Performing a genome-wide association study of Salmonella enterica serovar Typhi (S. Typhi) invasion, Alvarez et al. (1) identify a trait-associated SNP, rs8060947, in VAC14. rs8060947 is an expression quantitative trait locus for VAC14 RNA expression, and carriage of the A allele is associated with reduced VAC14 RNA and protein expression, and increased invasion of S. Typhi. VAC14-associated inhibition of S. Typhi invasion is mediated by a reduction in host cell membrane cholesterol. Carriage of the A allele at rs8060947 is associated with typhoid fever in Vietnamese individuals [cases = 496, controls = 500; P = 0.01, additive odds ratio (OR) = 1.38]. The authors further identify a SNP in high linkage disequilibrium with rs8060947 and rs8044133, located in a transcription factor binding site, which merits further investigation as the causative SNP.
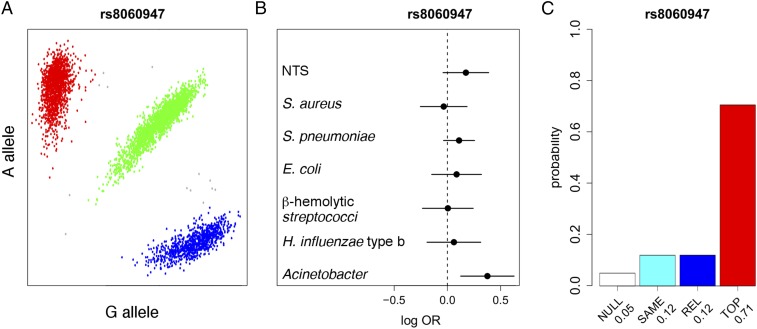
Alvarez et al. (1) note that cholesterol has been implicated in the invasion and pathogenesis of a wide range of pathogens (2–5). We therefore used previously published genome-wide association study data, describing susceptibility to bacteraemia secondary to diverse pathogens in Kenyan children (6), to explore whether genetic variation at the VAC14 locus is associated with susceptibility to invasive bacterial infections other than typhoid fever. In Kenyan children (cases = 1,536, controls = 2,677), rs8060947 (Fig. 1A) is significantly associated with all-cause bacteremia: Padditive = 0.02, OR = 1.11 [95% confidence interval (CI) 1.02–1.22]. In keeping with the effect observed in typhoid, carriage of the A allele at rs8060947 increases risk of bacteremia.
Fig. 1.
rs8060947 and major causes of bacteremia in Kenyan children. (A) Cluster plot of rs8060947 (Affymetrix SNP 6.0 chip) in Kenyan children (n = 4,924); minor allele frequency (MAF) = 0.44, Hardy–Weinberg equilibrium (HWE) P = 0.11. (B) rs8060947 association with major causes of bacteremia in Kenyan children; NTS, n = 180, Staphylococcus aureus, n = 175, S. pneumoniae, n = 426, E. coli, n = 151, β-hemolytic Streptococci, n = 146, Haemophilus influenza type b, n = 128, Acinetobacter species, n = 130. Log-transformed ORs and 95% CIs of rs8060947 association (additive model) are calculated by multinomial logistic regression, including four principal components of genome-wide genotyping data to account for population structure. (C) Posterior probabilities of models of association at rs8060947: NULL, no association with any pathogen; REL, related effects ∼ N(0,0.22) across all pathogens (correlation is ρ=0.96); SAME, the same effect ∼ N(0,0.22) across all pathogens (correlation is ρ = 1); TOP, a nonzero effect in bacteremia secondary to nontyphoidal Salmonella, E. coli, S. pneumoniae, and Acinetobacter species alone; this model (highlighted in red) is the most probable (Bayes factor c.f. NULL = 14). Genotype and phenotype data are derived from Rautanen et al. (6). Methods are as described in Rautanen et al. (6).
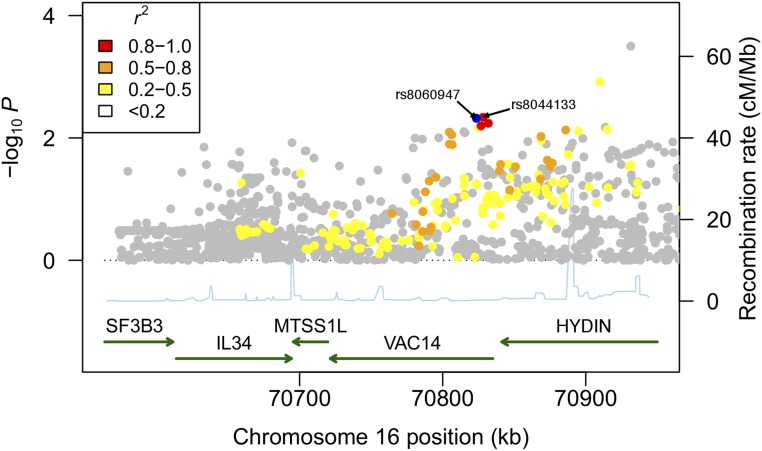
To understand whether risk of bacteremia conferred by rs8060947 is shared across pathogens causing bacteremia in Kenyan children, we conducted a Bayesian analysis comparing models of association at rs8060947 with the major causes of bacteremia in this population (Fig. 1B). The most probable model is one in which rs8060947 is associated with susceptibility to bacteraemia caused by nontyphoidal Salmonella (NTS), Streptococcus pneumoniae, Escherichia coli, and Acinetobacter species, but not bacteremia caused by other pathogens (Fig. 1C). We performed imputation-based mapping of the association at the VAC14 locus with bacteremia secondary to NTS, S. pneumoniae, E. coli, and Acinetobacter species (Fig. 2). In that analysis, there is evidence for association between bacteremia secondary to these four pathogens and both rs8060947 [Padditive = 4.76 × 10−3, OR = 1.17 (95% CI = 1.05–1.30)] and rs8044133 [Padditive = 4.58 × 10−3, OR = 1.17 (95% CI = 1.05–1.30)].
Fig. 2.
Association plot of bacteremia susceptibility at the VAC14 locus. SNPs are colored according to strength of linkage disequilibrium (r2) to rs8060947. Association statistics are calculated using an additive model including bacteremia cases secondary to NTS, E. coli, S. pneumoniae, and Acinetobacter species (n = 887) and shared, healthy controls (n = 2,677). rs8044133 is imputed (imputation information score = 0.996), MAF = 0.47, with no evidence of departure from HWE (P = 0.73). Common (MAF > 0.05), well-imputed SNPs (imputation information score > 0.4), with no evidence for departure from HWE (P > 1 × 10−10) were included in the analysis. Association statistics are calculated by logistic regression, including four principal components of genome-wide genotyping data to account for population structure. Genotype and phenotype data are derived from Rautanen et al. (6). Methods are as described in Rautanen et al. (6).
Alvarez et al. (1) demonstrate that genetic variation in VAC14 is a determinant of clinical typhoid fever in Vietnamese individuals. Our data expand on this observation, demonstrating that the same risk allele at rs8060947, with a comparable effect size, increases risk of bacteremia secondary to diverse pathogens in Kenyan children. This observation may reflect a previously unrecognized role for cholesterol in the pathogenesis of diverse bacterial pathogens, or a role for cholesterol in a shared risk factor for these pathogens in this population (e.g., malaria or HIV). Future studies will be required to further define the range of clinical diseases associated with VAC14, and to fine-map the genetic signal at VAC14.
Acknowledgments
The Kenyan Bacteraemia Study Group consists of the following. Principal Investigators: A.V.S.H. (Chair), T.N.W., J.A.G.S., and Stephen J. Chapman. Key Personnel: A.R., Tara C. Mills, Kirk Rockett, Anne W. Ndungu, Vivek Naranbhai, Alex W. Macharia, Sophie Uyoga, Carolyne Ndila, N.M., P.N., Shebe Mohammed, James A. Berkley, Isaiah Mwangi, S.M., Barnes S. Kitsao, Brett S. Lowe, Susan C. Morpeth, and Iqbal Khandwalla. The Kilifi DNA extraction Group: Alex W. Macharia, Sophie Uyoga, Herbert Opi, Carolyne Ndila, Emily Nyatichi, Prophet Ingosi, Barnes Kitsao, Clement Lewa, Johnstone Makale, Adan Mohamed, Kenneth Magua, Mary Njoroge, Gideon Nyutu, Ruth Mwarabu, Metrine Tendwa, and T.N.W.. The Kilifi Bacteraemia Surveillance Group: Ismail Ahmed, Samuel Akech, Alexander Balo Makazi, Mohammed Bakari Hajj, Andrew Brent, Charles Chesaro, Hiza Dayo, Richard Idro, Patrick Kosgei, Kathryn Maitland, Kevin Marsh, Laura Mwalekwa, Shalton Mwaringa, Charles Newton, Mwanajuma Ngama, Allan Pamba, Norbert Peshu, Anna Seale, Alison Talbert, and T.N.W.
The Wellcome Trust Case Control Consortium 2 consists of the following. Management Committee: Peter Donnelly (Chair), Ines Barroso (Deputy Chair), Jenefer M. Blackwell, Elvira Bramon, Matthew A. Brown, Juan P. Casas, Aiden Corvin, Panos Deloukas, Audrey Duncanson, Janusz Jankowski, Hugh S. Markus, Christopher G. Mathew, Colin N. A. Palmer, Robert Plomin, A.R., Stephen J. Sawcer, Richard C. Trembath, Ananth C. Viswanathan, and Nicholas W. Wood. Data and Analysis Group: Chris C. A. Spencer, Gavin Band, Céline Bellenguez, Colin Freeman, Garrett Hellenthal, Eleni Giannoulatou, M.P., Richard Pearson, Amy Strange, Zhan Su, Damjan Vukcevic, and Peter Donnelly. DNA, Genotyping, Data Quality Control, and Informatics Group: Cordelia Langford, Sarah E. Hunt, Sarah Edkins, Rhian Gwilliam, Hannah Blackburn, Suzannah J. Bumpstead, Serge Dronov, Matthew Gillman, Emma Gray, Naomi Hammond, Alagurevathi Jayakumar, Owen T. McCann, Jennifer Liddle, Simon C. Potter, Radhi Ravindrarajah, Michelle Ricketts, Matthew Waller, Paul Weston, Sara Widaa, Pamela Whittaker, Ines Barroso, and Panos Deloukas. Publications Committee: Christopher G. Mathew (Chair), Jenefer M. Blackwell, Matthew A. Brown, Aiden Corvin, and Chris C. A. Spencer.
Footnotes
The authors declare no conflict of interest.
2A complete list of Wellcome Trust Case-Control Consortium 2 and The Kenyan Bacteraemia Study Group can be found in the Acknowledgments.
Contributor Information
Collaborators: Adrian V S Hill, Thomas N Williams, J Anthony G Scott, Stephen J Chapman, Anna Rautanen, Tara C Mills, Kirk Rockett, Anne W Ndungu, Vivek Naranbhai, Alex W Macharia, Sophie Uyoga, Carolyne Ndila, Neema Mturi, Patricia Njuguna, Shebe Mohammed, James A Berkley, Isaiah Mwangi, Salim Mwarumba, Barnes S Kitsao, Brett S Lowe, Susan C Morpeth, Iqbal Khandwalla, Alex W Macharia, Sophie Uyoga, Herbert Opi, Carolyne Ndila, Emily Nyatichi, Prophet Ingosi, Barnes Kitsao, Clement Lewa, Johnstone Makale, Adan Mohamed, Kenneth Magua, Mary Njoroge, Gideon Nyutu, Ruth Mwarabu, Metrine Tendwa, Thomas N Williams, Ismail Ahmed, Samuel Akech, Alexander Balo Makazi, Mohammed Bakari Hajj, Andrew Brent, Charles Chesaro, Hiza Dayo, Richard Idro, Patrick Kosgei, Kathryn Maitland, Kevin Marsh, Laura Mwalekwa, Shalton Mwaringa, Charles Newton, Mwanajuma Ngama, Allan Pamba, Norbert Peshu, Anna Seale, Alison Talbert, Thomas N Williams, Peter Donnelly, Ines Barroso, Jenefer M Blackwell, Elvira Bramon, Matthew A Brown, Juan P Casas, Aiden Corvin, Panos Deloukas, Audrey Duncanson, Janusz Jankowski, Hugh S Markus, Christopher G Mathew, Colin NA Palmer, Robert Plomin, Anna Rautanen, Stephen J Sawcer, Richard C Trembath, Ananth C Viswanathan, Nicholas W Wood, Chris C A Spencer, Gavin Band, Céline Bellenguez, Colin Freeman, Garrett Hellenthal, Eleni Giannoulatou, Matti Pirinen, Richard Pearson, Amy Strange, Zhan Su, Damjan Vukcevic, Peter Donnelly, Cordelia Langford, Sarah E Hunt, Sarah Edkins, Rhian Gwilliam, Hannah Blackburn, Suzannah J Bumpstead, Serge Dronov, Matthew Gillman, Emma Gray, Naomi Hammond, Alagurevathi Jayakumar, Owen T McCann, Jennifer Liddle, Simon C Potter, Radhi Ravindrarajah, Michelle Ricketts, Matthew Waller, Paul Weston, Sara Widaa, Pamela Whittaker, Ines Barroso, Panos Deloukas, Christopher G Mathew, Jenefer M Blackwell, Matthew A Brown, Aiden Corvin, and Chris C A Spencer
References
- 1.Alvarez MI, et al. Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol. Proc Natl Acad Sci USA. 2017;114:E7746–E7755. doi: 10.1073/pnas.1706070114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hacke M, et al. Inhibition of Ebola virus glycoprotein-mediated cytotoxicity by targeting its transmembrane domain and cholesterol. Nat Commun. 2015;6:7688. doi: 10.1038/ncomms8688. [DOI] [PubMed] [Google Scholar]
- 3.Jutras I, Abrami L, Dautry-Varsat A. Entry of the lymphogranuloma venereum strain of Chlamydia trachomatis into host cells involves cholesterol-rich membrane domains. Infect Immun. 2003;71:260–266. doi: 10.1128/IAI.71.1.260-266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voisset C, et al. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J Biol Chem. 2005;280:7793–7799. doi: 10.1074/jbc.M411600200. [DOI] [PubMed] [Google Scholar]
- 5.Samuel BU, et al. The role of cholesterol and glycosylphosphatidylinositol-anchored proteins of erythrocyte rafts in regulating raft protein content and malarial infection. J Biol Chem. 2001;276:29319–29329. doi: 10.1074/jbc.M101268200. [DOI] [PubMed] [Google Scholar]
- 6.Rautanen A, et al. Kenyan Bacteraemia Study Group; Wellcome Trust Case Control Consortium 2 (WTCCC2); Kilifi Bacteraemia Surveillance Group Polymorphism in a lincRNA associates with a doubled risk of pneumococcal bacteremia in Kenyan children. Am J Hum Genet. 2016;98:1092–1100. doi: 10.1016/j.ajhg.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]