Abstract
ISSR (Inter simple sequence repeat) markers were used to assess the genetic diversity and population structure in 53 indigenous and exotic genotypes of gladiolus (Gladiolus hybridus Hort.). Molecular markers analysis showed PIC ranges from 0.42 (ISSR 861) to 0.99 (ISSR 855, ISSR 856 and ISSR 889) with an average 0.812, marker index ranged from 0.99 (ISSR 889) to 9.26 (ISSR 851) with an average 4.66 and resolving power of the primers ranged from 0.03 (ISSR 889) to 11.58 (ISSR 861) with an average value 3.80. The dendrogram based UPGMA clustering showed that all the 53 genotypes grouped into three main clusters. Nei’s gene diversity (Na) varied from 0.929 to 1.717, effective number of alleles (Ne) varied from 1.262 to 1.369, Shannon’s information index (I) ranged from 0.251 to 0.359 and gene diversity (He) was in the range from 0.167 to 0.229. Population structure analysis revealed three groups in which 32 genotypes were admixture types.
Keywords: Gladiolus, Molecular diversity, Inter simple sequence repeats, Population structure
Introduction
Gladiolus is a genus of perennial herbaceous bulbous flowering plants of high economic importance. Cultivated gladiolus (Gladiolus hybridus Hort.) is native of South Africa and belongs to the family Iridaceae. Most of the gladiolus (Gladiolus grandiflorus Hort.) are tetraploid (2n = 4x = 60), interspecific hybrids being cultivated for more than 260 years (Goldblatt 1996). The current number of species in the genus is 255 (Goldblatt and Manning 1998). Among these, 180 species are originally from South Africa (Duncan 1996, 2000; Goldblatt 1996; Goldblatt and Manning 1998; Goldblatt et al. 1993; Manning et al. 2002).
Several features like massive spikes with florets, brilliant colors, attractive shapes etc. makes it a queen of bulbous flowers. They are ideal for display, floral arrangement, interior decoration and making high quality bouquet (Lepcha et al. 2007). Among bulbous flowering plants, glaldiolus are ranked fifth to tulip (Tulip sp.), lily (Lilium sp.), fressia (Fressia sp.) and hippestrum (Hippestrum sp.) (Flower Council of Holland 2008). In the cut flower trade, they ranked fourth in the International market after the rose, carnation and chrysanthemum (Rathod et al. 2011). In India, commercial value of gladiolus has increased a lot because of its local and export market value.
Nowadays, most of the gladiolus cultivars are developed from inter-specific hybridization among several species. Hence, wide variation is exhibited among the cultivars for their growth, shape, spike length and floret colour. In India, efforts have been made on collection and evaluation of gladiolus germplasm in order to gather ample information for higher flower quality as well as flower yield and their contributing characters. Being ornamental, morphological characterization is highly recommended step in gladiolus for selection using flower, corm and vegetative traits etc. However, environmental influence on these traits limits their sole usage in selection. Improvement of traits like flower color, fragrance, disease resistance etc. is lagging due to inadequate exploration of diversity present within different species of gladiolus. There is much needed effort required for gladiolus improvement and to explore unexplored diversity present within large gladiolus germplasm. However, performance of particular gladiolus genotype depends on the climatic conditions in which they were grown (Swaroop and Janakiram 2010). This suggests large scale evaluation of wild and cultivated gladiolus genotypes under certain agro-climatic conditions to explore and utilize tremendous genetic diversity. It would be highly desirable to prioritize in breeding programmes, genotypes with broader agro-climatic adaptability. Therefore, large-scale diversity evaluation in gladiolus would help to exploit uncharacterized genetic components and to test their efficacy for developing genotypes with resistance for different abiotic and biotic stresses. In last three decades, numerous plant genetic diversity studies have been conducted using wide range of molecular markers including traditional markers like RAPDs, SSRs, AFLP or next generation sequencing (NGS) methods etc. (Mondini et al. 2009). Molecular markers aids in proper selection and characterization of plant germplasm. Among several markers available, Inter Simple Sequence Repeats (ISSR) markers are highly reproducible, polymorphic, informative and quick to use (Bornet and Branchard 2001). For any diversity study, it is advantageous to include original parents of the progeny genotypes tested in order to evaluate possible hybrid vigour and distribution of diversity among progeny genotypes. In the present study, authors assess the genetic diversity and population structure in 53 commercial gladiolus genotypes, including some parental genotypes, collected from different parts of country, using discriminating power of ISSR markers. To the best of author’s knowledge, present study is first report in which ISSR molecular markers have been used for diversity and population structure analysis in gladiolus.
Materials and methods
Plant material
In total, 53 genotypes of gladiolus were obtained from different sources namely: Directorate of Floricultural Research IARI, New Delhi, Indian Institute of Horticultural Research, (IIHR) Bangalore, Punjab Agricultural University (PAU), Ludhiana and National Botanical Research Institute (NBRI), Lucknow. Details like genotypes name, flower colour, origin, and pedigree is provided in Table 1.
Table 1.
Details of gladiolus genotypes studied
| S. no. | Name of variety | Origin | Flower colour | Parental cross or source | Collection centre |
|---|---|---|---|---|---|
| 1 | Punjab Glance | India | Orange with yellow blotch | Happy End × Yellow Stone | PAU, Ludhiana, Punjab |
| 2 | Punjab Flame | India | Carmine with red blotch | Sylvia × White Prosperity | PAU, Ludhiana, Punjab |
| 3 | Punjab Pink | India | Pink | Suchitra × White Prosperity | PAU, Ludhiana, Punjab |
| 4 | Punjab Glad-1 | India | Orange with yellow centre | Happy End × True Yellow | PAU, Ludhiana, Punjab |
| 5 | Pacifica | Exotic | Red group | Unknown | NBRI, Lucknow, UP |
| 6 | Orange Ginger | Exotic | Orange | Unknown | NBRI, Lucknow, UP |
| 7 | Prabha | India | Red group | Unknown | NBRI, Lucknow, UP |
| 8 | Sylvia | Exotic | Orange | Comm Koehl × Moorish Cherry | NBRI, Lucknow, UP |
| 9 | Aldebaran | Exotic | Yellow group | Unknown | NBRI, Lucknow, UP |
| 10 | Tiger Flame | Unknown | Red | Unknown | NBRI, Lucknow, UP |
| 11 | Victor | Exotic | Pink | Unknown | NBRI, Lucknow, UP |
| 12 | Pusa Shagun | India | Red | White Oak × Oscar | NBRI, Lucknow, UP |
| 13 | Regency | India | Red group | Ruffled Ebony × Ace of spades | NBRI, Lucknow, UP |
| 14 | Snow Princes | Exotic | White | Unknown | DFR, New Delhi |
| 15 | Inter Pearl | Unknown | Red group | Unknown | NBRI, Lucknow, UP |
| 16 | Yellow Stone | Exotic | Yellow | Unknown | DFR, New Delhi |
| 17 | Limoncilla | Exotic | Yellow lemon colour | Unknown | DFR, New Delhi |
| 18 | Pricilla | Exotic | Purple Pink | Diamond × Leana | DFR, New Delhi |
| 19 | Novalux | Exotic | Yellow group | Unknown | DFR, New Delhi |
| 20 | Gold Field | Exotic | Yellow | Unknown | DFR, New Delhi |
| 21 | Ocilla | Exotic | Cream white | Unknown | DFR, New Delhi |
| 22 | Punjab Dawn | India | Pink/peach | Suchitra × Melody | DFR, New Delhi |
| 23 | Chandni | India | Greenish white | Green Woodpecker × White Butterfly (1997) | DFR, New Delhi |
| 24 | Arka Darshan | India | Red purple with white blotch | Watermelon Pink × Shirley | IIHR, Bangalore, Karnataka |
| 25 | Arka Naveen | India | Purple–Violet group | Hybrid 74-39-1 × Tropic Sea | IIHR, Bangalore, Karnataka |
| 26 | Arka Shobha | India | Light Pink | Induced mutant from cv. wild Rose | IIHR, Bangalore, Karnataka |
| 27 | Jester Gold | Exotic | Yellow | Jester sport | DFR, New Delhi |
| 28 | Flavour Souvenir | Exotic | Yellow | Unknown | DFR, New Delhi |
| 29 | Forta Rosa | Exotic | Soft Pink | Unknown | DFR, New Delhi |
| 30 | Prince Margret Rosa | Exotic | Orange | Unknown | DFR, New Delhi |
| 31 | Arka Sagar | India | Pink with red and yellow blotch | Melody × Wild Rose | IIHR, Bangalore, Karnataka |
| 32 | Arka Poonam | India | Dresden Yellow | Geliber Herald’ × ’R.N. 121′ | IIHR, Bangalore, Karnataka |
| 33 | Arka Tilak | India | Red | Watermelon Pink × Lady John | IIHR, Bangalore, Karnataka |
| 34 | Arka Kum Kum | India | Red | Watermelon Pink × Lady John | IIHR, Bangalore, Karnataka |
| 35 | Arka Keshar | India | Yellow –Orange | Vink’s Glory × Sagar | IIHR, Bangalore, Karnataka |
| 36 | Arka Gold | India | Yellow with red blotch | Green Bay × Gold Medal-412 | IIHR, Bangalore, Karnataka |
| 37 | Pusa Sukanya | India | White with scarlet ring in the lip | Salmon Queen seedling selection | IARI, New Delhi |
| 38 | Wind Song | Exotic | Purple | Unknown | IARI, New Delhi |
| 39 | Hunting Song | Exotic | Scarlet red | Unknown | IARI, New Delhi |
| 40 | Friendship Sancere | Unknown | Snow white | Unknown | IARI, New Delhi |
| 41 | Shere Punjab | India | Pinkish orange | Suchitra × Melody | IARI, New Delhi |
| 42 | Arka Amar | India | Pink with White blotch | Watermelon Pink × Arka Aarti | IIHR, Bangalore, Karnataka |
| 43 | Pusa Suhagin | India | Florets ruby-red with barium yellow streak | Sylvia seedlings (1987) | IARI, New Delhi |
| 44 | Arun | India | Red | Sylvia × Fancy (1984) | IARI, New Delhi |
| 45 | Pusa Kiran | India | Florets are white with ray like red on throat | Ave open | IARI, New Delhi |
| 46 | Mohini | India | Floret colour red–purple | Ave × Christian Jane (2000) | IARI, New Delhi |
| 47 | Peter Pears | Exotic | Red–purple group | Salmoe × Maolete | IARI, New Delhi |
| 48 | Arka Aarti | India | Poppy-red with purple–red | Shirley × Melody | IARI, New Delhi |
| 49 | Legend Pink | Exotic | Pink | Unknown | IARI, New Delhi |
| 50 | Sancerre | Exotic | Snow White | Unknown | IARI, New Delhi |
| 51 | Novalux Yellow | India | Yellow group | unknown | IARI, New Delhi |
| 52 | American Beauty | Exotic | Pink/Peach | Unknown | IARI, New Delhi |
| 53 | White prosperity | Exotic | White | Unknown | IARI, New Delhi |
DNA extraction, PCR amplification and cluster analysis
Genomic DNA extraction, PCR amplification and gel electrophoresis was performed as described earlier (Kumar et al. 2009). A set of 19 ISSR primers (Jingang et al. 2008; Shufang et al. 2010) were initially tested for amplification with selected genotypes (Table 2). Out of 19 primers, 17 primers that showed consistent and good amplification were further used for PCR amplification with 53 genotypes. The data generated by 17 ISSR primers on 53 genotypes were scored in binary matrix format and used for different statistical analysis. The 0–1 matrix data was subjected to calculate pairwise genetic similarity using Jaccard’s coefficient (Jaccard 1908). The similarity matrix thus obtained was used to prepare dendrogram by unweighted pair group method of arithmetic averages (UPGMA) with the help of NTSYS-PC software version 2.02e (Rohlf 1993). Besides, PIC (polymorphism information content) (Botstein et al. 1980), marker index (MI) (Milbourne et al. 1997) and Resolving Power (Rp) (Prevost and Wilkinson 1999) were also calculated.
Table 2.
Details of the 17 ISSR primers used for diversity study on 53 gladiolus genotypes PIC (polymorphism information content), MI (marker index), RP (Resolving Power)
| Primer code | Sequence (5’–3’) | PIC | RP | MI | No. of alleles | Polymorphic alleles | Monomorphic alleles | Polymorphism percentage | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 22,203 | GAGAGAGAGAGAGAGAC | 0.82 | 3.73 | 4.92 | 6 | 6 | 0 | 100 |
| 2 | 22,204 | GAGAGAGAGAGAGAGAA | 0.59 | 6.37 | 2.98 | 6 | 5 | 1 | 83.33 |
| 3 | 22,207 | CTCTCTCTCTCTCTCTG | 0.65 | 3.99 | 2.60 | 4 | 4 | 0 | 100 |
| 4 | 22,209 | CACACACACACACACAA | 0.81 | 2.90 | 3.27 | 4 | 4 | 0 | 100 |
| 5 | 22,215 | TCTCTCTCTCTCTCTCC | 0.55 | 6.45 | 2.79 | 5 | 5 | 0 | 100 |
| 6 | 22,218 | ACACACACACACACACC | 0.89 | 4.48 | 7.14 | 8 | 8 | 0 | 100 |
| 7 | 22,219 | ACACACACACACACACG | 0.84 | 4.67 | 7.56 | 9 | 9 | 0 | 100 |
| 8 | 834 | AGAGAGAGAGAGAGAG(CT)T | 0.78 | 5.20 | 5.52 | 7 | 7 | 0 | 100 |
| 9 | 836 | AGAGAGAGAGAGAGAG(CT)A | 0.96 | 1.92 | 5.78 | 6 | 6 | 0 | 100 |
| 10 | 840 | GAGAGAGAGAGAGAGA(CT)T | 0.79 | 2.33 | 3.16 | 4 | 4 | 0 | 100 |
| 11 | 851 | GTGTGTGTGTGTGTGT(CT)G | 0.92 | 4.45 | 9.26 | 10 | 10 | 0 | 100 |
| 12 | 855 | ACACACACACACACAC(CT)T | 0.99 | 0.22 | 3.99 | 4 | 4 | 0 | 100 |
| 13 | 856 | ACACACACACACACAC(CT)A | 0.99 | 0.41 | 3.98 | 4 | 4 | 0 | 100 |
| 14 | 857 | ACACACACACACACAC(CT)G | 0.94 | 3.58 | 8.51 | 9 | 9 | 0 | 100 |
| 15 | 861 | ACCACCACCACCACCACC | 0.42 | 11.58 | 3.37 | 8 | 8 | 0 | 100 |
| 16 | 866 | CTCCTCCTCCTCCTCCTC | 0.87 | 2.26 | 3.48 | 4 | 4 | 0 | 100 |
| 17 | 889 | AGT)(CGT)(AGT)ACACACACACACAC | 0.99 | 0.03 | 0.99 | 1 | 1 | 0 | 100 |
| Average | 0.812 | 3.80 | 4.66 | 5.82 | 5.76 | – | 99.02 | ||
Population structure analysis
Gene diversity in population and subpopulation was measured by Nei (1973). Other genetic diversity estimates namely Na (number of different alleles), Ne (effective number of alleles) and I (Shannon’s information index) were also calculated. Further, in order to estimate the number of subpopulations in the gladiolus germplasm, population STRUCTURE analysis was done using program STRUCTURE version 2.2 (Pritchard et al. 2000). The membership of each genotypes was tested for K = 2 to K = 10 with admixture model. Three independent runs were assessed for each fixed K and each run consisted of 30,000 burn-in period and 1,00,000 MCMC (Markov Chain Monte Carlo) iterations. The optimal value of K was determined by examination of the ΔK statistic and L (K) (Evanno et al. 2005) using Structure Harvester program (Earl and vonHoldt 2012).
Result
DNA profiling using IISRs
In the present study, 17 ISSR primers generated a total of 99 alleles among 53 genotypes which varied from one to seventeen with an average of 5.8 alleles per primer pair. Out of 99 alleles generated by the ISSR primers, 98 alleles were polymorphic and one was monomorphic, thus generating 99.01% polymorphism (Table 2, Fig. 1). ISSR 851 primer gave highest number of alleles (10 alleles) followed by, ISSR 22,219 and ISSR 857 (9 alleles each) and ISSR primers 889 gave minimum number (1 alleles) (Table 2). The PIC value ranged from 0.42 (ISSR 861) to 0.99 (ISSR 855, ISSR 856 and ISSR889) with an average of 0.812, marker index ranged from 0.99 (ISSR 889) to 9.26 (ISSR, 851) with an average 4.66 and resolving power of the primers ranged from 0.03 (ISSR 889) to 11.58 (ISSR 861) with an average value of 3.80. Prevost and Wilkinson (1999) described the parameter resolving power (RP) as a measure of the discriminatory power of ISSR molecular markers. The values of resolving power in present study ranged from 0.03 to 11.58. Genetic similarities (GS) were calculated using the Nei–Li similarity co-efficient. Significant genetic variation was found among all the gladiolus genotypes with the GS value ranging from 0.11 to 1.00. Of the 53 pair wise combinations, the highest genetic similarity of 0.82 was found between the genotype Novalux Yellow and American Beauty, followed by 0.80 between genotype Shagun and Regency and minimum genetic similarity was observed between Punjab Glance to White Prosperity.
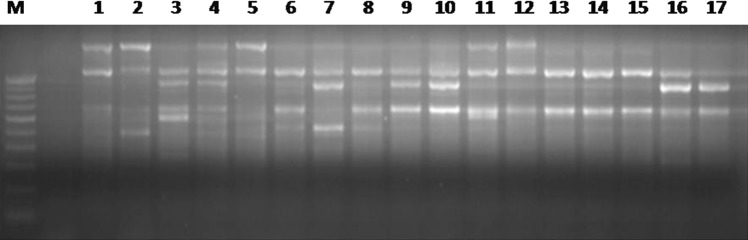
Fig. 1.
Representative gel image depicting PCR amplification of 17 gladiolus genotypes (1 Punjab Flame, 2 Punjab Pink, 3 Punjab Glad-1, 4 Pacifica, 5 Orange Ginger, 6 Prabha, 7 Sylvia, 8 Aldebaran, 9 Tiger Flame, 10 Victor, 11 Pusa Shagun, 12 Regency, 13 Snow Princes, 14 Inter Pearl, 15 Yellow Stone, 16 Limoncilla and 17 Pricilla) with primer ISSR 834. M represents 100 bp ladder
Cluster analysis and relationship among the genotypes
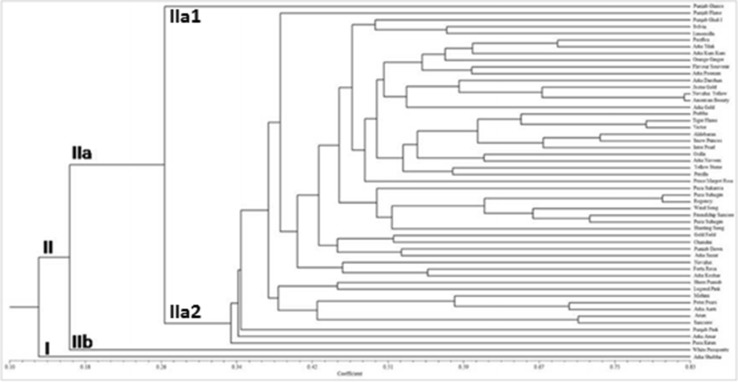
The UPGMA based clustering as depicted by dendrogram (Fig. 2) showed that all the 53 gladiolus genotypes were grouped into two major groups (Group I to II) at the coefficient of GS = 0.14 (Fig. 2). the group II contains maximum number of 52 genotypes which can be further divided into two subgroups, namely, IIa and IIb. The group I and IIb comprise of a single genotype each namely “Arka Shobha” and “White Prosperity” respectively. The sub group IIa1 contain single genotype namely “Punjab Glance” whose parents were “Happy End” and “Yellow Stone” (Table 1). The sub group IIa2 consist of 50 genotypes. Among these, five genotypes namely “Punjab Glance”, “Pusa Kiran”, “Arka Amar”, “Punjab Pink” and “Punjab Flame” were found to be the most distinct and present separately from other clustered genotypes.
Fig. 2.
Dendrogram of 53 gladilous genotypes generated by UPGMA clustering using Jaccards similarly matrix obtained from ISSR markers
Genetic diversity estimates
Among genetic diversity estimates, Na (number of different alleles) varied 0.929–1.717 with an average of 1.39, Ne (effective number of alleles) varied from 1.262 to 1.369 with an average of 1.24, similarly I (Shannon’s information index) ranged from 0.251 to 0.359 with an average 0.25 and He (Nei 1973) gene diversity was in the range from 0.167 to 0.229 with an average 0.15 (Table 3).
Table 3.
Different genetic diversity estimates for six populations (based on collection site) of gladiolus based on 99 loci
| Population (sample size) | Na | Ne | I | He |
|---|---|---|---|---|
| Pop1 (4) | 1.162 ± 0.095 | 1.262 ± 0.031 | 0.260 ± 0.026 | 0.167 ± 0.017 |
| Pop2 (10) | 1.343 ± 0.088 | 1.333 ± 0.038 | 0.295 ± 0.028 | 0.195 ± 0.020 |
| Pop3 (13) | 1.485 ± 0.084 | 1.326 ± 0.034 | 0.311 ± 0.026 | 0.200 ± 0.018 |
| Pop4 (10) | 1.717 ± 0.067 | 1.369 ± 0.034 | 0.359 ± 0.023 | 0.229 ± 0.017 |
| Pop5 (12) | 1.253 ± 0.097 | 1.319 ± 0.038 | 0.359 ± 0.028 | 0.186 ± 0.020 |
| Pop6 (4) | 0.929 ± 0.099 | 1.292 ± 0.037 | 0.251 ± 0.029 | 0.170 ± 0.020 |
| Mean | 1.39 ± 0.015 | 1.24 ± 0.005 | 0.25 ± 0.004 | 0.15 ± 0.003 |
Na number of different alleles, Ne effective number of alleles, I Shannon’s information index He Nei’s (1973) gene diversity
Population STRUCTURE of gladiolus populations
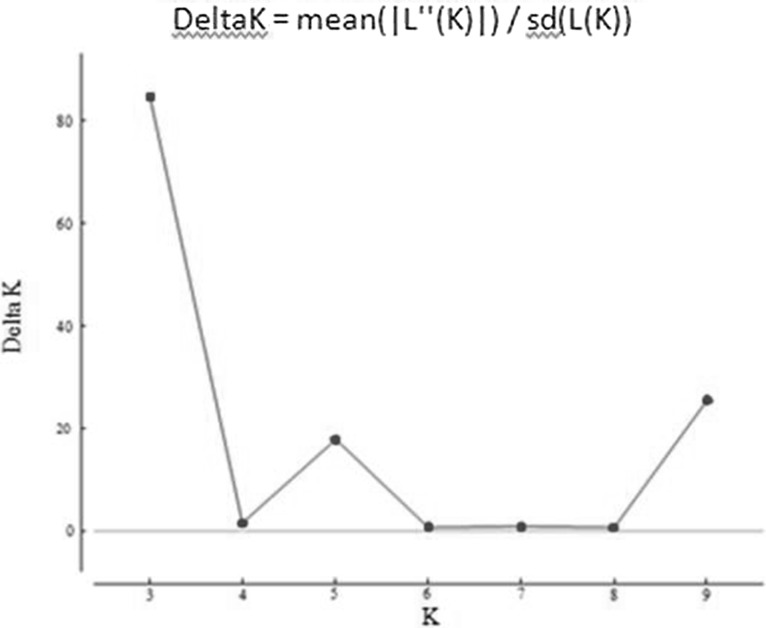
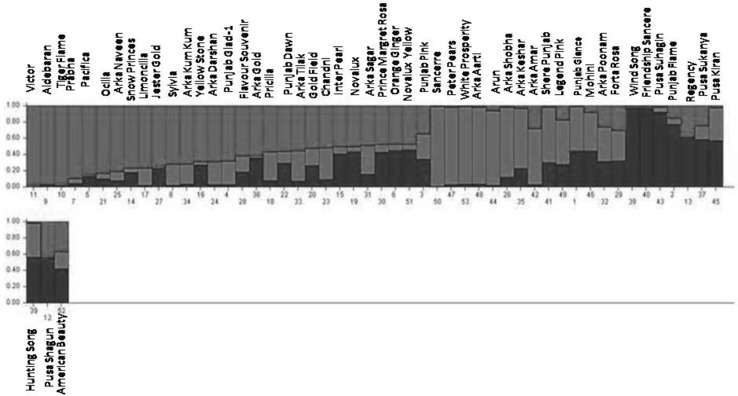
Model based STRUCTURE’ program was used to study the underlying population structure among the 53 gladiolus genotypes. The highest value ΔK was for K = 3, so the value of K = 3 was chosen for the final analysis of population structure (Fig. 3). Therefore, all 53 gladiolus genotypes were assigned to three clusters at K = 3 by their inferred genome fraction value. A genotype was assigned to a group if more than 75% of its genome fraction value is derived from that group (Fig. 4). Genotypes with membership probabilities < 0.75 were assigned to an admixed group. Out of 53 genotypes, 21 were assigned to three groups, and 32 genotypes were retained in the admixed clusters. Among three groups, red group contains 10 genotypes, green group comprised of seven (7) genotypes and finally blue group comprised of four (4) genotypes.
Fig. 3.
Highest ∆K value for the gladiolus genotype at K = 3
Fig. 4.
STRUCTURE plot of 53 gladiolus genotypes with K = 3. The different colour bars referred to three different genetic groups/pools respectively (color figure online)
Discussion
Molecular markers provide an effective tool to study the association and relationship among different genotypes. Several properties especially non responsive towards environmental, pleiotropic and epistatic effects make them highly advantageous and useful over traditional morphological or phenotypic markers (Mondini et al. 2009). Polymorphic genetic markers have wide potential applications in plant improvement programmes as a means for varietal and parentage identification, evaluation of polymorphic genetic loci affecting quantitative economic traits and genetic mapping. In the present study, authors selected ISSR markers for diversity evaluation because of their easy application and results interpretation. These markers are highly polymorphic and have been successfully used in many bulbous crops (Jingang et al. 2008; Kiani et al. 2012; Kameswari et al. 2014). In earlier studies, di-nucleotide based ISSR primers have been used in genotyping of some important boulbous flowering plant with high reproducibility and sufficient polymorphism (Kameswari et al. 2014). Rp values of markers suggested that the set of ISSR primers used were capable of distinguishing different gladiolus genotypes. Results observed in present study suggested the use of MI and PIC parameters to compare the information content of polymorphic ISSR and the use of Rp to select the most informative ISSR marker for best selection among different genotypes.
In the present study ISSR profile was used for diversity analysis and relationship among different gladiolus genotypes. The ISSR polymorphism obtained in the present study is 99% which is more than that reported by Jingang et al. (2008) who found 93% polymorphism using same ISSR markers in different gladiolus collection. Results showed that primers namely ISSR 851, ISSR 22,219, ISSR 857, ISSR 861 and ISSR 22,218 that have above average PIC, MI and Rp, were to be more efficient. Previously researchers have compared the genetic diversity in other floricultural bulbous crops, with pedigree information (Patra et al. 2008; Cui et al. 2014; Anderson et al. 2010). In the present study, genotype Arka Shobha, belonging to group I, is an induced mutant cultivar from wild rose and found to be most distinct genotype. Pedigree of White Prosperity from group II is unknown. Pusa Kiran had been developed from Ave open with white colored florets, Arka Amar is a cross of Water melon × Arka Aarti. Punjab Pink and Punjab Flame shared common parent i.e. White Prosperity. Punjab Pink has been developed by hybridizing Suchitra and White Prosperity while Punjab Flame is the progeny from Sylvia × White Prosperity cross. Clustering of some genotypes could be proved by their parental relationship. Two closely clustered genotypes namely “Punjab Down” and “Arka Sagar” share common parent i.e. genotype “Melody”. Punjab Down is a cross between Suchitra and Melody, while Arka Sagar is a cross between Melody and Wild Rose. Another genotype named “Arka Aarti” although shared common parent i.e. Melody but found to be far present in different subcluster. Likewise, Arka Tilak, Arka Kumkum and Arka Darshan shared common parent i.e. Watermelon Pink. These three genotypes are present close to each other. Arka Tilak and Arka Kumkum both originated from same parental cross i.e. Watermelon Pink x Lady John, while Arka Darshan originated from a cross between Watermelon Pink x Shirley. Likewise genotype namely “Pusa Suhagin” is present near to genotype named “Sylvia”. Pusa Suhagin is found to have been originated from Sylvia seedlings (Table 1). It has also been observed from the present study that majority of the exotic genotypes like Sylvia, Aldebaran, Limoncilla, Pricilla, Snow Princes, Ocilla, Yellow Stone and Prince Margret Rosa were found to be clustered together. From clustering pattern, it seems that up to some extent parental-progeny relationship exists among the genotypes and some related genotypes clustered together. The results were consistent with Pragya et al. (2010) and Ranjan et al. (2010) who had also observed the parental relationship among the gladiolus genotypes by using RAPD and AFLP markers.
STRUCTURE results showed the presence of mixed populations among three clusters with majority of the mixtures in cluster A. The mixed population could be attributed to various reasons like breeding/domestication history, high level of heterozygosity etc. It is believed that the cultivated gladiolus, Gladiolus grandiflorus now known as G. hybridus Hort. developed from a number of wild species viz G. Papilioentus, G. Netalensis, G. Opposityflorus, G. Papilio and G. Saundersii (Banard 1972; Imanishi 1989). Garden gladiolus varieties of today has emerged from diverse genetic parentage that are heteroploids ranging from 2n = 30 to 180 (Bhajantri and Patil 2013). Nowadays, most of the gladiolus genotypes are developed from inter-specific hybridization among several species. Thus modern genotypes are the results of complex inter specific crosses. They are heterozygous and tetraploid and the knowledge of the hereditary transmission of numerous characteristics is poor (Cantor and Chis 2009). Results of population structure analysis and UPGMA clustering partially corroborates each other. Not all but many genotypes were assigned to same cluster by population structure analysis and Jaccard’s similarity coefficients based dendrogram (Figs. 2, 4). For example genotypes namely Sancerre, White Prosperity, Arka Aarti, Peter Pears, Arun, Arka Amar, Arka Shobha etc. assigned to same cluster by population structure analysis and UPGMA clustering.
Implication for future gladiolus improvement programme
Improvement of floricultural crop like gladiolus depends on the evaluation and characterization of genetically diverse plant material. The genetic relationship among genotypes, observed in the present study could be useful for designing particular breeding programme specially selection of parents. Clustered presence of exotic genotypes in the present study indicates that these genotypes harbor different genetic components that could be useful for the improvement of indigenous and development of novel genotypes. Genotypes documented in the present study can be useful for breeders to broaden the genetic base. Genotypes namely Arka Shobha, Punjab Glance, White Prosperity, Pusa Kiran etc. could be used as parents in a particular breeding depending on their interesting desirable economic traits and hybridization potential.
Acknowledgements
The authors are grateful to Prf. Gaya Prasad, Hon’ble Vice Chancellor of the Sardar Vallabhbhai Patel University of Agriculture & Technology, Meerut, U.P. India for providing facilities and encouragement. Also, IIHR, Bangalore, PAU, Ludhiana, NBRI, Lucknow and IARI, New Delhi deserves thanks for providing gladiolus germplasm.
Contributor Information
Veena Chaudhary, Email: veena_chaudhary@yahoo.co.in.
Shailendra Sharma, Email: shgjus6@gmail.com.
References
- Anderson NO, Younis A, Sun Y. Inter simple sequence repeats distinguish genetic differences in easter lily ‘Nellie White’ clonal ramets within and among bulb growers over years. J Am Soc Hortic Sci. 2010;135(5):445–455. [Google Scholar]
- Banard TT (1972) On hybrid and hybridization. In: Lewis GJ, Obermeyer AA, Barnard TT (eds) Gladiolus a revision of the south african species. J of South Afric Bot, Supplement, 10: 304–310
- Bhajantri A, Patil VS. Studies on ethyl methane sulphonate (EMS) induced mutations for enhancing variability of gladiolus varieties (Gladiolus hybridus Hort.) in M1V2 generation. Karnataka J Agric Sci. 2013;26:403–407. [Google Scholar]
- Bornet B, Branchard M. Non-anchored inter simple sequence repeat (ISSR) markers reproducible and specific tool for genome fingerprinting. Plant Mol Biol Rep. 2001;22:427–432. [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Cantor M, Chis LM. New gladiolus cultivars homologated at USAMV Cluj-Napoca. Bull UASVM Hortic. 2009;66:504–509. [Google Scholar]
- Cui GF, Wu LF, Wang XN, Jia WJ, Duan Q, Ma LL, Jiang XL, Wang JH. Analysis of genetic relationships and identification of lily cultivars based on inter-simple sequence repeat markers. Genet Mol Res. 2014;13:5778–5786. doi: 10.4238/2014.July.29.5. [DOI] [PubMed] [Google Scholar]
- Duncan G (1996) Growing South African bulbous plants, National Botanical Institute. Cape Town, S. Africa. ISBN 1-874907-15-3
- Duncan G (2000) Grow bulbs: a guide to the species cultivation and propagation of South African Bulbs, National Botanical Institute, Kirstenbosch Gardening Series. Claremont, S. Africa. ISBN 0-900048-53-0
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Flower Council of Holland (2008) Facts and figures. http://www.flowercouncil.org/us/marketinformation/. Accessed 14 June 2009
- Goldblatt P (1996) Gladiolus in tropical Africa: systematics, biology & evoluation. Timber Press, Portland, Oregon. ISBN 10: 0-8819-233-38/0-88192-333-8
- Goldblatt P, Manning J (1998) Gladiolus in Southern Africa: timber press. Portland. ISBN 10: 1874950326
- Goldblatt P, Takei M, Razzaq ZA. Chromosome cytology in tropical African gladiolus (Iridaceae) Ann Mo Bot Gard. 1993;80:461–470. doi: 10.2307/2399794. [DOI] [Google Scholar]
- Imanishi H. Gladiolus. In: Mastsuo T, editor. Collected data of plant genetic resources. Tokyo: Kodansya. Scientific; 1989. pp. 1077–1080. [Google Scholar]
- Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaud Nat. 1908;44:223–270. [Google Scholar]
- Jingang W, Ying G, Daidi C, Shenkui L, Chuanpin Y. ISSR analysis of 26 general species of Gladiolus hybridus Hort. J Northeast Agric Univ. 2008;15(4):6–10. [Google Scholar]
- Kameswari PL, Girwani A, Radh-Rani K. Genetic diversity in tuberose (Polianthes tuberosa L.) using morphological and ISSR markers. Electron J Plant Breed. 2014;5(1):52–57. [Google Scholar]
- Kiani M, Memariani F, Zarghami H. Molecular analysis of species of Tulipa L. from Iran based on ISSR marker. Plant Syst Evol. 2012;298:1515–1522. doi: 10.1007/s00606-012-0654-0. [DOI] [Google Scholar]
- Kumar V, Sharma S, Sharma AK, Sharma S, Bhat KV. Comparative analysis of diversity based on morpho-agronomic traits and microsatellite markers in common bean. Euphytica. 2009;170:249–262. doi: 10.1007/s10681-009-9965-9. [DOI] [Google Scholar]
- Lepcha B, Nautiyal MC, Rao VK. Variability studies in gladiolus under mid hill conditions of Uttrakhand. J Ornam Hortic. 2007;10(3):169–172. [Google Scholar]
- Manning J, Goldblatt P, Snijman D (2002) The color encyclopedia of Cape Bulbs, Timber Press. Portland. ISBN 9780881925470
- Milbourne D, Meyer R, Bradshaw JE, Baird E, Bonar N, Provan J, Powell W, Waugh R. Comparison of PCR-based marker systems for the analysis of genetic relationships in cultivated potato. Mol Breed. 1997;3(2):127–136. doi: 10.1023/A:1009633005390. [DOI] [Google Scholar]
- Mondini L, Noorani A, Pagnotta MA. Assessing plant genetic diversity by molecular tools. Diversity. 2009;1:19–35. doi: 10.3390/d1010019. [DOI] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B, Acharya L, Mukherjee AK, Panda MK, Panda MC. Molecular characterization of ten cultivars of Canna lilies (Canna Linn.) using PCR based molecular markers (RAPDs and ISSRs) Int J Integr Biol. 2008;2:129–137. [Google Scholar]
- Pragya Bhat KV, Misra RL, Ranjan JK. Analysis of diversity and relationships among gladiolus cultivars using morphological and RAPD markers. Indian J Agric Sci. 2010;80(90):766–772. [Google Scholar]
- Prevost A, Wilkinson MJ. A new system of comparing PCR primers applied to ISSR finger printing of potato cultivars. Theor Appl Genet. 1999;98:107–112. doi: 10.1007/s001220051046. [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan P, Bhat KV, Mishra RL, Singh SK, Ranjan JK. Genetic relationship of gladiolus cultivars inferred from fluorescence based AFLP markers. Sci Hortic. 2010;123:562–567. doi: 10.1016/j.scienta.2009.11.013. [DOI] [Google Scholar]
- Rathod DM, Chawla SL, Ahur TR, Patel MA. Effect of planting time and chamicals on growth, flowering and yield of gladiolus (Gladiolus grandiflorus) cv. American beauty. J Ornam Hortic. 2011;14(1&2):24–27. [Google Scholar]
- Rohlf FJ (1993) NTSYS-PC: numerical taxonomy and multivariate analysis system. Version 2.02e, Exeter Software, Setauket, New York
- Shufang G, Huijuin FU, Jingang W. ISSR analysis of M1 generation of Gladiolus hybridus Hort. treated by EMS. J Northeast Agric Univ. 2010;17(2):22–26. [Google Scholar]
- Swaroop K, Janakiram T. Divergence studies in gladiolus. Indian J Hortic. 2010;67:352–355. [Google Scholar]