurs primarily along the diphosphoglyceryl backbone, cleaving the CL into two PA fragments (PA1/2), which are substituted with two FAs, respectively (FA1–4). Three major PA fragment types are formed (SI Appendix, Fig. S5). (B) MS2 scans of CL70:4 (1,428.1 m/z, Left) and CL72:5 (1,454.2 m/z, Right) are shown, along with their manual PA fragment annotation. For example, the three major PA34:2 fragments correspond to 671.6, 727.6, and 807.6 m/z. (C) The PA fragment m/z distributions overlap between the three fragmentation paths and hamper an automated unique annotation. (D) CL characterization of one RAW 264.7 sample including an absolute quantitative CL profile (Top), and PA composition data projected onto the carbon chain length (Middle) and double-bond count (Bottom) components, respectively. (E) PA and (F) FA abundance profiles for the same sample as in D.
A major problem was that manual and even semiautomatic filtering and interpretation of up to 1,450 MS/MS spectra per sample is not feasible within reasonable time frames. Therefore, we developed a robust structural modeling approach that allows us to perform such an analysis in an automated fashion. A major challenge was the partial overlap of the fragment space of the three major CL fragmentation paths (Fig. 2C), which hampers a clear and distinct fragment mapping. Thus, deconvolution of the fragment spectra into the contributions of the three fragmentation paths represents an important step for constructing quantitative CL fragment profiles. First, a fragmentation path annotation matrix is constructed from all theoretically possible CL side-chain combinations in a mass-resolution–dependent manner (Methods). This allows applying this approach also on instruments with limited mass resolution. Together with individual fragmentation path efficiencies, a theoretical fragment spectrum is constructed and the Euclidean distance to the measured spectra is minimized using a box-constrained limited-memory modification of the Broyden–Fletcher–Goldfarb–Shanno quasi-Newton algorithm (22). The resultant profile explains the FA distribution along the individual phosphatidic acid (PA) subunits of the CL molecules. This profile was then further used, for example, to determine CL carbon side-chain and double-bond distributions. Additionally, an individual FA profile for each sample was mathematically modeled (in analogy to the procedure described above) on the basis of the extracted PA data and the theoretically available FA space. This approach follows the principal logic of using MS1 data for CL quantification and MS2 data for their structural characterization (SI Appendix, Fig. S5). As quality control, the obtained FA and PA profiles were used to predict the respective higher structural layers of the CL molecules and were compared with measured profiles.
By applying this analysis strategy to fragment spectra data recorded for lipid extracts of murine RAW 264.7 cells, it was possible to obtain—additionally to a pure CL abundance profile (Fig. 2D, Top)—detailed structural composition data for each CL species. Projected onto the respective carbon chain length and the double-bond count components (Fig. 2D, Middle and Bottom), the results show that the CL composition in RAW 264.7 cells is highly symmetrical and utilizes mainly even-numbered FAs. It becomes apparent that more saturated CLs exhibit a highly symmetric double-bond distribution, while highly unsaturated CLs are more likely to be asymmetric, possibly due to one-sided substitution with a polyunsaturated fatty acid (PUFA). The detailed quantitative and structural insights into the CL compositions in cells can be expressed as cumulative profiles of CLs, PAs, and FAs for each sample (Fig. 2D, Top, and Fig. 2 E and F). Importantly, these results allow studying the structural compositions of individual CL species and predicting their underlying FA profiles. The cardiolipin CL72, for example, consists of ∼80% of two PA36 subunits, while the remaining CL72 mass is made up of mainly PA34/PA38, and to a smaller degree, of other combinations as well (SI Appendix, Fig. S6). This clearly impacts on estimations made for the FA pool composition incorporated into CLs. To cross-validate the generated FA profiles, their predictability for previously obtained PA profiles was tested. Pearson correlations of typically above 0.95 between the FA-predicted PA profile and the PA pattern extracted from fragment spectra indicate a strong agreement (SI Appendix, Fig. S7). Furthermore, the FA profiles were compared with matching published datasets, which were in good accordance with our results (SI Appendix, Fig. S8).
Impairment of CLs in Barth Syndrome.
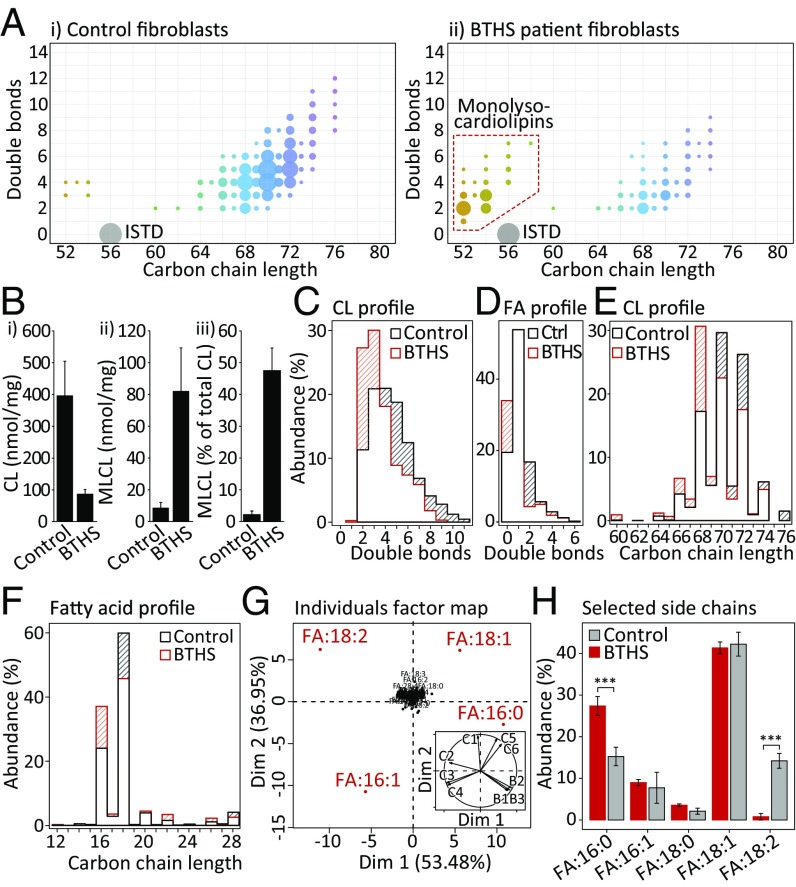
We used this method to analyze the impairment of CL compositions caused by dysfunction of Tafazzin in cultured BTHS patient fibroblasts. Compared with unaffected controls, we identified profoundly altered CL patterns (Fig. 3A), a strong depletion of CL mass to about 22% and a simultaneous 9.7-fold accumulation of MLCL species (Fig. 3B). A detailed species-specific structural analysis of CL profiles revealed the preferential depletion of double-bond–rich CLs (Fig. 3C), an effect that can also be observed on the PA level (SI Appendix, Fig. S9) and on the FA level (Fig. 3D). Similarly, a reduction of the CL carbon chain length, and their structural PA and FA components was observed (Fig. 3 E and F).
Fig. 3.
Impairment of CLs in BTHS patient fibroblasts. (A) Quantified CL composition observed in representative (i) control (n = 6) and (ii) BTHS patient fibroblasts (n = 3). Legend according to Fig. 1B. (B) BTHS cells exhibit (i) a reduced total CL content and increased (ii) absolute and (iii) relative MLCL levels. (C) The double-bond distribution in BTHS CLs is shifted toward a higher degree of saturation and (D) the same trend was observed at the FA level. Side-chain length distributions of (E) CLs and (F) FA in BTHS cells are enriched for lower carbon numbers. (G) Principal component analysis of FA profiles clearly separated BTHS cells from controls in dimension 1 (53.48% of variance, see Inset) and FA16:0, FA16:1, FA18:1, and FA18:2 were identified as major factors. (H) Two-way ANOVA was conducted on the influence of two variables (cell line, FA). The main effect for FA profiles resulted in an F-ratio of F(112, 1) = 167.6, P < 2e-16. Post hoc Bonferroni-corrected analysis revealed 2 of 112 possible fatty acids being significantly altered. We find a significant increase of FA16:0 (P = 2.11e-36) in BTHS cells while FA18:2 was strongly depleted (P = 3.24e-34). ***P < 0.001.
To identify the main targets of this effect, we conducted a principal component analysis on basis of the extracted FA compositions (Fig. 3G). Component 1 explained 53.48% of total variance and clearly separated controls from BTHS fibroblasts (Fig. 3G, Inset). The factors mainly responsible for this separation were identified as palmitic acid (FA16:0), oleic acid (FA18:1), and linoleic acid (FA18:2). While a relative depletion of FA18:2 was observed, FA16:0 was increased in the FA profiles (Fig. 3H). Importantly, our methodology is not only able to capture alterations of canonical CLs but also the fate of FAs in MLCL (SI Appendix, Fig. S10), as well as in oxidized CL species (SI Appendix, Fig. S11).
CL Heterogeneity and Reconfiguration in Human Cell Lines.
Next, we analyzed the CL composition in 16 different cultured human cell lines, grown under recommended growth conditions. Cell lines were of epithelial, lymphoblastic, endothelial, or myeloblastic origin obtained from different tissues of female or male donors (SI Appendix, Table S1). Interestingly, we observed clearly diverging CL profiles, which allowed grouping the cells lines into three clusters (SI Appendix, Figs. S12 and S13A), ranging from a group of cells with CLs harboring shorter FA side chains, over an intermediate group, to a group of cells with increased side-chain length. When analyzed for the influence of specific growth conditions, the only significant enrichment was found for DMEM-based high-glucose media in the intermediate cluster (P = 0.0048) (SI Appendix, Table S2). However, A431 and SK-N-SH were grown under similar conditions, but matched to the other two clusters. The cell line heterogeneity becomes apparent in view of the mean carbon chain length (SI Appendix, Fig. S13B), ranging from 67.5 to 71.3 (mean = 69.4), and double-bond count diverging between 3.9 and 5.5 per average CL (SI Appendix, Fig. S13C). Cell lines with longer-chained CLs also exhibited more double bonds. When comparing the FA profiles of the two most-extreme cell lines, U937 and K562 (SI Appendix, Fig. S13B), we found that the altered CL composition was not caused by incorporation of a new set of FAs, but was to a large extent the result of a shifted balance between palmitoleic acid (FA16:1) and linoleic acid (FA18:2) (SI Appendix, Fig. S13D).
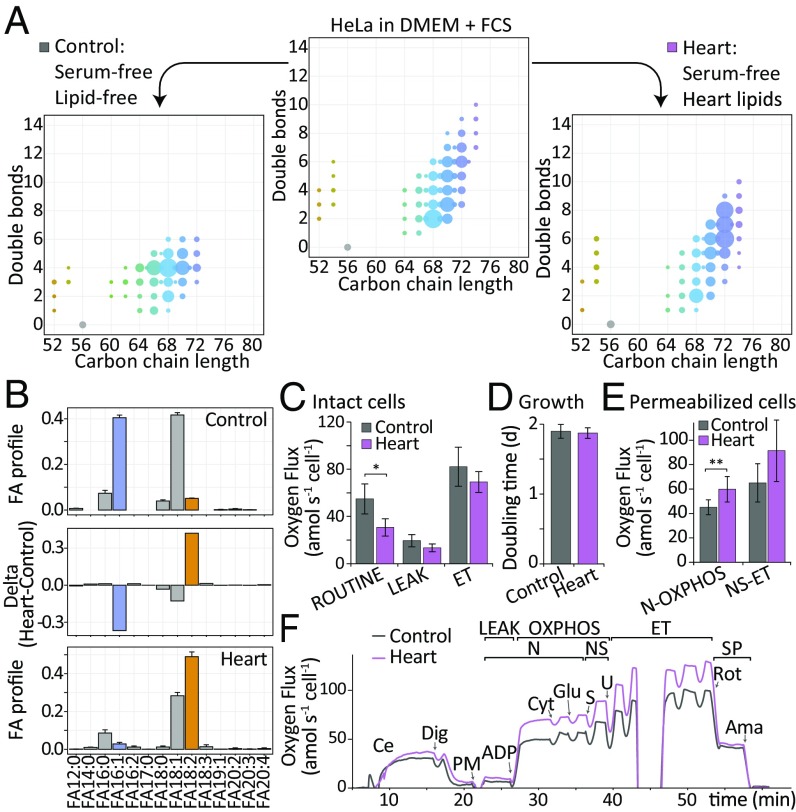
With the standard culture conditions described above, cells were exposed to serum, which could impose its intrinsic lipid compositions on the cellular phospholipid profiles and eventually impact their functions. Thus, we next studied the functional consequences of modulated lipid availability. HeLa cells were grown either in serum or lipid-free media. Cells were either forced to form fatty acids via de novo biosynthesis (control) or were supplemented with linoleic acid-rich pig heart lipid extracts (heart) (Fig. 4A). We analyzed the CL compositions generated by these treatments, which both showed to be highly different from the standard growth media (DMEM/FCS) (Fig. 4A). While CL profiles in the lipid-free control centered around CL68:4, the heart lipid supplementation shifted the CL compositions toward CL72:8. Our structural modeling analysis revealed that under lipid-free conditions the CL side chains were dominated by FA16:1 and FA18:1, whereas CLs of heart lipid-supplemented cells exhibited a high linoleic acid (FA18:2) content, which was indicative of a strong rearrangement of the mitochondrial phospholipid composition (Fig. 4B). In a next step, to study possible cooccurring alterations of core mitochondrial metabolic functions, we conducted high-resolution respirometry experiments in intact cells. Interestingly, tetralinoleoyl CL-rich cells had a significantly lower respiration in the presence of endogenous substrates (ROUTINE respiration), compared with controls (Fig. 4C). This effect was independent of cell growth rates, which were unaffected by the supplementation, arguing against a general inhibition of ATP production (Fig. 4D). To examine the functional origin of a potentially more-efficient respiration in cells supplemented by heart lipid, we performed a respirometric analysis in permeabilized cells and found that both conditions did not alter β-oxidation and complex IV activity. However, NADH-pathway capacity through complex I tended to be increased in heart lipid–treated cells in the presence of saturating substrate supply (SI Appendix, Fig. S14). Hence, we investigated this part of mitochondrial metabolism in more detail. Indeed the NADH-pathway capacity was significantly increased in heart lipid–treated cells (Pcor = 0.01, n = 8) (Fig. 4 E and F), in line with the reconfiguration of the mitochondrial CL side-chain substitution (Fig. 4A).
Fig. 4.
CL profile adaptation in response to lipid supplementation. (A) Quantified CL composition of HeLa cells grown in lipid and serum free (control, Left), standard (DMEM/FCS, Center), and pig heart lipids supplemented conditions (Heart, Right). One representative sample is shown (n = 4). Legend according to Fig. 1B. (B) FA composition in control (Top) and heart (Bottom) condition and their differential profile (Delta, Middle) (n = 4). (C) Oxygen flow in intact control and heart samples. (D) Doubling times of control and heart samples (n = 4). (E) Oxygen flow in permeabilized control and heart samples. NADH-pathway capacity (N-OXPHOS) was significantly increased upon heart lipid supplementation (n = 8, P = 0.01, Bonferroni-adjusted with m = 7). (F) Representative traces of oxygen flow for experiment shown in E used to calculate respiratory activities in different pathway-control states. Arrows indicate substrate-uncoupler-inhibitor titration steps: ADP, adenosine diphosphate; Ama, antimycin A; Ce, cells; Cyt, cytochrome c; Dig, digitonin; ET, maximal electron transfer capacity in presence of CCCP uncoupler (n = 4, Bonferroni-adjusted with m = 9); Glu, glutamate; LEAK, respiration after inhibition of ATP synthase; N, NADH-pathway; NS, convergent N- and succinate-pathway; PM, pyruvate and malate; Rot, rotenone; ROUTINE, cell respiration in presence of endogenous substrates; S, succinate; SP, succinate-pathway; U, uncoupler (CCCP). If not otherwise stated, data are shown as mean ± SD. *P < 0.05, **P < 0.01.
CL Diversity in Model Organisms.
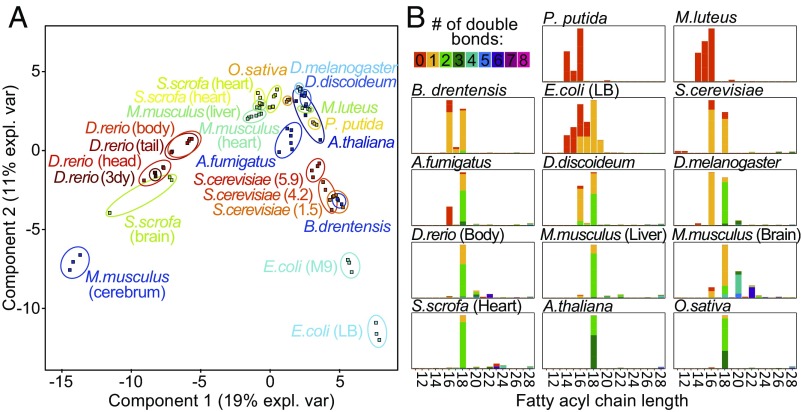
We assessed the structural variability of CLs in a selection of widely used model organisms (SI Appendix, Table S3). CLs were readily detectable with our analytical technique in all organisms tested. We observed highly diverse CL compositions, varying in an organism-, tissue-, and condition-dependent manner (SI Appendix, Fig. S15). The number of CL species accounting for 95% of total CL mass ranges between only 4 in Arabidopsis thaliana to 26 in Aspergillus fumigatus (see nCL95% values in SI Appendix, Fig. S15) and is indicative for this variability. Profiles were also highly diverse in terms of carbon chain length and degree of saturation. While in some bacteria only simple, short-chained, and completely saturated CLs were detected (Micrococcus luteus, Pseudomonas putida), zebrafish (Danio rerio) exhibited a complex pattern of long-chain and highly unsaturated CLs. We conducted a partial least-squares discriminant analysis (PLS-DA) that projected 19% and 11% of the variance in this CL dataset onto components 1 and 2, respectively, clearly separating the individual sample groups (Fig. 5A). Interestingly, some tissue and growth specificities contributed more to this separation than organism-specific differences.
Fig. 5.
CL compositions across the domains of life. (A) PLS-DA visualizes the differences among CL profiles recorded for species and conditions. Components 1 and 2 captured 19% and 11% of variability, respectively. Clear species and condition-specific differences were observed and samples grouped according to their biological replicates. (B) FA profiles calculated for CL profiles shown in A. Bars represent the mean profile abundance of each FA in all biological replicates measured for the respective condition. Double-bond number per FA is depicted in color code. Complete data can be found in Dataset S1 and illustrated in SI Appendix, Fig. S18.
An illustrative example for the conditional remodeling of CL compositions was the tissue-specificity observed in mouse and pig lipid extracts. Differences between these two species were dwarfed by the large tissue-specific variation of CL and FA profiles (SI Appendix, Text S3 and Fig. S16). In heart tissues, linoleoyl-rich CL species dominated, as expected. Although also in liver high FA18:2 levels were quantified, an increased FA18:1 and FA16:1 to FA18:2 ratio resulted in a broadening of the CL profile. This contrasts the FA profiles resolved for the brain, where only a small fraction of FA18:2 was found, in exchange for increased levels of FA20:4 and FA 22:6. A second example was that the Escherichia coli CL and FA patterns were strongly altered, when comparing the same strain grown in LB versus the minimal M9 medium (SI Appendix, Text S2 and Fig. S17). Furthermore, we observed distinct reconfigurations of the CL species during different growth phases of Saccharomyces cerevisiae (SI Appendix, Text S2 and Fig. S18). The underlying FA profiles were mainly based on FA16:1 and FA18:1. During logarithmic growth, CL side chains also contained their saturated counterparts, which, however, got depleted in later growth phase, leading to sharpened profiles centering at fourfold unsaturated CL species.
In general, the FA compositions incorporated into CLs resulting from analysis of this set of model organisms clearly followed the anticipated trend of an increased degree of unsaturation from bacteria toward higher eukaryotes (Fig. 5B). Procaryotic CLs contained shorter chain FAs, also with odd carbon chain length, and were typically highly saturated. The apparently complex CL patterns observed in brains of Mus musculus, Sus scrofa, and in different body regions of D. rerio was predominantly caused by high contents of the PUFAs FA20:4 and FA22:6 (SI Appendix, Fig. S19 and Dataset S1). In contrast, the highly unsaturated CL species in plants (A. thaliana, Oryza sativa) were based on an enrichment of α-linolenic acid (FA18:3).
Discussion
The structural variability of CLs observed in our study clearly demonstrates that the total CL mass alone, which is regularly used as mitochondrial marker (23), does by far not provide an accurate description of CL compositions. Even when utilizing MS2 fragment spectra for identification of major molecular lipid species, the results fall short of accurately describing the structural diversity of CL species, as demonstrated in SI Appendix, Fig. S5, where up to 20% of the CL72 mass alone would be incorrectly attributed to the tetraoleyl-CL molecular species. However, in several studies the recorded raw data would already contain the information required for structural characterization with our new data analysis strategy (16, 18, 21).
In contrast to other approaches (18, 24), we focus on single charged CL ions, which dominate under the present conditions. Furthermore, we operate the mass spectrometer in full scan mode with data-dependent acquisition of fragment spectra followed by targeted data extraction of CL features, rather than performing selected reaction monitoring-type experiments (24, 25). This comes along with the advantage of univocal identification of CLs orienting along internal references. Importantly, retrospect extraction of novel features is possible from previously recorded data and prevents incomplete data by extracting (baseline) information for lipids below the limit of detection. Dealing with the challenge of isobaric lipid species and fragments, our methodology solves a problem that persists also with high-resolution mass spectrometers. Nevertheless, increased sensitivities and reduced scan times allow recording additional MS2 data and performing MS3 experiments for all major CL species, which can connect the present FA profiles to an additional constraining experimental dataset (26).
The mechanisms that determine the exact molecular composition of CLs are so far only poorly understood. However, it is clear that the BTHS-linked transacylase Tafazzin plays a major role in the required remodeling process. In analogy to reports in the literature (e.g., ref. 27), we observed a strong reduction of CL mass in all BTHS patient cells. We further observed a preferential depletion of FA18:2 acyl chains compared with its more saturated counterparts. This is in good agreement with the augmented susceptibility of PUFAs to chemical (per)oxidation (28), suggesting that cellular oxidative damage preferentially targets double-bond–rich FA residues in CLs, while their saturated counterparts are more robust. Oxidative damage could therefore be a major driver behind CL depletion in BTHS patient fibroblasts and in CL homeostasis in general. In this context, Tafazzin could represent an essential part of a CL repair mechanism, after oxidized FAs are recognized and cleaved in a phospholipase-dependent manner.
A broad structural diversity of CLs was observed in different species and cell lines, where we also investigated the impact of environmental factors, growth conditions, and growth phases (Figs. 4 and 5 and SI Appendix, Fig. S13). These effects act in interdependence with cellular regulatory components of CL homeostasis, as for example shown by the cell lines U937 and K562, which were grown under similar conditions but exhibited different CL compositions with a mean carbon chain length deviation of ∼3.8 carbons. Importantly, the strong discrepancy between CL compositions in human cell lines and mammalian tissues advises caution when generalizing findings, especially in context of core CL functions in metabolism and apoptosis (9, 10, 29). This is also relevant when studying the connection between oxidative damage and CL remodeling. Linoleic acid (FA18:2) accounts for 10–20% of the CL mass in cultured cells (Figs. 3 and 4), whereas it constitutes a much larger fraction in heart and liver tissues (SI Appendix, Fig. S16C).
Our experiments show that lipid availability is a crucial factor for the emergence of specific CL compositions. When cells relied only on fatty acid de novo biosynthesis, CL pools were mainly composed of palmitoleic and oleic acid, whereas supplementation with heart lipid extracts generated a profile mimicking heart tissue (Fig. 4A and SI Appendix, Fig. S16). Importantly, these treatments did not only remodulate the mitochondrial lipid pool composition, but also had functional consequences for mitochondrial metabolic processes. We show that heart lipid treatment resulted in the activation of the NADH-linked mitochondrial pathway in permeabilized cells, which could be causative for the growth-independent reduction of ROUTINE oxygen consumption in intact cells (Fig. 4 C and E). Although the exact mechanistic links are still elusive, these observations fall in line with previous reports about the interplay between CL compositions and mitochondrial function (30), potentially via specific CL interactions with respiratory protein complexes (14) or its role in establishing efficient mitochondrial membrane structures (7, 31).
In summary, we comprehensively analyzed and structurally characterized the molecular mitochondrial CL composition in: (i) disease models, (ii) cell-culture models, (iii) CL modulation experiments, and (iv) a set of 13 species, and all data are openly available. By combining HPLC-MS/MS with a subsequent mathematical structural modeling pipeline for recorded fragment spectra, we were able to investigate CL variability in great molecular detail (Fig. 2). This will thus greatly expand our ability to study the underlying mechanistic principles responsible for establishing the large diversity of CL patterns that are observed in different species and under different conditions (SI Appendix, Fig. S15), and to understand their functional consequences.
Methods
Materials and methods are described briefly. Details in SI Appendix.
Sample Material and Respirometry.
The study was conducted in accordance with the Helsinki Declaration of 1975, as revised in 2000, and was approved as part of the Biobank for Rare Diseases by the Ethics Committee of the Medical University of Innsbruck, Austria (study no. UN4501), and experiments were performed after obtaining informed consent. If not stated otherwise, cells were grown under standard conditions (SI Appendix, Table S1) and biomass was homogenized in PBS. For lipid supplementation, S. scrofa heart lipid extracts were added to lipid-free Panserin 401 cell culture medium. High-resolution respirometry was performed in intact and permeabilized cells with the Oxygraph-2k (Oroboros Instruments).
Lipid Extraction and CL Analysis by LC-MS/MS.
Sample material was homogenized in water and lipids were extracted following the Folch method with CL(14:0)4 as internal standard (ISTD). Lipids were dissolved in HPLC starting condition separated by reversed-phase HPLC on a Dionex Ultimate 3000 HPLC (Thermo Fisher) and quantified with a Thermo Velos Pro Dual-Pressure Linear Ion Trap Mass Spectrometer.
Quantitative Data Analysis.
Data were analyzed in MZMine2 (32) and R (www.r-project.org/). Targeted peak integration was performed on baseline-corrected data. Quantification was achieved by external calibration, accounting for the ISTD response, and normalization to protein content.
MS/MS-Based Structural CL and FA Modeling.
Mathematical structural modeling was achieved by minimizing the Euclidean distance between a theoretical PA profile-dependent theoretical fragmentation model and the measured MS2 spectrum (Dataset S2). This minimization was achieved by using the box constraint, limited-memory modification of the BFGS quasi-Newton method “L-BFGS-B” (22). For each MS2 scan a modeled PA profile was obtained. To construct the final CL structural landscape, all PA profiles referring to one CL species were summed proportionally to their MS2 intensities and subsequently weighted them according to their respective MS1 levels. FA side-chain distributions were modeled in an analog manner, by linking and minimizing the analyzed PA compositions with theoretical PA profiles based on dual FA combinations within the available FA space (Dataset S3). FA profiles were either solved from cumulative PA profiles, or for PA compositions of single CL species.
Data Analysis.
Data visualization and statistical analysis was conducted using R.
Supplementary Material
Acknowledgments
The authors thank Petra Loitzl and Nina Madl for expert technical help; Prof. Hubertus Haas and Dr. Beatrix Lechner (Medical University of Innsbruck) for providing Aspergillus; Prof. Alexandra Lusser and Ines Schoberleitner (Medical University of Innsbruck) for providing Drosophila; Dr. Sandrine Dubrac (Medical University of Innsbruck) for providing fibroblasts; Daniele Seppi (Medical University of Innsbruck) for providing Dictyostelium; Prof. Dirk Meyer (University of Innsbruck) for providing zebrafish; and Dr. Sabine Podmirseg and Dr. Andreas Wagner (University of Innsbruck) for providing bacterial strains. This work was supported by an Erwin Schrödinger postdoctoral fellowship (Fonds zur Förderung der Wissenschaftlichen Forschung J3341) and a Medical University of Innsbruck Start grant of the Medical University of Innsbruck (to M.A.K.). G.O. was supported by an i-med Research Scholarship of the Medical University of Innsbruck.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Source data have been deposited to the EMBL-EBI MetaboLights database with the identifier MTBLS636. All quantified data files have been deposited in the Mendeley Data repository (dx.doi.org/10.17632/kntnfm8grc.1) and are accessible in a browsable format at www.humgen.at/cardiolipin/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719407115/-/DCSupplemental.
References
- 1.Holthuis JCM, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 2.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, Willers C, Kunji ERS, Crichton PG. Uncoupling protein 1 binds one nucleotide per monomer and is stabilized by tightly bound cardiolipin. Proc Natl Acad Sci USA. 2015;112:6973–6978. doi: 10.1073/pnas.1503833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 5.Acehan D, et al. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys J. 2011;100:2184–2192. doi: 10.1016/j.bpj.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:C33–C44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 7.Harner ME, et al. Aim24 and MICOS modulate respiratory function, tafazzin-related cardiolipin modification and mitochondrial architecture. eLife. 2014;3:e01684. doi: 10.7554/eLife.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luévano-Martínez LA, Forni MF, dos Santos VT, Souza-Pinto NC, Kowaltowski AJ. Cardiolipin is a key determinant for mtDNA stability and segregation during mitochondrial stress. Biochim Biophys Acta. 2015;1847:587–598. doi: 10.1016/j.bbabio.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalvez F, Gottlieb E. Cardiolipin: Setting the beat of apoptosis. Apoptosis. 2007;12:877–885. doi: 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]
- 10.Kagan VE, et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 11.Schlame M. Cardiolipin remodeling and the function of tafazzin. Biochim Biophys Acta. 2013;1831:582–588. doi: 10.1016/j.bbalip.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Ren M, Phoon CKL, Schlame M. Metabolism and function of mitochondrial cardiolipin. Prog Lipid Res. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Vreken P, et al. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun. 2000;279:378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, et al. Loss of protein association causes cardiolipin degradation in Barth syndrome. Nat Chem Biol. 2016;12:641–647. doi: 10.1038/nchembio.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barth PG, et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci. 1983;62:327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- 16.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: Focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal Chem. 2011;83:940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyurina YY, et al. LC/MS characterization of rotenone induced cardiolipin oxidation in human lymphocytes: Implications for mitochondrial dysfunction associated with Parkinson’s disease. Mol Nutr Food Res. 2013;57:1410–1422. doi: 10.1002/mnfr.201200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valianpour F, Wanders RJA, Barth PG, Overmars H, van Gennip AH. Quantitative and compositional study of cardiolipin in platelets by electrospray ionization mass spectrometry: Application for the identification of Barth syndrome patients. Clin Chem. 2002;48:1390–1397. [PubMed] [Google Scholar]
- 19.Minkler PE, Hoppel CL. Separation and characterization of cardiolipin molecular species by reverse-phase ion pair high-performance liquid chromatography-mass spectrometry. J Lipid Res. 2010;51:856–865. doi: 10.1194/jlr.D002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens J-C, et al. Lipidomic analysis of molecular cardiolipin species in livers exposed to ischemia/reperfusion. Mol Cell Biochem. 2015;400:253–263. doi: 10.1007/s11010-014-2282-1. [DOI] [PubMed] [Google Scholar]
- 21.Hsu F-F, et al. Structural characterization of cardiolipin by tandem quadrupole and multiple-stage quadrupole ion-trap mass spectrometry with electrospray ionization. J Am Soc Mass Spectrom. 2005;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Byrd RH, Lu P, Nocedal J, Zhu C. A limited memory algorithm for bound constrained optimization. SIAM J Sci Comput. 1995;16:1190–1208. [Google Scholar]
- 23.Larsen S, et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol. 2012;590:3349–3360. doi: 10.1113/jphysiol.2012.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowron A, et al. Diagnosis of Barth syndrome using a novel LC-MS/MS method for leukocyte cardiolipin analysis. J Inherit Metab Dis. 2013;36:741–746. doi: 10.1007/s10545-012-9552-4. [DOI] [PubMed] [Google Scholar]
- 25.Garrett TA, Kordestani R, Raetz CRH. Quantification of cardiolipin by liquid chromatography-electrospray ionization mass spectrometry. Methods Enzymol. 2007;433:213–230. doi: 10.1016/S0076-6879(07)33012-7. [DOI] [PubMed] [Google Scholar]
- 26.Hsu F-F, Turk J. Toward total structural analysis of cardiolipins: Multiple-stage linear ion-trap mass spectrometry on the [M - 2H + 3Li]+ ions. J Am Soc Mass Spectrom. 2010;21:1863–1869. doi: 10.1016/j.jasms.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlame M, et al. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann Neurol. 2002;51:634–637. doi: 10.1002/ana.10176. [DOI] [PubMed] [Google Scholar]
- 28.Xu L, Davis TA, Porter NA. Rate constants for peroxidation of polyunsaturated fatty acids and sterols in solution and in liposomes. J Am Chem Soc. 2009;131:13037–13044. doi: 10.1021/ja9029076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlame M, Greenberg ML. Biosynthesis, remodeling and turnover of mitochondrial cardiolipin. Biochim Biophys Acta. 2016;1862:3–7. doi: 10.1016/j.bbalip.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vergeade A, et al. Cardiolipin fatty acid remodeling regulates mitochondrial function by modifying the electron entry point in the respiratory chain. Mitochondrion. 2016;28:88–95. doi: 10.1016/j.mito.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim T-H, et al. Bid-cardiolipin interaction at mitochondrial contact site contributes to mitochondrial cristae reorganization and cytochrome C release. Mol Biol Cell. 2004;15:3061–3072. doi: 10.1091/mbc.E03-12-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.