Abstract
Bixa orellana L., popularly known as annatto, produces several secondary metabolites of pharmaceutical and industrial interest, including bixin, whose molecular basis of biosynthesis remain to be determined. Gene expression analysis by quantitative real-time PCR (qPCR) is an important tool to advance such knowledge. However, correct interpretation of qPCR data requires the use of suitable reference genes in order to reduce experimental variations. In the present study, we have selected four different candidates for reference genes in B. orellana, coding for 40S ribosomal protein S9 (RPS9), histone H4 (H4), 60S ribosomal protein L38 (RPL38) and 18S ribosomal RNA (18SrRNA). Their expression stabilities in different tissues (e.g. flower buds, flowers, leaves and seeds at different developmental stages) were analyzed using five statistical tools (NormFinder, geNorm, BestKeeper, ΔCt method and RefFinder). The results indicated that RPL38 is the most stable gene in different tissues and stages of seed development and 18SrRNA is the most unstable among the analyzed genes. In order to validate the candidate reference genes, we have analyzed the relative expression of a target gene coding for carotenoid cleavage dioxygenase 1 (CCD1) using the stable RPL38 and the least stable gene, 18SrRNA, for normalization of the qPCR data. The results demonstrated significant differences in the interpretation of the CCD1 gene expression data, depending on the reference gene used, reinforcing the importance of the correct selection of reference genes for normalization.
Electronic supplementary material
The online version of this article (10.1007/s12298-018-0528-1) contains supplementary material, which is available to authorized users.
Keywords: Normalizer genes, Endogenous genes, Quantitative real time, Stability, Gene expression
Introduction
Bixa orellana, popularly known as annatto, achiote or urucu, is native to tropical America and belongs to the Bixaceae family (Moreira et al. 2015). This plant species is economically and culturally important since it is the sole source of the pigment bixin, a dicarboxylic monomethyl ester apocarotenoid that confers red color to seeds and it is widely used as a traditional dye and as an additive in several industries (Giuliano et al. 2003). In the food industry, annatto dye is used for colouring an assortment of foods, including pastas, cheeses, sausages, snacks and ice creams. Due to its color spectra obtained from intense red and low toxicity, annatto is the second most consumed among natural dyes, after caramel, and is comparatively inexpensive (Mercadante and Pfander 1998). Another important market for annatto dye is the condiment industry for the manufacture of colorants, spices and other products used in gastronomy (Mercadante et al. 1997). Annatto is also used for body care products (e.g. creams, lotions and shampoos) in the cosmetics industry and for dyeing leather (Giuliano et al. 2003). In addition to bixin, other secondary metabolites produced by annatto include several phenolic compounds, in particular flavonoids, which together with bixin confer antioxidant potential to annatto seeds (Moreira et al. 2014).
Due to the importance of bixin, several studies have investigated the expression of genes that are potentially involved in the biosynthesis of carotenoids and bixin, using RT-PCR and qPCR techniques (Soares et al. 2011; Rodríguez-Ávila et al. 2011a, b; Cárdenas-Conejo et al. 2015). The qPCR reaction is sensitive, accurate and yields rapid results, representing a favorite technique for analyzing gene expression in different organisms, tissues and experimental conditions (Derveaux et al. 2010) and for elucidating complex gene networks that control the metabolic pathways (Kanakachari et al. 2016). However, the accuracy of the results generated by qPCR depends on the precise normalization of the transcripts using stably expressed reference genes, which in turn allows the elimination of possible non-biological variations and experimental errors (Dheda et al. 2004). The use of non-stable reference genes may compromise the analysis of the target genes normalized by them (Gutierrez et al. 2008).
Reference genes, also known as endogenous or normalizer genes, are believed to be those with constant expression in all tissues of the organism, at any stage of development, or undergoing any type of treatment. However, this universalization does not seem to exist completely in practice, which generates the need to verify the stability among different tissues and treatments under evaluation in the qPCR (Andersen et al. 2004).
In light of the considerations above, it is necessary to validate reference genes in order to increase the reliability of the expression data of target genes. The reference genes commonly used in plants are those coding for actin, ubiquitin, tubulin, elongation factors, 18S ribosomal RNA (18SrRNA), 60S ribosomal proteins (RPLs) and 40S ribosomal proteins (RPSs) (Gutierrez et al. 2008; Borowski et al. 2014). In B. orellana, gene coding for 18SrRNA has been the only reference gene used for normalizing gene expression data (Soares et al. 2011; Rodríguez-Ávila et al. 2011a, b; Cárdenas-Conejo et al. 2015). However, no studies published to date have examined the stability of this reference gene, as well as other candidates for reference genes in this plant species. The selection of reference genes will benefit future studies of gene expression in B. orellana.
Various statistical algorithms are able to evaluate the stability of expression of reference genes in a group of samples. Some of the most commonly used algorithms include NormFinder (Andersen et al. 2004), geNorm (Vandesompele et al. 2002), BestKeeper (Pfaffl et al. 2004), ΔCt method (Silver et al. 2006) and RefFinder (Xie et al. 2012). They select the most appropriate gene or gene combination based on the calculation of expression stability for normalization. The objective of this study was to analyze the expression stability of four candidate reference genes in B. orellana, by calculating their stability and validating their reliability to normalize the expression profiles of a target gene coding for carotenoid cleavage dioxygenase 1 (CCD1), potentially involved in the biosynthesis of bixin. The results provide information for the proper choice of reference genes in B. orellana.
Materials and methods
Plant material
Flower buds, flowers, mature leaves and immature seeds at different stages of development of two biological replicates of annatto plants (Bixa orellana L., var. Embrapa 37) were harvested in the Sempre Viva Farm, located at BR-367, in the municipality of Porto Seguro (Bahia, Brazil). Immature seeds at four different developmental stages were collected based on the fruit capsule diameter (mm)/length (mm), as follows: S1 (1st stage of seed development; 7.3/9.1), S2 (2nd stage of seed development; 10.6/13.6), S3 (3rd stage of seed development; 14.1/22.2) and S4 (4th stage of seed development; 19.1/31.9). All samples were collected and immediately submerged in liquid nitrogen (N2) and maintained at − 80 °C for subsequent analysis.
RNA isolation and cDNA synthesis
Total RNA was isolated using the RNAqueous® kit (Ambion Inc., Austin, TX, USA), together with the Plant RNA Isolation Aid reagent (Ambion). Samples were subjected to ultrasonication (2 pulses of 5 s each, amplitude of 70%, with intervals of 10 s) in an ultrasonic processor (Ultrasonic processor Gex 130, Cole-Parmer, Vernon Hill, IL, USA), during the process of RNA isolation. DNA residues were eliminated using the Turbo DNA-free™ kit (Ambion). All of these procedures followed the manufacturer’s instructions. The integrity of RNA was analyzed on a GelRed™-stained 1% agarose gel. RNA concentration was quantified using the Qubit® RNA BR assay kit by Qubit® fluorometer (Life Technologies, Grand Island, NY, USA). The RNA purity was confirmed by the ratio of the readings at 260 and 280 nm in the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). The concentration and quality of total RNA used for cDNA synthesis are shown in Table S1. First-strand cDNA was synthesized using the First Strand cDNA Synthesis kit (Fermentas, Waltham, MA, USA), according to the manufacturer’s instructions. The cDNA concentration was quantified in the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), diluted to a concentration of 10 ng μL−1 and stored at − 20 °C until further use.
Selection of candidate reference genes and primer design
Candidate reference genes coding for 40S ribosomal protein S9 (RPS9), histone H4 (H4), 60S ribosomal protein L38 (RPL38) and 18S ribosomal RNA (18SrRNA) were selected from an EST database of B. orellana (Soares et al. 2011). The candidates for reference genes in B. orellana were selected based on their reported stabilities in other expression studies in different plant species (Kundu et al. 2013; Figueiredo et al. 2013; Wei et al. 2013; Kanakachari et al. 2016; Shivhare and Lata 2016). 18SrRNA, RPS9 and RPL38 are involved in the formation of the ribosome structure, which in turn plays an essential role in the synthesis of proteins. In eukaryotes, the ribosomes consist of a structure formed by ribosomal RNA (rRNA) and proteins arranged in two subunits, the large (60S) one and the small (40S) one. The 40S subunit is composed of the 18S rRNA and 33 ribosomal proteins, among them RPS9. This subunit has a region over which the tRNA pairs with complementary three-base codon in the mRNA. The 60S subunit is composed of 5S, 28S and 5.8S rRNAs and 49 ribosomal proteins, among them RPL38. This subunit is responsible for catalyzing the peptide bonds between the amino acids. On the other hand, H4-type histone is able to bind to DNA to form the nucleosome, a basic unit of DNA packaging in eukaryotes. This structure is composed of eight histone proteins, two of each H2A, H2B, H3 and H4, forming a core of histones around which DNA is wrapped, besides the histone H1 that links the histone cores helping to compact the nucleosomes.
Their cDNA sequences and the respective primer annealing regions are shown in Table S2. The target gene used in the validation of the candidate reference genes was a dioxygenase cleavage of carotenoid type 1 (BoCCD1) of B. orellana, selected from the NCBI database (accession # EF493214.1). Primers were designed using the Primer3 website (version 0.4.0; available from http://bioinfo.ut.ee/primer3–0.4.0/) and their sequences and amplicon characteristics are shown in Table 1.
Table 1.
Candidate reference and target genes, primer and amplicon characteristics, and amplification efficiency (E) obtained by qPCR
| Gene | Description | Primer sequences (5′- 3′) | Amplicon (bp) | E (%) | Tm (°C) |
|---|---|---|---|---|---|
| H4 | Histone class H4 | F-TGACAGCATCGCGAATCACA R-GTGAAGAGAATCAGCGGCCT |
85 | 85.47 | 57.1 57.3 |
| RPS9 | 40S ribosomal protein S9 | F-ACTTCTCTCTCACCAGTCCGT R-GCCTTCTTCATGGAGGCCTT |
82 | 84.24 | 57.3 57.6 |
| RPL38 | 60S ribosomal protein L38 | F-ACTTCCTTCTCACCGCCAGA R-CAGCGGACCTTGAACTTGAC |
85 | 83.77 | 58.3 57.2 |
| 18SrRNA | 18S ribosomal RNA | F-CGCAACACCGGCATAAGG R-CCGATCGGCCATTTTGG |
55 | 81.01 | 57.0 57.2 |
| CCD1 | Carotenoid cleavage dioxygenase type 1 | F-CAGGGAAAACAAGGATCGAA R-AGGCACAAAAACAGCCTCTG |
92 | 82.70 | 52.7 56.0 |
F forward, R reverse
Quantitative real-time PCR (qPCR) and statistical analysis
All the qPCR reactions were run on a Stratagene Mx 3005P apparatus (Agilent Technologies, Santa Clara, CA, USA) using the 5 × HOT FIREPol® EvaGreen® qPCR Supermix kit (Solis BioDyne, Tartu, Estonia), according to the manufacturer’s instructions. The qPCR reactions were carried out in triplicate for two biological replicates, in a reaction volume of 20 μL [4 μL syber green, 1 μL (5 mM) each primer, 5 μL sterile Milli-Q water and 10 μL (10 ng) cDNA sample]. The amplification conditions were performed using the following steps: activation of Taq DNA Polymerase at 95 °C for 12 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s and extension at 72 °C for 30 s. The non-template control (NTC) reactions were included in the analysis in triplicate. In total, 180 cDNA samples were used in the qPCR analysis.
For the purpose of validation of the reference genes, the comparative cycle threshold (Ct; 2−ΔΔCt) method was applied to calculate the fold-change of the BoCCD1 target gene in technical triplicates and two biological replicates. Statistical analysis of gene expression was carried out using the BioEstat 5.3 software (Universidade Federal do Pará, Brazil) and the statistical differences were assessed based on the analysis of variance (ANOVA) and means separation by the Student’s t test, with a critical value of P ≤ 0.05. The amplification efficiency (E) of each primer was calculated using the Miner 2.2 software (Zhao and Fernald 2005).
Expression stability analyses for the reference genes
Expression stability of the candidate reference genes was analyzed using NormFinder, geNorm, BestKeeper, ΔCt method and RefFinder. NormFinder evaluates a set of candidate genes for normalization in qPCR from raw expression data. This program analyzes the stability of expression globally, as well as intragroup. The gene or combination of genes with the lowest stability value is the most suitable for normalization (Andersen et al. 2004). The geNorm calculates an expression stability measure (M) for each candidate gene in a given set of samples, and selects these genes by progressively eliminating less stable genes from the analysis; the reference gene with the least variation is the most stable. In addition, the software indicates the minimum number of genes required for normalization of the data, evaluating the pairwise variation (Vn/n + 1) between normalization factors (NFn and NFn + 1, n ≥ 2). The value Vn/n + 1 below 0.15 suggests that an additional reference gene is not required for normalization (Vandesompele et al. 2002). BestKeeper ranks the stability of the candidates on the basis of the Ct standard deviation (SD) of all samples for a given reference gene, as well as their coefficient of variation (CV). The smaller these values, the more stable expression of candidate reference gene (Pfaffl et al. 2004). The ΔCt method was employed by comparing relative expression of pairs of genes within each sample. The Ct variation of these genes implies their stability (Silver et al. 2006). RefFinder (https://omictools.com/reffinder-tool) is a comprehensive reference gene classification tool based on different software programs, including NormFinder, GeNorm, BesKeeper and ΔCt method (Xie et al. 2012).
Results
Candidate reference genes selection
Expression analysis of candidate reference genes
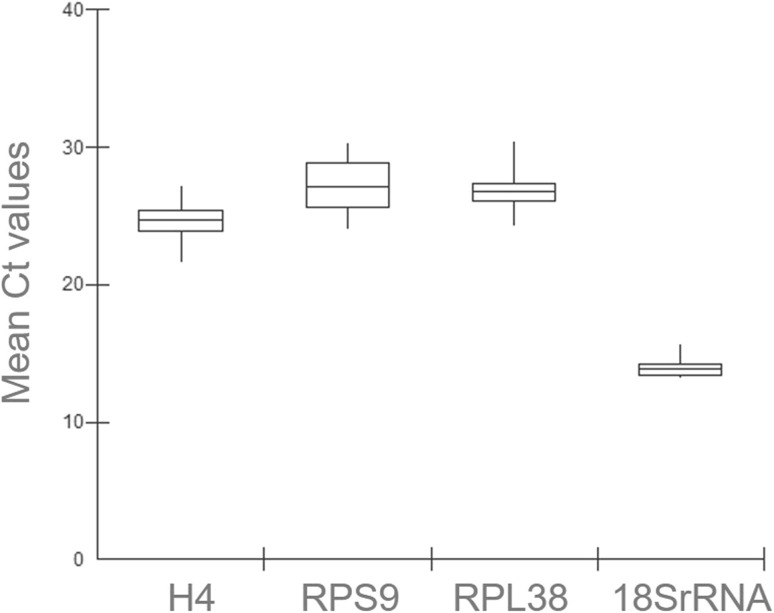
Raw expression levels of the candidate reference genes across the different tissues (flower buds, flowers, leaves and seeds at different developmental stages) were determined by Ct analysis (Table S3 and Fig. S1). Equal amounts of cDNA samples from seeds at the S2 and S3 developmental stages were pooled together, and designated as S2 + S3 seeds, in order to simplify the analysis. Gene-specific amplification of each of the four candidate reference genes in the different samples was confirmed by the appearance of a single, dominant peak in the qPCR dissociation curve analyses, confirming the specificity of the primers used in the qPCR reactions (Fig. S2). The mean values of Ct showed variations for each gene among the different samples: Ct varied from 13.21 to 15.57 for 18SrRNA, from 24.03 to 30.25 for RPS9, from 24.29 to 30.41 for RPL38, and from 21.58 to 27.12 for H4 (Fig. 1). 18SrRNA showed more transcript abundance and, hence, the lowest Ct values (average of 14.02) in all samples, followed by H4 (average of 24.56). RPS9 and RPL38 showed a relatively less abundance of transcripts, with average Ct of 27.21 and 26.97 respectively. The global Ct variation ranged from 13.21 (for 18SrRNA) to 30.41 (for RPL38). The Ct values with SD for all genes in all tissues and developmental stages are shown in Table S4.
Fig. 1.
Box-plot representation of the expression levels of four candidate reference genes (H4, RPS9, RPL38 and 18SrRNA) among the different samples. The lines inside the box show the median values surrounded by lower and upper boxes indicating the first and third quartile. The vertical lines (whiskers) indicate value ranges
Expression stability of candidate reference genes
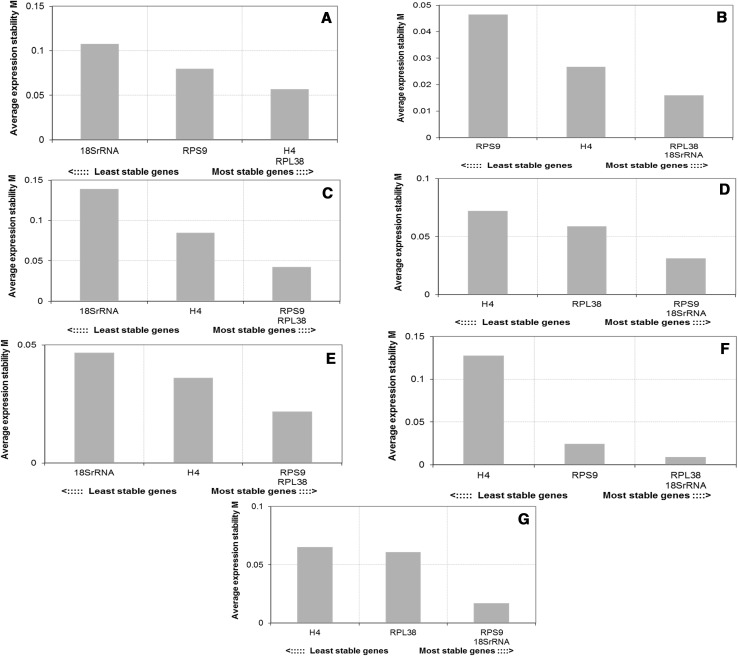
Expression stability of the four candidates for reference genes in B. orellana was analyzed using five statistical tools, geNorm, NormFinder, BestKeeper, ΔCt method and RefFinder. For each method and gene, a ranking of stability values was calculated for both individual and all samples together. In the geNorm analysis, the M-value was used to represent the average stability of the expression. A lower M-value indicates more stable expression. The M-values for all the candidate reference genes analyzed in the different samples were below the geNorm default threshold of 1.5 (Fig. 2), indicating that the four chosen candidate reference genes have expression stability. When evaluating all the samples together, the combination of H4 and RPL38 showed the higher stability value and 18SrRNA, the lower one (Fig. 2a). In flower buds, the most stable combination was RPL38 and 18SrRNA and the least stable gene was RPS9 (Fig. 2b). RPL38 and RPS9 were the most stable combination in flowers and S2 + S3 seeds, while 18SrRNA was the least stable gene (Figs. 2c, e). The best stability values in S1 seeds and leaves were obtained for the combination of RPS9 and 18SrRNA (Figs. 2d, g), while in S4 seeds, the best values were obtained for the combination of RPL38 and 18SrRNA (Fig. 2f). H4 was the least stable gene in these tissues (Figs. 2d, f, g).
Fig. 2.
Gene expression stability of four candidate reference genes (H4, RPS9, RPL38 and 18SrRNA) and the best combination of two genes obtained by geNorm. a All tissues. b Flower buds. c Flowers. d S1 seeds. e S2 + S3 seeds. f S4 seeds. g Leaves. Mean expression stability (M) was calculated following stepwise exclusion of the least stable gene across all the treatment groups
The geNorm analysis of pairwise variation (Vn/n + 1) to establish the optimum number of normalizer genes indicated that two reference genes were sufficient for normalization among the analyzed samples, with no effect on the normalization factor by adding additional reference genes (V2/3 = 0.000, V3/4 = 0.000).
Based on the values of SD and CV calculated by BestKeeper, the candidate reference genes were ranked in the different samples (Table 2). In the analysis of all tissues, 18SrRNA was the most stable, followed by RPL38, and H4 was the least stable among the four candidate reference genes. In the analysis of individual tissues, RPS9 showed the best stability values in five out of the six tissues analyzed (leaves, flower buds, flower, S1 and S4 seeds). 18SrRNA was the most stable gene in S2 + S3 seeds and H4 remained as the least stable gene in each individual tissue.
Table 2.
Stability ranking for the different samples obtained by BestKeeper
| Sample | Gene | BestKeeper | ||
|---|---|---|---|---|
| Ranking | SD | CV | ||
| All tissues | H4 | 4 | 2.52 | 10.26 |
| RPS9 | 3 | 1.90 | 6.97 | |
| RPL38 | 2 | 1.71 | 6.36 | |
| 18SrRNA | 1 | 0.63 | 4.5 | |
| Leaves | H4 | 4 | 1.64 | 6.05 |
| RPS9 | 1 | 0.06 | 0.21 | |
| RPL38 | 3 | 1.58 | 5.18 | |
| 18SrRNA | 2 | 0.12 | 0.85 | |
| Flower buds | H4 | 4 | 1.40 | 5.55 |
| RPS9 | 1 | 0.06 | 0.20 | |
| RPL38 | 3 | 0.81 | 3.12 | |
| 18SrRNA | 2 | 0.54 | 4.03 | |
| Flowers | H4 | 4 | 2.36 | 9.28 |
| RPS9 | 1 | 0.43 | 1.61 | |
| RPL38 | 2 | 1.02 | 3.75 | |
| 18SrRNA | 3 | 1.20 | 7.72 | |
| S1 seeds | H4 | 4 | 1.88 | 7.80 |
| RPS9 | 1 | 0.02 | 0.09 | |
| RPL38 | 3 | 1.50 | 5.43 | |
| 18SrRNA | 2 | 0.24 | 1.79 | |
| S2 + S3 seeds | H4 | 4 | 1.20 | 5.54 |
| RPS9 | 2 | 0.52 | 2.18 | |
| RPL38 | 3 | 0.82 | 3.38 | |
| 18SrRNA | 1 | 0.04 | 0.25 | |
| S4 seeds | H4 | 4 | 3.95 | 16.53 |
| RPS9 | 1 | 0.13 | 0.51 | |
| RPL38 | 3 | 0.62 | 2.36 | |
| 18SrRNA | 2 | 0.34 | 2.43 | |
SD standard deviation, CV coefficient of variation
Data in bold indicate the best gene for the sample group analyzed
NormFinder ranks the reference genes on the basis of their stability value (SV), where lower values indicate more stable gene expression. RPL38 showed the lowest SV in all tissues and also in individual tissues such as flower buds, leaves and S1 and S4 seeds (Table 3). 18SrRNA also showed a lower SV in S4 seeds. RPS9 exhibited the lowest SV in flowers and S2 + S3 seeds. The least stable genes were H4 in all tissues and S1 and S4 seeds, 18SrRNA in flower and leaves, RPL38 in S2 + S3 seeds and RPS9 in flower buds (Table 3). NormFinder also provides the better combination of two genes for normalization of the samples (Table 3). However, only for leaves (RPS9 and RPL38), S1 seeds (H4 and RPS9) and S2 + S3 seeds (RPS9 and RPL38), stability values of the combination were lower than those of the first ranked individual gene.
Table 3.
Stability values for the different samples obtained by NormFinder
| Sample | H4 | RPS9 | RPL38 |
18S
rRNA |
Best combination of two genes | Stability value for best combination of two genes |
|---|---|---|---|---|---|---|
| All tissues | 0.073 | 0.050 | 0.034 | 0.062 | RPS9 and RPL38 | 0.040 |
| Leaves | 0.036 | 0.029 | 0.025 | 0.039 | RPS9 and RPL38 | 0.003 |
| Flower buds | 0.026 | 0.034 | 0.002 | 0.013 | H4 and RPS9 | 0.008 |
| Flowers | 0.078 | 0.011 | 0.023 | 0.097 | H4 and 18SrRNA | 0.014 |
| S1 seeds | 0.045 | 0.039 | 0.020 | 0.031 | H4 and RPS9 | 0.005 |
| S2 + S3 seeds | 0.030 | 0.009 | 0.031 | 0.009 | RPS9 and RPL38 | 0.002 |
| S4 seeds | 0.114 | 0.054 | 0.033 | 0.033 | H4 and RPS9 | 0.046 |
Data in bold indicate the best candidate reference gene or their combination for the sample group analyzed
The ΔCt method showed the RPL38 was the more stable for all samples, except for S2 +S3 and S4 seeds, in which RPS9 and 18SrRNA were the most stable, respectively (Table 4). On the other hand, H4 was the least stable for flower buds, S1 and S4 seeds, whereas 18SrRNA was the least stable for all tissues, leaves, flowers and S2 + S3 seeds (Table 4).
Table 4.
Stability ranking for the different samples obtained by the ΔCt method
| Sample | Gene | ΔCt method | |
|---|---|---|---|
| Ranking | Average of SD | ||
| All tissues | H4 | 2 | 2.43 |
| RPS9 | 3 | 2.45 | |
| RPL38 | 1 | 2.09 | |
| 18SrRNA | 4 | 2.6 | |
| Leaves | H4 | 3 | 1.23 |
| RPS9 | 2 | 1.20 | |
| RPL38 | 1 | 1.18 | |
| 18SrRNA | 4 | 1.25 | |
| Flower buds | H4 | 4 | 1.04 |
| RPS9 | 3 | 1.00 | |
| RPL38 | 1 | 0.61 | |
| 18SrRNA | 2 | 0.61 | |
| Flowers | H4 | 3 | 2.48 |
| RPS9 | 2 | 1.56 | |
| RPL38 | 1 | 1.54 | |
| 18SrRNA | 4 | 2.62 | |
| S1 seeds | H4 | 4 | 1.41 |
| RPS9 | 3 | 1.30 | |
| RPL38 | 1 | 1.14 | |
| 18SrRNA | 2 | 1.15 | |
| S2 + S3 seeds | H4 | 3 | 0.81 |
| RPS9 | 1 | 0.56 | |
| RPL38 | 2 | 0.56 | |
| 18SrRNA | 4 | 0.91 | |
| S4 seeds | H4 | 4 | 3.84 |
| RPS9 | 3 | 1.66 | |
| RPL38 | 2 | 1.49 | |
| 18SrRNA | 1 | 1.50 | |
SD standard deviation
Data in bold indicate the best gene for the sample group analyzed
Finally, we compared and ranked the four candidates based on the comprehensive analysis tool RefFinder, which integrates the outputs of NormFinder, GeNorm, BestKeeper and ΔCt method. According to these results, RefFinder ranked RPL38 as the most stable and 18SrRNA as the least stable gene among all the samples (Table 5).
Table 5.
Stability ranking for all tissue samples obtained by the RefFinder using the output data of ΔCt, BestKeeper, NormFinder and GeNorm methods
| Method | Ranking | |||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| ΔCt | RPL38 | H4 | RPS9 | 18SrRNA |
| BestKeeper | 18SrRNA | RPL38 | RPS9 | H4 |
| NormFinder | RPL38 | RPS9 | H4 | 18SrRNA |
| GeNorm | H4 | RPL38 | RPS9 | 18SrRNA | |
| Recommended comprehensive ranking Geomean of ranking values |
RPL38 | H4 | RPS9 | 18SrRNA |
| 1.19 | 2.21 | 2.71 | 2.83 | |
Data in bold indicate the best gene
Validation of the selected reference genes
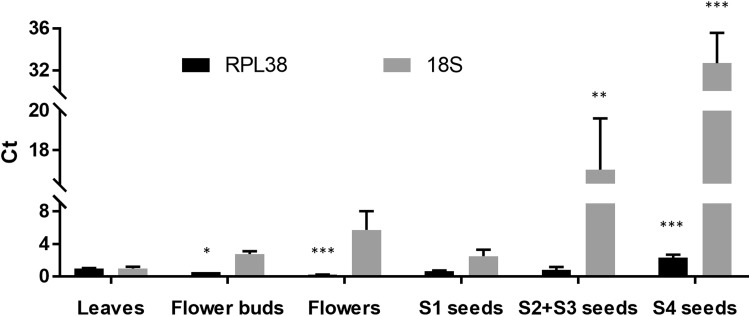
In order to validate the reliability of the candidate reference genes, the most stable RPL38 and the least stable 18SrRNA were used to normalize the expression profiles of the target gene BoCCD1 in the different tissues. When using RPL38 as reference gene, the BoCCD1 gene expression was significantly downregulated in flower buds and flowers, upregulated in S4 seeds, and relatively constant in S1 and S2 + S3 seeds in comparison with the expression levels in leaves, which was used as a calibrator (Fig. 3). On the other hand, when 18SrRNA was used as a reference gene, significant differences in the expression levels of BoCCD1 were detected only for S2 + S3 and S4 seeds, which exhibited values much higher than those found when RPL38 was used as a normalizer (Fig. 3). Taken together, the results revealed significant differences in the BoCCD1 expression profiles depending on the reference genes, whether stable or unstable, used for normalization of the qPCR data.
Fig. 3.
Relative quantification of CCD1 expression in diverse tissue samples of B. orellana using different candidate reference genes for normalization of qPCR data. Significantly different from leaves (calibrator) according to Student’s t-test at levels of P ≤ 0.05 (*), P ≤ 0.01 (**) and P ≤ 0.001 (***)
Discussion
The selection of stable reference genes may vary across different species, genotypes, experimental conditions, tissues and stages of development. In this work, the reference genes or their combinations that were considered stable by the different algorithms (geNorm, NormFinder, BestKeeper, ΔCt method and RefFinder) varied according to the tissue and developmental stage of seeds. Variations in the ranking of stability of the candidate reference genes in the different samples were also observed among the different tools. These variations are expected to occur due to the differences in statistical algorithms used for each program. Therefore, it is advisable to evaluate the stability of each candidate reference gene or their combinations based on the global results of the three programs in order to overcome their individual limitations and to provide a more reliable evaluation of the data (Kanakachari et al. 2016).
The Ct variation of the candidate reference genes in the different tissues analyzed in the present study was of 2–6 cycles (Fig. 1), with 18SrRNA showing the lowest values. In principle, this result could demonstrate its higher stability in comparison with the other candidate reference genes tested. However, 18SrRNA was barely ranked as the most stable gene by the different algorithms used in the analysis of stability. It was only ranked as the most stable gene in all tissues and S2 + S3 seeds, by the BestKeeper, and in S4 seeds, by the NormFinder and ΔCt method, or when combined with other reference genes in samples of flower buds, leaves, S1 and S4 seeds, by the geNorm. On the other hand, it was considered the least stable gene in flower and leaf samples, by the NormFinder, in all tissues, flowers and S2 + S3 seeds, by the geNorm, and also in flower, flower bud, leaves and S2 + S3 seeds, by the ΔCt method. 18SrRNA was also considered the least stable gene in different tissues of Bupleurum chinense (Dong et al. 2011), in lettuce under abiotic stresses (Borowski et al. 2014), in strawberry under osmotic stress (Galli et al. 2015) and in eggplant at different stages of fruit development (Kanakachari et al. 2016), but it was the most stable gene in different organs and developmental stages of eggplant (Gantasala et al. 2013) and Descurainia sophia (Xu et al. 2016). Since 18SrRNA has been the only reference gene used in B. orellana (Soares et al. 2011; Rodríguez-Ávila et al. 2011a, b; Cárdenas-Conejo et al. 2015), caution must be taken when considering to use it in future studies of gene expression in this plant species, since it was ranked as the least stable gene in our study. Besides, 18SrRNA was the candidate reference gene that showed the lowest Ct values (Fig. 1), indicating that its transcripts are more abundant in the different tissues than those of the other candidate reference genes analyzed. Such an abundance of 18SrRNA transcripts has been also widely reported in other studies, with Ct varying between the 12th and 15th cycles (Wei et al. 2013; Galli et al. 2015; Xu et al. 2016; Kanakachari et al. 2016), and it may constitute an additional problem for the normalization of rare transcripts such as those of genes related to the secondary metabolism.
H4 was considered in most analyses as the least stable among all the candidate reference genes tested in the present study. It ranked last in BestKeeper’s stability ranking for all the samples. According to geNorm, it was also ranked last in samples of leaves and S1 and S4 seeds, and only in samples of all tissues showed better stability when combined with RPL38. In the NormFinder, it appeared as the most unstable gene in samples of all tissues and S1 and S4 seeds, and it was not ranked first in any of the individual analyses. In the ΔCt method, it was ranked as the least stable in all tissues, flower bud, S1 and S4 seeds. Besides, H4 did not appeared in the first position in the ranking in any of the samples. However, H4 was considered as the most stable gene in a study in strawberry under osmotic stress (Galli et al. 2015).
Genes coding for 60S and 40S ribosomal proteins have been widely used in the analyses of candidate reference genes in plants. RPS9 was the most stable gene by BestKeeper in samples of flower buds, flowers, leaves and S1 and S4 seeds, and ranked as the second most stable gene in S2 + S3 seed samples and the third one in samples of all tissues. According to NormFinder, RPS9 was the most stable gene in samples of flowers and S2 + S3 seeds, and as one of the genes in the best combination for all the samples, except flowers. In the geNorm, RPS9 formed the most stable gene pairs in flowers, leaves, S1 and S2 + S3 seeds, and it was the most unstable in flower buds. Similarly, RPL38 also formed the best combination pairs in most samples by the geNorm, including with RPS9 in flower and S2 + S3 seed samples. In the ΔCt method, RPS9 was the most stable only in the S2 + S3 seeds. By the BestKeeper, RPL38 ranked second in flowers and all tissues, and third in the other samples analyzed. According to NormFinder, it was the most stable gene in most samples (all tissues, leaves, flower buds, S1 and S4 seeds), and together with RPS9, it formed the most stable combination for all tissues, leaves and S2 + S3 seeds. By the ΔCt method, RPL38 was the most stable in flower buds, flowers, leaves, S1 and S4 seeds. Considering all the data, RPL38 was ranked as the most stable candidate reference gene using the RefFinder. In other studies, the ranking of stability of genes coding for ribosomal proteins was variable, ranging from most stable (Dong et al. 2011; Figueiredo et al. 2013; Borowski et al. 2014) to most unstable (Shivhare and Lata 2016) in different plant tissue samples.
BoCCD1 was selected as a target gene to validate the reliability of the candidate reference genes analyzed, since its sequence and mRNA expression were previously characterized in B. orellana (Rodríguez-Ávila et al. 2011a). BoCCD1 was shown to be a homolog of CCD1s from Arabidopsis thaliana, Coffea canephora and Vitis vinifera, and to have the highly conserved RPE65 domain responsible for the catalytic activity of CCDs. Previous RT-PCR analysis showed that BoCCD1 expression was lower in flowers, immature fruits and mature seeds, but it was higher in leaves, flower buds and immature seeds (Rodríguez-Ávila et al. 2011a). A remarkable increase in the mRNA expression of BoCCD1 was observed in the last developmental stage of immature seeds, which was coordinated with the accumulation of carotenoids, mainly bixin, in seeds (Rodríguez-Ávila et al. 2011a). Therefore, BoCCD1 was considered a good target gene for validation of the candidate reference genes in this study. Interestingly, our results showed significant differences in the BoCCD1 expression profiles depending on the reference genes used for normalization, whether stable or unstable. The main divergences were observed in samples of flower buds and flowers, in which the BoCCD1 expression was downregulated using RPL38, but relatively constant using 18srRNA as reference genes, in comparison with the expression levels in leaves (Fig. 3). Besides, despite the fact that the two normalizers detected a significant upregulation of BoCCD1 in S4 seeds, its levels of relative expression varied considerably using the unstable 18SrRNA. These results clearly indicate that the incorrect use of reference genes without previous validation may introduce significant bias in the analysis and lead to misinterpretation of data. Similar results were also reported in other plant species using stable or unstable normalizers (Guénin et al. 2009; Dong et al. 2011; Figueiredo et al. 2013; Borowski et al. 2014; Galli et al. 2015, Kanakachari et al. 2016; Shivhare and Lata 2016).
Conclusion
In conclusion, the results obtained in the present study indicate RPL38 as the most stable reference gene in the different tissues and developmental stages of seeds in B. orellana. 18SrRNA was the least stable. These results support the selection of suitable reference genes for future gene expression studies in this important plant species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by research grants from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brasília, Brazil), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasília, Brazil) and UESC (Universidade Estadual de Santa Cruz, Ilhéus, Bahia, Brazil). We gratefully acknowledge the Ph.D. scholarship provided by CAPES Foundation to VSM.
Authors’ contributions
VSM conducted the experiments. VSM, VLFS, RJSS and AOS analyzed the data. VSM drafted the manuscript. VLFS, WCO and MGCC supported the project and designed the experiments. VLFS and MGCC revised the manuscript. All authors read and approved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Research involving human participants and/or animals
The authors declare that the present study does not involve any human participants and/or animals.
Informed consent
The authors declare that the present study does not involve any informed consent.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12298-018-0528-1) contains supplementary material, which is available to authorized users.
References
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Borowski JM, Galli V, da Silva Messias R, Perin EC, Buss JH, Silva SDDA, Rombaldi CV. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta. 2014;239:1187–1200. doi: 10.1007/s00425-014-2041-2. [DOI] [PubMed] [Google Scholar]
- Cárdenas-Conejo Y, Carballo-Uicab V, Lieberman M, Aguilar-Espinosa M, Comai L, Rivera-Madrid R. De novo transcriptome sequencing in Bixa orellana to identify genes involved in methylerythritol phosphate, carotenoid and bixin biosynthesis. BMC Genom. 2015;16:877. doi: 10.1186/s12864-015-2065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time. Methods. 2010;50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112–114. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- Dong L, Sui C, Liu Y, Yang Y, Wei J, Yang Y. Validation and application of reference genes for quantitative gene expression analyses in various tissues of Bupleurum chinense. Mol Biol Rep. 2011;38:5017–5023. doi: 10.1007/s11033-010-0648-3. [DOI] [PubMed] [Google Scholar]
- Figueiredo A, Loureiro A, Batista D, Monteiro F, Várzea V, Pais MS, Gichuru EK, Silva MC. Validation of reference genes for normalization of qPCR gene expression data from Coffea spp. hypocotyls inoculated with Colletotrichum kahawae. BMC Res Notes. 2013;6:388. doi: 10.1186/1756-0500-6-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli V, Borowski JM, Perin EC, da Silva Messias R, Labonde J, IdosS Pereira, Silva SD, Rombaldi CV. Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene. 2015;554:205–214. doi: 10.1016/j.gene.2014.10.049. [DOI] [PubMed] [Google Scholar]
- Gantasala NP, Papolu PK, Thakur PK, Kamaraju D, Sreevathsa R, Rao U. Selection and validation of reference genes for quantitative gene expression studies by real-time PCR in eggplant (Solanum melongena L) BMC Res Notes. 2013;6:312. doi: 10.1186/1756-0500-6-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Rosati C, Bramley PM. To dye or not to dye: biochemistry of annatto unveiled. Trends Biotechnol. 2003;21:513–516. doi: 10.1016/j.tibtech.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot. 2009;60:487–493. doi: 10.1093/jxb/ern305. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2008;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- Kanakachari M, Solanke AU, Prabhakaran N, Ahmad I, Dhandapani G, Jayabalan N, Kumar PA. Evaluation of suitable reference genes for normalization of qPCR gene expression studies in brinjal (Solanum melongena L.) during fruit developmental stages. Appl Biochem Biotechnol. 2016;178:433–450. doi: 10.1007/s12010-015-1884-8. [DOI] [PubMed] [Google Scholar]
- Kundu A, Patel A, Pal A. Defining reference genes for qPCR normalization to study biotic and abiotic stress responses in Vigna mungo. Plant Cell Rep. 2013;32:1647–1658. doi: 10.1007/s00299-013-1478-2. [DOI] [PubMed] [Google Scholar]
- Mercadante AZ, Pfander H. Carotenoids from annatto: a review. Recent Res Devel Agric Food Chem. 1998;2:79–91. [Google Scholar]
- Mercadante AZ, Steck A, Pfander H. Isolation and identification of new apocarotenoids from annatto (Bixa orellana) seeds. J Agric Food Chem. 1997;45:1050–1054. doi: 10.1021/jf960412k. [DOI] [Google Scholar]
- Moreira VS, Rebouças TNH, de Moraes MOB, São José AR, da Silva MV. Atividade antioxidante de urucum (Bixa orellana L.) in natura e encapsulado. Rev Iberoam Tecnol Postcosecha. 2014;15:201–209. [Google Scholar]
- Moreira PA, Lins J, Dequigiovanni G, Veasey EA, Clement CR. The domestication of annatto (Bixa orellana) from Bixa urucurana in Amazonia. Econ Bot. 2015;69:127. doi: 10.1007/s12231-015-9304-0. [DOI] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ávila NL, Narváez-Zapata JA, Aguilar-Espinosa M, Rivera-Madrid R. Regulation of pigment-related genes during flower and fruit development of Bixa orellana. Plant Mol Biol Rep. 2011;29:43–50. doi: 10.1007/s11105-010-0207-z. [DOI] [Google Scholar]
- Rodríguez-Ávila NL, Narvaez-Zapata JA, Ramírez-Benítez JE, Aguilar-Espinosa ML, Rivera-Madrid R. Identification and expression pattern of a new carotenoid cleavage dioxygenase gene member from Bixa orellana. J Exp Bot. 2011;62:5385–5395. doi: 10.1093/jxb/err201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivhare R, Lata C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci Rep. 2016;6:23036. doi: 10.1038/srep23036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares VL, Rodrigues SM, de Oliveira TM, De Queiroz TO, Lima LS, Hora-Júnior BT, Gramacho KP, Micheli F, Cascardo JCM, Otoni WC, Gesteira AS, Costa MGC. Unraveling new genes associated with seed development and metabolism in Bixa orellana L. by expressed sequence tag (EST) analysis. Mol Biol Rep. 2011;38:1329–1340. doi: 10.1007/s11033-010-0234-8. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(research0034):1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Miao H, Zhao R, Han X, Zhang T, Zhang H. Identification and testing of reference genes for sesame gene expression analysis by quantitative real-time PCR. Planta. 2013;237:873–889. doi: 10.1007/s00425-012-1805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- Xu X, Liu X, Chen S, Li B, Wang X, Fan C, Wang G, Hanwen N. Selection of relatively exact reference genes for gene expression studies in flixweed (Descurainia sophia) by quantitative real-time polymerase chain reaction. Pestic Biochem Physiol. 2016;127:59–66. doi: 10.1016/j.pestbp.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.