Abstract
Modulation of expression of noncoding RNAs is an important aspect of the oncogenic activities of high-risk human papillomavirus (HPV) E6 and E7 proteins. While HPV E6/E7-mediated alterations of microRNAs (miRNAs) has been studied in detail there are fewer reports on HPV-mediated dysregulation of long noncoding RNAs (lncRNAs). The cervical carcinoma expressed PCNA regulatory (CCEPR) lncRNA is highly expressed in cervical cancers and expression correlates with tumor size and patient outcome. We report that CCEPR is a nuclear lncRNA and that HPV16 E6 oncogene expression causes increased CCEPR expression through a mechanism that is not directly dependent on TP53 inactivation. CCEPR depletion in cervical carcinoma cell lines reduces viability, while overexpression enhances viability. Moreover, we determined that CCEPR is a nuclear lncRNA. In contrast to what was published and inspired its designation, there is no evidence for PCNA mRNA stabilization, and hence CCEPR likely functions through a different mechanism.
Keywords: long non-coding RNA, PCNA, cervical cancer, cell viability, viral oncogenesis
INTRODUCTION
High-risk human papillomaviruses (HPVs) cause approximately 5% of all human cancers including almost all cervical carcinomas as well as a large fraction of other anogenital tract and oropharyngeal carcinomas (Cubie, 2013). E6 and E7 are the only viral proteins that are consistently expressed in the tumors, and their expression is required for cancer initiation and maintenance (Moody and Laimins, 2010). The two best studied cellular targets of HPV16 E6 and E7 are the TP53 and RB1 tumor suppressors, respectively. However, there is compelling evidence that the high-risk HPV E6 and E7 proteins also target other cellular factors and over the years, many additional cellular protein targets of HPV E6 and E7 have been identified (Roman and Munger, 2013; Vande Pol and Klingelhutz, 2013).
Only a small fraction (<2%) of the transcribed human genome correspond to protein-coding mRNAs (Djebali et al., 2012). Most of the transcribed RNAs are not translated, and are members of various classes of noncoding RNAs. One such class are microRNAs (miRNAs) and several studies have investigated the modulation of cellular miRNA expression by HPV E6/E7 (Gunasekharan and Laimins, 2013; Harden et al., 2017; Melar-New and Laimins, 2010; Wang et al., 2014). Another class are the long non-coding RNAs (lncRNAs). These are defined as transcripts longer than 200 nucleotides with limited coding potential of <100 amino acids. LncRNAs can specifically interact with proteins, DNA and RNA and function as scaffolds for multiprotein complexes, guiding such complexes to specific portions of the genome, binding and affecting the stability of cognate mRNA partners or serving as microRNA sponges (Long et al., 2017; Wang and Chang, 2011). LncRNAs are dysregulated in many diseases and can drive important cancer phenotypes by acting as tumor suppressors or oncogenes (Gutschner and Diederichs, 2012; Marchese et al., 2017; Wapinski and Chang, 2011).
Three microarray studies have interrogated lncRNA expression in cervical tumor samples and at different cancer stages and have suggested that lncRNAs may contribute to the development and progression of cervical carcinoma (Chen et al., 2015; Gibb et al., 2012; Sun et al., 2014). However, the molecular mechanism(s) that drive the expression of these lncRNAs and the potential pathophysiological contributions of specific lncRNAs to HPV carcinogenesis remain largely unknown. Moreover, these studies do not provide insights whether the observed aberrant lncRNA expression represents a direct consequence of HPV E6 and/or E7 expression or whether these changes occur as a consequence of or in response to cellular mutations that accumulate during malignant progression.
The cervical carcinoma expressed PCNA regulatory lncRNA gene (CCEPR; aka cervical carcinoma high-expressed lncRNA 1, CCHE1) is located on chromosome 10 and encodes a 2502 nucleotide RNA. As the designation implies, it was found to be highly expressed in cervical cancer tissues. CCEPR expression levels were shown to correlate with larger tumor size and serve as a predictor for poor prognosis of cervical cancer patients (Chen et al., 2017). It was reported that CCEPR overexpression might contribute to cellular hyperproliferation of cervical cancer cells by binding and stabilizing PCNA mRNA (Yang et al., 2015). Additionally, high level CCEPR expression was also detected in other cancer types that are not associated with HPV infections, including hepatocellular and urothelial bladder carcinomas and high-level expression may also serve as a predictor of poor prognosis with these tumors, as well (Peng and Fan, 2016; Zhan et al., 2017). These results suggest that CCEPR may be an oncogenic lncRNA.
Here, we report that increased CCEPR expression is driven by the HPV E6 oncogene, through a mechanism that is not directly dependent on TP53 inactivation. While our results are consistent with earlier results that showed that CCEPR expression modulates proliferation of cervical cancer cell lines, we did not detect any correlation of CCEPR and PCNA levels. Moreover, CCEPR is a predominantly nuclear lncRNA, suggesting that it may function through mechanisms that are independent of PCNA mRNA stabilization.
MATERIALS AND METHODS
Cell culture
Primary human foreskin keratinocytes (HFKs) were prepared from a pool of 3 to 5 neonatal foreskins as previously described (McLaughlin-Drubin et al., 2008). HFKs were maintained in keratinocyte-serum-free media (KSFM) supplemented with human recombinant epidermal growth factor 1-53 and bovine pituitary extract (Invitrogen). HFK populations were generated by retroviral infection with the corresponding LXSN based retroviruses followed by G418 selection (500 μg/ml) (Halbert et al., 1991). The LXSN based vector for expression of a dominant negative C-terminal TP53 minigene (dn-p53) was a kind gift from Moshe Oren’s group (Gottlieb et al., 1994). All experiments were performed with donor and passage matched HFKs that were passaged less than 8 times. CaSki, C33A and HeLa cells (ATCC) were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Cell fractionation
Cell fractionation was performed using the “Rapid, Efficient And Practical” (REAP) method for subcellular fractionation (Suzuki et al., 2010). Briefly, cells were washed with ice-cold phosphate buffered saline (PBS) and triturated with ice cold 0.1 % NP40 containing PBS five times. Cell were pelleted for 10 seconds at 13,000 x g and the supernatant, which contains the cytoplasmic fraction, was collected. The nuclear pellet was re-suspended in ice-cold 0.1 % NP40 containing PBS and triturated only once. After a 10 second spin at 13,000 x g, the supernatant was discarded and the nuclear pellet was collected. Both cytoplasmic and nuclear fractions were mixed with RNA lysis buffer from the Quick-RNA MiniPrep kit (Zymo Research) for RNA isolation.
Antibodies, plasmids, transfections, and lentiviral transduction
The following primary antibodies used were used for western blotting: TP53 (OP43, Calbiochem, 1:1,000), RB1 (Ab-5, Millipore, 1:100) and actin (Ab-1501, Millipore, 1:1,000). A horseradish peroxidase conjugated anti-mouse antibody (NA931, GE Healthcare Life Sciences, 1:10,000) was used for detection by enhanced chemiluminescence and images were acquired on a Syngene ChemiXX6 imager equipped with Genesys software version 1.5.5.0. Transient transfections were performed using Lipofectamine 3000 (Thermo Fisher) according to the manufacturer’s protocol. The cells were transfected with lentiviral shRNA expression vectors with the following target sequences, shRNA-CCEPR1: 5′-GGCGAGCATGTTTGTTGTTTA-3′ (Yang et al., 2015), shRNA-CCEPR2: 5′-GTGAGAAATGAGCGGATTACC-3′, and a non-targeting control shRNA: 5′-CCTAAGGTTAAGTCGCCCTCG-3′ (Bryant et al., 2010; Shen et al., 2011). A vector for expression of the full length CCEPR lncRNA from the SR alpha promoter (composed of the simian virus 40 (SV40) early promoter and the R segment and part of the U5 sequence (R-U5′) of the long terminal repeat of human T-cell leukemia virus type 1) was purchased from the EST consortium in Japan, cDNA clone ORF AK055418/FLJ30856. The obtained plasmid was verified for expression of full length CCEPR by sequencing.
RNA isolation and real-time quantitative PCR analysis
Total RNA was extracted using the Quick-RNA MiniPrep (Zymo Research). cDNA was reverse transcribed using the Quantitect Reverse Transcription Kit (Qiagen). Quantitative PCR (qPCR) was performed in triplicate on a StepOne Plus (Applied Biosystems) thermocycler using SYBR Green PCR Master Mix (Applied Biosystems) reagents. Primer sequences used for RT-qPCR analysis in this study are as follows: CCEPR: 5′-AAGGTCCCAGGATACTCGC-3′ (forward) and 5′-GTGTCGTGGACTGGCAAAAT-3′ (reverse) (Yang et al., 2015); PCNA: 5′-TTAAATTGTCACAGACAAGTAATGTCG-3′ (forward) and 5′-TGGCTTTTGTAAAGAAGTTCAGGTAC-3′ (reverse) (Bustin et al., 2001); MALAT1: 5′-GACGGAGGTTGAGATGAAGC-3′ (forward) and 5′-ATTCGGGGCTCTGTAGTCCT-3′ (reverse) (Tripathi et al., 2010); GAPDH: 5′-GATTCCACCCATGGCAAATCC-3′ (forward) and 5′-TGGGATTTCCATTGATGACAAG-3′ (reverse) (McLaughlin-Drubin et al., 2011). Primers for CCEPR were tested for exponential amplification and CCEPR qPCR amplicons were verified by sequencing. Expression data was quantified using the ΔΔCT method and normalized to expression of the GAPDH as the housekeeping gene.
Cell viability determination
Two days post transfection, media was removed, and 10 μg/ml resazurin (Sigma) was added to assess redox fitness. Cells were incubated with dye for one hour and then sample fluorescence was read in triplicate using 560 nm excitation and 590 nm emission filters on a Synergy H1 microplate reader (BioTek).
Quantification of cell numbers
Four days post transfection, cells were fixed overnight with 10% (w/v) trichloroacetic acid, washed and incubated in 0.4% Sulforhodamine B (SRB) (Sigma) (Vichai and Kirtikara, 2006) in 1% acetic acid for ten minutes. After washing with 1% acetic acid five times plates were air-dried and SRB dye was eluted in 10 mM Tris buffer, pH 10.5. Absorbance readings were taken at 492 nm on a Synergy H1 microplate reader (BioTek).
RESULTS
CCEPR expression is mainly driven by HPV16 E6 oncogene
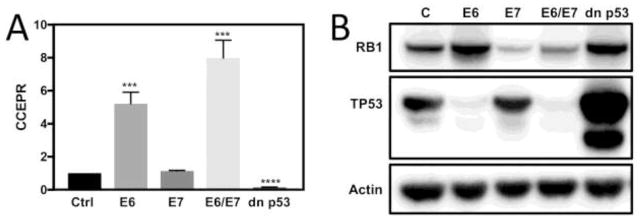
It has been reported that CCEPR expression is elevated in cervical cancer tissues and cell lines (Chen et al., 2017; Yang et al., 2015). It was unknown, however, whether increased CCEPR expression is driven by the HPV E6 and/or E7 oncoproteins or whether expression increases late during carcinogenesis and is not directly linked to E6/E7 expression. To investigate this issue, we used quantitative reverse-transcription polymerase chain reaction (qRT-PCR) technology to determine CCEPR levels in donor and passage matched primary human foreskin keratinocyte (HFK) cultures with ectopic expression of HPV16 E6 and/or E7 oncogene expression. These experiments showed that CCEPR levels were increased in HPV16 E6/E7 and HPV16 E6 expressing HFKs but not in HPV16 E7 expressing HFKs. Hence, E6 is the main driver of CCEPR expression in HPV oncogene expressing cells, and E7 co-expression may further enhance CCEPR levels (Figure 1A). Expression of HPV16 E6 and E7 was assessed indirectly by analyzing TP53 and RB1 levels. As expected, RB1 levels were lower in E7 expressing cells and TP53 levels were lower in E6 expressing cells (Figure 1B).
Figure 1.
CCEPR levels as determined by qRT-PCR in HFKs with stable co-expression of HPV16 E6/E7, individual expression of HPV16 E7, HPV16 E6 or a dominant negative TP53 minigene (dnp53) (A). Results are expressed relative to control vector transduced HFKs (Ctrl). Means ± SEM calculated from a single representative HFKs population, each performed in triplicate are shown. *** p<0.001 (Student’s t test). Validation of HPV16 E6 and/or E7 and dn p53 expression in HFKs by western blot analysis of RB1 and TP53. Actin is shown as a loading control (B).
HPV16 E6 mediated CCEPR upregulation is not a direct consequence of TP53 inactivation
The TP53 tumor suppressor is a major cellular target of high-risk HPV E6 oncoproteins. Given that CCEPR is also overexpressed in non-HPV associated hepatocellular and urothelial bladder carcinomas where TP53 is frequently mutated, we next investigated if high-level CCEPR expression was caused by TP53 inactivation. CCEPR levels were determined by qRT-PCR in HFK populations with expression of the dn p53 minigene (Gottlieb et al., 1994). Unlike HPV16 E6 expression, inactivation of TP53 did not cause increased CCEPR expression; in fact, CCEPR expression was lower in dominant negative TP53 expressing HFKs than in control vector transduced HFKs. Hence, CCEPR upregulation by HPV16 E6 expression is not a direct consequence of TP53 inactivation (Figure 1A). as well as the dominant negative TP53 (dn p53) minigene (Gottlieb et al., 1994). As expected, dn p53 expression in HFKs caused increased p53 levels, presumably by stabilizing and inactivating endogenous wild type TP53 (Gottlieb et al., 1994) (Figure 1B).
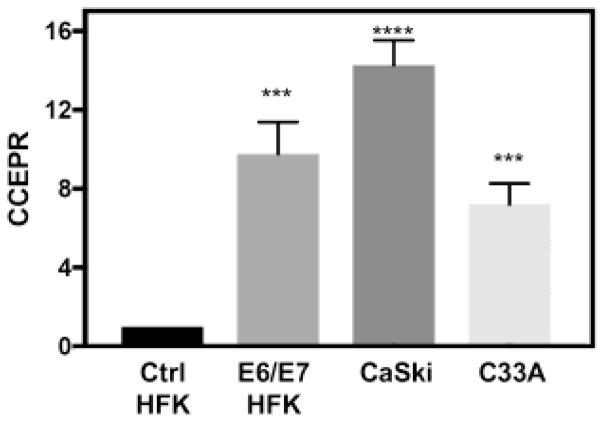
CCEPR levels in HPV16 E6/E7 expressing cells are similar to cervical cancer cell lines
To determine whether HPV16 E6/E7 expression increases CCEPR to levels similar to those in HPV16 positive cervical carcinoma cells, we compared CCEPR expression in HPV16 E6/E7 expressing HFKs and HPV16 positive CaSki cervical cancer cells by qRT-PCR. These experiments showed HPV16 E6/E7 expression increases CCEPR levels in HFKs to levels similar to CaSki cells (Figure 2). Moreover, given that CCEPR is also expressed in non-HPV-associated cancers, we determined CCEPR levels in HPV negative C33A cervical carcinoma cells and found that CCEPR levels were expressed higher levels in these cells than in HFKs (Figure 2).
Figure 2.
CCEPR levels as determined by qRT-PCR in HPV16 E6/E7 expressing HFKs (E6/E7 HFK), HPV16 positive CaSki and HPV negative C33A cervical carcinoma cells. Results are normalized relative to control HFKs (Ctrl HFK). Means ± SEM, each performed in triplicate are shown. ****p<0.0001, ***p<0.001 (Student’s t test).
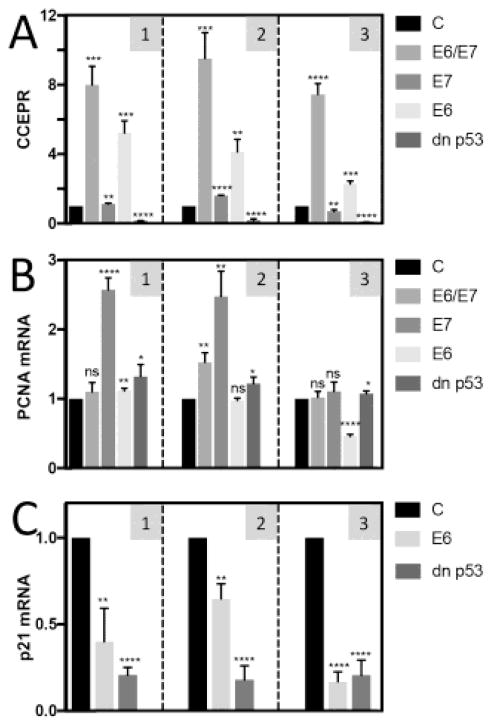
PCNA mRNA levels do not correlate with CCEPR
A previous study reported a significant correlation of CCEPR and PCNA mRNA levels in cervical cancer tissues, and suggested that CCEPR overexpression may cause enhanced proliferation by PCNA mRNA binding and stabilization (Yang et al., 2015). We, therefore, determined CCEPR (Figure 3A) and PCNA mRNA levels (Figure 3B) in three independently derived HFK populations each with stable expression of HPV16 E6 and/or E7, or dn p53 by qRT-PCR. Our results showed that while CCEPR and PCNA mRNA levels each were high in HPV16 E6/E7 expressing HFKs, PCNA mRNA levels were increased in E7 expressing cells where CCEPR levels were low, similar to control HFKs, whereas HPV16 E6 expressing cells, which expressed high levels of CCEPR, had low PCNA levels similar to control HFKs. Moreover, TP53 expressing which contained lower CCEPR levels than control vector transduced HFKs showed no concomitant decreased in PCNA mRNA. To document TP53 inhibition by HPV16 E6 and dn p53 expression, we determined mRNA expression of the prototypical TP53 responsive CDKN1A (p21CIP1) gene. As expected, CDKN1A mRNA levels were significantly lower both in HPV16 E6 and dn p53 expressing HFKs (Figure 3C). Hence our results show that there is no correlation between CCEPR and PCNA mRNA levels.
Figure 3.
Levels of CCEPR (A) PCNA mRNA (B) as determined by qRT-PCR in HFKs with stable co-expression of HPV16 E6/E7, individual expression of HPV16 E7, HPV16 E6 or a dominant negative TP53 mutant (dn p53). Validation of TP53 inactivation in HFKs expressing HPV16 E6 or dn p53 by analyzing expression of the TP53 responsive CDKN1A (p21) levels (C). Results shown are normalized to control vector transduced HFKs (C). Means ± SEM calculated from each of the three independently derived HFKs populations, performed in triplicate are shown. Data shown in panel 1 correspond to the data shown in Figure 1.
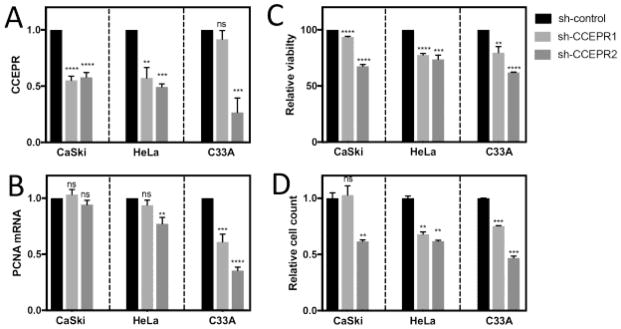
Depletion of CCEPR inhibits viability of HPV positive and HPV negative cervical cancer lines
Depletion of CCEPR has been reported to inhibit HeLa cervical cancer cell proliferation and decrease PCNA levels (Yang et al., 2015). Since the previous study only used a single CCEPR targeting siRNA and our results did not provide any evidence for CCEPR regulating PCNA mRNA levels, we tested the effect of depleting CCEPR in HPV18 positive HeLa, HPV16 positive CaSki and HPV negative C33A cervical cancer cells using two independent CCEPR targeting shRNAs. A non-targeting control shRNA was transfected as a negative control. CCEPR depletion was validated using qRT-PCR (Figure 4A). We also analyzed PCNA mRNA levels and consistent with our results shown in Figure 3, CCEPR depletion did not trigger a consistent, concomitant decrease in PCNA mRNA levels (Figure 4B). As a biological read out, cellular viability was assessed by reduction of resazurin at two days post shRNA transfection (Figure 4C). To confirm that the observed differences in resazurin reduction reflect differences in cellular viability versus changes in redox metabolism, we also determined cell numbers by Sulforhodamine B (SRB) staining at four days post shRNA transfection (Figure 4D). Consistent with a previous study in HeLa cells (Yang et al., 2015) our experiments showed a decrease in viability and cell numbers in response to CCEPR depletion in each of the cell lines tested.
Figure 4.
Levels of CCEPR (A) and PCNA mRNA (B) as determined by qRT-PCR in HPV16 positive CaSki, HPV18 positive HeLa and HPV-negative cervical cancer cell lines transiently transfected with two separate CCEPR shRNA vectors. RNAs were harvested at two days post transfection, and results are expressed relative to cells transfected with a control non-targeting shRNA vector. Cell viability at two days post transfection was assessed by resazurin assays (C). Cell numbers were determined by Sulforhodamine B (SRB) protein stain assay at four days post transfection (D). Bars represent averages ± SEM of single representative experiments, each performed in triplicate. ****p<0.0001, ***p<0.001, **p<0.01, ns = non-significant (Student’s t test). Similar results were observed in three independent experiments.
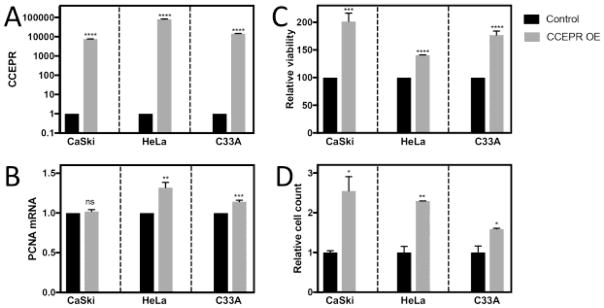
CCEPR overexpression in cervical cancer lines does not cause increased PCNA mRNA expression
Next, we tested whether transiently overexpressing CCEPR in the cervical cancer cell lines would have the opposite effect of CCEPR depletion. HPV18 positive HeLa, HPV16 positive CaSki and HPV negative C33A cervical carcinoma lines were each transfected with full a length CCEPR expression plasmid or a control vector and ectopic CCEPR expression was documented by qRT-qPCR (Figure 5A). Even at these very high levels of CCEPR expression there was no consistent, significant increase in PCNA mRNA levels, (Figure 5B) even though we detected an increase in cell viability (Figure 5C) and cell numbers (Figure 5D) at two and four days post transfection, respectively. These results show that even very high, CCEPR overexpression, while enhancing cell viability and causing an increase in cell numbers, did not cause a consistent, concomitant increase in PCNA levels. In combination with the results shown in Figures 3 and 4, these experiments show that CCEPR can modulate cell viability, but that this likely independent of modulation of PCNA mRNA levels.
Figure 5.
Levels of CCEPR (A) and PCNA mRNA (B) as determined by qRT- PCR in HPV16 positive CaSki, HPV18 positive HeLa and HPV negative C33A cervical cancer cells transiently transfected with either control plasmid or a full length CCEPR expression vector. RNAs were harvested at two days post transfection. were, and results are expressed relative to control vector transfected cells. Cell viability at two days post transfection was assessed by resazurin assays (C). Cell numbers were determined by Sulforhodamine B (SRB) protein stain assay at four days post transfection (D). Bars represent averages ± SEM of single representative experiments, each performed in triplicates. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05 (Student’s t test). Similar results were observed in three independent experiments.
CCEPR is a nuclear lncRNA
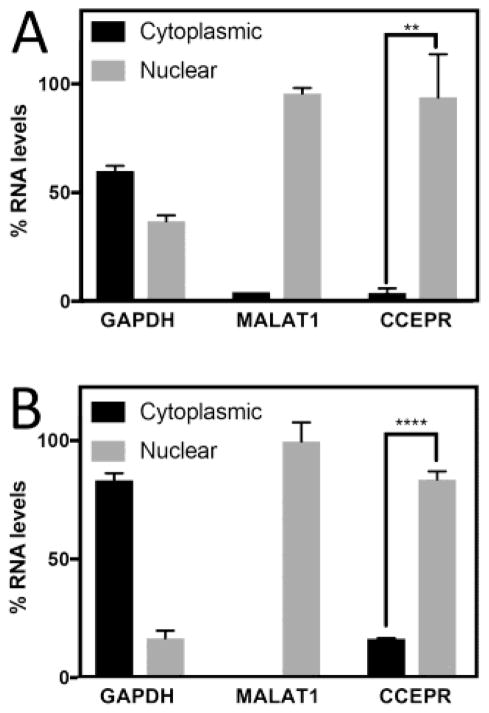
Determination of the subcellular localization can provide insights regarding potential biological activities and biochemical targets of lncRNAs (Chen, 2016; Fatica and Bozzoni, 2014; Zhang et al., 2014). Nuclear lncRNAs often function as epigenetic regulators and/or affect nuclear mRNA metabolism, while cytoplasmic lncRNAs have been implicated in the regulation of the stability and translation of mRNA targets (Long et al., 2017; Wang and Chang, 2011). Given that our results do not support the model that CCEPR modulates PCNA mRNA stability and abundance, we determined CCEPR levels by qRT-PCR in nuclear and cytoplasmic fractions of the HPV18 positive HeLa cervical carcinoma line (Figure 6A) as well as HPV16 E6/E7 expressing HFKs (Figure 6B). The known nuclear lncRNA MALAT1 (Bernard et al., 2010; Tripathi et al., 2010) and GAPDH mRNA (found in both nucleus and cytoplasm) served as controls for RNA detection in the nuclear and cytoplasmic fractions, respectively. These experiments revealed that similar to MALAT1, CCEPR is predominantly nuclear lncRNA (Figure 6).
Figure 6.
Subcellular localization of CCEPR as determined by qRT–PCR in cytoplasmic and nuclear fractions of the of HPV18 positive HeLa cervical carcinoma cells (A) and HPV16 E6/E7 expressing HFKs (B). Means ± SEM, each performed in triplicate are shown. MALAT1 and GAPDH are used as controls. ****p<0.0001, **p<0.01 (Student’s t test). Similar results were also obtained with HPV16 positive CaSki cervical carcinoma cells.
DISCUSSION
High-risk HPV E6 and E7 oncoproteins dysregulate a variety of host cellular processes to induce host cell transformation. The ability of HPV E6 and E7 to modulate expression of various coding genes and microRNAs have been well documented. Despite a few reports of aberrant lncRNA expression profiles in cervical cancer tissues (Chen et al., 2015; Gibb et al., 2012; Sun et al., 2014), the molecular mechanisms that drive the expression of these lncRNAs and whether and how they may contribute to cervical carcinogenesis remains largely unknown.
LncRNAs are emerging as a critical component of the cancer transcriptome, and have been reported to play a role in variety of cellular processes through their interactions with proteins, DNA and RNA (Long et al., 2017; Wang and Chang, 2011). We focused on CCEPR because its expression has been analyzed in detail in cervical carcinoma tissue and expression was shown to increase during cervical cancer development correlate with tumor size (Chen et al., 2017). However, it was not known whether increased CCEPR expression was a direct consequence of HPV E6 and/or E7 expression. Here, we show that HPV16 E6/E7 expression in primary human foreskin keratinocytes causes increased CCEPR expression to levels that are similar to those detected in HPV16 positive CaSki cervical carcinoma cells. Moreover, we show that the HPV16 E6 oncoprotein is the major driver of CCEPR expression. CCEPR levels were even higher in E6/E7 expressing cells, suggesting that E7 may further enhance CCEPR levels, although we cannot rule out that E6 expression may be higher in the E6/E7 co-expressing HFKs. The finding that the HPV negative C33A cervical carcinoma cell line also highly expresses CCEPR is consistent with the model that the pathway targeted by HPV16 E6 that triggers increased CCEPR expression may also be defective, presumably by mutation, in the HPV negative C33A cervical carcinoma line and presumably other in other HPV negative cancers, including hepatocellular and urothelial bladder carcinomas that have also been reported to highly express CCEPR. Given that the TP53 tumor suppressor is a major cellular target of the cancer associated HPV E6 proteins that TP53 is mutated in C33A cells as well as many other cancers including and hepatocellular and urothelial bladder carcinomas, we hypothesized that HPV16 E6 my drive CCEPR expression via TP53 inactivation. Somewhat surprisingly, we found that expression of a dominant negative TP53 mutant that inactivates TP53 by forming non-functional tetramers (Gottlieb et al., 1994), decreased rather than increased CCEPR expression. Hence, HPV16 E6 drives CCEPR expression through a pathway that not directly dependent on TP53, but that this pathway may also be dysfunctional in HPV negative cancers.
CCEPR has been reported to bind to and stabilize PCNA mRNA thereby regulating proliferation (Yang et al., 2015). Consistent with this study, we observed that CCEPR depletion not only inhibits the viability of HPV18 positive HeLa but also of HPV16 positive CaSki, and HPV negative C33A cervical cancer cell lines, whereas overexpressing CCEPR enhanced viability of these cell lines. Viability measurements reflected changes in cell numbers, indicating that CCEPR can modulate cell proliferation, which supports the previously described pro-proliferative role of CCEPR in cervical cancer cell lines. In contrast, however, our results did not provide any evidence for a correlation between CCEPR and PCNA mRNA levels as was noted previously (Yang et al., 2015). Indeed, and as expected and presumably as a consequence of E2F transcription factor activation (Cheng et al., 1995), high-level PCNA mRNA expression in HPV16 E6/E7 expressing cells was mostly driven by E7, which did not increase CCEPR levels. Conversely, whereas HPV16 E6 expression in HFKs increased CCEPR levels, it did not cause an increase in PCNA mRNA levels. Similarly, expression of the dominant negative TP53 mutant, while decreasing CCEPR levels did not affect PCNA mRNA levels. Furthermore, perturbation of CCEPR expression in cervical cancer cell lines did not cause consistent, opposite alterations in PCNA mRNA levels. Hence, PCNA mRNA stabilization is not be the primary mechanism by which CCEPR modulates cellular viability and proliferation. It will be interesting to determine the molecular mechanisms by which CCEPR modulates cellular viability.
The subcellular localization can give important hints regarding the biological activity of a given lncRNA (Chen, 2016; Fatica and Bozzoni, 2014; Zhang et al., 2014). We determined that CCEPR was predominantly nuclear. Prevailing models suggest that binding and altering the stability of cognate mRNA partners is mostly achieved by cytoplasmic lncRNAs, whereas nuclear lncRNAs often play roles in nuclear organization, epigenetic regulation of gene expression or alternative splicing (Long et al., 2017; Wang and Chang, 2011). Hence it is likely that CCEPR primarily functions in the nucleus by interacting with proteins, chromatin and/or RNA, rather than by modulating PCNA mRNA levels in the cytoplasm. One important future endeavor will be to identify the molecular targets of CCEPR to understand its mechanism of action. The study of lncRNAs in human diseases including cancer is an emerging field with enormous, largely untapped potential as these molecules may not just be cancer biomarkers but represent novel targets for drug development (Vitiello et al., 2015).
Highlights.
The human papillomavirus E6 protein drives expression of the cervical carcinoma expressed PCNA regulatory (CCEPR) long noncoding RNA (lncRNA).
CCEPR induction is independent of TP53 inactivation
CCEPR regulates cellular proliferation
CCEPR levels do not correlate with PCNA levels
Acknowledgments
We thank Drs. Amy Yee, Philip Hinds, Peter Juo and members of the Munger Lab for stimulating discussions, suggestions and advice and the two anonymous reviewers for their helpful suggestions. Supported by PHS grant CA066980 (KM). SS was a Dean’s Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. Embo j. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Li SR, Dorudi S. Expression of the Ca2+-activated chloride channel genes CLCA1 and CLCA2 is downregulated in human colorectal cancer. DNA Cell Biol. 2001;20:331–338. doi: 10.1089/10445490152122442. [DOI] [PubMed] [Google Scholar]
- Chen J, Fu Z, Ji C, Gu P, Xu P, Yu N, Kan Y, Wu X, Shen R, Shen Y. Systematic gene microarray analysis of the lncRNA expression profiles in human uterine cervix carcinoma. Biomed Pharmacother. 2015;72:83–90. doi: 10.1016/j.biopha.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang CX, Sun XX, Wang C, Liu TF, Wang DJ. Long non-coding RNA CCHE1 overexpression predicts a poor prognosis for cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21:479–483. [PubMed] [Google Scholar]
- Cheng S, Schmidt-Grimminger DC, Murant T, Broker TR, Chow LT. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- Cubie HA. Diseases associated with human papillomavirus infection. Virology. 2013;445:21–34. doi: 10.1016/j.virol.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Gibb EA, Becker-Santos DD, Enfield KS, Guillaud M, Niekerk D, Matisic JP, Macaulay CE, Lam WL. Aberrant expression of long noncoding RNAs in cervical intraepithelial neoplasia. Int J Gynecol Cancer. 2012;22:1557–1563. doi: 10.1097/IGC.0b013e318272f2c9. [DOI] [PubMed] [Google Scholar]
- Gottlieb E, Haffner R, von Ruden T, Wagner EF, Oren M. Down-regulation of wild-type p53 activity interferes with apoptosis of IL-3-dependent hematopoietic cells following IL-3 withdrawal. Embo j. 1994;13:1368–1374. doi: 10.1002/j.1460-2075.1994.tb06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekharan V, Laimins LA. Human papillomaviruses modulate microRNA 145 expression to directly control genome amplification. J Virol. 2013;87:6037–6043. doi: 10.1128/JVI.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden ME, Prasad N, Griffiths A, Munger K. Modulation of microRNA-mRNA Target Pairs by Human Papillomavirus 16 Oncoproteins. MBio. 2017:8. doi: 10.1128/mBio.02170-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Crum CP, Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci U S A. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Huh KW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J Virol. 2008;82:8695–8705. doi: 10.1128/JVI.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212–5221. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacother. 2016;83:450–455. doi: 10.1016/j.biopha.2016.06.056. [DOI] [PubMed] [Google Scholar]
- Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, Kaelin WG., Jr Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene. Cancer Discov. 2011;1:222–235. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang SB, Jin ZJ, Sun SH, Wang F, Li W. Long noncoding RNA-EBIC promotes tumor cell invasion by binding to EZH2 and repressing E-cadherin in cervical cancer. PLoS One. 2014;9:e100340. doi: 10.1371/journal.pone.0100340. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. REAP: A two minute cell fractionation method. BMC Res Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445:115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- Vitiello M, Tuccoli A, Poliseno L. Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol (Dordr) 2015;38:17–28. doi: 10.1007/s13402-014-0180-x. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang HK, Li Y, Hafner M, Banerjee NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, Tuschl T, Zheng ZM. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proc Natl Acad Sci U S A. 2014;111:4262–4267. doi: 10.1073/pnas.1401430111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 2015;36:7615–7622. doi: 10.1007/s13277-015-3465-4. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Li Y, Guan B, Chen X, Chen Z, He A, He S, Gong Y, Peng D, Liu Y, Cai Z, Li X, Zhou L. Increased expression of long non-coding RNA CCEPR is associated with poor prognosis and promotes tumorigenesis in urothelial bladder carcinoma. Oncotarget. 2017;8:44326–44334. doi: 10.18632/oncotarget.17872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX, Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547:1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]