Abstract
All known orthopoxviruses, including ectromelia virus (ECTV), contain a gene in the E3L family. The protein product of this gene, E3, is a double-stranded RNA-binding protein. It can impact host range and is used by orthopoxviruses to combat cellular defense pathways, such as PKR and RNase L. In this work, we constructed an ECTV mutant with a targeted disruption of the E3L open reading frame (ECTVΔE3L). Infection with this virus resulted in an abortive replication cycle in all cell lines tested. We detected limited transcription of late genes but no significant translation of these mRNAs. Notably, the replication defects of ECTVΔE3L were rescued in human and mouse cells lacking PKR. ECTVΔE3L was nonpathogenic in BALB/c mice, a strain susceptible to lethal mousepox disease. However, infection with ECTVΔE3L induced protective immunity upon subsequent challenge with wild-type virus. In summary, E3L is an essential gene for ECTV.
Keywords: ectromelia virus, mousepox virus, double-stranded RNA, dsRNA, E3L gene, innate immune evasion, host range gene
Introduction
Ectromelia virus (ECTV; also referred to as “mousepox virus”) is a double-stranded DNA virus in the Poxviridae family of the genus orthopoxvirus. ECTV is a natural pathogen of mice and primarily infects new hosts through abrasions in the skin (Fenner, 1947). Following initial replication at the site of infection, the virus spreads to multiple organs and eventually disseminates to the skin where characteristic pock lesions can manifest in certain mouse strains (Esteban and Buller, 2005; Fenner and Mortimer, 2006).
The formation of double-stranded RNA (dsRNA) occurs during the replication cycles of numerous viral infections (Weber et al., 2006). For this reason, dsRNA is an important pathogen-associated molecule that initiates an anti-viral immune response. With respect to poxviruses, it has been known for some time that vaccinia virus (VACV) forms abundant amounts of dsRNA, especially during intermediate and late time points of the infection cycle (i.e., after DNA replication; “post-replicative”) (Boone et al., 1979; Colby and Duesberg, 1969; Colby et al., 1971; Duesberg and Colby, 1969). Other orthopoxviruses, such as cowpox virus, also form dsRNA albeit to a varying degree relative to VACV (Frey et al., 2017). The mechanism of dsRNA formation has been discussed elsewhere (Arndt et al., 2016; Frey et al., 2017). Briefly, it has been demonstrated using VACV as a model that dsRNA forms as a result of several factors: minimal intergenic space, convergent transcription from opposite DNA strands, and inefficient termination of transcription, particularly during post-replicative time points (Xiang et al., 2000; 1998). These events lead to the production of mRNAs with the capacity for complementary base pairing, thus allowing for dsRNA formation in the cytoplasm.
There are two important anti-viral pathways in cells that are induced by the presence of viral dsRNA: PKR and 2’5’-OAS/RNase L. Upon activation, both pathways can result in the termination of viral protein synthesis, thus reducing viral dissemination within the host organism. Upon recognition of dsRNA, PKR phosphorylates the alpha subunit of eukaryotic initiation factor 2 (eIF2α), which results in translational termination (Proud, 2005). RNase L non-specifically cleaves cytoplasmic RNA molecules, including viral transcripts, which inhibits protein synthesis at the level of RNA stability (Chakrabarti et al., 2011). Because dsRNA is a potent trigger of anti-viral immunity, many viruses have evolved strategies to evade detection (Langland et al., 2006; Seet et al., 2003). This study focused on the immune evasion gene E3L, which produces a protein able to blunt anti-viral responses that become induced by viral dsRNA.
The E3L gene is conserved among a large fraction of the members of the subfamily Chordopoxvirinae (Bratke et al., 2013; Haller et al., 2014; Myskiw et al., 2011). It encodes a protein product (termed E3) with an amino-terminal Z-DNA-binding domain (Kim et al., 2003) and a carboxy-terminal dsRNA-binding domain (Chang and Jacobs, 1993). This protein was originally discovered from extracts of VACV-infected cells due to its ability to bind to Poly(I:C), a synthetic dsRNA analog (Chang et al., 1992; Watson et al., 1991). E3 has been shown to inhibit activation of PKR and RNase L, which is likely due in large part to its ability to sequester dsRNA (Chang et al., 1992; Frey et al., 2017; Langland et al., 2006; Rivas et al., 1998; Xiang et al., 2002; Zhang et al., 2008). Moreover, the dsRNA-binding domain is necessary for the virulence of VACV upon infection of mice (Brandt and Jacobs, 2001). E3L can be an important host range gene since its deletion renders some poxviruses unable to replicate in cells that are normally permissive for infection (Beattie et al., 1996; Rahman et al., 2013).
In this study, we demonstrate the critical importance of E3L to the ability of ECTV to replicate in cultured cells and cause disease in mice. An ECTV mutant (ECTVΔE3L) lacking an intact E3L open reading frame was replication-incompetent in all tested mammalian cell lines and avirulent in a mouse strain that is highly susceptible to lethal infection with wild-type ECTV. Interestingly, ECTVΔE3L induced protective immunity in these mousepox-susceptible mice when used as a vaccine prior to challenge with wild-type virus. This report is the first to characterize ECTVΔE3L and to define the role of E3L in the replication capacity and pathogenicity of mousepox virus.
Materials and methods
Cells and culture methods
The following cell lines were used in this study: BS-C-1 (ATCC# CCL-26), HeLa (ATCC# CCL-2), and L-929 (ATCC# CCL-1). These cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) supplemented with 5% fetal bovine serum (FBS; Gemini BioProducts), and penicillin-streptomycin (Gemini BioProducts). The DMEM contained 4mM L-glutamine, 1 mM sodium pyruvate, and 4.5 mg/mL D-glucose from the manufacturer. All cells were maintained at 37°C in a 5% CO2 incubator and sub-cultured when they reached approximately 80–90% confluency. BS-C-1 cells constitutively expressing the E3 protein of VACV were also utilized (termed BS-C-1+E3L cells; generously supplied by Dr. Stefan Rothenburg). The E3 protein produced in these cells has mCherry fused at the C-terminus to aid in visualization using fluorescence microscopy. The BS-C-1+E3L cells were maintained in the continuous presence of 500 µg/mL Geneticin/G418 (Gemini BioProducts) to preserve high E3 levels, which were consistently ≥90% positive as determined by flow cytometry (Fig. 1A). These cells fully rescue the replication of VACVΔE3L to wild-type levels (data not shown). Wild-type and PKR knockout mouse embryonic fibroblasts (MEFs) transformed with SV40 large T antigen were kindly provided by Dr. Robert Silverman (Jha et al., 2011; Zhou et al., 1999). These cells were grown in RPMI medium (Invitrogen) supplemented with 10% FBS and penicillin-streptomycin. HeLa cells in which PKR expression was stably knocked-down by RNAi or cells expressing a control siRNA were kindly provided by Dr. Charles Samuel (Zhang et al., 2008; Zhang and Samuel, 2007). These cells were maintained in DMEM+5% FBS containing 1 µg/mL puromycin (Invivogen) to maintain stable knockdown of PKR. We independently confirmed the absence of detectable PKR in the MEFs and HeLa cells via Western blotting before conducting experiments with these cells (data not shown).
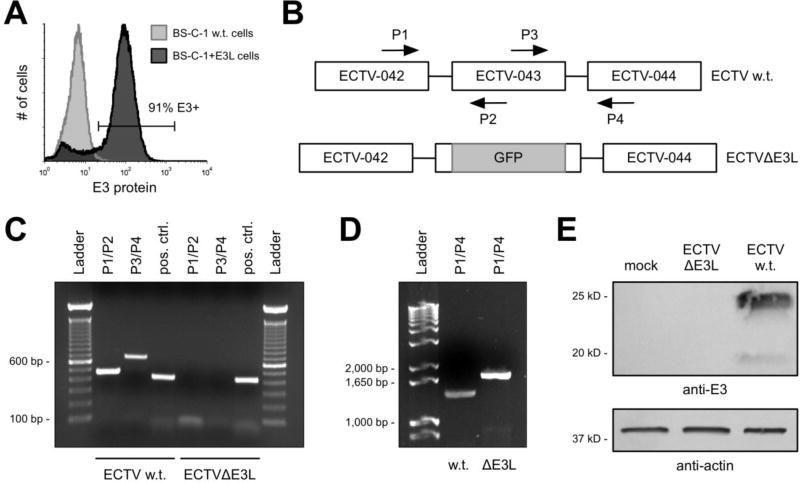
Figure 1. Construction and confirmation of ECTVΔE3L.
(A) Uninfected w.t. BS-C-1 and BS-C-1+E3L cells were stained intracellularly for E3 protein and analyzed using flow cytometry. The positive gate was set based upon staining in the w.t. cells, which are negative for E3. (B) Representation (not drawn to scale) of the genome of w.t. ECTV in the region of the E3L gene (ECTV-043) and the approximate location where the GFP cassette was inserted to disrupt the coding region of E3L. The arrows depict the binding locations of PCR primers (P). (C and D) Using the primers shown in (B), PCR reactions were performed to confirm deletion of the predicted sequence from the E3L locus. The positive control (pos. ctrl.) reaction was carried out using primers (not depicted) to amplify a shared sequence located elsewhere in the genome. (E) BS-C-1 cells were infected (MOI=5) for 18 hrs prior to isolating total protein. A Western blot was performed to detect E3 from these cellular extracts. Separate lanes of the SDS-PAGE gel were loaded with equal amounts of total protein to detect actin, which was used as a loading control.
Viruses
The following viruses were used during the course of this work: ECTV wild-type (Moscow strain; gift of Dr. Laurence Eisenlohr), ECTV expressing GFP [Moscow background; gift of Dr. Luis Sigal (Fang et al., 2008)], ECTVΔE3L, VACV wild-type (Western Reserve strain; gift of Dr. Laurence Eisenlohr), and VACVΔE3LΔK3L (Copenhagen background; gift of Dr. Stefan Rothenburg). VACVΔE3LΔK3L was constructed using standard homologous recombination techniques and expresses green fluorescent protein (GFP) (Brennan et al., 2014). ECTVΔE3L was constructed in a similar manner. Approximatley 90% of the E3L open reading frame was replaced with the coding sequence for GFP under the transcriptional control of the poxviral p7.5 early/late promoter. After the infection/transfection stage, a GFP-positive plaque was isolated and subsequently underwent six rounds of passage in BS-C-1+E3L cells to ensure that 100% of plaques stably expressed GFP. As a control, we also constructed a revertant virus (ECTV E3L REV) in which the native E3L open reading frame was recombined back into ECTVΔE3L. To create the revertant virus, we amplified the region surrounding the E3L locus (~500 bp upstream and downstream) from w.t. ECTV. After purification of the PCR product, we performed a similar procedure as described above to generate the ECTV E3L REV. After the initial infection/transfection stage, a GFP-negative plaque was isolated that subsequently underwent several rounds of passaging on normal BS-C-1 cells. We then performed sequencing of the E3L gene to confirm that the wild-type sequence was present in the ECTV E3L REV (data not shown). VACVΔE3LΔK3L and ECTVΔE3L were titrated using BS-C-1+E3L cells. All other viruses were titrated using normal BS-C-1 cells. Virus was released from infected cells by three cycles of freezing in liquid nitrogen followed by thawing in a 37°C water bath. In some experiments, cytosine β-D-arabinofuranoside (AraC; Sigma-Aldrich) was used at a final concentration of 50 µg/mL to block viral DNA replication. This drug was added at the time of infection.
Mice
Female C57BL/6 and BALB/c mice aged 6 to 8 weeks were obtained from Jackson Laboratories. Immunizations with ECTVΔE3L or VACV were carried out using i.p. injection in 300 µL total volume of 1× PBS. Some infections, as described in the results, were carried out via injection of virus in 30 µL total volume of 1x PBS into the right hind footpad. Experimental procedures involving mice were carried out in accordance with regulations from the National Institutes of Health, the Association for Assessment and Accreditation of Laboratory Animal Care International, and the United States Department of Agriculture. Animal protocols were approved by the Institutional Animal Care and Use Committees (IACUC) at Thomas Jefferson University (Philadelphia, PA) [former institution of L.C. Eisenlohr] and the Children’s Hospital of Philadelphia Research Institute (Philadelphia, PA) [current institution of L.C. Eisenlohr]. All mouse work was carried out in a humane manner and virus infections were performed under isoflurane anesthesia. Mice were sacrificed upon losing 20% of their pre-infection body weight.
Enzyme-linked immunospot (ELISPOT) assay
ELISPOT procedures were carried in a similar manner as described previously (Siciliano et al., 2014). On the indicated days post-infection, spleens were isolated and processed (including red blood cell lysis) to create single-cell suspensions. Three mice were used for each condition and splenocytes were pooled prior to plating and stimulation. T-cells were activated using bone marrow-derived dendritic cells (BMDCs) that had been incubated in the presence of the indicated peptides (2 µg/mL final concentration). BMDCs were grown and prepared in a similar manner as described previously (Hersperger et al., 2012; Siciliano et al., 2014). All cells were cultured in the presence of RPMI (Invitrogen) supplemented with 10% FBS. In some experiments, CD4+ T-cells were negatively selected using the Dynabeads Untouched Mouse CD4 Cells kit (Invitrogen) according to the manufacturer’s instructions. The antibodies used to capture and detect mouse IFN-γ and the AEC substrate development reagents were purchased from BD Biosciences and used according to the manufacturer’s instructions.
Serum antibody levels
Serum was collected from immunized mice by cheek bleeding and analyzed for anti-poxvirus IgG using a standard ELISA protocol. Briefly, serum was serially diluted (1:500, 1:1,000, 1:2,000, 1:4,000, 1:8,000, and 1:16,000) in 1x PBS supplemented with 1% low-IgG bovine serum albumin (BSA; Gemini Bio Products). Each dilution (plated in triplicate) was then incubated in high binding EIA/RIA plates (Corning) that had been pre-coated with 12,500 PFU VACV (Western Reserve strain) per well. Plates were washed with PBS+0.01% Tween (PBST) and incubated with peroxidase labeled anti-mouse IgG (H+L) (Vector Laboratories) at 1:200 dilution in PBS/BSA as above. Plates were developed using ABST Peroxidase Substrate (KPL) and read at 405 nm.
Fluorescence microscopy
Fluorescence microscopy was carried out using a Zeiss Axiostar plus epifluorescence microscope and images were captured with an Optronics camera system. All images were manipulated (brightness, contrast, etc.) in an equivalent fashion using ImageJ software (version 1.50g). Cells were grown in four-well chamber slides (Nunc Lab-Tek II; Thermo Scientific), infected with either ECTV or VACV (MOI=10), and fixed at the indicated time points with 5% formalin for 10 min. at room temperature. Cell surface expression of B5 and intracellular levels of dsRNA were assessed in a similar manner as a prior report (Frey et al., 2017). For dsRNA detection, cells were fixed, permeabilized in acetone (Fisher Scientific) at −20° C, and incubated with anti-dsRNA monoclonal antibody (clone J2; Scicons). After washing, the secondary antibody goat anti-Mouse IgG2a Alexa Fluor 568 (Invitrogen) was applied. Glass coverslips were then mounted using ProLong Gold antifade reagent with 4’-6-diamidino-2-phenylindole (DAPI; Invitrogen) and allowed to cure overnight prior to visualization. For surface B5 staining, unpermeabilized cells were fixed and then incubated with an anti-B5 monoclonal antibody (clone VMC-22; BEI resources NR-553). The same secondary antibody and mounting procedure was performed as described above for dsRNA detection. To visualize actin, fixed and permeabilized cells were incubated in the presence of Alexa Fluor 488-phalloidin (Invitrogen) before carrying out the previously outlined mounting procedure.
Flow cytometry
Flow cytometry data were collected using a FACSCalibur instrument (BD Biosciences) equipped with red and blue lasers. A minimum of 75,000 total cells were collected for each sample. The acquired data were analyzed using FCS Express 4 Flow Cytometry (De Novo Software; version 6.04.0027). To measure intracellular levels of E3, cells were first harvested via 0.25% trypsin (Invitrogen) digestion and washed once with 1x PBS containing 1% BSA (Gemini BioProducts). Cells were then fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions. After permeabilization, monoclonal antibodies were added to the cells to detect levels of E3 protein (BEI resources NR-4547) or dsRNA (clone J2; Scicons). After 40 minutes of incubation at room temperature, the cells were washed twice with Perm/Wash Buffer (BD Biosciences) followed by the addition of anti-mouse IgG2a APC (eBioscience) for 30 minutes. A final wash with 1x Perm/Wash Buffer was performed prior to the addition of a 2% paraformaldehyde (Electron Microscopy Sciences) solution in 1x PBS. In experiments examining the spread of virus, 5×105 BS-C-1 w.t. or BS-C-1+E3L cells were seeded into six-well plates. They were infected (MOI=0.005) the next day with GFP-expressing ECTV or ECTVΔE3L. After 24, 48, 72, and 96 hours, the cells were harvested using trypsin digestion and immediately analyzed. Virus spread in the culture was quantified by measuring the percentage of total cells that were positive for GFP expression at each time point.
RNase L activity assay
Cells were plated in 6-well culture plates to be 80–90% confluent at the time of infection. Cells were either infected (MOI=5) or mock treated the following day. Infections were carried out in DMEM+2% FBS for one hour in minimal volume followed by replacement with standard media (DMEM+5% FBS). Total RNA was isolated using the PureLink RNA Mini kit (Invitrogen) following the manufacturer’s instructions. RNA electrophoresis was performed using a “bleach gel” (Aranda et al., 2012), which contained 1% agarose and 1% bleach (Clorox; 6% sodium hypochlorite) in 1x TBE buffer. Each lane contained 4 µg of RNA. The gel was stained with ethidium bromide (2 µg/ml) to visualize the 28S and 18S rRNA bands and any possible RNA cleavage products.
Real-time PCR
BS-C-1 cells were plated in 6-well culture plates one day prior and in sufficient numbers so as to be 80–90% confluent at the time of infection. Cells were either infected (MOI=5) or mock treated. Infections were carried out in DMEM+2% FBS for one hour in minimal volume followed by replacement with standard media (DMEM+5% FBS). Total RNA was isolated at the indicated times post-infection using the PureLink RNA Mini kit (Invitrogen) following the manufacturer’s instructions. The relative levels of F17R and H4L transcripts were quantified as previously described (Frey et al., 2017). GAPDH was used as the internal control and for standardization among samples in a similar manner as Arndt, et al. (Arndt et al., 2016). Predesigned primers to detect GAPDH transcripts from rhesus monkey were purchased from Bio-Rad (qMccCID0018506).
Reporter plasmid
A plasmid was constructed using the GeneArt Synthesis service (Invitrogen) containing the gene for mCherry. The promoter for the VACV gene A14L, a known late gene (Yang et al., 2013), was placed immediately upstream of the start codon for mCherry. The promoter for the ECTV A14L ortholog differs by a single nucleotide from the VACV sequence (data not shown). BS-C-1 cells were transfected first and then infected (MOI=5) with the indicated viruses two hours later. The reporter gene plasmid was introduced into cells using the Lipofectamine 3000 kit (Invitrogen) according the manufacturer’s instructions. Fluorescence was measured using flow cytometry or microscopy at the indicated time points post-infection.
Western blotting
Cells were plated in 6-well culture plates to be 80–90% confluent at the time of infection. Cells were either infected (MOI=5) or mock treated the following day. Infections were carried out in DMEM+2% FBS for one hour in minimal volume followed by replacement with standard media (DMEM+5% FBS). For eIF2α detection, total protein was collected at 6 hours post-infection using cell extraction buffer (Invitrogen) supplemented with Halt™ Protease and Phosphatase Inhibitor Cocktail (Invitrogen; 100x concentrate) and EDTA (Invitrogen; 5 mM final concentration). A total of 100 µg of protein for each sample was separated using 12% Mini-PROTEAN TGX gels (Bio-Rad) and transferred onto a 0.45µm polyvinyl difluoride (PVDF) membrane. Membranes were blocked using StartingBlock™ T20 (in TBS) blocking buffer (Invitrogen). For E3 and A3 protein detection, cells were infected for 18 hrs prior to isolating total protein as described above. The following primary antibodies were used: rabbit anti-phospho (Ser51) eIF2α (clone S.674.5; Invitrogen), mouse anti-total eIF2α (clone EIF2-alpha; Invitrogen), mouse anti-beta actin (clone AC-15; Sigma-Aldrich), mouse anti-VACV E3 (clone 3015B2; gift of Dr. Stuart Isaacs [(Weaver et al., 2007)]), and rabbit anti-VACV A3 (gift of Dr. Bernard Moss). Detection using the appropriate secondary antibody conjugated to alkaline phosphatase (AP) was performed according to the manufacturer’s instructions (Bio-Rad).
Results
Creation and verification of a mutant ECTV lacking a functional E3L gene
In order to study the importance of the E3 protein to the replication of ECTV, we constructed a mutant form of the virus (ECTVΔE3L) with a disrupted E3L gene [this gene is also known as ECTV-043 (Chen et al., 2003); E3L is from VACV nomenclature]. Using standard homologous recombination techniques (Johnston and McFadden, 2004), we replaced approximately 90% of the open reading frame of E3L with the gene for GFP under the transcriptional control of the p7.5 early/late promoter. Initially, we used w.t. BS-C-1 cells for the infection/transfection stage and subsequent passages of GFP-positive plaques. However, GFP expression was not stable from passage to passage and we failed to isolate any pure ECTVΔE3L after several attempts. Therefore, we made use of BS-C-1 cells engineered to stably express the E3 protein of VACV (Hand et al., 2015) (Fig. 1A) and were successful at creating ECTVΔE3L in this complementing cell line.
To confirm disruption of the E3L locus, we constructed PCR primers such that one primer of each pair was internal to the region of E3L replaced by the GFP gene (Fig. 1B). As shown in Figure 1C, we did not detect a PCR product for the mutant virus but did observe bands of the expected length for w.t. virus. Using the outermost primers (P1 and P4 in Fig. 1B), we observed a PCR product for both viruses, as expected (Fig. 1D). The band in the ECTVΔE3L lane was approximately 300 bp longer than in the lane corresponding to w.t. ECTV because the inserted p7.5 promoter (~100 bp) and GFP coding region (720 bp) is larger than the E3L gene (573 bp). Therefore, at the genetic level it appeared that the E3L locus was successfully replaced by the GFP gene. To confirm loss of E3 at the protein level, we performed a Western blot from extracts of virus-infected cells. As shown in Figure 1E, we detected robust levels of E3 from BS-C-1 cells infected with w.t. ECTV but did not observe any signal from cells infected with ECTVΔE3L.
The yield of ECTVΔE3L was slightly reduced relative to w.t. ECTV
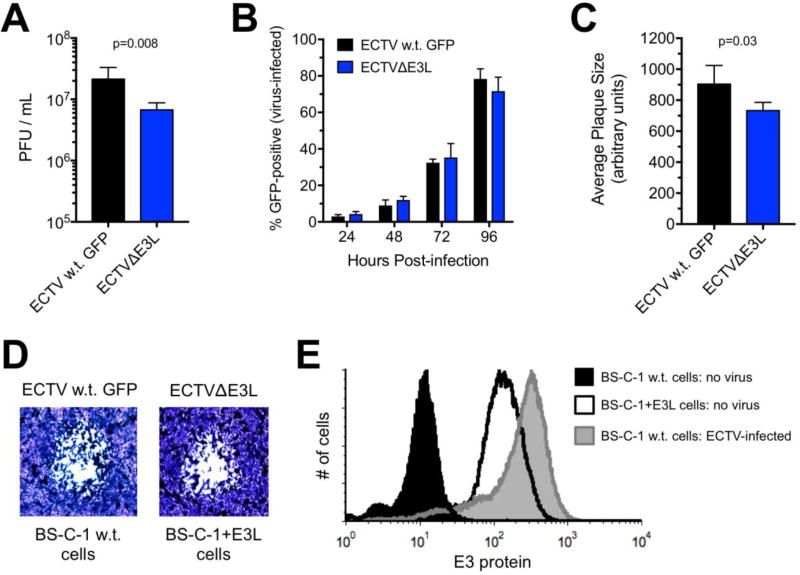
Having successfully created a mutant ECTV lacking the E3L gene, we next examined the replication capacity of this virus relative to w.t. ECTV expressing GFP (to control for the presence of the GFP gene). We performed a single-step growth analysis to compare virus yield at 24 hrs post-infection. We already established from the passaging of virus during the construction of ECTVΔE3L that the mutant cannot grow in normal BS-C-1 cells. Therefore, we infected equal numbers of normal BS-C-1 and +E3L cells with w.t. ECTV and ECTVΔE3L, respectively. As shown in Figure 2A, there was a small but statistically significant reduction in the replication capacity of ECTVΔE3L relative to w.t. virus. However, both viruses spread (as assessed using GFP expression) throughout a cell monolayer in an equivalent manner after being initially infected with a low MOI (Fig. 2B). With respect to the plaque size, w.t. ECTV produced plaques that were slightly larger than ECTVΔE3L (Fig. 2C). Representative images of plaques produced by both viruses are shown in Figure 2D. During the course of these experiments, we noticed that the amount of E3 protein in BS-C-1+E3L cells was less than the levels found in normal BS-C-1 cells infected with w.t. ECTV (Fig. 2E). Therefore, the slight attenuation of ECTVΔE3L described above may be the result of the lower levels of E3 in the BS-C-1+E3L cells relative to the levels of this protein observed during natural infection. Moreover, not every single BSC1+E3L cell produced E3 at all times (Fig. 1A). Finally, it should be noted that we compared the virus yield from equal numbers of normal BS-C-1 and +E3L cells infected with w.t. ECTV (MOI=5). The amount of virus produced was slightly higher from the BS-C-1+E3L cells (average of three trials: 1.8×106 PFU/mL; standard deviation: 2.0×105) relative to normal BS-C-1 cells (average of three trials: 1.1×106 PFU/mL; standard deviation: 1.6×105) but the difference was not statistically different. Therefore, the “extra” E3 in the BS-C-1+E3L cells did not have a dramatic effect on the replication ability of w.t. ECTV.
Figure 2. Growth comparison between wild-type ECTV and ECTVΔE3L.
(A) Equal numbers of normal BS-C-1 and +E3L cells were infected (MOI=5) with w.t. ECTV and ECTVΔE3L, respectively. At 24 hrs post-infection, virus was harvested from cells and quantified using a standard plaque assay. The bars depict the average of four separate wells and the error bars represent the standard deviations. The data are representative of two independent experiments. Statistical analysis [performed using GraphPad Prism software (version 7.0c)] was carried out using the Mann-Whitney test. (B) Normal BS-C-1 or BS-C-1+E3L cells were initially infected with a low amount (MOI=0.01) of w.t. ECTV expressing GFP or ECTVΔE3L, respectively. The capacity of each virus to spread within the well was quantified using flow cytometry. The graph depicts the levels of GFP at the indicated times post-infection. The bars depict the average of four separate wells at each time point and the error bars represent the standard deviations. The data are representative of two independent experiments. (C) Cells were infected as in (B) using a MOI of 0.001. Plaques were visualized at day five post-infection using a 1% crystal violet solution (in 20% methanol/1x PBS). The diameter of individual plaques [representative phase contrast images shown in (D) for each virus] were measured using ImageJ software (version 1.50g) at the widest point of the area of observable cytopathic effect. A total of 40 plaques for each virus across two separate experiments were quantified and plotted. The bars depict the average and the error bars represent the standard deviations. Statistical analysis was performed as above. (E) Levels of intracellular E3 protein were analyzed using flow cytometry in the following conditions: uninfected normal BS-C-1 cells, normal BS-C-1 cells infected (MOI=5) with w.t. ECTV for 18 hrs, and uninfected BS-C-1+E3L cells. The data are representative of two independent experiments.
ECTVΔE3L displayed an abortive infection in normal BS-C-1 cells but productive replication is rescued in BS-C-1+E3L cells
Using immunofluorescence, we next examined various infection events at the cellular level. We first infected normal BS-C-1 cells with w.t. ECTV expressing GFP and ECTVΔE3L. Both viruses have the GFP gene under the transcriptional control of the poxvirus p7.5 early/late promoter. As expected based on our prior data, there was no detectable GFP in cells infected with ECTVΔE3L (Fig. 3A). We used DAPI staining to visualize the appearance of viral DNA in the cytoplasm of infected cells. These “virus factories” were clearly discernable in cells infected with w.t. ECTV (large areas of extra-nuclear staining; Fig. 3A). While most cells infected with ECTVΔE3L contained a factory, they were significantly smaller relative to w.t. virus (Fig. 3A). Additionally, we did not detect signs of apoptosis, such as nuclear fragmentation (Hornemann et al., 2003), in any condition. The lack of discernible apoptosis induction was confirmed by staining infected cells with an antibody that specifically detects activated caspase 3. No active caspase 3 signal was observed in any condition except for the positive control (data not shown).
Figure 3. ECTVΔE3L infection of w.t. BS-C-1 cells resulted in an abortive replication cycle.
Normal BS-C-1 (A) or BS-C-1+E3L (B) cells were infected (MOI=10) with the indicated viruses. At 24 hrs post-infection, cells were examined for GFP expression, stained with DAPI to visualize virus factories (arrows) and cell nuclei (N), stained for B5 at the plasma membrane, or stained for actin. Examples of actin tails are indicated by the arrow heads. Representative images are depicted; each row does not necessarily show the same field of view.
We also stained cells to measure the viral protein B5 at the cell surface, which is a late (i.e., post-DNA replication) infection event since treatment of cells with AraC prohibited positive staining (data not shown). We observed robust levels of B5 at the plasma membrane of w.t. ECTV-infected cells but failed to detect any surface B5 after infection with ECTVΔE3L (Fig. 3A). Additionally, we examined the formation of actin tails, which is another late infection event (Lynn et al., 2012). These are induced by poxviruses to aid in viral spread from cell-to-cell (Roberts and G. L. Smith, 2008). We did not detect actin tail formation in ECTVΔE3L-infected cells, whereas they were readily detectable after infection with w.t. virus (Fig. 3A). We observed comparable results as outlined above (i.e., abortive infection) upon ECTVΔE3L infection of the following cell lines: 293T, HeLa [human], Vero [African green monkey], L-929, NIH-3T3, and DC2.4 [mouse] (data not shown). We chose these cell lines because they are derived from different mammalian species and are all permissive for w.t. ECTV. Finally, we also examined the above parameters in BS-C-1+E3L cells. As shown in Figure 3B, GFP expression, factory formation, cell surface B5, and the presence of actin tails were similar between w.t. ECTV- and ECTVΔE3L-infected cells.
Higher levels of dsRNA were detected in ECTVΔE3L-infected cells compared to w.t. ECTV
Next, we employed a commercially available monoclonal antibody (J2) that binds specifically to double-stranded RNA (dsRNA). This antibody has been used to detect duplex RNA in cells infected by certain viruses (Weber et al., 2006). Using this antibody, we recently reported that ECTV formed significantly lower amounts of dsRNA during infection of BS-C-1 cells compared to VACV (Frey et al., 2017). Other groups have also used the J2 antibody to measure dsRNA formation in the context of poxvirus infection (Arndt et al., 2016; Burgess and Mohr, 2015; Liu and Moss, 2016; Willis et al., 2011). Since E3 contains a dsRNA-binding domain and has been shown experimentally to bind to a dsRNA analog (Chang et al., 1992; Watson et al., 1991), we reasoned that duplex RNA may be more readily detectable in cells infected with ECTVΔE3L relative to w.t. ECTV. To this point, there is published evidence suggesting that the presence of E3 likely reduces the binding efficiency of J2 to its epitope on dsRNA (Frey et al., 2017). As shown in Figure 4A to 4C, we observed higher levels of dsRNA in BS-C-1 cells infected with ECTVΔE3L compared to w.t. ECTV, a finding that is consistent with our hypothesis. Treatment of cells at the time of infection with AraC abolished detectable dsRNA staining (Fig. 4A). Therefore, in the absence of E3, viral dsRNA was no longer shielded by this protein; thus, the epitope was presumably more accessible to the J2 antibody. Similar results were also observed in HeLa and L-929 cells (data not shown), which we used as representative human- and murine-derived cells, respectively.
Figure 4. Higher amounts of dsRNA were detected during infection with ECTVΔE3L compared to w.t. ECTV.
(A) BS-C-1 cells were infected (MOI=10) with the indicated viruses for 24 hrs prior to staining for dsRNA (red) and cell nuclei/virus factories (DAPI; blue). Images (400x total magnification) were merged using ImageJ software and are representative of two independent trials. AraC was added at the time of infection where indicated. (B) BS-C-1 cells were infected as above prior to staining for intracellular dsRNA using flow cytometry. The depicted data are representative of two independent trials. (C) BS-C-1 cells were infected as above for various lengths of time prior to measuring dsRNA positivity using flow cytometry. The bars depict the average and the error bars represent the standard deviations of three independent trials for each time point.
ECTVΔE3L activates PKR but not RNase L in cells derived from multiple species
Since dsRNA was detected in the majority of ECTVΔE3L-infected cells (Fig. 4), we hypothesized that PKR and/or RNase L would become stimulated in these cells. We used an antibody that recognizes eIF2α after it has been phosphorylated at serine-51 by activated PKR. As shown in Figure 5A, we found evidence of PKR activation in BS-C-1, HeLa, and L-929 cells after infection with ECTVΔE3L. However, we did not observe a change in the phosphorylation state of eIF2α after infection with w.t. ECTV relative to mock-treated cells (Fig. 5A). Next, we assessed RNase L activation by employing an assay that detects rRNA cleavage products (Silverman et al., 1983; Wreschner et al., 1981). We used VACVΔE3LΔK3L as a control since it has been shown previously to induce RNase L activation in BS-C-1 cells (Frey et al., 2017). As expected, we observed rRNA breakdown following infection of BS-C-1 cells with VACVΔE3LΔK3L (Fig. 5B). Conversely, we did not detect RNase L activation following infection with ECTVΔE3L (Fig. 5B). Interestingly, neither virus induced rRNA cleavage in HeLa or L-929 cells (Fig. 5B), which suggests that there may be varying thresholds of RNase L activation in different cell types.
Figure 5. PKR was the primary anti-viral pathway responsible for inhibiting the replication cycle of ECTVΔE3L.
(A) The indicated cell lines were infected (MOI=5) for 6 hrs prior to isolating total protein. A Western blot was performed to detect the indicated proteins from these cellular extracts. Since we could not strip the membrane and re-probe, separate lanes of a SDS-PAGE gel were loaded with equal amounts of total protein and run concurrently. These data are representative of three independent trials. (B) Cells were infected as above with the indicated viruses. Total RNA was isolated at 16 hrs post-infection and separated using a 1% bleach TBE gel. Each lane contained 4 µg of RNA. The data are representative of two independent trials. (C) The indicated cells were infected (MOI=10) for 18 hrs prior to measuring GFP expression using flow cytometry. The depicted data are representative of two independent trials.
Prior work (Rahman et al., 2013; Zhang et al., 2008) – along with our data – point to PKR as the primary anti-viral pathway that blunts the replication cycle of E3L-null poxviruses (Fig. 3A). Therefore, we compared the replication capacity of ECTVΔE3L relative to w.t. ECTV in PKR competent and deficient cells. We used PKR−/− MEFs (mouse) and PKR knockdown HeLa cells (human) for these experiments. As shown in Figure 5C, the absence of PKR restored the replication of ECTVΔE3L to wild-type levels in both cell types. In these experiments, we used GFP expression as a proxy for productive replication capacity. Similar results were also obtained using single-cycle growth curves (data not shown).
Late genes are transcribed but not translated in ECTVΔE3L-infected cells
The immunofluorescence data in Figure 3 showed that ECTVΔE3L was unable to complete its replication cycle within infected cells. Cytoplasmic DAPI staining revealed that ECTVΔE3L began DNA replication but was not able to complete the process. The virus factories were present but significantly smaller than those found during w.t. infection. Since expression of viral intermediate and late genes relies on prior DNA replication (Broyles, 2003; Vos and Stunnenberg, 1988), we turned our attention to the examination of “post-replicative” gene transcription. The dsRNA staining in Figure 4 indicated that intermediate and/or late gene transcription occurred in ECTVΔE3L-infected cells because the presence of AraC precluded dsRNA detection. Moreover, prior research has shown that dsRNA is mostly formed at late time points of the VACV infection cycle (Boone et al., 1979; Colby and Duesberg, 1969; Duesberg and Colby, 1969). Therefore, we determined if the levels of post-replicative transcripts varied between ECTVΔE3L and w.t. virus by performing real-time PCR analysis of two late genes, H4L and F17R (VACV nomenclature). As shown in Figure 6A, the levels of these transcripts were significantly reduced following infection with ECTVΔE3L. In summary, the data in Figure 4 and Figure 6A, clearly demonstrate that late gene transcripts were present in ECTVΔE3L-infected cells but they were at significantly lower levels compared to w.t. virus.
Figure 6. Transcription of late genes was detected but their translation was severely blunted during the replication cycle of ECTVΔE3L.
(A) BS-C-1 cells were infected (MOI=5) with ECTVΔE3L or w.t. ECTV. Total RNA was isolated at the indicated time points. Real-time reverse-transcriptase PCR was performed using gene-specific primers for F17R and H4L. The data points represent duplicate reactions from two independent infections. Statistical analysis [performed using GraphPad Prism software (version 7.0c)] was carried out using the Mann-Whitney test. * denotes a p value <0.05. (B) BS-C-1 cells were transfected and then infected (MOI=5) with the indicated viruses two hours later. Fluorescence was measured using flow cytometry at the indicated time points after adding virus. The bars depict the average and the error bars represent the standard deviations of three wells per virus per time point. The depicted data are representative of two independent trials. (C) BS-C-1 cells were infected (MOI=10) for 18 hrs prior to isolating total protein. A Western blot was performed to detect A3 from these cellular extracts. Separate lanes of the SDS-PAGE gel were loaded with equal amounts of total protein to detect actin, which was used as a loading control. AraC was added at the time of infection where indicated. The depicted data are representative of two independent trials. (D) Wild-type or PKR−/− MEFs were transfected/infected as in (B). Fluorescence was observed 18 hrs after adding virus. Roughly equal numbers of cells are in each image (all at 400x total magnification) as determined by DAPI staining (data not shown). (E) Wild-type and PKR−/− MEFs were infected (MOI=10) with the indicated viruses. At 24 hrs post-infection, cells were examined for GFP expression, stained with DAPI to visualize virus factories and cell nuclei, and stained for B5 at the plasma membrane. Representative images (all at 100x total magnification) from two independent trials are shown. The DAPI and B5 images show the same field of view.
We next determined if translation of late transcripts could be detected within ECTVΔE3L-infected cells. To address this question, we used a reporter construct in which the gene for mCherry was placed under the control of a known late gene promoter. We initially performed a time course experiment to measure mCherry expression in cells that were transfected with the reporter plasmid and subsequently infected. As shown in Figure 6B, we failed to detect fluorescence in cells that had been infected with ECTVΔE3L whereas w.t. ECTV yielded a clear mCherry signal beginning at 8 hrs post-infection. Fluorescence was absent in w.t. ECTV-infected cells treated with AraC (data not shown), which confirmed that expression of mCherry was restricted to the predicted post-replicative time point. To confirm our findings from the reporter assays, we attempted to detect expression of the A3 protein (VACV nomenclature), which is a late gene product (Assarsson et al., 2008; Rosel and Moss, 1985). In a Western blot assay, the levels of A3 within ECTVΔE3L-infected cells were substantially lower than the amount detected for w.t. virus but similar to AraC-treated w.t. infection (Fig. 6C). Therefore, the presence of late gene products during ECTVΔE3L infection was virtually undetectable. To establish the role of PKR-mediated translational arrest in this process, we conducted a similar infection/transfection experiment (as in Fig. 6B) in w.t. and PKR−/− MEFs. We observed equivalent levels of mCherry fluorescence (Fig. 6D) and A3 protein (data not shown) in PKR−/− MEFs infected with either virus. Moreover, the presence of B5 at the plasma membrane was fully restored in the PKR −/− MEFs (Fig. 6E). In summary, it appears that PKR is the host factor principally responsible for curtailing the replication cycle of ECTVΔE3L.
ECTVΔE3L is non-pathogenic in mousepox-susceptible mice and stimulates T-cell responses
We next examined the in vivo characteristics of ECTVΔE3L. To determine if the mutant virus retained any virulent properties, we infected BALB/c mice in a hind footpad with 10,000 PFU. This is a relatively high challenge dose in this mouse strain, which experiences a lethal outcome following infection with low amounts of w.t. ECTV (Xu et al., 2008). Footpad inoculation is often used in an attempt to mimic the natural dermal route of transmission (Fenner, 1947). As a control, we also created a revertant virus (ECTV E3L REV) in which the native E3L sequence was returned to the ECTVΔE3L virus. As shown in Figure 7A, infections with w.t. ECTV and ECTV E3L REV resulted in 100% mortality – as expected – whereas all mice infected with ECTVΔE3L survived. Swelling at the site of inoculation was severe for C57BL/6 mice infected with the revertant virus but did not manifest following ECTVΔE3L infection (Fig. 7B). The assessment of swelling was performed in C57BL/6 mice, a mousepox-resistant strain, because it can be difficult to assess changes in footpad thickness in susceptible mice that succumb quickly to the infection.
Figure 7. ECTVΔE3L was nonpathogenic in vivo but induced detectable T-cell responses.
(A) Groups of BALB/c mice (n=5 per group) were infected with 10,000 PFU of the indicated viruses in the right hind footpad and monitored for mortality. (B) Groups of C57BL/6 mice (n=5 per group) were infected as above. Swelling of the infected footpad was measured using digital calipers on the indicated days post-infection. Data points indicate the average value at each time point and the error bars represent the standard deviation. (C) C57BL/6 mice were infected as above. On day 10 post-infection, splenocytes were isolated and pooled (n=3 per group) prior to isolating CD4+ T-cells. An ELISPOT assay was performed to quantify CD4+ T-cells responses to individual 15-mer peptides derived from both early and post-replicative viral gene products (Assarsson et al., 2008). The dashed line indicates the threshold (defined as three times the standard deviation of the DMSO control) for positive responses. The names of viral proteins from which each peptide is derived are indicated using VACV nomenclature. The numbers in parentheses indicate the amino acid position at the beginning of each 15-mer peptide (amino acid #1 is at the N-terminus of the protein). The depicted data are representative of two independent trials. The bars depict the average and the error bars represent the standard deviation. SFC, spot forming cells. (D) An ELISPOT assay was performed as above except that total splenocytes (pooled, n=3 per group) were stimulated with the immunodominant MHC class I-restricted epitope derived from the VACV B8 protein (amino acids #20–27). The bars depict the average and the error bars represent the standard deviation. SFC, spot forming cells.
We also employed C57BL/6 mice to assess the generation of ECTV-specific T-cell responses during acute infection. We used a panel of previously identified I-Ab-restricted epitopes [(Siciliano et al., 2014) and unpublished results] to quantify CD4+ T-cell responses using an IFN-γ ELISPOT assay. We observed positive responses following infection with both ECTVΔE3L and ECTV E3L REV but the overall magnitude of these responses was reduced for the former (Fig. 7C). Additionally, we noticed that all of the positive responses following infection with ECTVΔE3L were derived from early genes (Fig. 7C). Conversely, positive responses induced by ECTV E3L REV were distributed across antigens expressed at multiple time points of the viral replication cycle (Fig. 7C). We also quantified the response raised against the immunodominant H-2Kb (MHC class I)-restricted epitope from the B8R protein of VACV (Tscharke, 2005), which is an early viral gene product (Symons et al., 2002). Both viruses induced detectable B8R-specific CD8+ T-cells but – similar to the CD4+ T-cell data – the magnitude was lower following ECTVΔE3L infection compared to the revertant virus (Fig. 7D).
ECTVΔE3L induces protective immunity in mousepox-susceptible mice
Since we established that ECTVΔE3L induces detectable immune responses, we investigated whether these could protect mice against lethal mousepox disease. We immunized mice with either a low (1×104 PFU) or high (1×105 PFU) dose of ECTVΔE3L and then challenged with two different amounts of w.t. ECTV thirty days later. We used 1×104 PFU VACV (Western Reserve strain) as a positive control (Hersperger et al., 2014). After challenge with 1,000 PFU w.t. ECTV in previously immunized BALB/c mice, we did not observe mortality in any group (Fig. 8A). However, the mock-vaccinated (saline) mice displayed significant weight loss while the ECTVΔE3L and VACV groups maintained stable body weights (Fig. 8A). Next, we challenged another group of immunized mice with 3,000 PFU w.t. ECTV, which is a commonly used infection dose (Fang and Sigal, 2006; Hersperger et al., 2012) and the highest amount allowed by our current IACUC protocol. The saline group was the only condition to experience mortality (Fig. 8B). Every group except for the VACV-immunized mice experienced some degree of morbidity but the saline condition displayed the most severe loss of body weight (Fig. 8B). As outlined in Table 1, the majority of mice in the saline group experienced several clinical signs of illness, such as pock lesions, whereas these disease signs were minimal in the ECTVΔE3L–immunized groups. Since all mice were challenged in a hind limb, we also measured the infected footpad to detect swelling. The saline condition displayed the greatest increase in footpad thickness and the swelling remained the highest of any group at day 25 post-challenge in the surviving animals (Fig. 8C). The swelling in the other groups was in the process of resolving by the same time point (Fig. 8C).
Figure 8. ECTVΔE3L elicited protective immunity against lethal mousepox disease in BALB/c mice.
(A and B) Groups of BALB/c mice (n=5 per group) were immunized via i.p. injection with VACV (1×104 PFU), two different doses of ECTVΔE3L (1×104 PFU or 1×105 PFU), or mock-immunized using 1x PBS (saline). After 30 days, mice were challenged with the indicated amounts of w.t. ECTV in the right hind footpad and monitored for mortality and weight loss. Data points indicate the average value at each time point and the error bars represent the standard deviation. +, death event in the saline group. (C) Footpad thickness was measured using digital calipers for all mice (n=5 per group) challenged with 3,000 PFU w.t. ECTV. The bars depict the average and the error bars represent the standard deviation. A one-way ANOVA test revealed that the groups were significantly different from each other. Further statistical analysis of column pairs was carried out using the Mann-Whitney test. All statistical tests were performed using GraphPad Prism software (version 7.0c). n.s., not significant. (D) Serum was isolated one day prior to challenge for all mice that received 3,000 PFU w.t. ECTV as in (B). A standard ELISA was performed to measure levels of anti-poxvirus IgG. Each bar indicates a different serum dilution; from left to right: 1:500, 1:1,000, 1:2,000, 1:4,000, 1:8,000, and 1:16,000. The bars depict the average and the error bars represent the standard error of the mean of the values obtained from individual mice (n=5 per group). The depicted data are representative of two independent trials.
Table 1.
Observations related to mousepox disease following challenge with 3,000 PFU of wild-type ECTV.
| Visible pock lesions on skin |
Persistent Footpad swelling* |
Persistent scab/exudate on infected footpad% |
|
|---|---|---|---|
| Saline | 4/5 | 3/3# | 5/5 |
| 1×104 ECTVAE3L | 1/5 | 5/5 | 2/5 |
| 1×105 ECTVAE3L | 0/5 | 2/5 | 1/5 |
| VACV | 0/5 | 0/5 | 0/5 |
defined as footpad thickness greater than or equal to 3.0 mm at day 25 post-challenge
two mice did not survive to this time point
observed at any point during the post-challenge period
Finally, we measured the pre-challenge immune responses generated by the VACV or ECTVΔE3L immunizations. In contrast to C57BL/6 mice, we did not possess a panel of MHC class I- or II-restricted epitopes for BALB/c mice, so we could not quantify T-cell responses. However, we did measure poxvirus-specific IgG from the sera of immunized animals one day prior to challenge. As shown in Figure 8D, all groups except for the saline condition demonstrated detectable virus-specific antibodies with the VACV group displaying the highest titers. In summary, ECTVΔE3L induced protective adaptive immune responses despite being replication incompetent.
Discussion
In this study, we characterized a novel ECTV mutant lacking the ability to produce a functional protein product from the E3L gene. We find that E3L is an essential gene for the productive replication of ECTV in all tested cells. To date, most of the work done on E3L has focused on VACV and various E3L mutants of this virus. Here, we hoped to expand our knowledge of this gene by examining its importance to another orthopoxvirus.
An E3L gene encoding a protein with Z-DNA- and dsRNA-binding domains, which are present in the ECTV and VACV versions of E3L, have been identified in almost all sequenced parapoxviruses, orthopoxviruses, and clade II poxviruses (Bratke et al., 2013). Using VACV as a model, E3 has been shown to be an important host range factor. For example, VACVΔE3L replicates poorly in HeLa (human), Vero (non-human primate), and L-929 (mouse) cells (Beattie et al., 1996; Chang et al., 1995; Langland and Jacobs, 2002). Conversely, loss of E3L has minimal impact on the replication of VACV in BHK-21 (Syrian hamster), CEF (chicken), or RK-13 (rabbit) cells (Beattie et al., 1995; Chang et al., 1995; Langland and Jacobs, 2002).
We infected multiple cell lines derived from several mammalian species that are permissive for w.t. ECTV. Interestingly, we found that ECTVΔE3L could not productively infect any of these cells. We were unable to detect GFP expression or late gene protein production even at 48 hrs post-infection with ECTVΔE3L (data not shown). Therefore, E3L is somewhat dispensable for VACV depending on the cell type but appears to be completely essential for ECTV in all of the cell lines we tested. E3L may be vital to the replication of ECTV because it lacks a functional K3L gene. The protein product of K3L has been demonstrated to inhibit PKR by acting as an eIF2α mimic (Davies et al., 1992). The ECTV K3L open reading frame contains several mutations that result in a truncated protein predicted to be non-functional (Bratke et al., 2013; Chen et al., 2003; V. P. Smith and Alcami, 2002). Thus, without K3L to aid E3L, it seems likely that ECTV has become wholly reliant on the latter in order to complete its replication cycle within cells that contain functional PKR.
DAPI staining revealed the presence of viral factories within ECTVΔE3L-infected cells but they were severely blunted in size compared to those found within w.t. virus-infected cells. Therefore, the mutant virus is able to commence genome replication but this process cannot be completed. Previous studies have shown that the transcription of late genes relies on prior DNA replication – hence the term “post-replicative” transcripts (Broyles, 2003; Vos and Stunnenberg, 1988). Multiple lines of evidence from our work show that mRNA synthesis of late genes does occur during the replication cycle of ECTVΔE3L; however, the overall levels of these mRNAs are greatly reduced relative to w.t. virus. Thus, it seems that transcription of late genes does not require the finalization of viral genome replication prior to the initiation of their synthesis. Additionally, we find that translation of late mRNAs is inhibited due to a PKR-mediated block during infection with ECTVΔE3L. In summary, our data suggest that certain intermediate and/or late protein products may be essential for the completion of viral genome replication, which would result in large viral factories as visualized by DAPI staining.
Given that ECTVΔE3L is replication-defective in vitro, we hypothesized that it would be non-pathogenic in BALB/c mice, a strain that is susceptible to mousepox disease. As predicted, there were no signs of mortality or weight loss in this mouse strain after infection with ECTVΔE3L via the footpad or i.p. routes. Therefore, this ECTV mutant appears to be completely avirulent in vivo. Interestingly, footpad infection with ECTVΔE3L failed to yield detectable swelling of the limb, which indicates that a robust local inflammatory response did not manifest. Despite the lack of clinical signs of infection and inflammation, a single inoculation with a relatively low amount of ECTVΔE3L induced virus-specific adaptive immune responses. However, the B- and T-cell responses elicited by ECTVΔE3L were of a lower magnitude relative to w.t. ECTV (in C57BL/6 mice) or VACV (in BALB/c mice). Nevertheless, these immune responses protected BALB/c mice from death after challenge with w.t. ECTV. Both ECTVΔE3L immunization groups experienced some weight loss after challenge, but the high dose ECTVΔE3L group was protected against all other clinical signs of mousepox to a greater extent than immunization with a 10-fold lower dose. In future studies, we will examine different doses, multiple immunizations, and inoculation routes to determine if higher magnitude immune responses can be generated by ECTVΔE3L. In the process, we will also learn if complete protection against signs of mousepox disease, including weight loss, can be achieved. Lastly, our findings also raise an interesting possibility that at least a portion of the pathology associated with mousepox infection of susceptible mice may be immune-mediated. It also appears that the removal viral PKR inhibitors from poxvirus vaccine vectors may be a beneficial strategy to dampen immune-mediated pathology while still eliciting protective immune responses (Bravo Cruz et al., 2017).
Historically, replication-competent w.t. VACV was employed to eradicate smallpox virus but its use as a vaccine vector has since been curtailed due to safety concerns in humans. This led to the development of highly attenuated poxviruses with improved safety profiles, especially in immunocompromised individuals and those contraindicated to receive w.t. VACV. Most of these weakened or replication-incompetent poxviruses have been various versions of VACV (Jacobs et al., 2009), such as MVA and NYVAC, but some work has also been done with an avipoxvirus called ALVAC (Franchini et al., 2004; Paoletti et al., 1994). The deletion of E3L from VACV or MVA yields highly attenuated viruses that are replication-deficient in many cells, including those derived from humans (Chang et al., 1995; Ludwig et al., 2006). These E3L-null mutants can induce protective immunity against subsequent challenge with homologous or heterologous viruses (Denzler et al., 2011; Jentarra et al., 2008; Volz et al., 2018). Our work here indicates that ECTVΔE3L could be considered as another option for an attenuated vaccine vector. Its lack of replication competence in vitro and virulence in BALB/c mice suggests that it would be nonpathogenic in humans and other animals. Wild-type ECTV is already known to be pathogenic only in mice (Buller et al., 1986). Therefore, ECTVΔE3L would likely be further attenuated in non-murine hosts compared to w.t. virus.
Upon closer examination of ECTVΔE3L at the cellular level, we find that early events of the viral replication cycle occur but post-replicative protein production is blocked. An ELISPOT assay quantifying epitope-specific CD4+ T-cells revealed that positive responses were principally restricted to early gene products. It seems that the late gene products in the initial inoculum (present in the injected virions) were insufficient to elicit T-cell responses. It is possible that a higher dose of ECTVΔE3L or multiple immunizations would have induced T-cell responses against post-replicative proteins. However, we have evidence suggesting that these approaches would also not yield meaningful CD4+ T-cell responses to late gene products (K.S. Forsyth, manuscript in preparation). Therefore, de novo protein synthesis and possibly endogenous antigen processing (Miller et al., 2015) appear to be important to the priming of poxvirus-specific CD4+ T-cells. If ECTVΔE3L were to be used as a vaccine vector, it seems that the expression of a recombinant antigen would need to be placed under the transcriptional control of an early viral promoter.
In conclusion, we report that ECTV relies on E3 to complete a productive replication cycle. We detect significantly more dsRNA within cells infected with ECTVΔE3L relative to w.t. virus. This exposed, E3-unbound dsRNA activates PKR, which blocks late protein production and, hence, the completion of genome replication and virion assembly. Despite ECTVΔE3L being replication-defective and nonpathogenic in mousepox-susceptible animals, this mutant virus is able to induce protective immunity in immunized mice challenged with w.t. ECTV. Future work may reveal w.t. ECTV or ECTVΔE3L to be attractive vaccine vectors.
Highlights.
Ectromelia virus (ECTV) requires E3L to replicate in cultured cells derived from multiple mammalian species.
ECTV lacking E3L (ECTVΔE3L) does not complete genome replication and displays little to no translation of late genes.
The replication defects of ECTVΔE3L were rescued in human and mouse cells lacking PKR, which is an important host anti-viral protein.
ECTVΔE3L was nonpathogenic in BALB/c mice, a strain susceptible to lethal mousepox disease.
When used a vaccine, ECTVΔE3L induced protective immunity upon subsequent challenge with wild-type ECTV.
Acknowledgments
As indicated in the methods section, some reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository. This work was supported in part by NIH grant R01 AI110542 and the Albright Creative Research Experience program, which provides internal grants at Albright College.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aranda PS, LaJoie DM, Jorcyk CL. Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt WD, White SD, Johnson BP, Huynh T, Liao J, Harrington H, Cotsmire S, Kibler KV, Langland J, Jacobs BL. Monkeypox virus induces the synthesis of less dsRNA than vaccinia virus, and is more resistant to the anti-poxvirus drug, IBT, than vaccinia virus. Virology. 2016;497:125–135. doi: 10.1016/j.virol.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Greenbaum JA, Sundström M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proceedings of the National Academy of Sciences. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E, Denzler KL, Tartaglia J, Perkus ME, Paoletti E, Jacobs BL. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. Journal of Virology. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E, Kauffman EB, Martinez H, Perkus ME, Jacobs BL, Paoletti E, Tartaglia J. Host-range restriction of vaccinia virus E3L–specific deletion mutants. Virus Genes. 1996;12:89–94. doi: 10.1007/BF00370005. [DOI] [PubMed] [Google Scholar]
- Boone RF, Parr RP, Moss B. Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. Journal of Virology. 1979;30:365–374. doi: 10.1128/jvi.30.1.365-374.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt TA, Jacobs BL. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. Journal of Virology. 2001;75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratke KA, McLysaght A, Rothenburg S. A survey of host range genes in poxvirus genomes. Infect. Genet. Evol. 2013;14:406–425. doi: 10.1016/j.meegid.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo Cruz AG, Han A, Roy EJ, Guzmán AB, Miller RJ, Driskell EA, O’Brien WD, Shisler JL. Deletion of the K1L gene results in a vaccinia virus that is less pathogenic due to muted innate immune responses, yet still elicits protective immunity. Journal of Virology. 2017;91:e00542–17. doi: 10.1128/JVI.00542-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan G, Kitzman JO, Rothenburg S, Shendure J, Geballe AP. Adaptive gene amplification as an intermediate step in the expansion of virus host range. PLoS Pathog. 2014;10:e1004002. doi: 10.1371/journal.ppat.1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles SS. Vaccinia virus transcription. J. Gen. Virol. 2003;84:2293–2303. doi: 10.1099/vir.0.18942-0. [DOI] [PubMed] [Google Scholar]
- Buller RM, Potter M, Wallace GD. Variable resistance to ectromelia (mousepox) virus among genera of Mus. Curr. Top. Microbiol. Immunol. 1986;127:319–322. doi: 10.1007/978-3-642-71304-0_38. [DOI] [PubMed] [Google Scholar]
- Burgess HM, Mohr I. Cellular 5'-3' mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe. 2015;17:332–344. doi: 10.1016/j.chom.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. Journal of Interferon & Cytokine Research. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Jacobs BL. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- Chang HW, Uribe LH, Jacobs BL. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. Journal of Virology. 1995;69:6605–6608. doi: 10.1128/jvi.69.10.6605-6608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Danila MI, Feng Z, Buller RML, Wang C, Han X, Lefkowitz EJ, Upton C. The genomic sequence of ectromelia virus, the causative agent of mousepox. Virology. 2003;317:165–186. doi: 10.1016/S0042-6822(03)00520-8. [DOI] [PubMed] [Google Scholar]
- Colby C, Duesberg PH. Double-stranded RNA in vaccinia virus infected cells. Nature. 1969;222:940–944. doi: 10.1038/222940a0. [DOI] [PubMed] [Google Scholar]
- Colby C, Jurale C, Kates JR. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. Journal of Virology. 1971;7:71–76. doi: 10.1128/jvi.7.1.71-76.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MV, Elroy-Stein O, Jagus R, Moss B, Kaufman RJ. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. Journal of Virology. 1992;66:1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler KL, Schriewer J, Parker S, Werner C, Hartzler H, Hembrador E, Huynh T, Holechek S, Buller RM, Jacobs BL. The attenuated NYCBH vaccinia virus deleted for the immune evasion gene, E3L, completely protects mice against heterologous challenge with ectromelia virus. Vaccine. 2011;29:9691–9696. doi: 10.1016/j.vaccine.2011.09.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg PH, Colby C. On the biosynthesis and structure of double-stranded RNA in vaccinia virus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 1969;64:396–403. doi: 10.1073/pnas.64.1.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban DJ, Buller RML. Ectromelia virus: the causative agent of mousepox. J. Gen. Virol. 2005;86:2645–2659. doi: 10.1099/vir.0.81090-0. [DOI] [PubMed] [Google Scholar]
- Fang M, Lanier LL, Sigal LJ. A role for NKG2D in NK cell-mediated resistance to poxvirus disease. PLoS Pathog. 2008;4:e30. doi: 10.1371/journal.ppat.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Sigal LJ. Direct CD28 costimulation is required for CD8+ T cell-mediated resistance to an acute viral disease in a natural host. J. Immunol. 2006;177:8027–8036. doi: 10.4049/jimmunol.177.11.8027. [DOI] [PubMed] [Google Scholar]
- Fenner F. Studies in infectious ectromelia in mice; natural transmission; the portal of entry of the virus. Aust J Exp Biol Med Sci. 1947;25:275–282. doi: 10.1038/icb.1947.39. [DOI] [PubMed] [Google Scholar]
- Fenner F, Mortimer P. Classic paper: Fenner on the exanthemata. Rev. Med. Virol. 2006;16:353–363. doi: 10.1002/rmv.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini G, Gurunathan S, Baglyos L, Plotkin S, Tartaglia J. Poxvirus-based vaccine candidates for HIV: two decades of experience with special emphasis on canarypox vectors. Expert Rev Vaccines. 2004;3:S75–88. doi: 10.1586/14760584.3.4.s75. [DOI] [PubMed] [Google Scholar]
- Frey TR, Lehmann MH, Ryan CM, Pizzorno MC, Sutter G, Hersperger AR. Ectromelia virus accumulates less double-stranded RNA compared to vaccinia virus in BS-C-1 cells. Virology. 2017;509:98–111. doi: 10.1016/j.virol.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller SL, Peng C, McFadden G, Rothenburg S. Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 2014;21:15–40. doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand ES, Haller SL, Peng C, Rothenburg S, Hersperger AR. Ectopic expression of vaccinia virus E3 and K3 cannot rescue ectromelia virus replication in rabbit RK13 cells. PLoS ONE. 2015;10:e0119189. doi: 10.1371/journal.pone.0119189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersperger AR, Siciliano NA, DeHaven BC, Snook AE, Eisenlohr LC. Epithelial immunization induces polyfunctional CD8+ T cells and optimal mousepox protection. Journal of Virology. 2014;88:9472–9475. doi: 10.1128/JVI.01464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersperger AR, Siciliano NA, Eisenlohr LC. Comparable polyfunctionality of ectromelia virus- and vaccinia virus-specific murine T cells despite markedly different in vivo replication and pathogenicity. Journal of Virology. 2012;86:7298–7309. doi: 10.1128/JVI.00038-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornemann S, Harlin O, Staib C, Kisling S, Erfle V, Kaspers B, Hacker G, Sutter G. Replication of modified vaccinia virus ankara in primary chicken embryo fibroblasts requires expression of the interferon resistance gene E3L. Journal of Virology. 2003;77:8394–8407. doi: 10.1128/JVI.77.15.8394-8407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs BL, Langland JO, Kibler KV, Denzler KL, White SD, Holechek SA, Wong S, Huynh T, Baskin CR. Vaccinia virus vaccines: past, present and future. Antiviral Research. 2009;84:1–13. doi: 10.1016/j.antiviral.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentarra GM, Heck MC, Youn JW, Kibler K, Langland JO, Baskin CR, Ananieva O, Chang Y, Jacobs BL. Vaccinia viruses with mutations in the E3L gene as potential replication-competent, attenuated vaccines: scarification vaccination. Vaccine. 2008;26:2860–2872. doi: 10.1016/j.vaccine.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha BK, Polyakova I, Kessler P, Dong B, Dickerman B, Sen GC, Silverman RH. Inhibition of RNase L and RNA-dependent protein kinase (PKR) by sunitinib impairs antiviral innate immunity. Journal of Biological Chemistry. 2011;286:26319–26326. doi: 10.1074/jbc.M111.253443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JB, McFadden G. Technical knockout: understanding poxvirus pathogenesis by selectively deleting viral immunomodulatory genes. Cellular Microbiology. 2004;6:695–705. doi: 10.1111/j.1462-5822.2004.00423.x. [DOI] [PubMed] [Google Scholar]
- Kim Y-G, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.3201/eid1204.051181. [DOI] [PubMed] [Google Scholar]
- Langland JO, Jacobs BL. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology. 2002;299:133–141. doi: 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- Liu R, Moss B. Opposing roles of double-stranded RNA effector pathways and viral defense proteins revealed with CRISPR-Cas9 knockout cell lines and vaccinia virus mutants. Journal of Virology. 2016;90:7864–7879. doi: 10.1128/JVI.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H, Suezer Y, Waibler Z, Kalinke U, Schnierle BS, Sutter G. Double-stranded RNA-binding protein E3 controls translation of viral intermediate RNA, marking an essential step in the life cycle of modified vaccinia virus Ankara. J. Gen. Virol. 2006;87:1145–1155. doi: 10.1099/vir.0.81623-0. [DOI] [PubMed] [Google Scholar]
- Lynn H, Horsington J, Ter LK, Han S, Chew YL, Diefenbach RJ, Way M, Chaudhri G, Karupiah G, Newsome TP. Loss of Cytoskeletal Transport during Egress Critically Attenuates Ectromelia Virus Infection In Vivo. Journal of Virology. 2012;86:7427–7443. doi: 10.1128/JVI.06636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Ganesan APV, Luckashenak N, Mendonca M, Eisenlohr LC. Endogenous antigen processing drives the primary CD4+ T cell response to influenza. Nature Medicine. 2015;21:1216–1222. doi: 10.1038/nm.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myskiw C, Arsenio J, Hammett C, van Bruggen R, Deschambault Y, Beausoleil N, Babiuk S, Cao J. Comparative analysis of poxvirus orthologues of the vaccinia virus E3 protein: modulation of protein kinase R activity, cytokine responses, and virus pathogenicity. Journal of Virology. 2011;85:12280–12291. doi: 10.1128/JVI.05505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti E, Tartaglia J, Taylor J. Safe and effective poxvirus vectors--NYVAC and ALVAC. Dev. Biol. Stand. 1994;82:65–69. [PubMed] [Google Scholar]
- Proud CG. eIF2 and the control of cell physiology. Semin. Cell Dev. Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Liu J, Chan WM, Rothenburg S, McFadden G. Myxoma Virus Protein M029 Is a Dual Function Immunomodulator that Inhibits PKR and Also Conscripts RHA/DHX9 to Promote Expanded Host Tropism and Viral Replication. PLoS Pathog. 2013;9:e1003465. doi: 10.1371/journal.ppat.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas C, Gil J, Mělková Z, Esteban M, Díaz-Guerra M. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- Roberts KL, Smith GL. Vaccinia virus morphogenesis and dissemination. Trends in Microbiology. 2008;16:472–479. doi: 10.1016/j.tim.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Rosel J, Moss B. Transcriptional and translational mapping and nucleotide sequence analysis of a vaccinia virus gene encoding the precursor of the major core polypeptide 4b. Journal of Virology. 1985;56:830–838. doi: 10.1128/jvi.56.3.830-838.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, Sypula J, Nazarian SH, Lucas A, McFadden G. Poxviruses and immune evasion. Annu. Rev. Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- Siciliano NA, Hersperger AR, Lacuanan AM, Xu R-H, Sidney J, Sette A, Sigal LJ, Eisenlohr LC. Impact of distinct poxvirus infections on the specificities and functionalities of CD4+ T cell responses. Journal of Virology. 2014;88:10078–10091. doi: 10.1128/JVI.01150-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. rRNA cleavage as an index of ppp(A2’p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. Journal of Virology. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VP, Alcami A. Inhibition of Interferons by Ectromelia Virus. Journal of Virology. 2002;76:1124–1134. doi: 10.1128/JVI.76.3.1124-1134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons JA, Tscharke DC, Price N, Smith GL. A study of the vaccinia virus interferon-gamma receptor and its contribution to virus virulence. J. Gen. Virol. 2002;83:1953–1964. doi: 10.1099/0022-1317-83-8-1953. [DOI] [PubMed] [Google Scholar]
- Tscharke DC. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. Journal of Experimental Medicine. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz A, Jany S, Freudenstein A, Lantermann M, Ludwig H, Sutter G. E3L and F1L Gene Functions Modulate the Protective Capacity of Modified Vaccinia Virus Ankara Immunization in Murine Model of Human Smallpox. Viruses 10. 2018;21 doi: 10.3390/v10010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos JC, Stunnenberg HG. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 1988;7:3487–3492. doi: 10.1002/j.1460-2075.1988.tb03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JC, Chang HW, Jacobs BL. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology. 1991;185:206–216. doi: 10.1016/0042-6822(91)90768-7. [DOI] [PubMed] [Google Scholar]
- Weaver JR, Shamim M, Alexander E, Davies DH, Felgner PL, Isaacs SN. The identification and characterization of a monoclonal antibody to the vaccinia virus E3 protein. Virus Res. 2007;130:269–274. doi: 10.1016/j.virusres.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. Journal of Virology. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis KL, Langland JO, Shisler JL. Viral double-stranded RNAs from vaccinia virus early or intermediate gene transcripts possess PKR activating function, resulting in NF-kappaB activation, when the K1 protein is absent or mutated. Journal of Biological Chemistry. 2011;286:7765–7778. doi: 10.1074/jbc.M110.194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschner DH, James TC, Silverman RH, Kerr IM. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2'p)nA) in interferon-treated cells. Nucleic Acids Res. 1981;9:1571–1581. doi: 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Condit RC, Vijaysri S, Jacobs B, Williams BRG, Silverman RH. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. Journal of Virology. 2002;76:5251–5259. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Latner DR, Niles EG, Condit RC. Transcription elongation activity of the vaccinia virus J3 protein in vivo is independent of poly(A) polymerase stimulation. Virology. 2000;269:356–369. doi: 10.1006/viro.2000.0242. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Simpson DA, Spiegel J, Zhou A, Silverman RH, Condit RC. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. Journal of Virology. 1998;72:7012–7023. doi: 10.1128/jvi.72.9.7012-7023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Cohen M, Tang Y, Lazear E, Whitbeck JC, Eisenberg RJ, Cohen GH, Sigal LJ. The orthopoxvirus type I IFN binding protein is essential for virulence and an effective target for vaccination. Journal of Experimental Medicine. 2008;205:981–992. doi: 10.1084/jem.20071854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Maruri-Avidal L, Sisler J, Stuart CA, Moss B. Cascade regulation of vaccinia virus gene expression is modulated by multistage promoters. Virology. 2013;447:213–220. doi: 10.1016/j.virol.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Jacobs BL, Samuel CE. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. Journal of Virology. 2008;82:840–848. doi: 10.1128/JVI.01891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Samuel CE. Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. Journal of Virology. 2007;81:8192–8200. doi: 10.1128/JVI.00426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Paranjape JM, Der SD, Williams BR, Silverman RH. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]