Abstract
Zika virus (ZIKV) belongs to the positive-sense single-stranded RNA-containing Flaviviridae family. Its recent outbreak and association with human diseases (e.g. neurological disorders) have raised global health concerns, and an urgency to develop a therapeutic strategy against ZIKV infection. However, there is no currently approved antiviral against ZIKV. Here we present a comprehensive overview on recent progress in structure–function investigation of ZIKV NS5 protein, the largest non-structural protein of ZIKV, which is responsible for replication of the viral genome, RNA capping and suppression of host interferon responses. Structural comparison of the N-terminal methyltransferase domain and C-terminal RNA-dependent RNA polymerase domain of ZIKV NS5 with their counterparts from related viruses provides mechanistic insights into ZIKV NS5-mediated RNA replication, and identifies residues critical for its enzymatic activities. Finally, a collection of recently identified small molecule inhibitors against ZIKV NS5 or its closely related flavivirus homologues are also discussed.
Keywords: Viral replication, Flavivirus, Non-structural protein 5, RNA capping, Pathogen–host interaction, Drug discovery, Antiviral inhibitors
Introduction
Viruses of the Flaviviridae family are widespread vector-borne pathogens, causing large epidemics and tens of thousands of deaths every year. The family is comprised of three genera—Pestivirus, Hepacivirus, and Flavivirus—with over 70 viruses, including the human pathogens dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), tick-borne encephalitis virus (TBEV), Japanese encephalitis virus (JEV), hepatitis C virus (HCV), and Zika virus (ZIKV) [1]. ZIKV emerged as a major global pathogen after large outbreaks occurred in Micronesia (2007), French Polynesia (2013), and Brazil (2015). In response to the accelerated global transmission of ZIKV, the World Health Organization (WHO) declared a public health emergency of international concern [2, 3]. In addition to its rapid transmission, these outbreaks have been associated with severe neurological disorders such as microcephaly [4] and Guillain–Barré syndrome (GBS) [5].
Like other flaviviruses, ZIKV is a small, enveloped virus with non-segmented ~ 10–12 kb single-stranded positive-sense RNA genome that is capped at the 5′ end, and lacks a 3′ polyA tail. ZIKV enters host cells via receptor-mediated endocytosis [6], with AXL, a gene that encodes a receptor tyrosine kinase, as a candidate receptor [7, 8]. The range of permissible cell types and the host-cell receptors that mediate ZIKV entry are still being investigated. Recent in vitro studies have shown that AXL and other receptors in the TYRO3-ALXL-MERTK (TAM) family permit ZIKV entry in human skin cells [9] and that AXL is highly expressed in developing human cerebral cortexes [8]. However, deletion of TAM receptors in mice does not reduce ZIKV replication in vivo, suggesting that these receptors may not play a role in ZIKV infection, or that several redundant entry receptors exist for ZIKV [10, 11]. Following endocytosis, the envelope protein undergoes a low pH-dependent conformational change that results in fusion of the viral membrane with the host membrane, leading to release of the capsid-bound genome (nucleocapsid) into the cytoplasm [12]. The genome is then dissociated from the capsid, translated as a single polyprotein, and replicated within endoplasmic reticulum (ER)-derived vesicle packets. The polyprotein is co- and post-translationally cleaved by both viral and host proteases to generate three structural (capsid, pre-membrane, and envelope) and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins [13].
Immature virions are assembled in the lumen of the ER and trafficked to the Golgi apparatus where select proteins are modified by glycosylation (Fig. 1) [14, 15]. Subsequent maturation of virions occurs through proteolytic cleavage of prM by a yet unknown host furin-like protease and homodimerization of the envelope protein in the trans-Golgi network [16, 17]. In addition to mature infectious viral particles, this process generates non-infectious subviral particles, which lack the capsid protein and the RNA genome. Both mature infectious viral progeny and subviral particles are released from the host cells through exocytosis (Fig. 1).
Fig. 1.
Schematic view of the ZIKV replication cycle. The replication cycle of ZIKV is similar to other known flaviviruses. Its E proteins are involved in the attachment of the virus to receptors on the host membrane. Subsequently, the virus enters the cell via endocytosis. Viral genomic RNA is released into cytoplasm after fusion of viral and host membranes. The single-stranded RNA (ssRNA) is first translated to a polyprotein, which is then cleaved into several viral proteins. Viral genomes replicate on the endoplasmic reticulum (ER) surface. After assembly at the ER, the virus buds with the help of the host ESCRT (endosomal sorting complexes required for transport) machinery. Through Golgi apparatus, its prM protein is cleaved, which indicates the maturation of virions. Finally, the ZIKV exits the cell via exocytosis
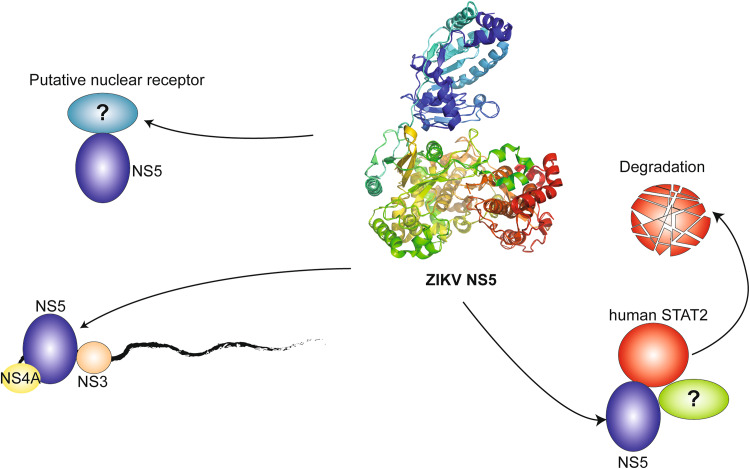
The role of ZIKV NS5 in viral replication and virus–host interaction
The viral NS5 protein is the largest (~ 100 kDa) and most conserved ZIKV protein, with 94% sequence identity between the two major ZIKV lineages: Asian and African [3]. NS5 is comprised of two domains, an N-terminal methyltransferase domain and an RNA-dependent RNA polymerase (RdRP) domain at the C-terminal end [18–20]. It performs three essential roles in the viral life cycle: genome replication [21, 22] and capping [23], and interferon suppression [24, 25] (Fig. 2). In addition, recent evidence suggested that it might also play a role in modulating the activity of cellular spliceosome [26].
Fig. 2.
The activity of ZIKV NS5 in host cells. On the ER surface, ZIKV NS5 forms a replication complex with other non-structural proteins of ZIKV (e.g., NS3 and NS4A) to mediate viral replication [18–20]. In the cytoplasm, through interaction with STAT2 and a yet unknown E3 ubiquitin ligase, ZIKV NS5 promotes proteasome-mediated protein degradation of human STAT2. It has been observed that a significant amount of ZIKV NS5 enters the nucleus, likely affecting the function of nuclear receptors or other nuclear activities
Genome replication
The RdRP domain of ZIKV NS5 (NS5-RdRP), like its counterparts in other members of Flaviviridae family, generates positive- and negative-sense copies of the RNA genome via a de novo mechanism [27–29], that is, it uses RNA as a template but does not require a primer to elongate nascent RNA. The NS5-RdRP-mediated replication process is thought to involve three distinct conformational states [27, 30–32]. In the first state, pre-initiation, NS5 is poised to receive initial NTPs, but the RNA exit tunnel is blocked. In the second state, initiation, the secondary structure of the 3′ and 5′ untranslated regions (UTRs) and the cyclization of the genome permit NS5 to bind to the 3′ end of the viral RNA template. Meanwhile, ATP and GTP molecules enter the active site of NS5-RdRP to form Watson–Crick pairs with the conserved C and U bases at the 3′ end of the viral template RNA. Subsequently, a ribose–phosphate bond is generated by nucleophilic attack by the activated alcohol group of the adenine ribose on the guanine α-phosphate. These two nucleotides serve as the initial dinucleotide primer [33]. In the third state, elongation, the RNA exit tunnel of NS5 opens up, ensuring processive RNA polymerization. It is likely that additional viral factors are also involved in facilitating ZIKV RNA replication, as observed in other flaviviruses. For example, ZIKV NS3 encodes a helicase domain, which is essential to unwind the double-stranded (ds) RNA intermediate formed during genome synthesis [34–38]. Additionally, the NS4B protein was reported to be involved in membrane alterations and anchoring of the viral replication complex on the cellular membrane [39].
Capping
The methyltransferase activity possessed by the N-terminal methyltransferase (MTase) domain of NS5 is required for the final steps in generating the type-I 5′ cap using S-adenosyl-methionine (SAM) as the methyl donor [40–42]. Based on in vitro evidence, the MTase domain mediates methylation of the N7 atom of the cap guanine (G0) and the 2′-O atom of the adenine ribose (the first nucleotide of the viral genome) through a two-step reaction [41]. In addition, the N-terminal MTase serves as a guanylyltransferase [41], which uses GTP as a substrate to form a covalent NS5–GMP intermediate, followed by transfer of the GMP from NS5 to the end of an acceptor RNA transcript. Notably, formation of the NS5–GMP intermediate can be stimulated by the NS3 helicase [41].
Interferon suppression
Type I interferons (IFN-α and -β) play a critical role in controlling viral infection [43–46]. To establish infection, flaviviruses have developed various immune evasion mechanisms to suppress the type I IFN antiviral responses [47–55]. Of particular note, both the DENV and ZIKV NS5 proteins inhibit the type I interferon responses by binding the host signal transducer and activator of transcription 2 (STAT2) and targeting it for proteasome-dependent degradation [49, 54–56]. DENV NS5 also interacts with the host E3 ubiquitin-protein ligase (UBR4), a component of the host proteasome that is required for DENV NS5-mediated STAT2 degradation [55]. In addition to recruiting UBR4, the DENV NS5 protein must be matured proteolytically at its N terminus for STAT2 degradation to occur [56]. Domain mapping revealed that the first ten amino acids of DENV NS5 are required for STAT2 degradation, but not for STAT2 binding [56], consistent with the fact that these residues are essential for UBR4 interaction [55]. The STAT2-binding site of DENV NS5 is suggested to be located in the region between the MTase and RdRP domains [55, 56]. However, the exact amino acids required for STAT2 interaction have yet to be elucidated. In contrast to DENV, N-terminal proteolytic processing of ZIKV NS5 is not required for STAT2 degradation, and, while it has been demonstrated that ZIKV NS5-mediated STAT2 degradation is proteasome dependent, it is not UBR4 dependent, and the precise component of the proteasome involved in ZIKV NS5-mediated STAT2 degradation has not been identified [49]. These observations indicate that ZIKV NS5 uses a mechanism distinct from that of DENV NS5 to degrade STAT2. Resolution of the ZIKV NS5–STAT2 interaction at the amino acid level, and the identification of the required host proteasome component, will contribute to the elucidation of the unique degradation mechanism employed by ZIKV NS5. This knowledge will also make it possible to generate an attenuated ZIKV strain for use in a vaccine that is unable to inhibit the host antiviral response, and may contribute to the development of antiviral therapeutics that target the NS5 protein.
Structural study of ZIKV NS5
Overall structure of ZIKV NS5
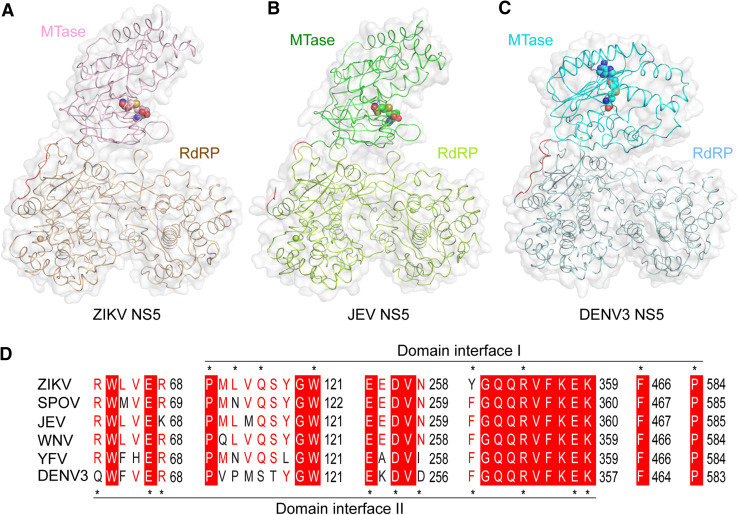
Recent evidence has led to the notion that the tandem organization of the MTase and RdRP subdomains of the flavivirus NS5 facilitates its sequential, RNA-templated activities [57, 58]. In particular, the presence of the MTase domain enhances the binding affinity of NS5 for RNA template and incoming nucleotides, thereby promoting the RdRP-mediated replication initiation and elongation [58]. However, the mechanism by which the two subdomains cooperate in RNA replication or capping remains undetermined. Nevertheless, the crystal structures of full-length NS5 from JEV [57], DENV3 [59] and ZIKV [18–20] have recently been determined, providing direct insights into the domain orientation of flavivirus NS5. The crystal structure of ZIKV NS5 reveals that the N-terminal MTase domain and C-terminal RdRP domain stack against each other, resulting in a buried surface area of 1400 Å2. Remarkably, this conformational state of ZIKV NS5 shows high resemblance with that of JEV NS5 (Fig. 3a), superposition of which gives a root-mean-square deviation (RMSD) of 0.63 Å over 872 Cα atoms, suggesting that the conformation of NS5 is conserved within the Flavivirus genus. Consistent with this suggestion, the amino acid residues at the domain interfaces are highly conserved (Fig. 3b), including a sequence motif (motif F, see below) that likely engages in recognition of template RNA as well as incoming NTPs [32]. In contrast, in the crystal structures of RdRP alone, this motif either becomes disordered [60, 61] or adopts different conformation [20], implying that its function is modulated by the inter-domain contact. On the other hand, the domain orientation of ZIKV NS5 differs significantly from that of DENV3 NS5 (Fig. 3a), despite that the individual domains superimpose well between the two proteins. The MTase domain of DENV3 NS5 interacts with the C-terminal RdRP domain through a distinct interface (Fig. 3a), resulting in a less extended conformation and an RMSD of 6.06 Å over 844 Cα atoms when superimposed with ZIKV NS5. Intriguingly, the residues at the domain interface of DENV3 NS5 are also highly conserved (Fig. 3b), suggesting functional relevance of this alternative conformation. The existence of multiple conformations of flavivirus NS5 is consistent with previous small angle X-ray scattering analysis of DENV3 NS5, which indicates the presence of a conformational ensemble of flavivirus NS5 in solution [62]. In support of this notion, mutating the evolutionarily conserved residues at the domain interface resulted in enhanced RdRP activity of DENV3 NS5, but impaired viral replication and infectivity, implying that the conformational dynamics of NS5 may regulate its activity in viral replication [59]. The exact functional implication of these alternative conformations awaits future biochemical and cellular investigations.
Fig. 3.
Structural comparison of full-length ZIKV NS5, JEV NS5 and DENV3 NS5. a Structural overview of ZIKV NS5 (light pink and wheat), JEV NS5 (green and light green) and DNEV3 NS5 (aquamarine and light blue) under transparent surface. The domain linkers are colored in red. The bound SAH molecules are shown in sphere representation. b The residues located at the two alternative domain interfaces, marked by asterisks, are conserved throughout evolution. Identical residues are labeled in white and highlighted in red. Similar residues are labeled in red
Structure of the ZIKV NS5 MTase domain
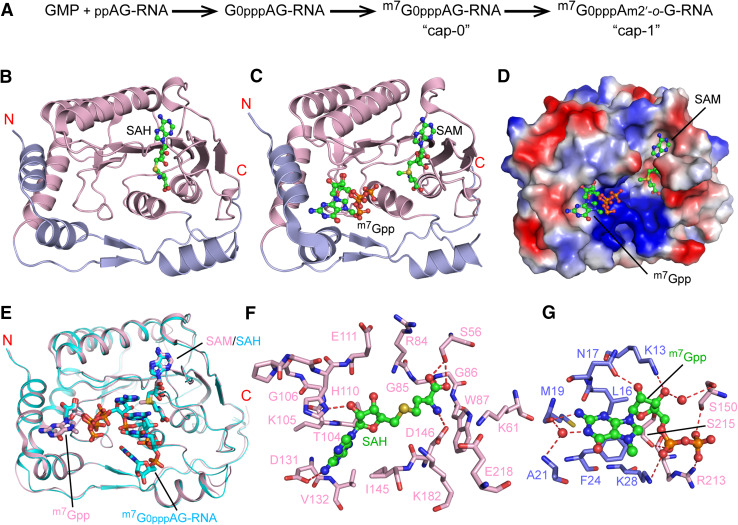
As described above, the MTase domain of flavivirus NS5 mediates the formation of type-1 RNA cap at the 5′-end of the nascent +RNA strand, involving a guanylyltransferase reaction that transfers GMP to the 5′-end of viral RNA, and two MTase reactions that methylate the G0 residue at N7 position (cap-0) and the 2′-O of A1 (cap-1) [63] (Fig. 4a). Capping of the viral RNA is essential for maintaining stability, and increases viral polyprotein translation efficiency [64]. The structures of the MTase domain of ZIKV NS5 [60, 65–68], similar to what was observed for the other flavivirus NS5s [69], reveal a dominant Rossmann fold comprised of a seven-stranded β-sheet sandwiched by two α-helices from one side and another α-helix from the other side, harboring a single binding site for cofactor SAM (Fig. 4b). In addition, two appendages from the N- and C-terminal ends join to the Rossmann fold, together forming the substrate-binding sites (Fig. 4b–d). Structural superposition of the ZIKV NS5 MTase bound to both m7Gpp and SAM with the DENV3 MTase bound to cofactor byproduct S-adenosyl-homocysteine (SAH) and m7G0pppAG-RNA revealed well-aligned interaction sites for SAM, m7Gpp, and the 2′-O group of A1 (Fig. 4e), suggesting that the MTase domain maintains a pre-configured conformation for enzymatic catalysis. The SAH-binding site of ZIKV NS5–MTase involves residues T104–E111 and D131–V132, which interact with the adenosine moiety, and residues S56, K61, R84–W98, I145–D146, K182 and E218 that make contacts with the homocysteine group (Fig. 4f). On the other hand, binding of m7Gpp mainly involves residues in the N-appendage, in which residues L16, M18, A21, F24 and K28 form a hydrophobic cave harboring the guanylyl ring, while residues K13, N17, K28, S150, R213 and S215 interact with the diphosphate group or the sugar ring through direct or water-mediated hydrogen bonds (Fig. 4g). These co-factor- and substrate-binding sites together define the druggable sites within ZIKV NS5.
Fig. 4.
Structural analysis of ZIKV NS5–MTase domain. a Enzymatic reactions mediated by ZIKV NS5–MTase. b Ribbon representation of ZIKV NS5–MTase (PDB: 5TMH) bound to SAH. The Rossmann fold is colored in light pink, and the N- and C-terminal extensions are colored in slate. The SAH molecule is shown in ball-and-stick representation. Ribbon (c) and surface electrostatic (d) representations of ZIKV NS5–MTase bound to both SAM and m7Gpp molecules (PDB 5KQS). e Structural overlay of SAM- and m7Gpp-bound ZIKV NS5–MTase (light pink, PDB 5KQS) and SAH- and m7G0pppAG-RNA-bound DENV3 NS5–MTase (cyan, PDB 5DTO). f Close-up view of the interaction between ZIKV NS5–MTase and SAH (PDB 5TMH). The hydrogen bonds are depicted as dashed lines. g Close-up view of the interaction between ZIKV NS5–MTase and m7Gpp (PDB 5KQS). The color schemes in b, f and g are the same as in a
Structure of the ZIKV NS5-RdRP domain
The flavivirus genome is strictly conserved with end sequences 5′-AG…CU-3′ [32]. Accordingly, the NS5-RdRP domain-mediated RNA replication always starts with synthesis of a pppAG dinucleotide [32]. Like with other viral RdRPs, flavivirus NS5-RdRP is comprised of Thumb, Palm and Fingers subdomains (Fig. 5a, b). Whereas the mechanism by which flavivirus NS5 initiates and elongates the RNA substrates remains poorly understood, recent evidence has suggested that these processes are regulated by various protein and RNA elements (e.g., stem loops at the 5′- and 3′-ends), to achieve virus-specific replication [70]. In accordance with these regulations, flavivirus NS5-RdRP contains a more compact active site, with narrow NTP- and RNA template-channels [71], in comparison with the RNA polymerases mediating primer-based RNA replication. For instance, structural studies of ϕ6 [72, 73], HCV [74, 75] and JEV [76] RdRPs have revealed that a loop protruding from the Thumb domain, termed priming loop, reaches out to the active site to stabilize the formation of a replication pre-initiation complex, while blocking the RNA exit channel. Subsequent transition from initiation state to elongation state led to a conformational transition of this loop [74], thereby allowing the RNA product to exit from the RdRP molecule. Given that the priming loop also exists within ZIKV NS5-RdRP [3], it is conceivable ZIKV NS5-RdRP employs a similar regulatory mechanism during the transition from the replication initiation stage to elongation stage [70, 71]. As illustrated in Fig. 5a, ZIKV NS5–RdRP-mediated RNA replication may start with a pre-initiation complex, in which the RdRP binds to ATP and GTP, the first two incoming nucleotides of RNA replication (Fig. 5a) [27, 76–79]. Next, a pair of divalent cations (Mn2+ or Mg2+) coordinates these initial nucleotides with the RdRP molecule to promote the synthesis of the 5′-AG-3′ dinucleotide, resulting in the initiation state of replication. Subsequent synthesis of longer RNA product leads to opening up of the RNA exit tunnel, allowing the RdRP to enter into the elongation state, in which the synthesized RNA processively elongates from the active site (Fig. 5a).
Fig. 5.
Structural analysis of ZIKV NS5-RdRP domain. a A model for the conformational transition of ZIKV NS5-RdRP during different stages of RNA replication, highlighting the regulatory role of the priming loop (slate). The active site is marked by red asterisk. The template and synthesized RNA strands are colored in blue and purple, respectively. b Structural overview of the ZIKV NS5-RdRP domain, with the N-terminal extension, Thumb, Fingers, Palm and Primer loop colored in orange, aquamarine, green, light pink and blue, respectively. The zinc ions are shown in purple spheres. c Structural overlay of ZIKV NS5-RdRP (wheat) and GTP-bound JEV NS5-RdRP (cyan), with the GTP binding sites highlighted in expanded view. The GTP-contact residues for JEV NS5-RdRP are labeled. The corresponding sites in ZIKV NS5-RdRP are labeled in parenthesis. The conserved motifs in ZIKV NS5-RdRP and JEV NS5-RdRP are colored in magenta and green, respectively. The GTP molecules are shown in stick representation. d Structural overlay of ZIKV NS5-RdRP (wheat, PDB 5TMH) and the replication initiation complex of HCV RdRP (green, PDB 4WTL), with the conserved motifs in ZIKV RdRP colored in magenta. The RNA molecule bound to HCV is shown in stick representation, and the Mn2+ ions are shown in salmon spheres. The potential repositioning of the priming loop of ZIKV NS5-RdRP during transition to the initiation state is indicated by a curved arrow. e, f Two expanded views of the HCV RdRP–RNA interaction. The RNA-contacting residues of HCV RdRP and their corresponding residues in ZIKV NS5-RdRP are colored in green and wheat, respectively. The hydrogen bonds are depicted as dashed lines. g Structural overlay of ZIKV NS5-RdRP (wheat, PDB 5TMH) and the replication elongation complex of HCV RdRP (blue, PDB 4WTG), with the conserved motifs in ZIKV RdRP colored in magenta. The potential repositioning of the priming loop of ZIKV NS5-RdRP required for transition to the elongation state is indicated by a curved arrow
The crystal structures of ZIKV NS5–RdRP, either in the isolated state [20, 60, 61] or in the context of full-length NS5 [18–20], have recently been determined. Similar to other viral RdRPs [71], the ZIKV NS5-RdRP adopts a capped right-hand fold that is further divided into the Palm, Fingers and Thumb subdomains (Fig. 5b), each of which harbors conserved motifs controlling RNA synthesis [70]. In addition, an N-terminal extension associates with the Fingers subdomain for potential functional regulation (Fig. 5b). The Palm subdomain serves as the catalytic center of the RdRP, containing conserved motifs with catalytic aspartic acids (motifs A and C) [77], mediating template binding, translocation and/or NTP specificity (motif B) [80, 81], nucleotide transfer (motif D) [80], and alignment of priming nucleotide ATP (motif E) (Fig. 5c) [77]. The Thumb subdomain presents the priming loop (Fig. 5b) that presumably promotes ATP-specific RNA initiation, as well as regulates the subsequent transition to the elongation phase [32]. The Fingers subdomain regulates the de novo RdRP activity through participation in the formation of the active site and NTP entry channel, with motif F involved in binding to the nascent base pair [82] and motif G involved in binding to RNA template [57]. All these motifs act in concert to form three tunnels, which ensure access to the template, the entrance of the incoming ribonucleoside triphosphate (NTP), and the exit of the newly synthesized RNA products. Like its homologues from JEV and DENV, the ZIKV NS5-RdRP contains two zinc ions, which might play a role in stabilizing the structural fold. Structural knowledge on the conformational transition of ZIKV NS5-RdRP between distinct replication states remains unavailable to date. Nevertheless, a structural comparison of ZIKV NS5-RdRP with the GTP-bound JEV RdRP [76], in which the GTP molecule partially occupies the space accommodating the first two nucleotides, sheds light onto the structural basis underlying the initial transition of ZIKV RdRP into a pre-initiation state (Fig. 5c). In essence, the priming loop of ZIKV NS5-RdRP appears to be in a position that is well poised for NTP recognition, with residues T796, W797 and S798 likely hydrogen bonded with the β,γ-phosphate groups of the initial NTP (Fig. 5c). By contrast, a large conformational adjustment is suggested for motif F, which would move toward the NTP molecule, with residues K462 and R473 interacting with the β-phosphate and base ring, respectively (Fig. 5c). In addition, recognition of the phosphate groups of the initial NTP may also involve residues R731 and R739 from the Thumb domain, recognition of the sugar group may involve D665 from motif C, and the base-specific recognition may involve residues D540 from motif A and S603 from motif B.
Structural comparison of ZIKV NS5-RdRP with the replication initiation complex of HCV NS5, with a 5′-UACC RNA template, a 5′-pGG RNA primer and an incoming UDP [74], provides further insights into the conformational transitions of ZIKV NS5 toward the initiation state (Fig. 5d). Notably, motifs A, B and C from the palm domain of ZIKV NS5-RdRP are positioned similar to the corresponding region of HCV NS5, with residues D535, D540, S603, N612, D665 and D666 well poised to coordinate Mn2+ ions or interact with the incoming nucleotide for enzymatic catalysis (Fig. 5e). Motif F of ZIKV NS5-RdRP is also well aligned with the corresponding region of HCV NS5 that interacts with the nascent Watson–Crick pair, likely involving residues K458 and R473 interacting with the incoming nucleotide and residues I475 and F477 packing against the pairing residue (Fig. 5e, f). Meanwhile, motif E residue S712 and Thumb residues R731 and R739 are positioned to donate hydrogen bonds to the backbone of the two primer nucleotides (Fig. 5e). On the other hand, it is apparent that the priming loop of ZIKV NS5-RdRP is positioned far deeper in the active site than the corresponding region of HCV NS5 (Fig. 5d), implying that formation of the initiation complex would require the priming loop to retract from the active site to accommodate the growing RNA product. Interestingly, the corresponding region in HCV NS5 (residues Y448–S450) stacks against the 3′-end of the template RNA (Fig. 5f), implying that the priming loop of ZIKV NS5-RdRP may play a similar role in stabilizing the initiation complex. Structural comparison with the initiation complex of HCV NS5 also supports the role of motifs B and G in escorting the template RNA, with motif B (residues G604–V606) making van der Waals contacts with sugar or base moieties of the template RNA and motif G (residues A408–A409) interacting with the backbone through main-chain hydrogen bonding interactions (Fig. 5f). Finally, structural comparison of ZIKV NS5-RdRP with the replication elongation complex of HCV NS5 with a self-complimentary 5′-CAAAAUUUU-3′ RNA duplex hints at additional conformational change of ZIKV NS5-RdRP accompanying the transition from the replication initiation state to the elongation state (Fig. 5g). Consistent with the current model [32] (Fig. 5a), this conformational rearrangement likely involves further retraction of the priming loop from the active site to accommodate the growing products of RNA synthesis.
Small molecule inhibitors for ZIKV NS5
The fact that viral NS5 is essential for viral replication but lacks a human counterpart makes NS5 an ideal drug target for potential therapeutics. In fact, the concept of NS5 as a drug target has been validated by the success of commercially available drugs targeting the viral polymerase of hepatitis C, which belongs to the same viral family as ZIKV [83]. Although there are currently no approved vaccines or antivirals for ZIKV disease, multiple strategies have been devised to develop inhibitors targeting ZIKV NS5 and its flavivirus homologues. Consequently, a variety of agents have been synthesized and evaluated for potential therapeutics, and re-purposing of some patented drugs was also attempted [84–96].
ZIKV NS5-RdRP inhibitors
ZIKV NS5-RdRP has been considered as a major drug target due to its virus-unique activity, which confers a potential advantage to development of inhibitors with fewer side effects [96–98]. In recent years, development of small nucleoside inhibitors (NIs) aiming to inhibit the activity of RdRP has proved to be a promising treatment approach [30, 99, 100]. Acting as substrate analogs, incorporation of these compounds into nascent RNA could result in chain termination or lethal replication [30]. For instance, a hepatitis C virus NS5B polymerase nucleoside inhibitor named sofosbuvir (Fig. 6a) was approved in the United States to cure the infection of hepatitis C virus, another member of the Flaviviridae family [101]. As a nucleotide analog, sofosbuvir could be metabolized to active triphosphate form and inhibit viral genome replication by performing as a chain terminator [102, 103]. Recent studies further demonstrated that sofosbuvir antagonized ZIKV genome replication in human cells in a concentration-dependent manner [89]. In addition, sofosbuvir treatment of ZIKV-infected mice deficient in type I IFN signaling pathway displayed higher survival rates than those of controls [89], which implied sofosbuvir might be a potential drug against ZIKV infection owing to its broad-spectrum anti-Flaviviridae activity [84, 104].
Fig. 6.
Structures of a list of potential inhibitors against ZIKV NS5. Small molecule inhibitors targeting NS5-RdRP (a–g), NS5–MTase (h, i) and the NS5–NS3 interaction (j). Tr triphenylmethyl
The high structural resemblance of RdRPs among Flaviviridae members [105] prompted efforts to re-purpose other previously identified NS5-targeting inhibitors for ZIKV NS5. For instance, 7-deaza-2′-C-methyl adenosine (7DMA) (Fig. 6b), which was previously identified as a potent inhibitor for HCV [106] and other flaviviruses [107, 108], exhibited consistent high potency against ZIKV in cell-based assays [88, 106, 109]. Likewise, another closely related NI, 2′-C-methyladenosine (2′CMA) (Fig. 6c), was also reported to reduce ZIKV infection pronouncedly [88]. A substitution of the 2′-C-methyl group for an ethynyl group further led to the generation of another candidate inhibitor, termed NITD008 (Fig. 6d), which exerted broad-spectrum antiviral effects on ZIKV, HCV and all the four serotypes of DENV [99]. Although direct pursuit of this inhibitor has been discontinued due to its toxicity in vivo [99], further modification of this inhibitor holds promise to yield an effective drug compound with less toxicity.
The combination of compound library screening with hit optimization has also led to discovery of an increasing number of antiviral inhibitors. For instance, a class of triphenylmethyl alkylated nucleoside analogs has been identified to antagonize DENV or YFV in vitro. Among them, the 3′,5′-bis-O-tritylated-5-chlorouridine compound (Fig. 6e) generated promising results, and has been selected for future development [110]. Given that these lipophilic compounds exert their function via inhibition of RNA replication [110], it is possible to extend the investigation of this class of inhibitors toward ZIKV NS5-mediated RNA replication.
In addition to the direct substrate-binding sites, allosteric regulation sites have started to gain attention for development of non-nucleoside inhibitors (NNIs) [111–113]. For instance, a fragment-based screening has recently led to the discovery of a cohort of NNIs targeting an allosteric pocket at the priming loop-proximate region of DENV NS5–RdRP, termed “N” pocket [112, 113]. In particular, two novel acyl-sulfonamide derivatives (Fig. 6f, g) targeting this pocket show high potency in DENV1–4 NS5-mediated replication initiation, as well as moderately inhibit the replication elongation process [112]. Importantly, our recent structural analysis has indicated that the inhibitor binding site for these compounds is conserved in ZIKV NS5-RdRP [19], which provides a new platform for future development of inhibitors for ZIKV disease.
ZIKV NS5–MTase inhibitors
The RNA capping function of ZIKV NS5–MTase makes it another attractive target for antiviral development. Two non-selective competitive inhibitors, sinefungin (Fig. 6h) and SAH, were previously identified to impede both N7 and 2′-O-methylation reactions of DENV NS5–MTase [114–116]. However, practical application of these two inhibitors is limited due to their cellular non-permeability and non-selectivity [105]. Later, structural characterization of DENV3, WNV and ZIKV NS5–MTase further identified a conserved hydrophobic pocket located near the SAM-binding site, which paved the way toward the development of highly specific inhibitors [60, 117, 118]. It is worth noting that a recent effort has identified such a SAM-competitive inhibitor, NSC 12155, which shows inhibitory activity toward NS5–MTase from WNV, DENV-2, DENV-3 and YFV in vitro, and high antiviral efficacy toward WNV, DENV-2 and JEV in cell-based assays [119]. It remains to be tested whether this compound also inhibits ZIKV. More recently, a high-resolution crystal structure of ZIKV NS5–MTase with a SAM analog (MS2042) (Fig. 6i) revealed that the 4-fluorophenyl moiety intruded into the RNA binding tunnel, suggesting that this compound might hinder the RNA methylation via occupation of the putative binding sites for the base and the 2′-OH groups of cap-0 adenosine [120]. One of the major challenges of this class of SAM analogs faces is that they may lack target specificity in cells, a recurring issues for all the NIs [99, 121]. The eukaryotic homologues of Flaviviridae NS5–MTase, such as DNA methyltransferases (DNMT) and RNA methyltransferases (RNMT), also use SAM as cofactors, thereby likely be non-specific targets of these compounds.
Potential inhibitors targeting the ZIKV NS5–NS3 complex
It is worth noting that exploration of potential inhibitors targeting other biological functions of NS5 is also underway. Previous studies have identified an allosteric pocket (cavity B) on DENV NS5 critical to DENV RNA synthesis and the formation of active NS5–NS3 complex [97], which has led to development of an allosteric inhibitor, termed 16i (Fig. 6j), which is capable of disrupting the interaction of DENV NS5–NS3 under in vitro conditions [122]. Considering the high structural similarity between ZIKV NS5 and DENV NS5 (Fig. 3a), this compound could potentially be re-purposed to target ZIKV NS5. Due to lack of structural knowledge and comprehensive characterizations of the NS5–NS3 interactions, it remains unclear how NS5 and NS3 cooperate in viral genome replication. Future structural and biochemical analyses of the ZIKV NS5–NS3 complex and their interactions with RNA will provide a novel framework for development of alternative antiviral therapeutic strategies against ZIKV.
Summary
The ZIKV NS5 protein plays a crucial role in genomic replication and RNA capping of ZIKV, as well as in interferon suppression. Recent progress in structural characterizations of ZIKV NS5 has provided an excellent opportunity for the development of inhibitors against ZIKV NS5. In particular, comparative structural analysis permits us to identify residues that are critical for different stages of ZIKV NS5-mediated RNA replication. Based on the structural similarities between ZIKV NS5 and its homologues from flaviviruses, it has become an attractive strategy to develop ZIKV NS5 inhibitors on the basis of the pool of small molecule inhibitors that were previously identified for the NS5 proteins from other Flaviviridae viruses. Whereas this process is still in the early stages, many inhibitors have demonstrated strong inhibitory effects on NS5-RdRP-mediated RNA replication or MTase-mediated RNA capping activities. The major challenges in clinical application of these inhibitors include their often-unpredictable cellular toxicity and the emergence of drug resistance. For instance, residues located in the catalytic or allosteric sites (e.g., “N” pocket) of the RdRP domain, the N7 and 2′-O-methylation sites of the MTase domain, and the NS3-interacting sites of NS5 might generate potential resistance profiles, as what have been observed in other viral models [106, 112, 123, 124]. Conceivably, a combined use of drugs that target different sites or functional stages of ZIKV NS5 might offer a solution in overcoming these challenges.
Acknowledgements
This work was supported by March of Dimes Foundation (1-FY15-345), Kimmel Scholar Award from Sidney Kimmel Foundation for Cancer Research and NIH (1R35GM119721) to J.S. This work is also partly funded by Trans fund of state of California (AB2664) to J.S. and R.H.
Footnotes
Boxiao Wang and Stephanie Thurmond contributed equally to this work.
Contributor Information
Rong Hai, Email: ronghai@ucr.edu.
Jikui Song, Email: jikui.song@ucr.edu.
References
- 1.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika Virus. N Engl J Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 3.Wang A, Thurmond S, Islas L, Hui K, Hai R. Zika virus genome biology and molecular pathogenesis. Emerg Microbes Infect. 2017;6:e13. doi: 10.1038/emi.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika virus and birth defects-reviewing the evidence for causality. N Engl J Med. 2016;374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 5.Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial AL, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra JC, Despres P, Fournier E, Mallet HP, Musso D, Fontanet A, Neil J, Ghawche F. Guillain–Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case–control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meertens L, Labeau A, Dejarnac O, Cipriani S, Sinigaglia L, Bonnet-Madin L, Le Charpentier T, Hafirassou ML, Zamborlini A, Cao-Lormeau VM, Coulpier M, Misse D, Jouvenet N, Tabibiazar R, Gressens P, Schwartz O, Amara A. Axl mediates ZIKA virus entry in human glial cells and modulates innate immune responses. Cell Rep. 2017;18:324–333. doi: 10.1016/j.celrep.2016.12.045. [DOI] [PubMed] [Google Scholar]
- 8.Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell. 2016;18:591–596. doi: 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, Cao-Lormeau VM, Choumet V, Briant L, Despres P, Amara A, Yssel H, Misse D. Biology of Zika virus infection in human skin cells. J Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Wang PR, Qu LB, Yi CH, Zhang FC, Tang XP, Zhang LG, Chen L. AXL is not essential for Zika virus infection in the mouse brain. Emerg Microbes Infect. 2017;6:e16. doi: 10.1038/emi.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastings AK, Yockey LJ, Jagger BW, Hwang J, Uraki R, Gaitsch HF, Parnell LA, Cao B, Mysorekar IU, Rothlin CV, Fikrig E, Diamond MS, Iwasaki A. TAM receptors are not required for Zika virus infection in mice. Cell Rep. 2017;19:558–568. doi: 10.1016/j.celrep.2017.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufmann B, Rossmann MG. Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes Infect. 2011;13:1–9. doi: 10.1016/j.micinf.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan C, Xie X, Muruato AE, Rossi SL, Roundy CM, Azar SR, Yang Y, Tesh RB, Bourne N, Barrett AD, Vasilakis N, Weaver SC, Shi PY. An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe. 2016;19:891–900. doi: 10.1016/j.chom.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guirakhoo F, Bolin RA, Roehrig JT. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology. 1992;191:921–931. doi: 10.1016/0042-6822(92)90267-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J Gen Virol. 1991;72(Pt 6):1323–1329. doi: 10.1099/0022-1317-72-6-1323. [DOI] [PubMed] [Google Scholar]
- 16.Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol. 2003;84:183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 17.Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Upadhyay AK, Cyr M, Longenecker K, Tripathi R, Sun C, Kempf DJ. Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr F Struct Biol Commun. 2017;73:116–122. doi: 10.1107/S2053230X17001601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Tan XF, Thurmond S, Zhang ZM, Lin A, Hai R, Song J. The structure of Zika virus NS5 reveals a conserved domain conformation. Nat Commun. 2017;8:14763. doi: 10.1038/ncomms14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao B, Yi G, Du F, Chuang YC, Vaughan RC, Sankaran B, Kao CC, Li P. Structure and function of the Zika virus full-length NS5 protein. Nat Commun. 2017;8:14762. doi: 10.1038/ncomms14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 22.Choi KH, Rossmann MG. RNA-dependent RNA polymerases from Flaviviridae. Curr Opin Struct Biol. 2009;19:746–751. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Dong H, Fink K, Zust R, Lim SP, Qin CF, Shi PY. Flavivirus RNA methylation. J Gen Virol. 2014;95:763–778. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- 24.Best SM. The many faces of the flavivirus NS5 protein in antagonism of type I interferon signaling. J Virol. 2017;91:e01970-16. doi: 10.1128/JVI.01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diamond MS. Mechanisms of evasion of the type I interferon antiviral response by flaviviruses. J Interferon Cytokine Res. 2009;29:521–530. doi: 10.1089/jir.2009.0069. [DOI] [PubMed] [Google Scholar]
- 26.De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, Mammi P, Mancini E, Yanovsky MJ, Andino R, Krogan N, Srebrow A, Gamarnik AV. The dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog. 2016;12:e1005841. doi: 10.1371/journal.ppat.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 28.Bressanelli S, Tomei L, Rey FA, De Francesco R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J Virol. 2002;76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao CC, Singh P, Ecker DJ. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 30.Malet H, Masse N, Selisko B, Romette JL, Alvarez K, Guillemot JC, Tolou H, Yap TL, Vasudevan S, Lescar J, Canard B. The flavivirus polymerase as a target for drug discovery. Antiviral Res. 2008;80:23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Morozova OV, Belyavskaya NA, Zaychikov EF, Kvetkova EA, Mustaev AA, Pletnev AG. Identification of RNA replicase subunits responsible for initiation of RNA synthesis of tick-borne encephalitis virus by affinity labelling. Biomed Sci. 1991;2:183–186. [PubMed] [Google Scholar]
- 32.Selisko B, Potisopon S, Agred R, Priet S, Varlet I, Thillier Y, Sallamand C, Debart F, Vasseur JJ, Canard B. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 2012;8:e1002912. doi: 10.1371/journal.ppat.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranjith-Kumar CT, Gutshall L, Kim MJ, Sarisky RT, Kao CC. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J Virol. 2002;76:12526–12536. doi: 10.1128/JVI.76.24.12526-12536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Clum S, You S, Ebner KE, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73:3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matusan AE, Pryor MJ, Davidson AD, Wright PJ. Mutagenesis of the Dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J Virol. 2001;75:9633–9643. doi: 10.1128/JVI.75.20.9633-9643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartelma G, Padmanabhan R. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology. 2002;299:122–132. doi: 10.1006/viro.2002.1504. [DOI] [PubMed] [Google Scholar]
- 37.Benarroch D, Selisko B, Locatelli GA, Maga G, Romette JL, Canard B. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004;328:208–218. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KH, Padmanabhan R. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J Biol Chem. 2005;280:27412–27419. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- 39.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15:2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 44.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuel MA, Diamond MS. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J Virol. 2005;79:13350–13361. doi: 10.1128/JVI.79.21.13350-13361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou A, Paranjape JM, Der SD, Williams BR, Silverman RH. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]
- 47.Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, Garcia-Sastre A. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe. 2010;8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, Boer E, Wolfinbarger JB, Bloom ME. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol. 2005;79:12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, Garcia-Sastre A, Khromykh AA, Best SM. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol. 2010;84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JM, Pisanelli G, Pham A, Ayllon J, Miorin L, Martinez-Romero C, tenOever BR, Garcia-Sastre A. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe. 2014;16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lubick KJ, Robertson SJ, McNally KL, Freedman BA, Rasmussen AL, Taylor RT, Walts AD, Tsuruda S, Sakai M, Ishizuka M, Boer EF, Foster EC, Chiramel AI, Addison CB, Green R, Kastner DL, Katze MG, Holland SM, Forlino A, Freeman AF, Boehm M, Yoshii K, Best SM. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe. 2015;18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- 55.Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, Garcia-Sastre A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu G, Gong P. Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013;9:e1003549. doi: 10.1371/journal.ppat.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potisopon S, Priet S, Collet A, Decroly E, Canard B, Selisko B. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res. 2014;42:11642–11656. doi: 10.1093/nar/gku666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Soh TS, Zheng J, Chan KW, Phoo WW, Lee CC, Tay MY, Swaminathan K, Cornvik TC, Lim SP, Shi PY, Lescar J, Vasudevan SG, Luo D. A crystal structure of the Dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog. 2015;11:e1004682. doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duan W, Song H, Wang H, Chai Y, Su C, Qi J, Shi Y, Gao GF. The crystal structure of Zika virus NS5 reveals conserved drug targets. EMBO J. 2017;36:919–933. doi: 10.15252/embj.201696241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Godoy AS, Lima GM, Oliveira KI, Torres NU, Maluf FV, Guido RV, Oliva G. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat Commun. 2017;8:14764. doi: 10.1038/ncomms14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bussetta C, Choi KH. Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry. 2012;51:5921–5931. doi: 10.1021/bi300406n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coloma J, Jain R, Rajashankar KR, Garcia-Sastre A, Aggarwal AK. Structures of NS5 methyltransferase from Zika virus. Cell Rep. 2016;16:3097–3102. doi: 10.1016/j.celrep.2016.08.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coutard B, Barral K, Lichiere J, Selisko B, Martin B, Aouadi W, Lombardia MO, Debart F, Vasseur JJ, Guillemot JC, Canard B, Decroly E. Zika virus methyltransferase: structure and functions for drug design perspectives. J Virol. 2017;91:e02217-16. doi: 10.1128/JVI.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang C, Feng T, Cheng J, Li Y, Yin X, Zeng W, Jin X, Guo F, Jin T. Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design. Biochem Biophys Res Commun. 2017;492:624–630. doi: 10.1016/j.bbrc.2016.11.098. [DOI] [PubMed] [Google Scholar]
- 68.Zhou H, Wang F, Wang H, Chen C, Zhang T, Han X, Wang D, Wu C, Xie W, Wang Z, Zhang L, Wang L, Yang H. The conformational changes of Zika virus methyltransferase upon converting SAM to SAH. Oncotarget. 2017;8:14830–14834. doi: 10.18632/oncotarget.14780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu L, Dong H, Chen H, Zhang J, Ling H, Li Z, Shi PY, Li H. Flavivirus RNA cap methyltransferase: structure, function, and inhibition. Front Biol (Beijing) 2010;5:286–303. doi: 10.1007/s11515-010-0660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Selisko B, Wang C, Harris E, Canard B. Regulation of Flavivirus RNA synthesis and replication. Curr Opin Virol. 2014;9:74–83. doi: 10.1016/j.coviro.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 73.Salgado PS, Makeyev EV, Butcher SJ, Bamford DH, Stuart DI, Grimes JM. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure. 2004;12:307–316. doi: 10.1016/j.str.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 74.Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, Fox D, 3rd, Wetmore DR, McGrath ME, Ray AS, Sofia MJ, Swaminathan S, Edwards TE. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 75.Lam AM, Edwards TE, Mosley RT, Murakami E, Bansal S, Lugo C, Bao H, Otto MJ, Sofia MJ, Furman PA. Molecular and structural basis for the roles of hepatitis C virus polymerase NS5B amino acids 15, 223, and 321 in viral replication and drug resistance. Antimicrob Agents Chemother. 2014;58:6861–6869. doi: 10.1128/AAC.03847-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Surana P, Satchidanandam V, Nair DT. RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state. Nucleic Acids Res. 2014;42:2758–2773. doi: 10.1093/nar/gkt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Selisko B, Dutartre H, Guillemot JC, Debarnot C, Benarroch D, Khromykh A, Despres P, Egloff MP, Canard B. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology. 2006;351:145–158. doi: 10.1016/j.virol.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 79.Nomaguchi M, Ackermann M, Yon C, You S, Padmanabhan R. De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters. J Virol. 2003;77:8831–8842. doi: 10.1128/JVI.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shu B, Gong P. The uncoupling of catalysis and translocation in the viral RNA-dependent RNA polymerase. RNA Biol. 2017;14:1314–1319. doi: 10.1080/15476286.2017.1300221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iglesias NG, Filomatori CV, Gamarnik AV. The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J Virol. 2011;85:5745–5756. doi: 10.1128/JVI.02343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsai CH, Lee PY, Stollar V, Li ML. Antiviral therapy targeting viral polymerase. Curr Pharm Des. 2006;12:1339–1355. doi: 10.2174/138161206776361156. [DOI] [PubMed] [Google Scholar]
- 84.Bullard-Feibelman KM, Govero J, Zhu Z, Salazar V, Veselinovic M, Diamond MS, Geiss BJ. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res. 2017;137:134–140. doi: 10.1016/j.antiviral.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cai L, Sun Y, Song Y, Xu L, Bei Z, Zhang D, Dou Y, Wang H. Viral polymerase inhibitors T-705 and T-1105 are potential inhibitors of Zika virus replication. Arch Virol. 2017;162:2847–2853. doi: 10.1007/s00705-017-3436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen L, Liu Y, Wang S, Sun J, Wang P, Xin Q, Zhang L, Xiao G, Wang W. Antiviral activity of peptide inhibitors derived from the protein E stem against Japanese encephalitis and Zika viruses. Antiviral Res. 2017;141:140–149. doi: 10.1016/j.antiviral.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 87.Delvecchio R, Higa LM, Pezzuto P, Valadao AL, Garcez PP, Monteiro FL, Loiola EC, Dias AA, Silva FJ, Aliota MT, Caine EA, Osorio JE, Bellio M, O’Connor DH, Rehen S, de Aguiar RS, Savarino A, Campanati L, Tanuri A. Chloroquine, an endocytosis blocking agent, inhibits Zika virus infection in different cell models. Viruses. 2016;8:E322. doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eyer L, Nencka R, Huvarova I, Palus M, Joao Alves M, Gould EA, De Clercq E, Ruzek D. Nucleoside Inhibitors of Zika Virus. J Infect Dis. 2016;214:707–711. doi: 10.1093/infdis/jiw226. [DOI] [PubMed] [Google Scholar]
- 89.Ferreira AC, Zaverucha-do-Valle C, Reis PA, Barbosa-Lima G, Vieira YR, Mattos M, Silva PP, Sacramento C, de Castro Faria HC, Neto L, Campanati A, Tanuri K, Bruning FA, Bozza PT, Souza TML. Sofosbuvir protects Zika virus-infected mice from mortality, preventing short- and long-term sequelae. Sci Rep. 2017;7:9409. doi: 10.1038/s41598-017-09797-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kamiyama N, Soma R, Hidano S, Watanabe K, Umekita H, Fukuda C, Noguchi K, Gendo Y, Ozaki T, Sonoda A, Sachi N, Runtuwene LR, Miura Y, Matsubara E, Tajima S, Takasaki T, Eshita Y, Kobayashi T. Ribavirin inhibits Zika virus (ZIKV) replication in vitro and suppresses viremia in ZIKV-infected STAT1-deficient mice. Antiviral Res. 2017;146:1–11. doi: 10.1016/j.antiviral.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu G, Bluemling GR, Collop P, Hager M, Kuiper D, Gurale BP, Painter GR, De La Rosa A, Kolykhalov AA. Analysis of ribonucleotide 5′-triphosphate analogs as potential inhibitors of Zika virus RNA-dependent RNA polymerase by using nonradioactive polymerase assays. Antimicrob Agents Chemother. 2017;61:e01967. doi: 10.1128/AAC.01967-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mumtaz N, Jimmerson LC, Bushman LR, Kiser JJ, Aron G, Reusken C, Koopmans MPG, van Kampen JJA. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antiviral Res. 2017;146:161–163. doi: 10.1016/j.antiviral.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 93.Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S, Brass AL. The IFITMs inhibit Zika virus replication. Cell Rep. 2016;15:2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- 94.Wang C, Yang SNY, Smith K, Forwood JK, Jans DA. Nuclear import inhibitor N-(4-hydroxyphenyl) retinamide targets Zika virus (ZIKV) nonstructural protein 5 to inhibit ZIKV infection. Biochem Biophys Res Commun. 2017;493:1555–1559. doi: 10.1016/j.bbrc.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 95.Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, Jacob F, Nguyen HN, Itkin M, Hanna C, Shinn P, Allen C, Michael SG, Simeonov A, Huang W, Christian KM, Goate A, Brennand KJ, Huang R, Xia M, Ming GL, Zheng W, Song H, Tang H. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zmurko J, Marques RE, Schols D, Verbeken E, Kaptein SJ, Neyts J. The viral polymerase inhibitor 7-Deaza-2′-C-methyladenosine is a potent inhibitor of in vitro Zika virus replication and delays disease progression in a robust mouse infection model. PLoS Negl Trop Dis. 2016;10:e0004695. doi: 10.1371/journal.pntd.0004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zou G, Chen YL, Dong H, Lim CC, Yap LJ, Yau YH, Shochat SG, Lescar J, Shi PY. Functional analysis of two cavities in flavivirus NS5 polymerase. J Biol Chem. 2011;286:14362–14372. doi: 10.1074/jbc.M110.214189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deng YQ, Zhang NN, Li CF, Tian M, Hao JN, Xie XP, Shi PY, Qin CF. Adenosine analog NITD008 is a potent inhibitor of Zika virus. Open Forum Infect Dis. 2016;3:ofw175. doi: 10.1093/ofid/ofw175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boldescu V, Behnam MAM, Vasilakis N, Klein CD. Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat Rev Drug Discov. 2017;16:565–586. doi: 10.1038/nrd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garcia LL, Padilla L, Castano JC. Inhibitors compounds of the flavivirus replication process. Virol J. 2017;14:95. doi: 10.1186/s12985-017-0761-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Keating GM, Vaidya A. Sofosbuvir: first global approval. Drugs. 2014;74:273–282. doi: 10.1007/s40265-014-0179-7. [DOI] [PubMed] [Google Scholar]
- 102.Murakami E, Tolstykh T, Bao H, Niu C, Steuer HM, Bao D, Chang W, Espiritu C, Bansal S, Lam AM, Otto MJ, Sofia MJ, Furman PA. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J Biol Chem. 2010;285:34337–34347. doi: 10.1074/jbc.M110.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HM, Niu C, Otto MJ, Furman PA. Discovery of a beta-d-2′-deoxy-2′-alpha-fluoro-2′-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem. 2010;53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- 104.Onorati M, Li Z, Liu F, Sousa AM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp AT, Han W, Pletikos M, Gao T, Zhu Y, Bichsel C, Varela L, Szigeti-Buck K, Lisgo S, Zhang Y, Testen A, Gao XB, Mlakar J, Popovic M, Flamand M, Strittmatter SM, Kaczmarek LK, Anton ES, Horvath TL, Lindenbach BD, Sestan N. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 2016;16:2576–2592. doi: 10.1016/j.celrep.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lim SP, Noble CG, Shi PY. The dengue virus NS5 protein as a target for drug discovery. Antiviral Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 106.Olsen DB, Eldrup AB, Bartholomew L, Bhat B, Bosserman MR, Ceccacci A, Colwell LF, Fay JF, Flores OA, Getty KL, Grobler JA, LaFemina RL, Markel EJ, Migliaccio G, Prhavc M, Stahlhut MW, Tomassini JE, MacCoss M, Hazuda DJ, Carroll SS. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob Agents Chemother. 2004;48:3944–3953. doi: 10.1128/AAC.48.10.3944-3953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eyer L, Valdes JJ, Gil VA, Nencka R, Hrebabecky H, Sala M, Salat J, Cerny J, Palus M, De Clercq E, Ruzek D. Nucleoside inhibitors of tick-borne encephalitis virus. Antimicrob Agents Chemother. 2015;59:5483–5493. doi: 10.1128/AAC.00807-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schul W, Liu W, Xu HY, Flamand M, Vasudevan SG. A dengue fever viremia model in mice shows reduction in viral replication and suppression of the inflammatory response after treatment with antiviral drugs. J Infect Dis. 2007;195:665–674. doi: 10.1086/511310. [DOI] [PubMed] [Google Scholar]
- 109.Eyer L, Smidkova M, Nencka R, Neca J, Kastl T, Palus M, De Clercq E, Ruzek D. Structure–activity relationships of nucleoside analogues for inhibition of tick-borne encephalitis virus. Antiviral Res. 2016;133:119–129. doi: 10.1016/j.antiviral.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 110.Chatelain G, Debing Y, De Burghgraeve T, Zmurko J, Saudi M, Rozenski J, Neyts J, Van Aerschot A. In search of flavivirus inhibitors: evaluation of different tritylated nucleoside analogues. Eur J Med Chem. 2013;65:249–255. doi: 10.1016/j.ejmech.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 111.Eltahla AA, Luciani F, White PA, Lloyd AR, Bull RA. Inhibitors of the hepatitis C virus polymerase; mode of action and resistance. Viruses. 2015;7:5206–5224. doi: 10.3390/v7102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lim SP, Noble CG, Seh CC, Soh TS, El Sahili A, Chan GK, Lescar J, Arora R, Benson T, Nilar S, Manjunatha U, Wan KF, Dong H, Xie X, Shi PY, Yokokawa F. Potent allosteric dengue virus NS5 polymerase inhibitors: mechanism of action and resistance profiling. PLoS Pathog. 2016;12:e1005737. doi: 10.1371/journal.ppat.1005737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yokokawa F, Nilar S, Noble CG, Lim SP, Rao R, Tania S, Wang G, Lee G, Hunziker J, Karuna R, Manjunatha U, Shi PY, Smith PW. Discovery of potent non-nucleoside inhibitors of dengue viral RNA-dependent RNA polymerase from a fragment hit using structure-based drug design. J Med Chem. 2016;59:3935–3952. doi: 10.1021/acs.jmedchem.6b00143. [DOI] [PubMed] [Google Scholar]
- 114.Lim SP, Wen D, Yap TL, Yan CK, Lescar J, Vasudevan SG. A scintillation proximity assay for dengue virus NS5 2′-O-methyltransferase-kinetic and inhibition analyses. Antiviral Res. 2008;80:360–369. doi: 10.1016/j.antiviral.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 115.Selisko B, Peyrane FF, Canard B, Alvarez K, Decroly E. Biochemical characterization of the (nucleoside-2′O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n) J Gen Virol. 2010;91:112–121. doi: 10.1099/vir.0.015511-0. [DOI] [PubMed] [Google Scholar]
- 116.Chung KY, Dong H, Chao AT, Shi PY, Lescar J, Lim SP. Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2′-O methyltransferase activity in dengue virus. Virology. 2010;402:52–60. doi: 10.1016/j.virol.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 117.Dong H, Liu L, Zou G, Zhao Y, Li Z, Lim SP, Shi PY, Li H. Structural and functional analyses of a conserved hydrophobic pocket of flavivirus methyltransferase. J Biol Chem. 2010;285:32586–32595. doi: 10.1074/jbc.M110.129197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong H, Liu B, Bodenreider C, Lee G, Ding M, Chan WL, Wang G, Jian YL, Chao AT, Lescar J, Yin Z, Vedananda TR, Keller TH, Shi PY. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem. 2011;286:6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brecher M, Chen H, Li Z, Banavali NK, Jones SA, Zhang J, Kramer LD, Li H. Identification and characterization of novel broad-spectrum inhibitors of the flavivirus methyltransferase. ACS Infect Dis. 2015;1:340–349. doi: 10.1021/acsinfecdis.5b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jain R, Butler KV, Coloma J, Jin J, Aggarwal AK. Development of a S-adenosylmethionine analog that intrudes the RNA-cap binding site of Zika methyltransferase. Sci Rep. 2017;7:1632. doi: 10.1038/s41598-017-01756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kohler JJ, Lewis W. A brief overview of mechanisms of mitochondrial toxicity from NRTIs. Environ Mol Mutagen. 2007;48:166–172. doi: 10.1002/em.20223. [DOI] [PubMed] [Google Scholar]
- 122.Vincetti P, Caporuscio F, Kaptein S, Gioiello A, Mancino V, Suzuki Y, Yamamoto N, Crespan E, Lossani A, Maga G, Rastelli G, Castagnolo D, Neyts J, Leyssen P, Costantino G, Radi M. Discovery of multitarget antivirals acting on both the dengue virus NS5–NS3 interaction and the host Src/Fyn kinases. J Med Chem. 2015;58:4964–4975. doi: 10.1021/acs.jmedchem.5b00108. [DOI] [PubMed] [Google Scholar]
- 123.Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, Cortese R, De Francesco R, Eldrup AB, Getty KL, Hou XS, LaFemina RL, Ludmerer SW, MacCoss M, McMasters DR, Stahlhut MW, Olsen DB, Hazuda DJ, Flores OA. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem. 2003;278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 124.Qing J, Luo R, Wang Y, Nong J, Wu M, Shao Y, Tang R, Yu X, Yin Z, Sun Y. Resistance analysis and characterization of NITD008 as an adenosine analog inhibitor against hepatitis C virus. Antiviral Res. 2016;126:43–54. doi: 10.1016/j.antiviral.2015.12.010. [DOI] [PubMed] [Google Scholar]