Abstract
Haloxylon ammodendron plays an important role in maintaining the structure and function of the entire ecosystem where it grows. No suitable reference genes have been reported in H. ammodendron plants to date. In this study, a total of 8 reference genes (18S, ACT1, ACT7, UBC18, TUA5, GAPDH, EF-1α and UBQ10) were selected from the available trancriptome database, and the expression stability of these 8 candidate genes was validated under different abiotic stress with three different statistical algorithms (geNorm, NormFinder and BestKeeper). The results produced from different models were in agreement with each other essentially: 18S and TUA5 were the most stable genes under drought stress, 18S, the most stable gene under heat stress and mechanical damage, ACT7 and UBC18, stable under salt stress while TUA5 and GAPDH expressed constantly under mechanical damage, and ACT1 expressed steadily under cold conditions. Expression profiles of several stress response genes, including FT-5, FT-9, DREB2A and DREB2C, were further confirmed with various candidate reference genes. None of the candidate genes showed a constant expression among all tested samples. Hence, it’s essential to use more than one reference gene in order to guarantee the accuracy of quantitative real-time PCR. The results of this study will contribute to the accuracy and reliability in transcripts quantification, which is of significance to transcription-based studies and applications in this important shrub H. ammodendron.
Electronic supplementary material
The online version of this article (10.1007/s12298-018-0520-9) contains supplementary material, which is available to authorized users.
Keywords: Haloxylon ammodendron, Reference genes, Abiotic stress, Expression stability
Introduction
The Haloxylon plants in Chenopodiaceae family are a kind of shrubs or small trees which best survive in fixed dune, semi-fixed dune, saline soil and Gobi desert in temperate and subtropical zones of Asia and Africa (25–48°10′N, 5–110°30′E) (Pyankov et al. 1999). The researchers have to date identified 11 species in Haloxylon plants, of which there are only two species (Haloxylon ammodendron and H. persicum) distributed in the desert and semi-desert areas of northwest China (Yu et al. 2012). H. ammodendron, known as the “Desert Guardian”, is one of the major species for desert afforestation in China. It plays an irreplaceable role in maintaining the structure and function of the ecosystem in arid areas (Sheng et al. 2005; Yu et al. 2012) due to its outstanding capacity to resist high temperature and drought. It also helps reducing the wind speed and improving the microclimate of the forest, thus facilitating the growth of other plants (Long et al. 2014). H. ammodendron has evolved many xeromorphic characteristics. For instance, its leaves are degenerated into scales and the annual succulent branches take over the function of assimilation (Yu et al. 2012). These type of branches are usually referred to as assimilation branches (Yu et al. 2012) (Supplementary Fig. 1).
The adaptation of H. ammodendron to the harsh environment such as drought and high temperature indicates its unique adaptability to these stresses, and some functional gene expression may play an important role in this case. With the help of transcriptome technique, many genes functioning in stress tolerance have been mined in H. ammodendron (Long et al. 2014). However, the molecular mechanism for such an adaptability of H. ammodendron to environmental stress has not been reported.
Quantitative real-time PCR (qRT-PCR) is a method commonly used to determine gene expression at the mRNA level with its high sensitivity, reproducibility and specificity (Bustin 2002; Derveaux et al. 2010). The use of the internal reference gene is considered as the most appropriate and standardized method as it may effectively correct the difference between the RNA starting amount and the reverse transcription efficiency (Bao et al. 2016; Zhang et al. 2016). Thus, it is critical to find suitable reference genes for normalizing the expression of stress resistance genes in H. ammodendron.
The ideal reference genes which can be used exclusively for different types of samples should be expressed stably in all cells under any physiological conditions (Ma et al. 2016; Li et al. 2017; Perez et al. 2017). However, in the plants of same species, no reference gene can be stably expressed with the change of experimental conditions, and a reference gene suitable under one experimental condition is not necessarily suitable under another experimental condition; and in the plants of different species, the same reference gene may be expressed differently under the same stress (Volkov et al. 2003). Therefore, it is very important to select the stable reference gene(s) under specific experimental conditions. The plant-originated reference genes most frequently used in qRT-PCR include actin (ACT) (Maroufi et al. 2010), tubulin (TUB) (Wan et al. 2010), ubiquitin (UBQ) (Chen et al. 2011), elongation factors (EF) (Yuan et al. 2014), ubiquitin-conjugating enzyme (UBC) (Ma et al. 2016), 18S ribosomal RNA (18S) (Amil-Ruiz et al. 2013) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Jain et al. 2006), etc.
Due to limited genome sequence information, no applicable reference genes have been explored from H. ammodendron yet. Based on our previous transcriptome analysis, 8 commonly used reference genes (18S, ACT1, ACT7, UBC18, TUA5, GAPDH, EF-1α and UBQ10) in H. ammodendron were cloned. In order to obtain the most suitable reference genes, expression assays were performed in diverse abiotic-stressed samples of H. ammodendron by using BestKeeper, geNorm and NormFinder. This study provides evidences on reference gene selection based on their expression levels under particular conditions, which will benefit the future studies related to H. ammodendron as well as other species of Haloxylon genus.
Materials and methods
Plant materials and treatments
Cultured 1-year seedlings of H. ammodendron from a nursery in Jimsar, Xinjiang were separately transplanted in a greenhouse [30 ± 2/20 ± 2 °C (day/night), 900–1260 μmol/m2 s] and ready for experimental use (Supplementary Fig. 1). Except for H. ammodendron seedlings treated under drought stress, all the other stressed H. ammodendron seedlings were sampled at 6 time points: 0, 4, 8 (or 6), 12, 24, and 72 h after treatments. After each stress treatment, the assimilation branches were cut off from stress treated H. ammodendron seedlings. For mechanical damage treatment, the assimilation branches of H. ammodendron seedlings were cut with a sterilized blade, and preserved immediately. For low temperature treatment, H. ammodendron seedlings were grown at 4 °C with 12/12 h day/night light cycle. During salt treatment, H. ammodendron seedlings were irrigated with 200 mM NaCl. For high temperature treatment, H. ammodendron seedlings were grown at 45 °C with 12/12 h day/night light cycle. For drought treatment, H. ammodendron seedlings were dried by decreasing soil moisture content to 15, 12, 9, 6 or 3% and then assimilation branches were sampled while the assimilation branches sampled at 20% soil moisture content were taken as the control. All assimilation branches were sampled from three different plants as biological replicates. Moreover, all the assimilation branch samples were snap frozen in liquid nitrogen and stored at − 80 °C for total RNA isolation.
Total RNA isolation and cDNA synthesis
200 mg of each assimilation branch sample was collected for total RNA isolation. Total RNA was isolated using Trizol (Life Technologies, USA). RNA concentration and purity were determined by spectrophotometer (Eppendorf, Germany). RNA integrity was confirmed by electrophoresis using 1.0% agarose gel. Total RNA isolated from assimilation branch samples were diluted at a concentration of 1 μg/μL and were used for the qRT-PCR experiments. The first-strand cDNA was synthesized from Dnase-free total RNA using M-MLV reverse transcriptase (Transgen, China).
Sequence retrieving and cloning of the candidate reference genes
Sequence information of 8 candidate genes (18S, ACT1, ACT7, UBC18, TUA5, GAPDH, EF-1α and UBQ10) was retrieved from our H. ammodendron transcriptome data. Based on sequence information, primers (Supplementary Table 1) for cloning full length coding domains were designed using the Primer Premier 5 software. Amplified full length coding domains of candidate genes were confirmed by sequencing. Sequences and accession numbers could be obtained from NCBI database (Table 1).
Table 1.
Details of candidate reference genes, primer sequences for qRT-PCR, products size and amplicon characteristics
| Gene symbol | Gene name | Accession no. | Primers for qRT-PCR (F/R) (5′–3′) | Cellular function | Amplicon length (bp) | Tm (°C) | PCR efficiency | Regression coefficient (R2) | References |
|---|---|---|---|---|---|---|---|---|---|
| ACT1 | Actin 1 | KM886609 | GAACCCCAAGGCTAACAGAGAAAA CTCACACCATCACCAGAGTCAAGA |
Cytoskeleton structure protein | 150 | 85.2 | 1.02 | 0.9988 | Maroufi et al. (2010) |
| 18S | 18S rRNA | KU685539 | CTCTGCCCGTTGCTCTGATGAT CCTTGGATGTGGTAGCCGTTTC |
Ribosomal RNA | 194 | 89.3 | 0.97 | 0.9991 | Amil-Ruiz et al. (2013) |
| TUA5 | Tubulin alpha-5 | KU685537 | CAACTGGCTTCAAATGTGGTATCA ATCTTCACGGGCTTCAGAAAACTC |
Cytoskeleton structure protein | 230 | 87.4 | 0.95 | 0.9960 | Wan et al. (2010) |
| ACT7 | Actin 7 | KU674357 | TTGAACCCCAAGGCTAACAGAG ACGACCAGCAAGATCCAAACGG |
Cytoskeleton structure protein | 222 | 87.2 | 1.05 | 0.9988 | Maroufi et al. (2010) |
| EF-1α | Elongation factors-1A | KU674358 | CGTGAAGGCGACTCGTTAATTG GCTTGGAGACGCTTGACAGGAC |
Translation eukaryotic factor | 190 | 84.2 | 1.14 | 0.9944 | Yuan et al. (2014) |
| GAPDH | 3-Phosphate-glyceraldehyde dehydrogenase | KU674359 | AATGCTATTGGCGGGAAGAGAAG CAAGGATGGAGTCGTATTTGAGG |
Glycolysis and gluconeogenesis | 205 | 86.9 | 1.10 | 0.9952 | Jain et al. (2006) |
| UBC18 | Ubiquitin conjugating enzyme 18 | KU685538 | GATGTCTTCTCATTTCCCCTTC ATCGCCGACTCTAAACCTCTAT |
Protein degradation | 204 | 83.2 | 1.11 | 0.9974 | Ma et al. (2016) |
| UBQ10 | Polyubiquitin 10 | KU674360 | CTGCTTATGGGTTGTCAGTTTTTA GAGCAAAATAGGCACAAACGAAAG |
Protein degradation | 192 | 81.0 | 0.98 | 0.9975 | Chen et al. (2011) |
Primer designing and qRT-PCR
Primers were designed by using Primer Premier 5. All primer sequences are listed in Supplemental Table 1 (for full-length cloning and qRT-PCR) and Table 1 (for qRT-PCR). The amplification efficiency (E) and correlation coefficients (R2) of the primers for qRT-PCR were calculated (Reddy et al. 2015). qRT-PCR reactions were carried out on 7500 Fast (ABI, USA), in 96-well optical reaction plates. Reactions were performed in a total volume of 20 μL, containing 2 μL of cDNA (1:10 dilution), 0.4 μL of each primer (10 mM), 10 μL of 2 × SYBR Select Master Mix (Transgen, China) and made to 20 μL with RNase-free H2O. The qRT-PCR cycling conditions were as follows: 95 °C for 5 min followed by 40 cycles of 15 s at 95 °C, annealing (at each pair of primer’s specific Tm) for 30 s and fluorescent signal recording for 30 s at 72 °C. The melting curve analysis was carried out after 40 cycles to verify the primer specificity by heating from 65 to 95 °C with fluorescence measured within 20 min. The relative expression levels were determined by 2−ΔΔCt method (Livak and Schmittgen 2001) in three biological replicates with three technical repeats.
Data analysis
A threshold fluorescence value of 0.1 was applied to analyze all amplification plots in order to obtain amplification cycle (Cq) values with the SDS version 1.1 software (ABI, USA). The value representing stability and suitability was evaluated respectively using geNorm (Vandesompele et al. 2002), NormFinder (Andersen et al. 2004) and BestKeeper (Pfaffl et al. 2004), as described in relevant reports.
Results
Identification of H. ammodendron reference genes
Eight candidate reference genes (18S, ACT1, ACT7, UBC18, TUA5, GAPDH, EF-1α and UBQ10) were chosen from the H. ammodendron transcriptome data and their full-length open reading frames were cloned and sequenced (Supplementary Fig. 2). Their detailed information, including GenBank accession number and putative function description are listed in Table 1.
For qRT-PCR analysis, total RNAs isolated from plant samples (including biological replicates) exhibited high quality. The A260/A280 ratios of extracted RNA samples ranged from 1.95 to 2.12, indicating that RNA samples were pure and protein-free. The amplification efficiencies (E), correlation coefficients (R2) and the Tm values of all amplification products are listed in Table 1. The specificity of primers had been confirmed by agarose gel electrophoresis and melting curve analysis (Supplementary Fig. 3). The amplification efficiencies (E) of reference genes ranged from 0.95 (TUA5) to 1.11 (UBC18). The correlation coefficient (R2) values varied from 0.9944 (EF-1α) to 0.9991 (18S) (Table 1). These results show that all primer pairs designed for qRT-PCR analysis were suitable.
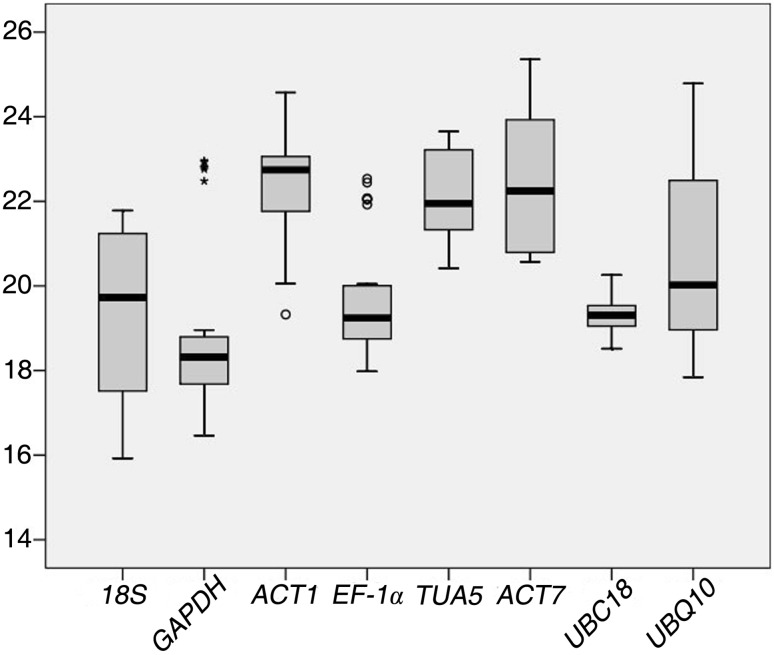
The results of expression stability validation showed that ACT7 had the highest mean Cq value (22.55) in all samples treated under stress (drought, heat, salt, cold and mechanical damage). On the other hand, the lowest mean Cq value was observed for GAPDH, followed by 18S and UBC18 (18.93, 19.28 and 19.32, respectively). Additionally, the most varied expression levels were observed for 18S, followed by UBQ10 and ACT7, as shown by the larger whisker taps and boxes comparing with the other genes. The least variation in expression levels was observed in UBC18, followed by EF-1α and GAPDH (Fig. 1), These results showed none of the candidate reference genes had constant expression levels across all the samples under the above-mentioned stresses (drought, heat, salt, cold and mechanical damage). Therefore, it is of necessity to validate suitable reference gene(s) for accurate normalization of gene expression under different stress conditions in H. ammodendron.
Fig. 1.
Expression levels of candidate reference genes among all samples: Lines across the boxes depict the medians. Boxes indicate the interquartile range. Whiskers represent 95% confidence intervals; Stars and circles indicate the presence of outliers
Expression stability of the candidate reference genes
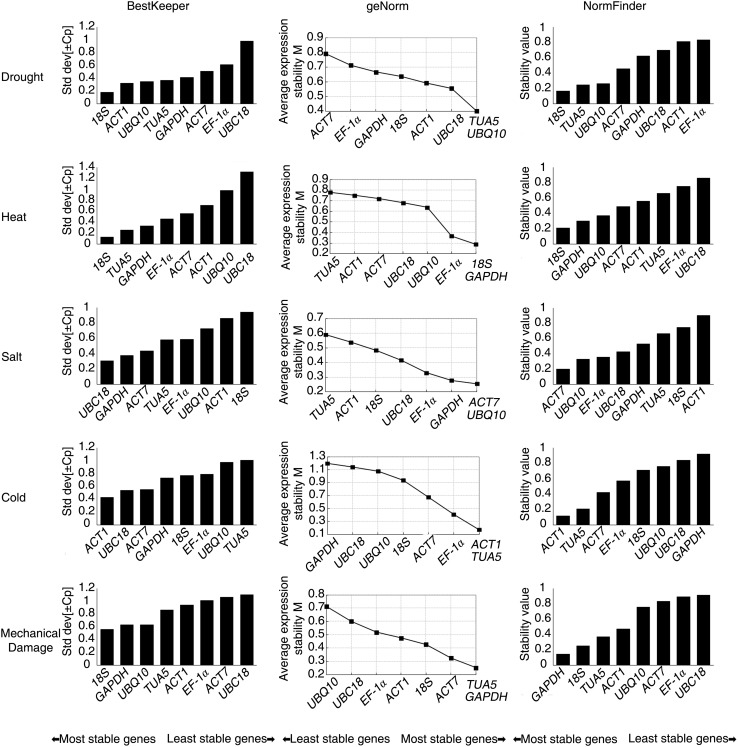
Tools are available to evaluate the expression stability of reference gene, such as geNorm (Vandesompele et al. 2002), NormFinder (Andersen et al. 2004) and BestKeeper (Pfaffl et al. 2004). In each sample subset, the 8 reference genes were ranked from the most stable to the least stable (Fig. 2). According to Bestkeeper, 18S was considered the most suitable gene under drought, heat stress and mechanical damage. Meanwhile, UBC18 and ACT1 were considered the most suitable genes under cold stress and salt stress. NormFinder analysis showed that 18S was the most suitable gene under drought and heat stress. ACT7, ACT1 and GAPDH were considered being stable under salt stress, cold stress and mechanical damage, respectively. Overall, the results shown in NormFinder were similar to those found in BestKeeper (Table 2). GeNorm analysis showed that the expression profiles of TUA5 and UBQ10 were the most constant under drought stress, 18S under heat stress, ACT7 and UBQ10 under salt stress, ACT1 and TUA5 under cold stress, and TUA5 and GAPDH under mechanical damage stress.
Fig. 2.
Expression stability of candidate reference genes in various stresses as calculated by BestKeeper, geNorm and NormFinder
Table 2.
Expression stability ranks of 8 candidate reference genes in different sets of stressed samples calculated using BestKeeper (BK), geNorm (GN) and NormFinder (NF) methods
| Gene | Drought | Heat | Salt | Cold | Mechanical damage | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BK | GN | NF | BK | GN | NF | BK | GN | NF | BK | GN | NF | BK | GN | NF | |
| 18S | 1 | 5 | 1 | 1 | 1 | 1 | 8 | 6 | 7 | 5 | 5 | 5 | 1 | 4 | 2 |
| ACT1 | 2 | 4 | 7 | 6 | 7 | 5 | 7 | 7 | 8 | 1 | 1 | 1 | 5 | 5 | 4 |
| UBQ10 | 3 | 2 | 3 | 7 | 4 | 3 | 6 | 2 | 2 | 7 | 6 | 6 | 3 | 8 | 5 |
| TUA5 | 4 | 1 | 2 | 2 | 8 | 6 | 4 | 8 | 6 | 8 | 2 | 2 | 4 | 1 | 3 |
| GAPDH | 5 | 6 | 5 | 3 | 2 | 2 | 2 | 3 | 5 | 4 | 8 | 8 | 2 | 2 | 1 |
| ACT7 | 6 | 8 | 4 | 5 | 6 | 4 | 3 | 1 | 1 | 3 | 4 | 3 | 7 | 3 | 6 |
| EF-1A | 7 | 7 | 8 | 4 | 3 | 7 | 5 | 4 | 3 | 6 | 3 | 4 | 6 | 6 | 7 |
| UBC18 | 8 | 3 | 6 | 8 | 5 | 8 | 1 | 5 | 4 | 2 | 7 | 7 | 8 | 7 | 8 |
Based on the expression stability analysis of candidate genes with algorithms geNorm, NormFinder and BestKeeper, we concluded the most stable genes under different stress conditions as follows, 18S and TUA5 under drought stress, 18S under heat stress, ACT7 and UBC18 under salt stress, ACT1 under cold stress, 18S, TUA5 and GAPDH under mechanical damage (Table 3).
Table 3.
Top 2 stable genes ranked by BestKeeper, geNorm and NormFinder
| Experimental sample sets | BestKeeper | geNorm | NormFinder |
|---|---|---|---|
| Drought | 18S | TUA5 | 18S |
| ACT1 | UBQ10 | TUA5 | |
| Heat | 18S | 18S | 18S |
| TUA5 | GAPDH | GAPDH | |
| Salt | UBC18 | ACT7 | ACT7 |
| GAPDH | UBQ10 | UBQ10 | |
| Cold | ACT1 | ACT1 | ACT1 |
| UBC18 | TUA5 | TUA5 | |
| Mechanical damage | 18S | TUA5 | GAPDH |
| GAPDH | GAPDH | 18S |
Optimal number of candidate genes for normalization
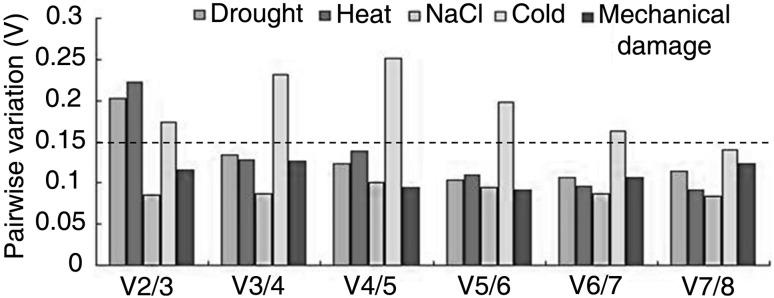
The optimal number of reference genes was determined by geNorm, which uses the step wise calculation of the pairwise variation (Vn/Vn + 1) between two sequential normalization factors with 0.15 as the cut-off value (Vandesompele et al. 2002). As shown in Fig. 3, the V2/3 value was lower than 0.15, indicating two reference genes were sufficient for normalizing gene expression under salt stress and mechanical damage. Three reference genes were sufficient for qRT-PCR analysis under drought and heat stress because the V2/3 value was higher than 0.15 and the V3/4 value lower than 0.15. Seven reference genes were needed for qRT-PCR analysis under cold stress because the V6/7 value was higher than 0.15 and the V7/8 value lower than 0.15.
Fig. 3.
Determination of the optimal number of reference genes for geNorm analysis. The pairwise variation (Vn/Vn + 1) was analyzed for the normalization factors NFn and NFn + 1 by geNorm program to determine (V < 0.15) the optimal number of reference genes. Threshold value (0.15) is displayed with a dash line
Validation of reference genes
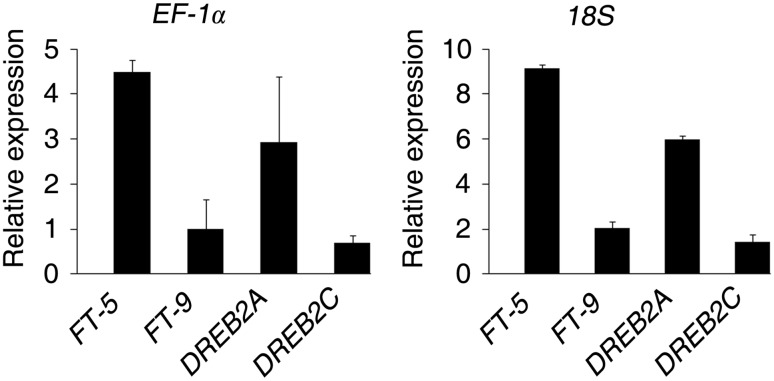
The expression of DREB genes was induced under drought stress in several plants such as Arabidopsis (Ding et al. 2013), rice (Oono et al. 2014), wheat (Shavrukov et al. 2016) and moso bamboo (Wu et al. 2015). Abiotic stresses also induce the expression of FT gene (Ryu et al. 2011, Manoharan et al. 2016), which regulates flower development and stomatal movement. Therefore, we evaluated relative expression level of FT-5, FT-9, DREB2A and DREB2C in H. ammodendron subjected to drought stress in order to determine the impact of different reference genes on normalization. According to NormFinder, the most stable gene (18S) and the least stable one (EF-1α) under drought stress were used as the reference gene control.
As shown in Fig. 4, when 18S gene is used for normalization, relative expression level of FT-5, FT-9, DREB2A and DREB2C were obviously high in comparison to EF-1α indicating that unsuitable reference gene led to different relative expression profiles. Moreover, these results indicate the importance of validating reference genes before applying them in experiments.
Fig. 4.
Expression profiles of FT-5, FT-9, DREB2A and DREB2C in H. ammodendron under drought stress. The best stable gene (18S) and the least stable reference gene (EF-1α) were used to normalize the expression data. Samples from the control treatment (0 h) were used as reference samples. Stressed samples were collected in 6 h after treatment. The relative expression of each gene was calculated as the mean fold change compared with the control samples in each experiment. Three independent biological samples with three technical replicates were used. At least three plants per treatment were used in each sample
Discussion
Haloxylon ammodendron, a perennial shrub in genus of Haloxylon Bunge (Chenopodiaceae), is distributed in northwest China and plays an important role in desert vegetation restoration. H. ammodendorn adapts to the harsh desert environment with its ability of efficient water utilization and adversity resistance (Yu et al. 2012). By using transcriptome technology, many genes related to environmental stress response have been explored in H. ammodendorn (Long et al. 2014). In order to understand the biological processes under environmental stresses, it is important to understand gene expression patterns. Among the several methods that help determining gene expression levels, qRT-PCR is the simplest one of high validity, accuracy and sensitivity. In addition, selection of suitable reference genes is vital to obtain reliable and accurate data for analyzing qRT-PCR, and the ideal reference genes should have relatively stable expression, irrespective of the types or physiological conditions of the sample (Crismani et al. 2006). Although, it is critical to highlight expression stability of reference genes, there is no single reference gene that has a stable expression under all conditions (Czechowski et al. 2005, Radonić et al. 2004; Wei et al. 2013). Therefore, the suitability of reference genes have a great influence on the reliability of the results.
In this study, we cloned 8 reference genes (18S, GAPDH, ACT1, EF-1Α, TUA5, ACT7, UBC18 and UBQ10) in H. ammodendorn and validated their expression stabilities. As we concluded, in H. ammodendron under drought stress, the most stable gene(s) were 18S and TUA5, under heat stress, 18S, under salt stress, ACT7 and UBC18, under cold stress, ACT1, and under mechanical damage 18S, TUA5 and GAPDH (Table 3). It is worth noting that much varied expression levels were observed for 18S and ACT7, as shown by the larger whisker taps and boxes comparing with the other genes (Fig. 1), indicating expression levels of 18S and ACT7 were highly unstable across all types of stress (drought, heat, salt, cold and mechanical damage). In other words, although expression levels of 18S and ACT7 were seriously affected by some types of stress (for 18S, salt and cold stresses; for ACT7, drought, heat, cold and mechanical damage stresses), 18S was still the optimal reference gene under drought, heat and mechanical damage stresses, while ACT7 was the optimal one in H. ammodendorn under salt and cold stresses as concluded above. These results showed that among the 8 reference genes tested in this study, there was no ideal reference gene(s) suitable to all stress conditions, each reference gene was only applicable to one or several stress conditions. Notably, a suitable reference gene should show a constant expression level in all samples under the same stress condition (Piron Prunier et al. 2016).
Moreover, previous research in soybean proved that 18S was not suitable as a reference gene under stresses due to its extremely high abundance. But 18S was one of the most suitable reference genes to study somatic embryo genesis or stimuli reaction of longan (Lin and Lai 2010; Wu et al. 2016) and papaya (Zhu et al. 2012). Under most other conditions, 18S was either one of the most or least stable genes (Wu et al. 2016). In this study, 18S ranked as the most stable gene under drought, heat stress or mechanical damage (Tables 2, 3), suggesting its applicability as a reference gene under these abiotic stresses in H. ammodendron. In addition, 18S gene has an expression level dramatically higher than other reference genes (Fig. 1). Wu et al. (2016) concluded that 18S was not an appropriate reference gene for normalization in qRT-PCR studies involving target genes of mid-or low-level expression. Therefore, 18S is suitable for normalization of high-level gene expression under drought, heat stress and mechanical damage in H. ammodendron.
Among all tested reference genes, GAPDH attracts specific attention because it ranked first or second as the most stable gene with three algorithms BestKeeper, geNorm and NormFinder (Tables 2, 3). Furthermore, GAPDH had less variation in expression levels than 18S, suggesting its higher stability. Hence GAPDH should be an appropriate reference gene in substitution for 18S for normalization in qRT-PCR studies involving target genes with mid-or low-level expression in H. ammodendron in the future. H. ammodendorn, the main pioneer plant and constructive species in desert areas, can tolerant high desert surface layer temperature (HDSLT) (more than 95 °C) (Yu et al. 2012). However, the ability of tolerance to environmental stresses such as HSLT stress is much poor in H. ammodendron seedlings, suggesting HDSLT stress tolerance is acquired by heat acclimation in H. ammodendron, and its molecular mechanism has not been reported. 18S and GAPDH will become the top two suitable reference genes for normalization in qRT-PCR concerning HSLT stress tolerance in H. ammodendorn.
Many evidences have shown that greater accuracy of qRT-PCR experiments in plants involves the application of more than one reference gene (Vandesompele et al. 2002; Le et al. 2012). As for the geNorm program, minimal reference genes are applied to calculate the pairwise variation Vn/Vn + 1 between two sequential normalization factors, in order to determine the necessity of adding other reference genes. A large variation indicates the necessity of adding another reference gene for calculation for more accurate normalization (Vandesompele et al. 2002). According to this criterion, the calculation result of V2/3 value is lower 0.15, which is the cut-off value subject to the subset of salt and damage stresses, suggesting that no more genes are needed to normalize gene expression in these samples (Fig. 3). However, three reference genes are suggested when performing qRT-PCR analysis using drought and heat stressed samples (Fig. 3). In another sample of the V6/7 value under cold stress, even though the value reaches the cut-off value of 0.15, the use of seven reference genes may be reconsidered for cost effectiveness (Fig. 3).
In conclusion, in this study we evaluated 8 candidate reference genes for the normalization of gene expression in H. ammodendron under drought, heat, salt, cold and mechanical damage stresses. The results suggested different sets of reference genes should be applied depending on stress conditions. The expression profiles of FT-5, FT-9, DREB2A and DREB2C confirmed the importance of validating reference genes in order to obtain accurate qRT-PCR results. This study will contribute to the accuracy and reliability in transcripts quantification, which provides the significance to transcription-based studies and applications in this important shrub H. ammodendron and other close relatives of the Haloxylon.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Phenotypic characteristics of one-year old seedlings of H. ammodendron. (a) General view of an untreated one-year old seedling growing in a tube filled with sand. (b) Assimilation branches separated from an untreated one-year old seedlings (Bar = 2 cm) (JPEG 218 kb)
Amplification of full length coding domains of ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10. M:DL 2000 DNA marker. Unit: bp (JPEG 722 kb)
Specificity of qRT-PCR amplicons. (a) 1.8% agarose gel electrophoresis showing amplification of a single product at the expected size for each reference gene. M represents 100 bp and 250 bp DNA Ladder. Lane 1-8 indicate ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10, respectively. (b) Dissociation curves with single peak were generated from all amplicons (JPEG 1060 kb)
Primers for cloning full length coding domains (ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10) and qRT-PCR (FT-5, FT-9, DREB2A and DREB2C) (DOCX 39 kb)
Funding
This work was funded by the National Natural Science Foundation of China (No. 31260181).
Compliance with ethical standards
Conflict of interest
The authors have no conflicts of interest to declare.
Footnotes
Bo Wang and Huihui Du have contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1007/s12298-018-0520-9) contains supplementary material, which is available to authorized users.
Contributor Information
Hua Zhang, Phone: +86 25 84395324, Email: hazelzhang@163.com.
Hao Ma, Phone: +86 25 84395324, Email: Lq-ncsi@njau.edu.cn.
References
- Amil-Ruiz F, Garrido-Gala J, Blanco-Portales R, Folta KM, Muñoz-Blanco J, Caballero JL. Identification and validation of reference genes for transcript normalization in strawberry (Fragaria × ananassa) defense responses. PLoS ONE. 2013;8:e70603. doi: 10.1371/journal.pone.0070603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Can Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bao W, Qu Y, Shan X, Wan Y. Screening and validation of housekeeping genes of the root and cotyledon of Cunninghamia lanceolata under abiotic stresses by using quantitative real-time PCR. Int J Mol Sci. 2016;17:1198. doi: 10.3390/ijms17081198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RTPCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Chen IC, Hill JK, Ohlemüller R, Roy DB, Thomas CD. Rapid range shifts of species associated with high levels of climate warming. Science. 2011;333:1024–1026. doi: 10.1126/science.1206432. [DOI] [PubMed] [Google Scholar]
- Crismani W, Baumann U, Sutton T, Shirley N, Webster T, Spangenberg G, Langridge P, Able JA. Microarray expression analysis of meiosis and microsporogenesis in hexaploid bread wheat. BMC Genomics. 2006;7:267. doi: 10.1186/1471-2164-7-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derveaux S, Vandesompele J, Hellemens J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Ding Y, Liu N, Virlouvet L, Riethoven JJ, Fromm M, Avramova Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013;13:229. doi: 10.1186/1471-2229-13-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Le DT, Aldrich DL, Valliyodan B, Watanabe Y, Ha CV, Nishiyama R, Guttikonda SK, Quach TN, Gutierrez-Gonzalez JJ, Tran LS, Nguyen HT. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS ONE. 2012;7:e46487. doi: 10.1371/journal.pone.0046487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang L, Zhang Y, Wang G, Song D, Zhang Y. Selection and validation of appropriate reference genes for quantitative real-time PCR normalization in staminate and perfect flowers of andromonoecious Taihangia rupestris. Front Plant Sci. 2017;8:729. doi: 10.3389/fpls.2017.00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Lai ZX. Reference genes selection for qPCR analysis during somatic embryo genesis in longan tree. Plant Sci. 2010;178:359–365. doi: 10.1016/j.plantsci.2010.02.005. [DOI] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long Y, Zhang J, Tian X, Wu S, Zhang Q, Zhang J, Dang Z, Pei XW. De novo assembly of the desert tree Haloxylon ammodendron (C. A. Mey.) based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genomics. 2014;15:1111. doi: 10.1186/1471-2164-15-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R, Xu S, Zhao Y, Xia B, Wang R. Selection and validation of appropriate reference genes for quantitative Real-Time PCR Analysis of gene expression in Lycoris aurea. Front Plant Sci. 2016;7:536. doi: 10.3389/fpls.2016.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan RK, Han JS, Vijayakumar H, Subramani B, Thamilarasan SK, Park JI, Nou IS. Molecular and functional characterization of FLOWERING LOCUS T homologs in Allium cepa. Molecules. 2016;21:217. doi: 10.3390/molecules21020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroufi A, Van Bockstaele E, De Loose M. Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol. 2010;11:15. doi: 10.1186/1471-2199-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Yazawa T, Kawahara Y, Kanamori H, Kobayashi F, Sasaki H, Mori S, Wu J, Handa H, Itoh T, Matsumoto T. Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice. PLoS ONE. 2014;9:e96946. doi: 10.1371/journal.pone.0096946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez LJ, Rios L, Trivedi P, D’Souza K, Cowie A, Nzirorera C, Webster D, Brunt K, Legare JF, Hassan A, Kienesberger PC, Pulinilkunnil T. Validation of optimal reference genes for quantitative real time PCR in muscle and adipose tissue for obesity and diabetes research. Sci Rep. 2017;7:3612. doi: 10.1038/s41598-017-03730-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: Bestkeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Piron Prunier F, Chouteau M, Whibley A, Joron M, Llaurens V. Selection of valid reference genes for reverse transcription quantitative PCR analysis in Heliconius numata (Lepidoptera: Nymphalidae) J Insect Sci. 2016;16:50. doi: 10.1093/jisesa/iew034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyankov VI, Black CC, Jr, Artyusheva EG, Voznesenskaya EV, Ku MSB, Edwards GE. Features of photosynthesis in Haloxylon species of Chenopo-diaceae that are dominant plants in Central Asia desert. Plant Cell Physiol. 1999;40:125–134. doi: 10.1093/oxfordjournals.pcp.a029519. [DOI] [Google Scholar]
- Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Reddy PS, Reddy DS, Sharma KK, Bhatnagar-Mathur P, Vadez V. Cloning and validation of reference genes for normalization of gene expression studies in pearl millet [Pennisetum glaucum (L.) R. Br.] by quantitative real-time PCR. Plant Gene. 2015;1:35–42. doi: 10.1016/j.plgene.2015.02.001. [DOI] [Google Scholar]
- Ryu JY, Park CM, Seo PJ. The floral repressor BROTHER OF FT AND TFL1 (BFT) modulates flowering initiation under high salinity in Arabidopsis. Mol Cells. 2011;32:295–303. doi: 10.1007/s10059-011-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavrukov Y, Baho M, Lopato S, Langridge P. The TaDREB3 transgene transferred by conventional crossings to different genetic backgrounds of bread wheat improves drought tolerance. Plant Biotechnol J. 2016;14:313–322. doi: 10.1111/pbi.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Zheng W, Pei K, Ma K. Genetic variation within and among populations of a dominant desert tree Haloxylon ammodendron (Amaranthaceae) in China. Ann Bot. 2005;96:245–252. doi: 10.1093/aob/mci171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov RA, Panchuk II, Schöffl F. Heat-stress-dependency and developmental modulation of gene expression: the potential of house-keeping genes as internal standards in mRNA expression profiling using real-time RT-PCR. J Exp Bot. 2003;54:2343–2349. doi: 10.1093/jxb/erg244. [DOI] [PubMed] [Google Scholar]
- Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399:257–261. doi: 10.1016/j.ab.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Wei L, Miao H, Zhao R, Han X, Zhang T, Zhang H. Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR. Planta. 2013;237:873–889. doi: 10.1007/s00425-012-1805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Lv H, Li L, Liu J, Mu S, Li X, Gao J. Genome-wide analysis of the AP2/ERF transcription factors family and the expression patterns of DREB genes in Moso Bamboo (Phyllostachys edulis) PLoS ONE. 2015;10:e0126657. doi: 10.1371/journal.pone.0126657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhang H, Liu L, Li W, Wei Y, Shi S. Validation of reference genes for RT-qPCR studies of gene expression in preharvest and postharvest Longan Fruits under different experimental conditions. Front Plant Sci. 2016;7:780. doi: 10.3389/fpls.2016.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Ren C, Zhang J, He X, Ma L, Chen Q, Qu Y, Shi S, Zhang H, Ma H. Effect of high desert surface layer temperature stress on Haloxylon ammodendron (C.A. Mey.) Bunge. Flora. 2012;207:572–580. doi: 10.1016/j.flora.2012.06.008. [DOI] [Google Scholar]
- Yuan M, Lu Y, Zhu X, Wan H, Shakeel M, Zhan S, Jin BR, Li J. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS ONE. 2014;9:e86503. doi: 10.1371/journal.pone.0086503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zeng Y, Yi X, Zhang Y. Selection of suitable reference genes for quantitative RT-PCR normalization in the halophyte Halostachys caspica under salt and drought stress. Sci Rep. 2016;6:30363. doi: 10.1038/srep30363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Li X, Chen W, Chen J, Lu W, Chen L, Fu D. Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE. 2012;7:e44405. doi: 10.1371/journal.pone.0044405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotypic characteristics of one-year old seedlings of H. ammodendron. (a) General view of an untreated one-year old seedling growing in a tube filled with sand. (b) Assimilation branches separated from an untreated one-year old seedlings (Bar = 2 cm) (JPEG 218 kb)
Amplification of full length coding domains of ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10. M:DL 2000 DNA marker. Unit: bp (JPEG 722 kb)
Specificity of qRT-PCR amplicons. (a) 1.8% agarose gel electrophoresis showing amplification of a single product at the expected size for each reference gene. M represents 100 bp and 250 bp DNA Ladder. Lane 1-8 indicate ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10, respectively. (b) Dissociation curves with single peak were generated from all amplicons (JPEG 1060 kb)
Primers for cloning full length coding domains (ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10) and qRT-PCR (FT-5, FT-9, DREB2A and DREB2C) (DOCX 39 kb)