CONSPECTUS
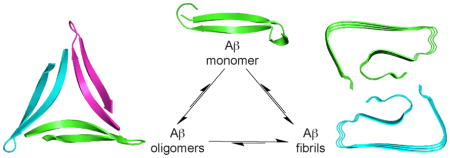
In the more than a century since its identification, Alzheimer’s disease has become the archetype of amyloid diseases. The first glimpses of the chemical basis of Alzheimer’s disease began with the identification of “amyloid” plaques in the brain in 1892 and extended to the identification of proteinaceous fibrils with “cross-β” structure in 1968. Further efforts led to the discovery of the β-amyloid peptide Aβ as a 40- or 42-amino acid peptide that is responsible for the plaques and fibrils. At this point, a three-decade long marathon began to elucidate the structure of the fibrils and identify the molecular basis of Alzheimer’s disease. Along the way, an alternative model began to emerge in which small aggregates of Aβ, called “oligomers”, rather than fibrils, are the culprits that lead to neurodegeneration in Alzheimer’s disease. This Account describes what is known about the structures of the fibrils and details our research group’s efforts to understand the structural, biophysical, and biological properties of the oligomers in amyloid diseases.
β-Sheets are the building blocks of amyloid fibrils and oligomers. Amyloid fibrils generally consist of extended networks of parallel β-sheets. Amyloid oligomers appear to be more compact enclosed structures, some of which are thought to be composed of antiparallel β-sheets comprising β-hairpins. β-Hairpins are special because their twisted shape, hydrophobic surfaces, and exposed hydrogen-bonding edges impart a unique propensity to form compact assemblies. Our laboratory has developed macrocyclic β-sheets that are designed to mimic β-hairpins formed by amyloidogenic peptides and proteins. The β-hairpin mimics contain two β-strand peptide fragments linked together at their N- and C-termini by two δ-linked ornithine turn mimics to create a macrocycle. An N-methyl group is installed on one of the β-strands to prevent uncontrolled aggregation. These design features facilitate crystallization of the β-hairpin mimics and determination of the X-ray crystallographic structures of the oligomers that they form.
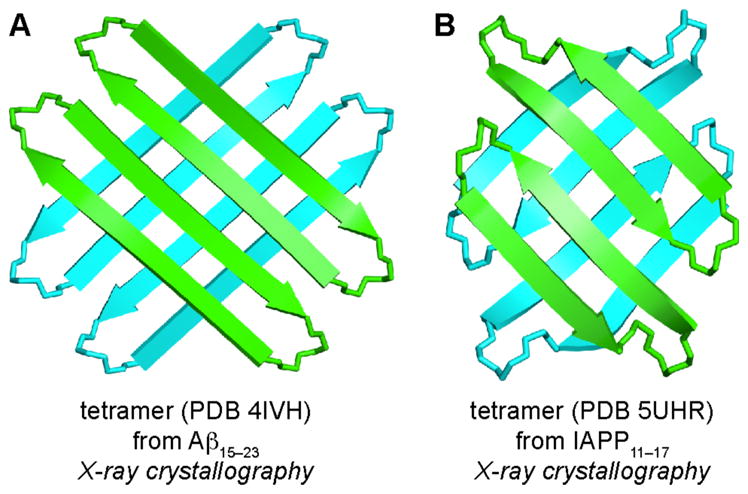
During the past few years, our laboratory has elucidated the X-ray crystallographic structures of oligomers formed by β-hairpin mimics derived from Aβ, α-synuclein, and β2-microglobulin. Out of these three amyloidogenic peptides and proteins, the Aβ β-hairpin mimics have provided the most insight into amyloid oligomers. Our studies have revealed a previously undiscovered mode of self-assembly, whereby three Aβ β-hairpin mimics assemble to form a triangular trimer. The triangular trimers are remarkable, because they contain two largely hydrophobic surfaces that pack together with other triangular trimers to form higher-order oligomers, such as hexamers and dodecamers. Some of the dodecamers pack in the crystal lattice to form annular porelike assemblies. Some of the β-hairpin mimics and triangular trimers assemble in solution to form oligomers that recapitulate the crystallographically observed oligomers. These oligomers exhibit toxicity toward neuronally derived cells, recapitulating the toxicity of the oligomers formed by full-length amyloidogenic peptides and proteins. These findings are significant, because they address a gap in understanding the molecular basis of amyloid diseases. We anticipate that these studies will pave the way for developing diagnostics and therapeutics to combat Alzheimer’s disease, Parkinson’s disease, and other amyloid diseases.
CONSPECTUS GRAPHIC
INTRODUCTION
In amyloid diseases, amyloidogenic peptides and proteins self-assemble into oligomers and fibrils (Table 1). Amyloid oligomers have emerged as important contributors to the pathogenesis of amyloid diseases.1,2 High-resolution structures of the toxic amyloid oligomers have eluded researchers since their discovery, constituting a significant gap in understanding amyloid diseases. Over the past few years, our laboratory has used X-ray crystallography to identify undiscovered modes of self-assembly of macrocyclic β-hairpin mimics containing sequences from amyloidogenic peptides and proteins. These assemblies provide insights into the structures of the elusive amyloid oligomers and may help shed light on the molecular basis of amyloid diseases. This Account summarizes the accomplishments thus far in elucidating the structures of amyloid fibrils and oligomers at high resolution.
Table 1.
Amyloidogenic peptides and proteins discussed in this Account and their associated diseases.
| disease | peptide/protein |
|---|---|
| Alzheimer’s disease | β-amyloid peptide Aβ |
| Parkinson’s disease | α-synuclein (α-Syn) |
| Huntington’s disease | huntingtin |
| amyotrophic lateral sclerosis (ALS) | superoxide dismutase 1 (SOD1) |
| Creutzfeld-Jakob disease | human prion protein (hPrP) |
| type-2 diabetes | islet amyloid polypeptide (IAPP) |
| dialysis-related amyloidosis | β2-microglobulin (B2M) |
STRUCTURAL ELUCIDATION OF AMYLOID FIBRILS
Aβ is the most extensively characterized of the more than thirty known amyloidogenic peptides and proteins and has served as the archetype for studying the structures of amyloid assemblies. X-ray diffraction measurements initially revealed that amyloid plaques produced a “cross-β” pattern, indicating that the proteinaceous components of the plaques have a “pleated sheet” conformation and providing a glimpse into the molecular structures of amyloid fibrils.3 The determination of the sequences of the 40- and 42-amino acid alloforms of Aβ (Aβ40 and Aβ42) provided the next piece of the puzzle. Solid-state NMR spectroscopy (ss-NMR) and X-ray diffraction of fibrils established that the fibrils are composed of an extended network of in-register parallel β-sheets, with the β-strands running perpendicular to the long axes of the fibrils and the hydrogen bonds running parallel. These studies produced the first molecular models of amyloid fibrils.
Elucidating the full three-dimensional structures of Aβ40 and Aβ42 fibrils has required further effort over the course of decades, because the structures of amyloid fibrils are difficult to determine by traditional high-resolution structure determination techniques. ss-NMR has thus far proven the most fruitful technique in fibril structure determination. Extensive ss-NMR studies by Tycko and coworkers unlocked the molecular structures of Aβ40 fibrils and revealed a rich structural polymorphism of the fibrils. ss-NMR studies by the research groups of Riek, Ishii, and Griffin elucidated the structure of a fibril polymorph of Aβ42. Eisenberg and coworkers pioneered X-ray crystallography of single micro-crystals composed of peptide fragments from key regions of amyloidogenic peptides and proteins, providing additional insights into how amyloid fibrils pack. Recently, cryo-electron microscopy (cryo-EM) has begun to emerge as a promising new technique for amyloid fibril structure determination. The following sections present the structures of fibrils of full-length amyloidogenic peptides and proteins, with a focus on structures that have been deposited in the Protein Data Bank (PDB) and are publically available to download (Table S1).
Aβ40 Fibrils
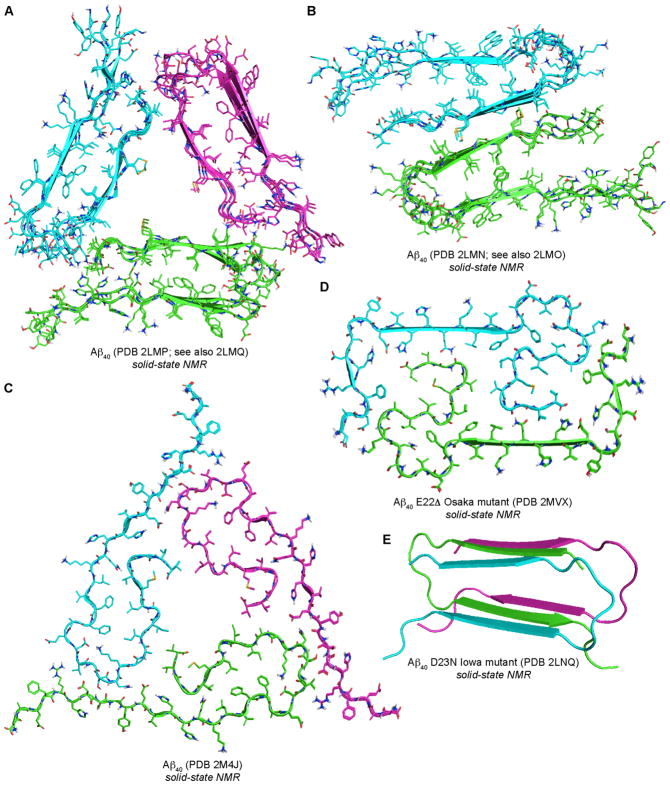
Tycko and coworkers are at the forefront of Aβ40 fibril structure determination and have developed methods for generating homogeneous fibrils from chemically synthesized Aβ40. These methods yielded two types of Aβ40 fibrils that have been reported thus far at high resolution: one contains three-fold symmetrical fibrils that have a twisted morphology; the other contains two-fold symmetrical fibrils that have a striated ribbon morphology.4,5 In the two-fold symmetrical fibrils, the two protofilament subunits stack on top of one another to create a four-layered β-sheet (Figure 1B). In the three-fold symmetrical fibrils, the three protofilament subunits arrange in a triangular fashion (Figure 1A). The structures of the protofilament subunits of both fibril types are very similar, each being composed of extended networks of layered, in-register parallel β-sheets. Central residues 11–22 and C-terminal residues 30–39 comprise two β-strands connected by a loop containing residues 23–29. The N-terminal residues 1–10 are unstructured.
Figure 1.
Structures of Aβ40 fibrils.
Tycko and coworkers have extended these techniques to determine the structure of Aβ40 fibrils from brain tissue using fibrils isolated from Alzheimer’s disease brain to seed fibrils of isotopically labeled Aβ40.6 ss-NMR revealed a three-fold symmetrical fibril (Figure 1C), similar to the three-fold symmetrical fibrils described above (Figure 1A). In this fibril structure, residues 12–19 form a β-strand which is connected to residues 30–40 by a loop comprising residues 21–29. Residues 30–40 are buried in the hydrophobic core of the fibril.
A small percentage of Alzheimer’s disease cases are associated with mutations in Aβ.7 Tycko and coworkers determined that the Aβ40 D23N Iowa mutant forms metastable fibrils composed of antiparallel β-sheets.8 The fibrils contain a double-layer antiparallel β-sheet in a single cross-β protofilament with β-strands comprising residues 16–23 and 30–36 connected by a loop comprising residues 24–29 (Figure 1E). Meier and coworkers determined the structure of fibrils formed by the Aβ40 E22Δ Osaka mutant.9 This structure revealed two-fold symmetrical fibrils with convoluted protofilament subunits that create a tightly packed core filled exclusively with hydrophobic residues (Figure 1D).
Aβ42 Fibrils
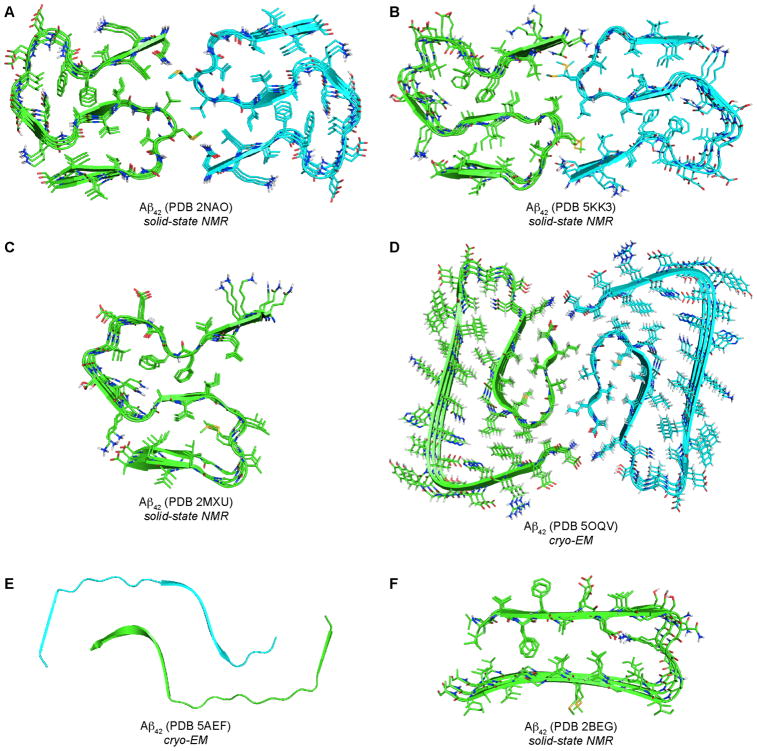
The research groups of Riek, Griffin, and Ishii have independently determined the structure of an Aβ42 fibril polymorph using ss-NMR.10,11,12 The protofilament subunits of the Aβ42 fibrils have a convoluted S shape, which differs substantially from the shape of the protofilament subunits in the Aβ40 fibrils (Figures 2A, B, and C). The structures from Riek and Griffin identify the fibrils as containing two protofilament subunits and being twofold symmetrical. The interior of each protofilament subunit contains packed hydrophobic cores, while the solvent-exposed exterior primarily contains charged and polar residues. A salt bridge between the side chain of Lys28 and the Ala42 carboxylate group provides additional stability and may explain why Aβ42 forms different fibrils than Aβ40. Schröder, Willbold, and coworkers recently determined the structure of a different Aβ42 fibril polymorph using cryo-EM (Figure 2D).13 Unlike previous Aβ fibril structures, the N-terminus is ordered and is part of the cross-β structure of the fibril.
Figure 2.
Structures of Aβ42 fibrils.
α-Syn and Tau Fibrils
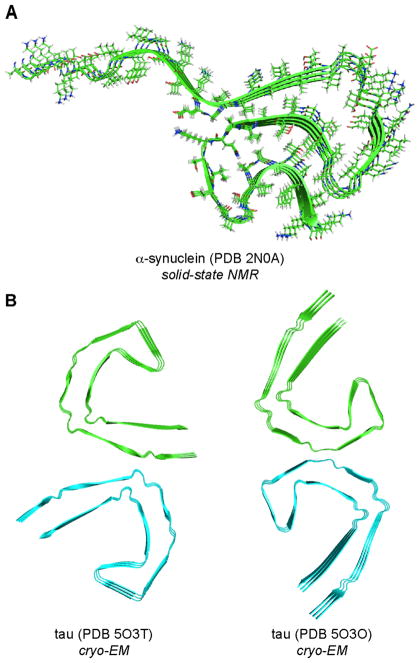
Rienstra and coworkers determined the structure of α-Syn fibrils generated from recombinantly expressed α-Syn.14 This structure revealed a single protofilament in which the packed core is composed of in-register parallel β-sheets (Figure 3A). Scheres and coworkers used cryo-EM to determine the structures of two different types of tau fibrils that were isolated from Alzheimer’s disease brains.15 Both consist of a pair of identical filaments encompassing tau306–378 and are composed of in-register parallel β-sheets (Figure 3B). The two types of fibrils differ in how the filaments pack together.
Figure 3.
Structures of an α-synuclein fibril (A) and two different paired filaments of the tau protein (B).
Additional Insights into Amyloid Fibril Structures
Eisenberg and coworkers have determined the X-ray crystallographic structures of fibrils formed by peptide fragments from amyloidogenic peptides and proteins.16,17,18 These structures revealed that the peptide fragments often form extended networks of in-register parallel β-sheets, which pack together through hydrophobic interactions. Eisenberg and coworkers used these peptide fragment structures to construct models of fibrils of full-length amyloidogenic peptides and proteins, providing a complementary approach to ss-NMR.
Over the last three decades, structural studies of amyloid fibrils have provided insights into the molecular basis of amyloid diseases. Through these studies, two structural patterns have emerged. Early work revealed flat, extended networks of laminated, layered β-sheets. Subsequent studies have shown more complex structures with convoluted shapes containing densely packed cores. The structures of amyloid fibrils are remarkable within the realm of structural biology, because they are composed of repeats of monomer subunits that extend in a single dimension. The extended networks in fibrils provide stark contrast with the enclosed structures of globular proteins, even though the same stabilizing forces that govern globular protein folding also direct amyloid fibril assembly.
STRUCTURAL ELUCIDATION OF AMYLOID MONOMERS
Amyloidogenic peptide and protein monomers are often classified as “intrinsically disordered proteins”, meaning that in their monomeric state the proteins lack stable secondary structural elements. The lack of well-defined structures makes meaningful characterization of the monomers by NMR spectroscopy and X-ray crystallography challenging. Characterization of smaller amyloidogenic peptides, such as Aβ, α-Syn, and IAPP is further complicated by the propensities of the peptides to aggregate. Interaction with other entities, such as another monomer, oligomer, fibril, or lipid membrane can shift the population of states and facilitate the formation of stable secondary structures. Table S2 summarizes key structures of monomeric amyloidogenic peptides and proteins that have been deposited in the PDB.
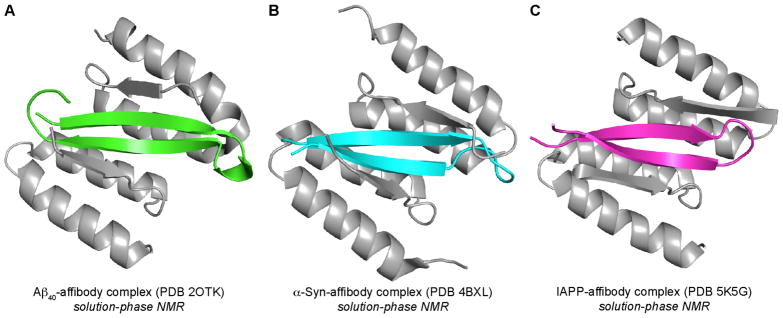
Folding of the monomers into β-hairpins is thought to have particular significance in the oligomerization of amyloidogenic peptides and proteins. β-Hairpins are special because their twisted shape, hydrophobic surfaces, and exposed hydrogen-bonding edges impart a unique propensity to form compact assemblies. Härd, Hoyer, and coworkers have reported three structures of β-hairpins formed by monomeric Aβ40, α-Syn, and IAPP (Figure 4).19,20,21 In each study, the monomeric peptide is bound to an affibody designed to stabilize the β-hairpin, and solution-phase NMR spectroscopy is used to elucidate the structure of the β-hairpin-affibody complex. These structures revealed the regions in Aβ, α-Syn, and IAPP that adopt β-strand conformations and the alignment of the β-strands that comprise the β-hairpin.
Figure 4.
Structures of β-hairpin-affibody complexes of Aβ (A), α-Syn (B), and IAPP (C).
STRUCTURAL ELUCIDATION OF AMYLOID OLIGOMERS
As researchers worked to understand amyloid fibrils, increasing evidence began to emerge that other, smaller soluble oligomers play a crucial role in the pathogenesis of amyloid diseases. Amyloid oligomers are heterogeneous in structure, stability, and stoichiometry, making it challenging to elucidate their structures by NMR spectroscopy or X-ray crystallography. Amyloid oligomers are thought to be different in structure from fibrils. Some oligomers are thought to be composed of antiparallel β-sheets comprising β-hairpins.22,23,24,25 Currently, no high-resolution structures of amyloid oligomers formed by full-length amyloidogenic peptides and proteins are available in the PDB. Excluding our own work, only three structures of amyloid oligomers composed of peptide fragments have been reported (Table S3).
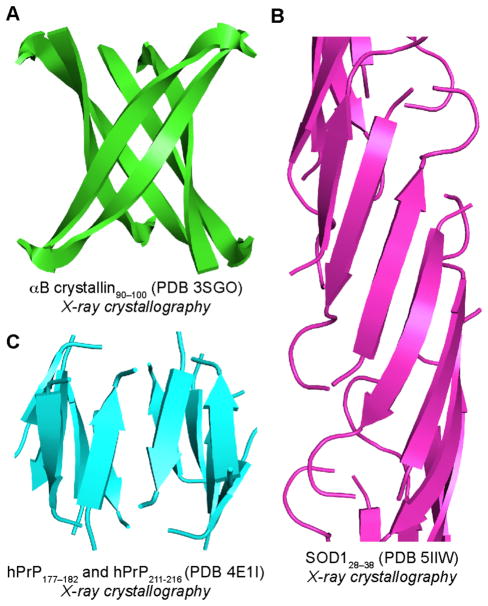
To better understand the structures of amyloid oligomers, researchers have turned to studying peptide fragments from regions of amyloidogenic peptides and proteins that are important in assembly. Eisenberg and coworkers determined the X-ray crystallographic structure of a β-barrel-like oligomer, termed a cylindrin, formed by an 11-residue peptide fragment from αB crystallin (Figure 5A).26 The cylindrin oligomer is composed of six β-strands that form a twisted antiparallel β-sheet that closes back on itself to form a cylinder. Recently, Eisenberg and coworkers determined the X-ray crystallographic structure of a corkscrew-like oligomer formed by an 11-residue peptide fragment derived from superoxide dismutase 1 (SOD1, Figure 5B).27 Surewicz and coworkers determined the X-ray crystallographic structure of a hexameric oligomer formed by a disulfide-stabilized β-sheet fragment from human prion protein (hPrP, Figure 5C).28 The hPrP oligomer is composed of three four-stranded antiparallel β-sheets that pack to form a hydrophobic core. These three structures illustrate occurrences of antiparallel β-sheets in amyloid oligomers and demonstrate the importance of hydrogen bonding and packed hydrophobic cores in oligomer stabilization.
Figure 5.
Structures of oligomers formed by peptide fragments derived from αB crystallin (A), SOD1 (B), and hPrP (C).
Our laboratory has pioneered elucidation of amyloid oligomer structures through X-ray crystallography of macrocyclic peptides containing fragments from amyloidogenic peptides and proteins. Our general approach involves stabilizing two β-strands in an antiparallel β-sheet conformation by linking the β-strands together at their N- and C-termini with two delta-linked ornithine turn units (δOrn) to form a macrocycle. To control aggregation of the peptide we use an unnatural group to block the hydrogen-bonding edge of one of the β-strands. The X-ray crystallographic structures of these macrocyclic peptides reveal that the peptides assemble hierarchically to form oligomers. The structures of these oligomers provide structural models for oligomers of full-length amyloidogenic peptides and proteins and illustrate their hierarchical assembly from smaller subunits. We then investigate the biophysical and biological properties of the oligomers observed crystallographically. These studies guide the design of new macrocyclic peptides that incorporate additional features from the full-length amyloidogenic peptides and proteins and also provide structural insights into the molecular basis of amyloid diseases (Chart 1). We envision that these studies will guide the identification of targets for drug discovery and facilitate the development of probes such as antibodies and fluorophores.
Chart 1.
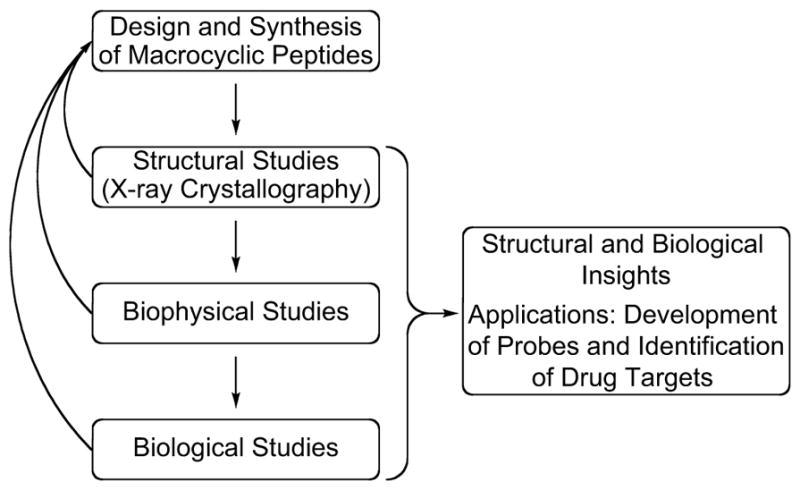
Our laboratory’s approach to studying amyloid oligomers.
Over the last ten years, our laboratory has designed macrocyclic peptides with the goal of mimicking the β-sheet structures formed by amyloidogenic peptides and proteins. Early work involved using the tripeptide mimic Hao to template β-sheet formation and block uncontrolled aggregation. These macrocyclic β-sheet peptides contain a single β-strand from Aβ, tau, B2M, or IAPP and a template strand that contains Hao (Figure 6).29,30,31,32,33 The X-ray crystallographic structures of these Hao-containing macrocyclic β-sheet peptides revealed that the peptides form flatter β-sheets that assemble to form hydrogen-bonded dimers, which sandwich together to form cruciform tetramers (Figure 7). Although the Hao-containing macrocyclic β-sheet peptides provided valuable insights into the supramolecular assembly of β-sheets derived amyloidogenic peptides and proteins, the template strand contains the unnatural Hao moiety and is not designed to mimic the amyloidogenic peptide or protein.
Figure 6.
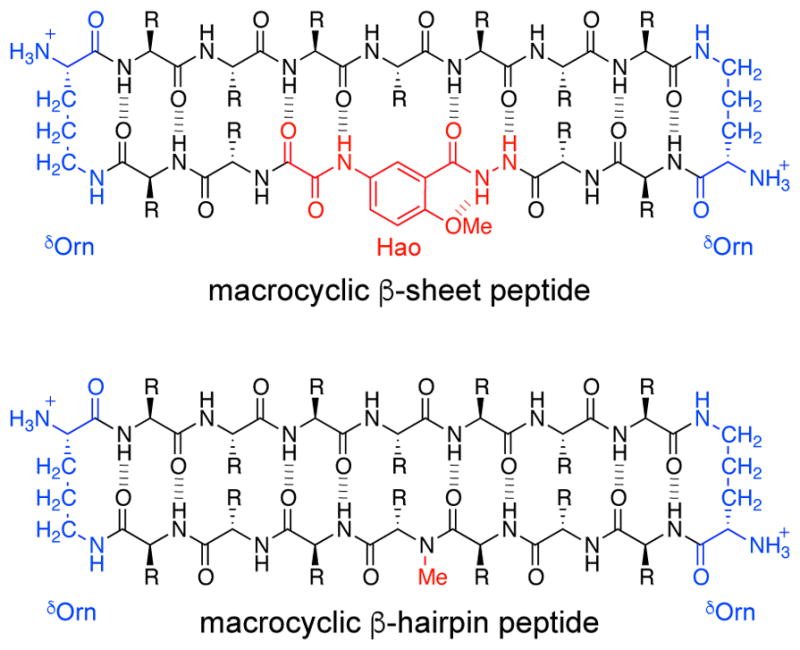
Chemical structures of a Hao-containing macrocyclic β-sheet peptide and a macrocyclic β-hairpin peptide.
Figure 7.
X-ray crystallographic structures of cruciform tetramers formed by Hao-containing macrocyclic β-sheet peptides derived from Aβ (A) and IAPP (B).
Inspired by the β-hairpin structures reported by Härd and Hoyer, former graduate student Ryan Spencer designed a macrocyclic peptide that better mimics β-hairpins formed by amyloidogenic peptides and proteins. These macrocyclic β-hairpin peptides contain two natural β-strand fragments linked together by two δOrn turn mimics (Figure 6). An N-methyl group on the amide backbone of one of the β-strands blocks uncontrolled aggregation by disrupting the ability of the peptide to form a continuous hydrogen-bonded β-sheet network. The design and study of these macrocyclic β-hairpin peptides represented a major breakthrough for our laboratory. The remainder of this Account will focus on our studies of macrocyclic β-hairpin peptides over the past five years. In these studies, X-ray crystallography has proven to be a fountainhead for elucidating the structures of the oligomers formed by these peptides.34
Structures of Oligomers Formed by Macrocyclic β-Hairpin Peptides Derived from Aβ17–36.35
We began our study of amyloid oligomers by designing two macrocyclic β-hairpin peptides that mimic an Aβ17–36 β-hairpin. In these macrocyclic β-hairpins, β-strands comprising Aβ17–23 and Aβ30–36 are linked together by two δOrn turn mimics to create peptides 1 and 2 (Figure 8). The δOrn turn mimic that links Asp23 to Ala30 replaces the Aβ24–29 loop. Peptide 1 contains an N-methyl group on Gly33; peptide 2 contains an N-methyl group on Phe20. To improve the solubility of these peptides, we replaced Met35 with the hydrophilic isostere ornithine.
Figure 8.
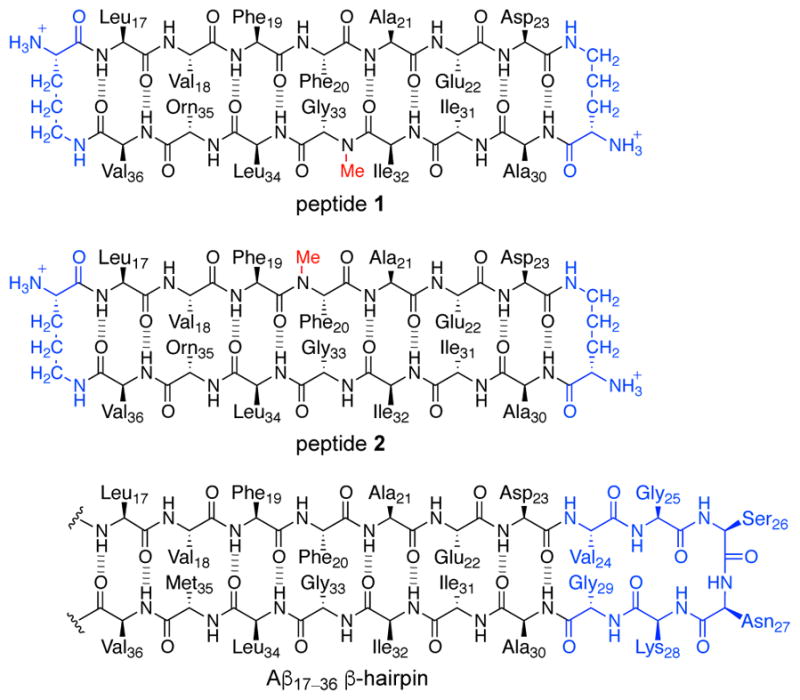
Chemical structures of peptides 1 and 2, illustrating their relationship to an Aβ17–36 β-hairpin.
The X-ray crystallographic structures of peptides 1 and 2 reveal that both peptides fold to form twisted antiparallel β-sheets that closely mimic the structure of the natural Aβ β-hairpin reported by Härd and Hoyer.19 The β-hairpin monomers formed by peptides 1 and 2 each assemble to form triangular trimers (Figure 9). The triangular trimers formed by peptides 1 and 2 are virtually identical, indicating that trimer formation is not guided by which β-strand contains the N-methyl group. In the triangular trimer formed by peptide 1, three ordered water molecules fill the center hole of the trimer, hydrogen bonding with each other and with the amide backbone of Phe20. In the triangular trimer formed by peptide 2, the three N-methyl groups on the Phe20 residues fill the center hole of the trimer, replacing the three ordered water molecules. The triangular trimers are stabilized by hydrophobic packing between amino acid side chains at the three corners of the trimer and by intermolecular hydrogen bonds between the amide backbones of adjacent monomers. The triangular trimers further assemble to form hexamers, in which two trimers pack face-to-face, and dodecamers, in which four trimers assemble in a tetrahedral fashion.
Figure 9.
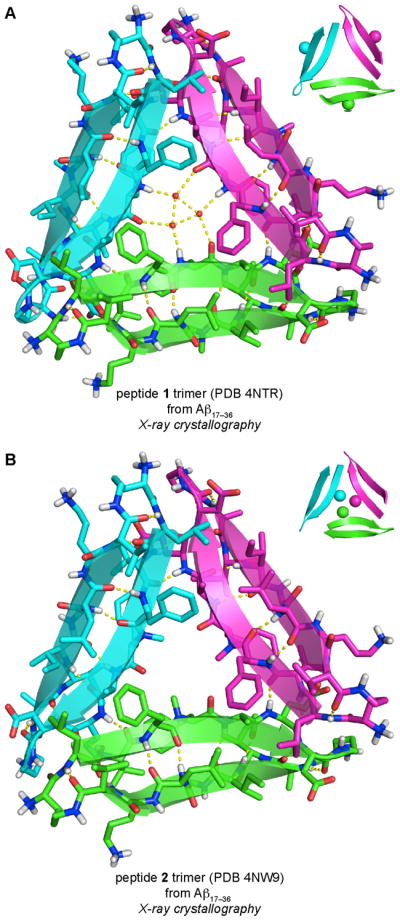
X-ray crystallographic structures of the triangular trimers formed by peptide 1 (A) and peptide 2 (B). In the insets, the N-methyl groups on each peptide are shown as spheres.
The X-ray crystallographic structures of the oligomers formed by peptides 1 and 2 transformed our laboratory’s perception about the supramolecular assembly of β-sheets, by revealing a richer more complex structural landscape for β-sheet assembly than had been previously observed. The triangular trimers differ from the flatter, edge-to-edge hydrogen-bonded β-sheets observed in the oligomers formed by the Hao-containing macrocyclic β-sheets. The triangular trimer motif appears to be a characteristic mode of assembly of β-hairpins. The foldon domain of bacteriophage T4 fibritin is composed of three β-hairpins that assemble to form a triangular trimer similar to the triangular trimers formed by peptides 1 and 2.36 Triangular trimers have also emerged as a common structural motif formed by other macrocyclic β-hairpin peptides developed by our laboratory and described below.
Structures of Oligomers Formed by a β-Hairpin Containing Aβ17–36. 37
In studying macrocyclic β-hairpin peptides, we have strived to design peptides that most closely mimic natural β-hairpins, while retaining the ability to crystalize the peptides and elucidate the X-ray crystallographic structures of the oligomers that they form. In peptides 1 and 2, the Aβ24–29 loop is replaced with a δOrn turn mimic, and Met35 is replaced with ornithine. To better mimic an Aβ17–36 β-hairpin, we reincorporated the Aβ24–29 loop and Met35 to create peptide 3 (Figure 10). Peptide 3 is a homologue of peptide 1 that contains 20 amino acids comprising the full length of an Aβ17–36 β-hairpin. To reinforce β-hairpin structure, we mutated residues Val24 and Gly29 to cysteine and formed an intramolecular disulfide bond. We had initially synthesized a variant of peptide 3 that contained the natural Val24 and Gly29 residues, but this peptide failed to grow crystals in any of the 864 crystallization conditions tested. We found that peptide 3 afforded crystals suitable for X-ray diffraction in the same crystallization conditions as peptides 1 and 2.
Figure 10.
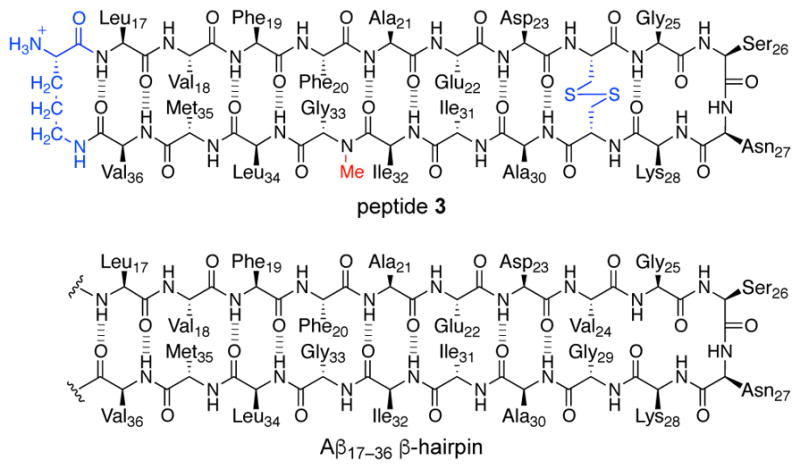
Chemical structure of peptide 3, illustrating its relationship to an Aβ17–36 β-hairpin.
The X-ray crystallographic structure of peptide 3 reveals that the peptide folds to form a twisted β-hairpin. The peptide 3 β-hairpin assembles in an identical fashion to peptides 1 and 2, to form a triangular trimer containing the same stabilizing interactions as the trimers formed by peptides 1 and 2 (Figure 11A). The peptide 3 trimer assembles hierarchically to form higher-order oligomers. Four trimers assemble in a tetrahedral fashion to form a dodecamer (Figure 11B), and five dodecamers pack together to form an annular porelike structure (Figure 11C). This study confirmed that the triangular trimers and dodecamers originally observed for peptides 1 and 2 can accommodate the Aβ24–29 loop. This study also revealed that dodecamers composed of triangular trimers can further assemble to form higher-order structures, such as annular pores.
Figure 11.
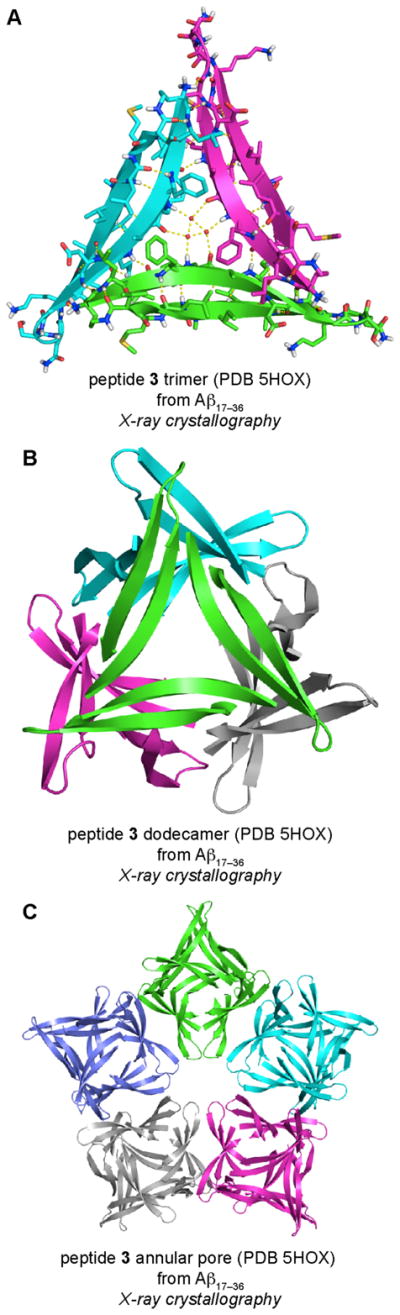
X-ray crystallographic structures of the trimer (A), dodecamer (B), and annular pore (C) formed by peptide 3.
The hierarchical assembly of peptide 3 to form trimers, dodecamers, and annular pores recapitulates aspects the Aβ oligomerization pathway thought to occur in the brain.2 Trimers and dodecamers of Aβ are thought to have special significance in the early stages of Alzheimer’s disease progression. 38 Aβ*56, a putative dodecamer isolated from the brains of Tg2576 transgenic mice, causes memory impairments when injected intracranially into rats. 39 Furthermore, annular pores isolated from Alzheimer’s disease brains appear similar in size and morphology to the annular pores formed by peptide 3.40 The structures of the oligomers formed by peptide 3 may help explain the significance of trimers, dodecamers, and annular pores formed by Aβ in Alzheimer’s disease.
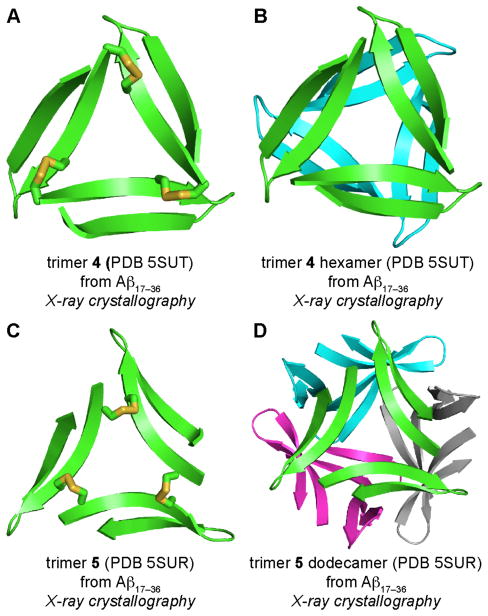
Stabilization, Assembly, and Toxicity of Trimers Derived from Aβ.41
Peptides 1, 2, and 3 appear to form oligomers at the millimolar concentrations of the X-ray crystallography experiments, but remain monomeric at the low micromolar concentrations of biological experiments. To better understand the biological significance of the triangular trimers formed by peptides 1 and 2, we created covalently stabilized versions of the trimers and performed biophysical and biological experiments on the stabilized trimers. We stabilized the trimers by engineering disulfide bonds at the three corners of each trimer to create trimers 4 and 5 (Figures 12A and C, and S1). Trimers 4 and 5 are identical, except that trimer 4 contains an N-methyl group on Gly33, while trimer 5 contains an N-methyl group on Phe20. The X-ray crystallographic structures of trimers 4 and 5 revealed that the two trimers assemble to form different higher-order oligomers. Trimer 4 assembles to form a sandwich-like hexamer (Figure 12B), while trimer 5 assembles to form a ball-shaped dodecamer (Figure 12D).
Figure 12.
X-ray crystallographic structures of trimers 4 and 5. (A) Trimer 4. (B) Trimer 4 hexamer. (C) Trimer 5. (D) Trimer 5 dodecamer.
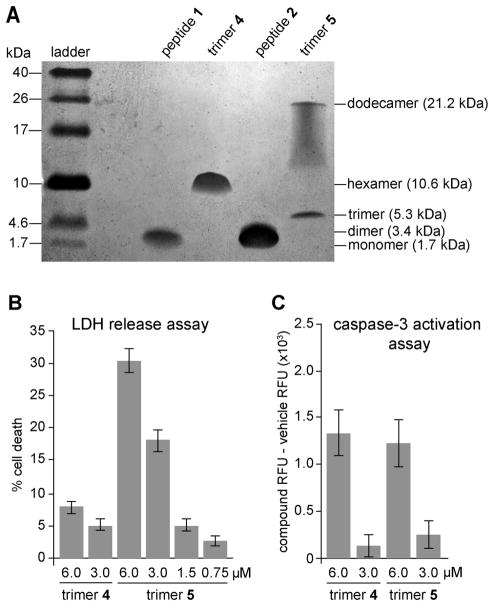
SDS-PAGE and size exclusion chromatography (SEC) show that trimers 4 and 5 assemble to form oligomers in solution that recapitulate the crystallographically observed hexamer and dodecamer (Figure 13A). LDH release assays and caspase-3 activation assays in the human neuroblastoma cell line SH-SY5Y show that trimers 4 and 5 are toxic (Figure 13B and C). Trimers 4 and 5 promote LDH release and caspase-3 activation at concentrations as low 6 μM, indicating toxicity toward cells. Dot blot assays indicate that the amyloid oligomer-specific antibody A11 recognizes trimers 4 and 5 as amyloid oligomers. These studies show that trimers 4 and 5 behave similarly to oligomers of full-length Aβ, thus providing evidence for the biological significance of the triangular trimer motif.
Figure 13.
Biophysical and biological studies of trimers 4 and 5. (A) Silver-stained SDS-PAGE gel. (B) LDH release assay. (C) Caspase-3 activation assay.
A Compact Dodecamer Formed by a Macrocyclic β-Hairpin Peptide Derived from Aβ16–36.42
β-Hairpins can adopt different registrations of β-strands. β-Hairpin registration is significant, because shifting the registration of β-strands changes both the pairings of the residues within the β-hairpin and the surfaces upon which the side chains are displayed. To explore the role that β-hairpin registration plays in the oligomerization of peptides derived from Aβ, we synthesized macrocyclic β-hairpin peptides 6, 7, and 8, which are designed to mimic β-hairpins from Aβ17–36, Aβ16–36, and Aβ15–36 (Figure 14). Each peptide contains an N-methyl group on Gly33 and p-iodophenylalanine (PheI) in place of Phe19 to facilitate X-ray crystallographic phase determination.
Figure 14.
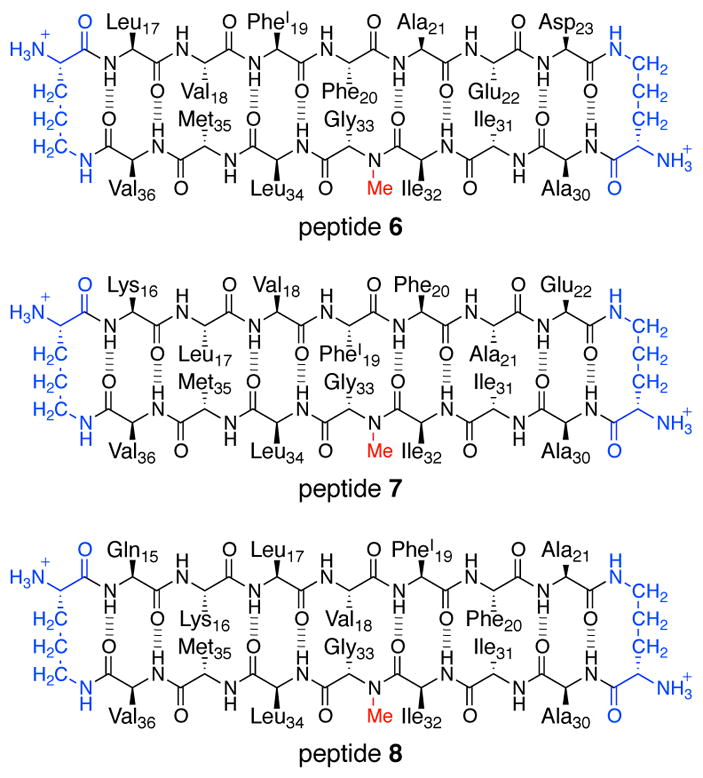
Chemical structures of peptides 6, 7, and 8, from Aβ.
The X-ray crystallographic structures of peptides 6, 7, and 8 reveal that the peptides assemble to form different structures (Figure 15). Peptide 6 assembles identically to the homologous peptide 1, whereby three β-hairpin monomers assemble to form a triangular trimer, which further assembles to form hexamers and dodecamers. Peptide 7 also assembles to form triangular trimers, which further assemble to form a dodecamer. The dodecamer formed by peptide 7 differs from the dodecamer formed by peptide 6, in that the peptide 7 dodecamer is more compact and forms a completely enclosed hydrogen-bonded β-sheet network. Peptide 8 does not assemble to form discrete oligomers, but rather forms a continuous network of out-of-register β-sheets, which resembles an amyloid fibril.
Figure 15.
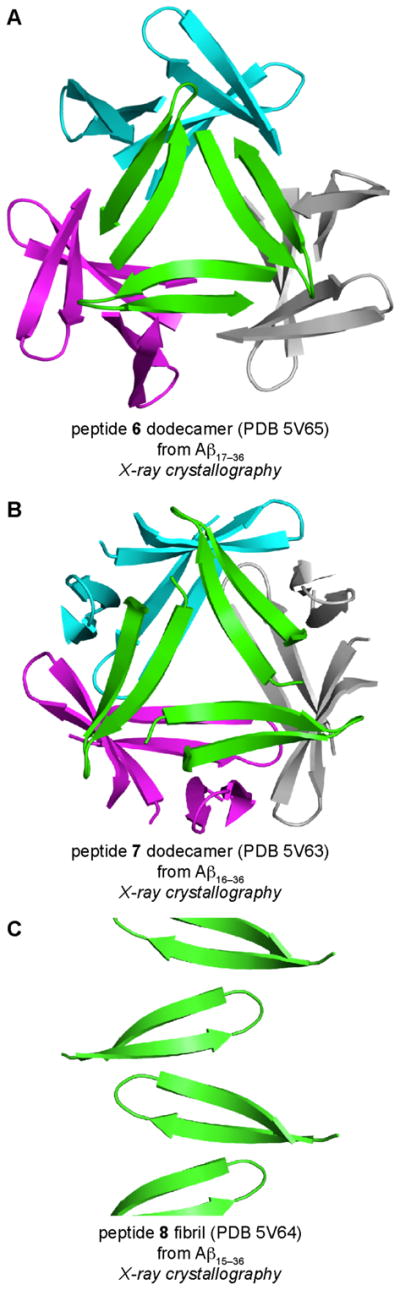
X-ray crystallographic structures of the dodecamers formed by peptides 6 (A) and 7 (B), and the fibrillar assembly formed by peptide 8 (C).
A Hexamer Formed by a Macrocyclic β-Hairpin Peptide Derived from Aβ16–36.43
To further explore the importance of β-hairpin registration in oligomer structure and assembly, we synthesized peptide 9. Peptide 9 is derived from an Aβ16–36 β-hairpin and contains Aβ16–22 and Aβ30–36 β-strands linked together by two δOrn turn mimics (Figure 16). The δOrn turn mimic that connects Glu22 to Ala30 replaces the Aβ23–29 loop. Peptide 9 also contains an N-methyl group on Phe19.
Figure 16.
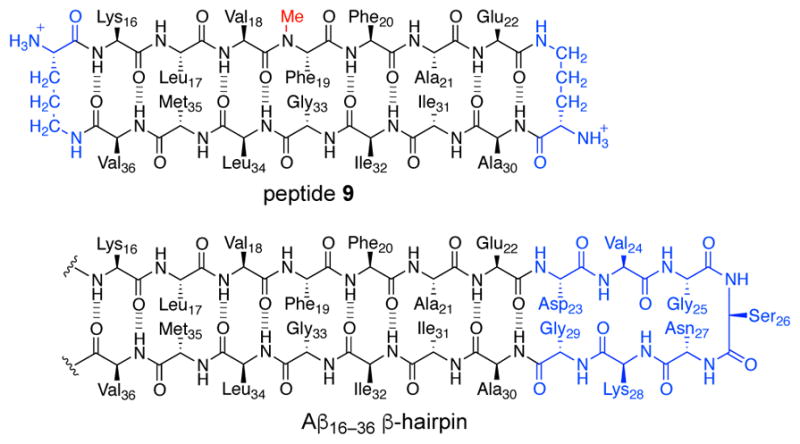
Chemical structure of peptide 9, illustrating its relationship to an Aβ16–36 β-hairpin.
The X-ray crystallographic structure of peptide 9 reveals that the peptide assembles to form a barrel-like hexamer that can be interpreted as either a dimer of triangular trimers or as a trimer of β-sheet dimers (Figure 17). The hexamer is stabilized by a densely packed hydrophobic core and a completely enclosed hydrogen-bonding network between the amide backbones of the monomer subunits. In SDS-PAGE, peptide 9 migrates at a molecular weight consistent with a hexamer, which recapitulates the hexamer observed crystallographically. Furthermore, peptide 9 is toxic toward SH-SY5Y cells at concentrations as low as 50 μM. These studies provide evidence that the peptide 9 hexamer behaves like an oligomer of full-length Aβ and demonstrate that β-hairpin registration may be important in the assembly and toxicity of oligomers formed by full-length Aβ β-hairpins.
Figure 17.
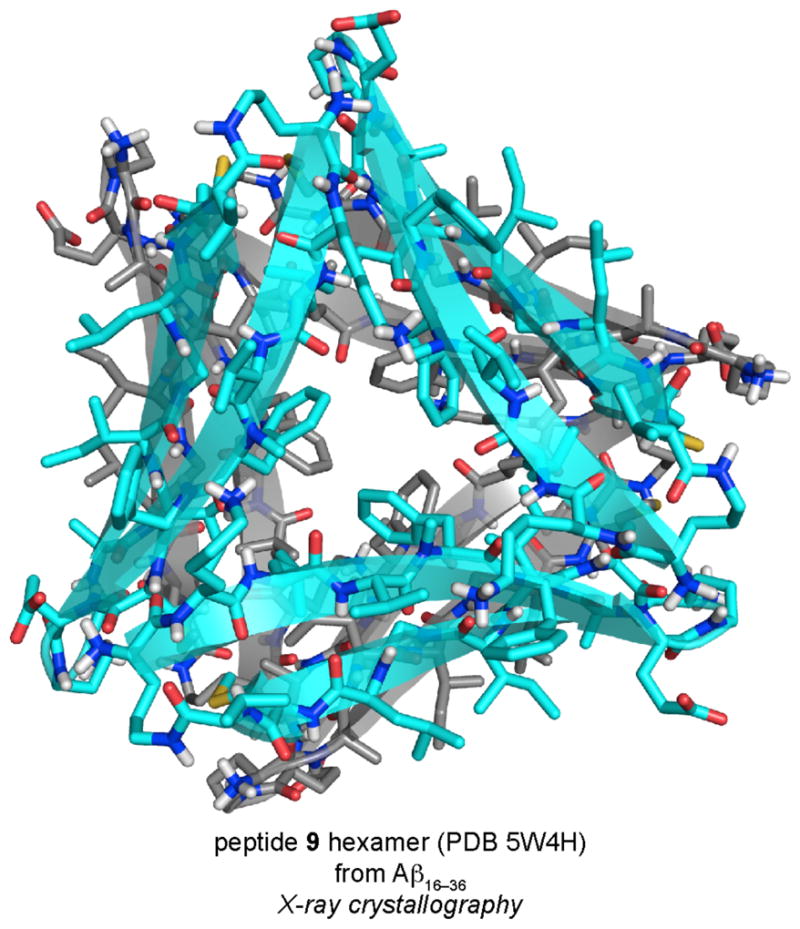
X-ray crystallographic structure of the hexamer formed by peptide 9.
Both peptides 7 and 9 are derived from an Aβ16–36 β-hairpin and differ only in the position of the N-methyl group and the presence or absence of a p-iodo group on Phe19. Despite their similarities, these peptides assemble to form different oligomers—peptide 7 forms a ball-shaped dodecamer and peptide 9 forms a barrel-like hexamer (Figures 15B and 17). This difference in assembly may result from the position of the N-methyl group. The N-methyl group on Gly33 in peptide 7 blocks formation of the barrel-like hexamer, promoting peptide 7 to assemble into the ball-shaped dodecamer. The formation of different oligomers is significant, because it demonstrates that macrocyclic β-hairpin peptides displaying almost identical amino acid side chains can fit together to form compact assemblies in more than one way. These findings may extend to the assembly of β-hairpins formed by full-length Aβ and provide molecular-level insights that could help explain the polymorphism and heterogeneity of Aβ oligomers.
Structures of Oligomers Formed by Macrocyclic β-Hairpin Peptides Derived from α-Syn36–55 and B2M63–69.44,45
β-Hairpins are a common structural motif in amyloid oligomers formed by other amyloidogenic peptides and proteins. Our laboratory has developed macrocyclic β-hairpin peptides derived from α-Syn and B2M and elucidated the structures of the oligomers these peptides form. The α-Syn-derived peptide 10 mimics the α-Syn β-hairpin observed by Hoyer and coworkers and contains α-Syn36–42 and α-Syn49–55 β-strands linked together with two δOrn turn mimics (Figure 18). Peptide 10 also contains an N-methyl group on Val52. To reinforce β-hairpin structure, Gly36 is mutated to alanine. To facilitate X-ray crystallographic phase determination, Tyr39 is mutated to p-iodophenylalanine. The X-ray crystallographic structure of peptide 10 reveals that the peptide assembles to form an asymmetric triangular trimer that further assembles to form a basket-shaped nonamer (Figure 19A). These structures provide insights into how a β-hairpin formed by full-length α-Syn might assemble to form oligomers.
Figure 18.
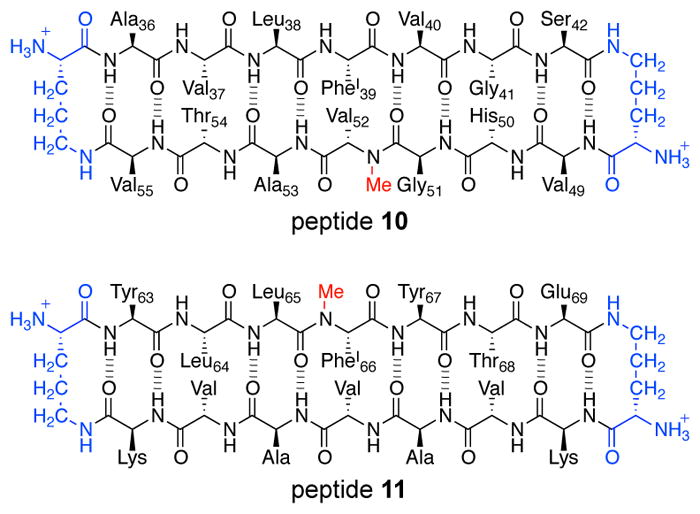
Chemical structures of peptides 10 and 11, from α-Syn and B2M.
Figure 19.
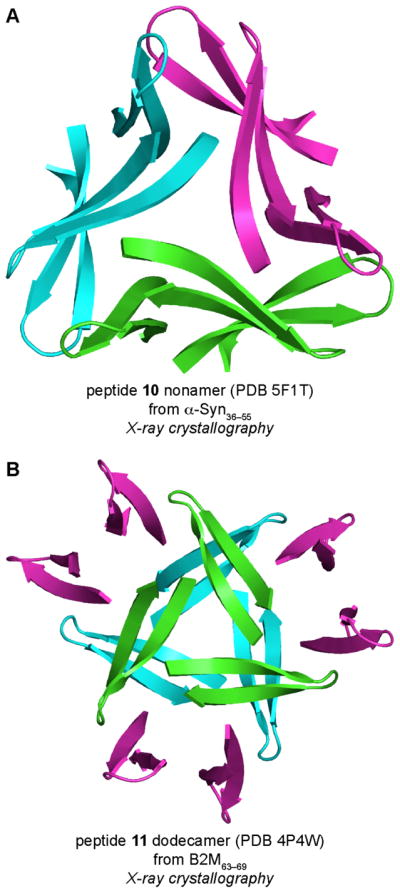
X-ray crystallographic structures of the nonamer formed by peptide 10 (A) and the dodecamer formed by peptide 11 (B).
The B2M-derived macrocyclic β-hairpin peptide 11 and related homologues only contain a single β-strand from B2M63–69 (Figure 18). The β-strand is incorporated into a macrocycle with an additional heptapeptide strand that is designed to template β-hairpin folding. The X-ray crystallographic structures of six different B2M-derived peptides demonstrate that small changes in the amino acid sequence of the additional heptapeptide strand can lead to different oligomeric assemblies, including hexamers, octamers, and dodecamers.45 Figure 19B shows the X-ray crystallographic structure of the dodecamer formed by peptide 11.
CONCLUSION
The formation of amyloid fibrils and oligomers has emerged as a widespread mode of protein folding and assembly central in amyloid diseases, such as Alzheimer’s disease, Parkinson’s disease, and type-2 diabetes. Understanding the amyloid state is one of the most important problems in structural biology and supramolecular chemistry. This problem is not only scientific, but is also critical to combatting the looming threat of neurodegenerative disorders in an aging population.
Although the structures of amyloid fibrils are now yielding to powerful techniques such as ss-NMR and cryo-EM, the structures of amyloid oligomers remain largely unknown. Elucidating the structures of amyloid oligomers is especially challenging, because the oligomers are heterogeneous and metastable. The use of constrained peptides in conjunction with X-ray crystallography is proving to be exceptionally fruitful in determining the structures of oligomers formed by amyloidogenic peptides and proteins. Thus far, this approach has yielded more than 30 structures that have been deposited in the PDB.
High-resolution structures of Aβ oligomers are desperately needed to better understand the molecular basis of Alzheimer’s disease. The X-ray crystallographic structures of the trimers, hexamers, and dodecamers formed by macrocyclic β-hairpin peptides offer a glimpse into how β-hairpins formed by full-length Aβ could fit together and provide high-resolution structural models of Aβ oligomers. These structures appear to be providing a meaningful window into the assembly of full-length Aβ. We are actively pursuing further studies of these structures and probing their role in Alzheimer’s disease.
To date, these X-ray crystallographic structures are the only high-resolution structural models of oligomeric assemblies of Aβ-derived peptides in the PDB. Complementary approaches are needed to shed additional light on the structures of amyloid oligomers. Other structural motifs may emerge from further studies. An enhanced understanding of the structural biology of amyloid oligomers should ultimately lead to diagnostics and therapies for Alzheimer’s disease and other amyloid diseases.
Supplementary Material
Acknowledgments
We thank the many people in our laboratory who have contributed to the research presented in this Account. We especially thank Ryan K. Spencer, Patrick J. Salveson, and Stan Yoo for their efforts in bringing these research projects to fruition, and the National Institutes of Health (Grant GM097562) and the National Science Foundation (CHE-1507840) for funding.
Biographies
Adam Kreutzer graduated from the University of Utah with a B.S. in Biology and M.S. in Oncological Sciences. He received his Ph.D. from the University of California, Irvine. He is currently a postdoctoral scholar at the University of California, Irvine. His research interests are in structural biology and biological probe development.
James Nowick is a Professor of Chemistry at the University of California, Irvine. He received his bachelors degree in Chemistry in 1985 from Columbia University and his Ph.D. in Organic Chemistry in 1990 from MIT. After an NSF postdoctoral fellowship in supramolecular chemistry at MIT, he began his independent career at UCI in 1991. He is currently Chair of the Department of Chemistry.
Footnotes
The authors declare no competing financial interest.
REFERENCES AND NOTES
- 1.Benilova I, Karran E, De Strooper B. The Toxic Aβ Oligomer and Alzheimer's Disease: An Emperor in Need of Clothes. Nat Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 2.Larson ME, Lesné SE. J Neurochem Soluble Aβ Oligomer Production and Toxicity. 2012;120:125–139. doi: 10.1111/j.1471-4159.2011.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.For leading references, see the Supporting Information.
- 4.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular Structural Basis for Polymorphism in Alzheimer's β-Amyloid Fibrils. Proc Natl Acad Sci US A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petkova AT, Yau WM, Tycko R. Experimental Constraints on Quaternary Structure in Alzheimer's β-Amyloid Fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular Structure of β-amyloid Fibrils in Alzheimer's Disease Brain Tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatami A, Monjazeb S, Milton S, Glabe CG. Familial Alzheimer's Disease Mutations within the Amyloid Precursor Protein Alter the Aggregation and Conformation of the Amyloid-β Peptide. J Biol Chem. 2017;292:3172–3185. doi: 10.1074/jbc.M116.755264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiang W, Yau WM, Luo Y, Mattson MP, Tycko R. Antiparallel β-sheet Architecture in Iowa-Mutant β-Amyloid Fibrils. Proc Natl Acad Sci US A. 2012;109:4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schütz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Güntert P, Böckmann A, Glockshuber R, Meier BH. Atomic-Resolution Three-Dimensional Structure of Amyloid β Fibrils Bearing the Osaka Mutation. Angew Chem Int Ed Engl. 2015;54:331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Böckmann A, Güntert P, Meier BH, Riek R. Atomic-Resolution Structure of a Disease-Relevant Aβ(1-42) Amyloid Fibril. Proc Natl Acad Sci US A. 2016;113:4976–4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, Griffin RG. Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Aβ(1-42) Fibril Structure Illuminates Self-Recognition and Replication of Amyloid in Alzheimer's Disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gremer L, Schölzel D, Schenk C, Reinartz E, Labahn J, Ravelli RBG, Tusche M, Lopez-Iglesias C, Hoyer W, Heise H, Willbold D, Schröder GF. Fibril Structure of Amyloid-β(1-42) by Cryoelectron Microscopy. Science. 2017 doi: 10.1126/science.aao2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, Wan W, Stubbs G, Schwieters CD, Lee VM, George JM, Rienstra CM. Solid-State NMR Structure of a Pathogenic Fibril of Full-Length Human α-Synuclein. Nat Struct Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SHW. Cryo-EM Structures of Tau Filaments from Alzheimer's Disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the Cross-β Spine of Amyloid-Like Fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Atomic Structures of Amyloid Cross-β Spines Reveal Varied Steric Zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 18.Colletier JP, Laganowsky A, Landau M, Zhao M, Soriaga AB, Goldschmidt L, Flot D, Cascio D, Sawaya MR, Eisenberg D. Molecular Basis for Amyloid-β Polymorphism. Proc Natl Acad Sci US A. 2011;108:16938–16943. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyer W, Grönwall C, Jonsson A, Ståhl S, Härd T. Stabilization of a β-Hairpin in Monomeric Alzheimer's Amyloid-β Peptide Inhibits Amyloid Formation. Proc Natl Acad Sci US A. 2008;105:5099–5104. doi: 10.1073/pnas.0711731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirecka EA, Shaykhalishahi H, Gauhar A, Akgül Ş, Lecher J, Willbold D, Stoldt M, Hoyer W. Sequestration of a β-Hairpin for Control of α-Synuclein Aggregation. Angew Chem Int Ed Engl. 2014;53:4227–4230. doi: 10.1002/anie.201309001. [DOI] [PubMed] [Google Scholar]
- 21.Mirecka EA, Feuerstein S, Gremer L, Schröder GF, Stoldt M, Willbold D, Hoyer W. β-Hairpin of Islet Amyloid Polypeptide Bound to an Aggregation Inhibitor. Sci Rep. 2016;6:33474. doi: 10.1038/srep33474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L, Edalji R, Harlan JE, Holzman TF, Lopez AP, Labkovsky B, Hillen H, Barghorn S, Ebert U, Richardson PL, Miesbauer L, Solomon L, Bartley D, Walter K, Johnson RW, Hajduk PJ, Olejniczak ET. Structural Characterization of a Soluble Amyloid β-Peptide Oligomer. Biochemistry. 2009;48:1870–1877. doi: 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- 23.Scheidt HA, Morgado I, Huster D. Solid-State NMR Reveals a Close Structural Relationship Between Amyloid-β Protofibrils and Oligomers. J Biol Chem. 2012;287:22822–22826. doi: 10.1074/jbc.M112.367474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doi T, Masuda Y, Irie K, Akagi K, Monobe Y, Imazawa T, Takegoshi K. Solid-state NMR Analysis of the β-Strand Orientation of the Protofibrils of Amyloid β-Protein. Biochem Biophys Res Commun. 2012;428:458–462. doi: 10.1016/j.bbrc.2012.10.096. [DOI] [PubMed] [Google Scholar]
- 25.Tay WM, Huang D, Rosenberry TL, Paravastu AK. The Alzheimer's Amyloid-β(1-42) Peptide Forms Off-Pathway Oligomers and Fibrils that are Distinguished Structurally by Intermolecular Organization. J Mol Biol. 2013;425:2494–2508. doi: 10.1016/j.jmb.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Atomic View of a Toxic Amyloid Small Oligomer. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sangwan S, Zhao A, Adams KL, Jayson CK, Sawaya MR, Guenther EL, Pan AC, Ngo J, Moore DM, Soriaga AB, Do TD, Goldschmidt L, Nelson R, Bowers MT, Koehler CM, Shaw DE, Novitch BG, Eisenberg DS. Atomic Structure of a Toxic Oligomeric Segment of SOD1 Linked to Amyotrophic Lateral Sclerosis (ALS) Proc Natl Acad Sci USA. 2017;114:8770–8775. doi: 10.1073/pnas.1705091114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apostol MI, Perry K, Surewicz WK. Crystal Structure of a Human Prion Protein Fragment Reveals a Motif for Oligomer Formation. J Am Chem Soc. 2013;135:10202–10205. doi: 10.1021/ja403001q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pham JD, Chim N, Goulding CW, Nowick JS. Structures of Oligomers of a Peptide from β-Amyloid. J Am Chem Soc. 2013;135:12460–12467. doi: 10.1021/ja4068854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Sawaya MR, Cheng PN, Zheng J, Nowick JS, Eisenberg D. Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic β-Sheet Mimics. J Am Chem Soc. 2011;133:6736–6744. doi: 10.1021/ja200222n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng PN, Liu C, Zhao M, Eisenberg D, Nowick JS. Amyloid β-Sheet Mimics that Antagonize Protein Aggregation and Reduce Amyloid Toxicity. Nat Chem. 2012;4:927–933. doi: 10.1038/nchem.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C, Zhao M, Jiang L, Cheng PN, Park J, Sawaya MR, Pensalfini A, Gou D, Berk AJ, Glabe CG, Nowick J, Eisenberg D. Out-of-Register β-Sheets Suggest a Pathway to Toxic Amyloid Aggregates. Proc Natl Acad Sci US A. 2012;109:20913–20918. doi: 10.1073/pnas.1218792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Kreutzer AG, Truex NL, Nowick JS. A Tetramer Derived from Islet Amyloid Polypeptide. J Org Chem. 2017;82:7905–7912. doi: 10.1021/acs.joc.7b01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer RK, Nowick JS. A Newcomers Guide to Peptide Crystallography. Israel J Chem. 2015;55:698–710. doi: 10.1002/ijch.201400179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer RK, Li H, Nowick JS. X-ray Crystallographic Structures of Trimers and Higher-Order Oligomeric Assemblies of a Peptide Derived from Aβ17–36. J Am Chem Soc. 2014;136:5595–5598. doi: 10.1021/ja5017409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao Y, Strelkov SV, Mesyanzhinov VV, Rossmann MG. Structure of Bacteriophage T4 Fibritin: A Segmented Coiled Coil and the Role of the C-Terminal Domain. Structure. 1997;5:789–798. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 37.Kreutzer AG, Hamza IL, Spencer RK, Nowick JS. X-ray Crystallographic Structures of a Trimer, Dodecamer, and Annular Pore Formed by an Aβ17–36 β-Hairpin. J Am Chem Soc. 2016;138:4634–4642. doi: 10.1021/jacs.6b01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lesné SE, Sherman MA, Grant M, Kuskowski M, Schneider JA, Bennett DA, Ashe KH. Brain Amyloid-β Oligomers in Ageing and Alzheimer's Disease. Brain. 2013;136:1383–1398. doi: 10.1093/brain/awt062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A Specific Amyloid-β Protein Assembly in the Brain Impairs Memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 40.Lasagna-Reeves CA, Glabe CG, Kayed R. Amyloid-β Annular Protofibrils Evade Fibrillar Fate in Alzheimer Disease Brain. J Biol Chem. 2011;286:22122–22130. doi: 10.1074/jbc.M111.236257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreutzer AG, Yoo S, Spencer RK, Nowick JS. Stabilization, Assembly, and Toxicity of Trimers Derived from Aβ. J Am Chem Soc. 2017;139:966–975. doi: 10.1021/jacs.6b11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salveson PJ, Spencer RK, Kreutzer AG, Nowick JS. X-ray Crystallographic Structure of a Compact Dodecamer from a Peptide Derived from Aβ16–22. Org Lett. 2017;19:3462–3465. doi: 10.1021/acs.orglett.7b01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kreutzer AG, Spencer RK, McKnelly KJ, Yoo S, Hamza IL, Salveson PJ, Nowick JS. A Hexamer of a Peptide Derived from Aβ16–36. Biochemistry. 2017 doi: 10.1021/acs.biochem.7b00831. ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salveson PJ, Spencer RK, Nowick JS. X-ray Crystallographic Structure of Oligomers Formed by a Toxic β-Hairpin Derived from α-Synuclein: Trimers and Higher-Order Oligomers. J Am Chem Soc. 2016;138:4458–4467. doi: 10.1021/jacs.5b13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spencer RK, Kreutzer AG, Salveson PJ, Li H, Nowick JS. X-ray Crystallographic Structures of Oligomers of Peptides Derived from β2-Microglobulin. J Am Chem Soc. 2015;137:6304–6311. doi: 10.1021/jacs.5b01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.