Abstract
It is a grand challenge to reveal the causal effects of DNA variants in complex phenotypes. Although statistical techniques can establish correlations between genotypes and phenotypes in Genome-Wide Association Studies (GWAS), they often fail when the variant is rare. The emerging Network-based Association Studies aim to address this shortcoming in statistical analysis, but are mainly applied to coding variations. Increasing evidences suggest that non-coding variants play critical roles in the etiology of complex diseases. However, few computational tools are available to study the effect of rare non-coding variants on phenotypes. Here we have developed a multiscale modeling variant-to-function-to-network framework VariFunNet to address these challenges. VariFunNet first predict the functional variations of molecular interactions, which result from the non-coding variants. Then we incorporate the genes associated with the functional variation into a tissue-specific gene network, and identify subnetworks that transmit the functional variation to molecular phenotypes. Finally, we quantify the functional implication of the subnetwork, and prioritize the association of the non-coding variants with the phenotype. We have applied VariFunNet to investigating the causal effect of rare non-coding variants on Alzheimer’s disease (AD). Among top 21 ranked causal non-coding variants, 16 of them are directly supported by existing evidences. The remaining 5 novel variants dysregulate multiple downstream biological processes, all of which are associated with the pathology of AD. Furthermore, we propose potential new drug targets that may modulate diverse pathways responsible for AD. These findings may shed new light on discovering new biomarkers and therapies for the prevention, diagnosis, and treatment of AD. Our results suggest that multiscale modeling is a potentially powerful approach to studying causal genotype-phenotype associations.
Keywords: single nucleotide polymorphism, RNA binding, transcription factor, systems biology, network robustness, complex disease
I. Introduction
The existing methods for genotype-phenotype association are mainly based on statistical or machine learning techniques. However, the power of these methods drops significantly when sample size is small or variants are rare [1, 2]. Furthermore, these approaches work as a black box without explicitly incorporating the information of biological mechanism. As a result, it is not as straightforward to interpret the causal association of non-coding region variants with phenotypes as coding region variants. Besides, limited analysis has been performed on multi-variants that work as a system. It is believed that occurrence of genetic diseases could be caused not only by individual primary variants in coding regions but also by modest effects of multi coding and non-coding variants collectively. Small effects of non-coding variants on regulatory functions could be ignored if analysis is not performed on systematic level. Systematic analysis and multi-scale modeling of coding variants by integrating tissue-specific gene-gene interactions or protein-protein interactions is proven fruitful [3–6].
In this paper, we have developed a multiscale modeling framework to establish causal functional impact of non-coding variants on the biological network through integrating heterogeneous data from GWAS, protein-DNA/RNA interaction, protein-protein interaction, and gene expression profile, and combining statistical and machine learning method, regulatory variants studies, and biological network analysis. We applied our pipeline to study the functional roles of non-coding variants and associated regulatory genes that are identified in Alzheimer Disease (AD). The appearance of GWAS and Next Generation Sequencing provide another tool for the identification of the genetic factors associated with AD. More than 20 genes have previously been proposed to be associated with AD [7]. Our method was validated by its ability to identify known risk genes, such as APOE, TOMMO40 and EGFR [7]. In addition, novel non-coding variants that may cause the dysregulation of genes such as CUX1, KHDRBS1, CNOT4, and SRSF10 were also identified as potential AD associated factors. Their down-stream regulated pathways have strong associations with AD, as supported by existing evidences. Furthermore, we propose potential new drug targets, particularly NTRK3 and CAMK2B, which may modulate diverse pathways responsible for the pathology of AD. Thus, our systematic biology approach that incorporates multi-scale models, from protein-DNA interactions in transcription level, protein-RNA interactions in translation level to gene network robustness at the system level, may shed new light on the functional roles of non-coding variants in AD in particular, and other multi-genic diseases in general.
II. Materials and Methods
A. Data set
Data used for our method application were acquired from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database [8, 9]. It was launched in 2004 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations. ADNI is the result from efforts of many co-investigators in a broad range of academic institutions and private corporations, and subjects have been recruited from more than 50 sites across the U.S. and Canada. Approximately 200 cognitively normal older individuals were followed for 3 years, 400 people with MCI were followed for 3 years and 200 people with early AD were followed for 2 years. Our study utilized ADNI1 GWAS genetic sequencing data and AD patients’ clinical data. More updated information could be found at www.loni.ucla.edu/ADNI.
B. The functional analysis of SNPs
54 SNPs were imputed to ANNOVAR [10, 11], with which SNPs position information in genes were annotated and flanking genes were indicated if it is in intergenic regions. Sequences upstream and downstream of these SNPs were retrieved from UCSC Genome Browser with hg19 as reference genome [12]. Each sequence’s length is 50nt. Regulatory analysis was performed with DeepBind software [13], with which binding scores of retrieved sequences with TF and RBP were predicted. 515 TF and 120 RBP human species were included in binding study. Each SNP’s two alleles differential binding scores of interactions with each regulatory protein, TF or RBP, were calculated. Then for each regulatory protein, two tailed student t-test (H0 : μ = 0 and Ha : μ ≠ 0) was performed on 53 differential binding scores that are corresponding to 53 SNPs (α = 0.05 and 52 degrees of freedom).
C. Analysis of gene network
The gene network of brain tissue was retrieved from Genome-scale Integrated Analysis of gene Networks (GIANT) [14]. We employed full gene network in brain, which consisted of 25689 genes, to conduct our analysis. GIANT network were based on gene-gene functional association. Gene-gene edges weights are proportional to confidence of gene-gene functional relationship and are ranged from 0 to 1.
The differential expressed genes of AD patients and normal controls came from Signaling Pathway Impact Analysis (SPIA) [15] and Common Significance Subnetworks Analysis [16] on gene expression datasets of healthy elders and AD downloaded from NCBI GEO Datasets-record of GSE5281. The dysregulated genes in these two subnetworks, including inflammation response subnetwork and calcium ion mechanism subnetwork, were involved into differentially expressed gene list. Besides, SPIA found dysregulated genes in KEGG AD pathway in hippocampus (HIP), middle temporal gyrus (MTG) and posterior cingulate cortex (PC). They also recognized perturbed genes in GABAergic synaptic pathway and glutamatergic synaptic pathway, which are two important neurotransmission system [15], in MTG, and in cytokine-cytokine receptor interactions, which are key components in inflammation process [17], in PC. These dysregulated genes were also combined into the differentially expressed gene list.
To identify pathways that link the genes associated with SNPs with the differentially expressed genes, we first map these genes onto the brain tissue-specific gene network. Then network-based approach Prize-Collection Steiner Tree (PCST) algorithm was applied to identify the subnetwork that connects genes.
D. Gene-set overrepresentation analysis
5 novel putative disease associated genes’ PCST subtrees, including RMBS3, CUX1, KHDRBS1, CNOT4, and SRSF10, were ranked on the top 21. Intermediate genes (excluding source and terminal nodes) in these subtrees were extracted and for gene-set overrepresentation analysis using ConsensusPathDB [18]. 13 pathway databases were integrated to this software, including KEGG, Reactome and WikiPathways. Enriched pathways passed the statistical significance threshold were listed in Supplementary Tables.
E. Calculation of Natural Connectivity
Natural connectivity is calculated as:
where λ1 ≥ λ2 ≥ …, ≥ λn denote a non-increasing ordering of the eigenvalues of adjacency matrix of the graph.
III. Results and Discussion
A. Majority of SNPs were positioned on non-coding regions
Quantitative trait association study was conducted on 620,901 single nucleotide polymorphisms (SNPs) using PLINK [19] (Figure 1) on a well-established dataset from ADNI [8, 9]. We used participants’ MMSE scores on the first-visit as the phenotypes. We selected top 54 SNPs, whose Bonferroni adjusted p-values were less than 1.0 and Benjamini & Yekutieli adjusted False Discovery Rate (FDR) values were less than 0.173. To uncover the functional roles of these 54 SNPs, we positioned them to human genome with ANNOVAR [10, 11]. This gene-based annotation was referred to Human Genome version 19 (hg19). Surprisingly, it turned out that 53 out of 54 SNPs are located in non-coding regions. This imposed a challenge to understand the functional impact of these SNPs because of very limited knowledge about the function of non-coding regions SNPs. We then proposed two ways to better understand their functions. Firstly, the flanking genes of SNPs were identified. The function of the flanking gene may shed light on the functional roles of these non-coding SNPs. Non-coding regions variants had no straightforward effect on expressed proteins sequence or structure, however they could be associated with proteins or transcripts expression level. In another word, they could vary individuals’ phenotypes through their regulatory functions, especially by the cis-regulation. Secondly, transcription factor (TF) and RNA binding protein (RBP) could bind with non-coding region DNA or RNA and then either promote or inhibit proteins and transcripts expressions. TF and RBP could also collectively regulate multiple transcripts expressions or protein expressions and thus give rise to phenotype variants.
Figure 1.
Schema of VariFunNet, a multi-scale modeling pipeline to study causal effect of non-coding SNPs on complex disease.
B. Identification and characterization of flanking genes
As proposed above, the flanking genes may provide a way to understand the mechanism of non-coding SNPs. ANNOVAR was used to find the flanking genes of these SNPs. In this way, totally 47 flanking genes were allocated. The most significantly associated SNP overlapped an intronic region of the most well studied AD related gene APOE. Thus, our strategy was supported by existing findings.
The second way that we proposed to study non-coding regions SNPs was to discover related TF and RBP through protein-DNA or protein–RNA interactions. TF and RBP could affect phenotype through interactions with multiple SNPs. The binding specificity of the interaction was predicted with DeepBind [13]. This software has been shown to outperform the widely used binding site prediction software, position weight matrices1 (PWMs) [13, 20]. The upstream and downstream DNA sequences of these 53 SNPs were generated using UCSC Genome Browser [12]. DeepBind was then used to generate scores for binding specificity of DNA sequences with 515 TFs. Scores differences for two different variants of each SNP were calculated. Two-tailed t-tests were performed on the score distribution of each SNP, and 27 statistical significant TFs were selected. Another 13 TFs were selected because binding scores differences of them with at least one of the 53 SNPs were higher than 4. These 40 TFs were then listed together.
C. Interpret selected genes with genetic network in system level
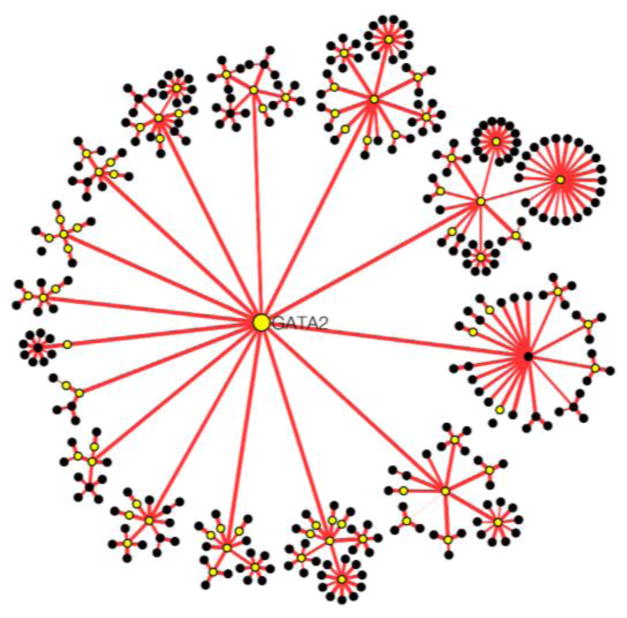
To better characterize the causal association of each selected SNP with AD, we integrated them into a GIANT [14]. Our method utilized PCST algorithm to find an optimal path from a gene that is associated with a SNP (source gene) to up- or down-regulated genes (terminal genes). 420 genes were used as terminal genes [15, 16, 21]. Each selected SNP-associated gene was used as source gene for the PCST. In the GIANT network, nodes denote all the identified genes in brain tissues. Each edge connects two functional related genes. The weight of each edge denotes the confidence for the functional relationship between two genes. The PCST algorithm can be used to identify a subtree that minimizes the sum of the total costs of edges included in the subtree plus the prizes of nodes not included in the subtree. In order to make the PCST work efficiently, we modified the network by deleting the edges whose weight is less than 0.3 (the edge weights ranges from 0.0 to 1.0). We mapped 1.0 minus edges weights from GIANT as the edge costs in our network in order to prioritize the edge that has high weight. We also assigned the prize of terminal node to be a fairly high value in order to contain as many terminal genes in the subtree as possible. The rest nodes have prize of 0.0. PCST could find out all the terminal genes that are related with source gene and exclude other genes from this subtree. Then the sums of total costs of edges included in the subtree plus the prizes of nodes not included in these subtrees were compared and ranked (Table 1). Lower sum value could demonstrate closer relationship of source gene with AD. Generated PCST trees were visualized with Cytoscape [22] (Figure 2).
Table 1.
Top Ranked PCST tree sums values associated with each source gene.
| Gene Symbol | PCST tree sums values | Gene Symbol | PCST tree sums values |
|---|---|---|---|
| EGFR | 654.461 | APOE | 841.9873 |
| HFE | 720.2541 | CUX1 | 891.3414 |
| PVRL2 | 722.942 | FMR1 | 906.7965 |
| RBMS3 | 752.6028 | TCF12 | 914.9009 |
| GATA2 | 753.7089 | TIA1 | 934.7990 |
| NFATC1 | 770.7396 | PTBP1 | 938.0070 |
| SP1 | 772.2513 | KHDRBS1 | 942.2435 |
| EGR1 | 781.6129 | CNOT4 | 956.1771 |
| NRG1 | 787.6241 | SRSF10 | 978.3997 |
| MEF2D | 800.2361 | TOMM40 | 987.009 |
| JUN | 825.4065 | POU2F1 | 1001.174 |
Figure 2.
Generated Prize-Collecting Steiner Tree for GATA2. Source gene GATA2 was labeled in the center. Black nodes denotes the terminal genes. Orange nodes denotes rest genes in the paths from source gene to terminal genes. PCST was visualized with Cytoscape.
D. Literature support of ranked genes
We manually inspected the association of top 21 ranked genes. 16 out of them in sorted gene list were found to be related to AD in the literature. For instance, Epidermal growth factor receptor (EGFR) has been reported to be potential drug target for β-amyloid peptides induced AD [23]. HFE has significant effects on iron and cholesterol metabolism, while both of these processes perform important roles in brain and are related to AD[24]. PVRL2 can mediate infection of herpes simplex. viruses (HSV), a virus found to be a risk factor for AD [25, 26]. GATA-2 activates transcription of Neuroglobin, which is implicated to reduce AD severity [27]. There is still a controversy about the effect of TOMM40 on AD, but TOMM40, as a mitochondrial transmembrane protein, is believed to be a risk gene in mitochondrial dysfunction in AD [28–30]. Other genes which are not listed here also has literature evidence for their relationship with AD.
E. Novel SNPs that are associated with AD
Five top-21 ranked SNPs that were linked to genes RMBS3, CUX1, KHDRBS1, CNOT4, and SRSF10 did not have direct evidences about their roles in AD. However, RMBS3 is shown to directly regulate SMAD2/3 [31]. It is also reported that TGFβ-SMAD2/3 signaling inhibition is one potential therapy for AD patients [32]. Thus RMBS3 can be associated with AD through regulating TGFβ-SMAD2/3 signaling. Besides, TGFβ signaling pathway is enriched when using Gene set overrepresentation analysis (GSOA) with a q-values of 0.0205. The GSOA is also on the PCST subtree of other four discovered genes, including CUX1, KHDRBS1, CNOT4, and SRSF10, using ConsensusPathDB [18]. In the CNOT4 PCST subtree, the most significantly enriched pathways are HIF-1-alpha transcription factor network, which is related to hypoxia [33], and circadian clock gene expression [34]. Increasing evidence suggests that both hypoxia and the disturbance of circadian cycle facilitate the pathogenesis of AD [35]. In the CUX1 PCST subtree, EGFR1 and FGF signaling pathway were two most significant enriched pathways. The modulation of EGFR [23] and FGF [36] have been reported to be potential therapy for the AD. The most significant pathways in KHDRBS1 PCST subtree were related to mRNA processing and splicing. In the SRSF10 PCST subtree, small nuclear ribonucleo proteins (snRNP) assembly that is related to mRNA splicing and metabolism of non-coding RNA, as well as E-cadherin adherens junction stabilization and expansion that are important for cell-cell adhension were the most significant enrichment. In summary, all five novel non-coding associations proposed in this study are supported by existing evidences.
F. Potential drug targets for the treatment of AD
We combine all pathways that are identified by the PCST into a sub-network that links genetic variants to molecular phenotypes of AD. The AD sub-network includes 1,326 genes and 6,289 directed edges. The node degree distribution in the AD sub-network follows a power-law distribution. Thus the AD sub-network has the robust-yet-fragile property of common biological network. Using the AD sub-network, it is possible for us to identify novel drug targets for the treatment of AD. Our rationale is that the potential drug target should maximally reduce the robustness of the AD subnetwork [37].. We quantify the impact of node inhibition on the network robustness using Natural Connectivity [38]. The Natural Connectivity corresponds to an “average” eigenvalue of a graph. Natural Connectivity has clear physical and structural meaning that can be tied to several connectivity properties of networks. In particular, it explicitly characterizes the redundancy and feedback of alternative paths in the network by quantifying the weighted number of closed walks of all lengths. It has been shown that the Natural Connectivity can more accurately quantify the network robustness than other connect metrics such as node degree and betweenness etc [39]. We calculate the impact of node inhibition on the Natural Connectivity using MIoBI algorithm [39]. The top 20 genes ranked by the MIoBI score are listed in Table 2. Interestingly, three protein kinases NTRK3, CAMK2B, and PARKAR1A are ranked at top 1, 3, and 12, respectively. These protein kinases are well known druggable targets. The association of NTRK3 with neurological disorders has been revealed recently [40]. CAMK2B is a tau kinase, playing a fundamental role in neurodegeneration and memory impairment in AD [41]. The role of PRKAR1A in AD is unknown. Further studies are needed to validate this hypothesis. Another interesting potential druggable target is GPCR protein GLP1R. It has been shown that GLP1R agonist reduces AD-associated tau hyperphosphorylation[42]. Due to heavy investments in the kinase and GPCR drug discovery, it is possible for us to repurpose FDA-approved drugs for the treatment of AD. Furthermore, as AD is a multi-genic complex disease, the conventional one-drug-one-target approach is less likely to be successful. Polypharmacology may be required to develop efficient AD therapies [43]. Protein kinases and GPCRs are excellent candidates for the targeted polypharmacology [44].
Table 2.
Top 20 critical nodes in the AD subnetwork determined by MIoBI. Potential kinase and GPCR drug targets are underlined.
| Gene ID | Gene Symbol | MIoBI Score |
|---|---|---|
| 4916 | NTRK3 | 0.323 |
| 1548 | CYP6A | 0.585 |
| 816 | CAMK2B | 0.622 |
| 6426 | SRSF1 | 0.713 |
| 56 | ACRV1 | 0.782 |
| 7534 | YWHAZ | 0.797 |
| 10772 | SRSF10 | 0.817 |
| 23369 | PUM2 | 0.864 |
| 9444 | QKI | 0.874 |
| 394 | ARHGAP5 | 0.901 |
| 6434 | TRA2B | 0.904 |
| 5573 | PRKAR1A | 0.943 |
| 6310 | ATXN1 | 0.966 |
| 2740 | GLP1R | 1.014 |
| 2334 | AFF2 | 1.015 |
| 6418 | SET | 1.019 |
| 2782 | GNB1 | 1.044 |
| 396 | ARHGDIA | 1.047 |
| 9436 | NCR2 | 1.054 |
| 860 | RNX2 | 1.061 |
Supplementary Material
Acknowledgments
Supported by Grant Number R56AG057555 from the National Institute of Aging (NIA) of the National Institute of Health (NIH).
Data used for our method application were acquired from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database [8, 9].
Contributor Information
Qiao Liu, Biochemistry, The Graduate Center, The City University of New York, New York, United States.
Chen Chen, School of Computing, Informatics and Decision Systems Engineering, Arizona State University, Tempe, United States.
Annie Gao, Princeton High School, Princeton, United States.
Hang Hang Tong, School of Computing, Informatics and Decision Systems Engineering, Arizona State University, Tempe, United States.
Lei Xie, Department of Computer Science Hunter College, The City University of New York, New York, United States.
V Reference
- 1.Clarke R, Ressom HW, Wang A, Xuan J, Liu MC, Gehan EA, et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008 Jan;8:37–49. doi: 10.1038/nrc2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickel PJ, Brown JB, Huang H, Li Q. An overview of recent developments in genomics and associated statistical methods. Philos Trans A Math Phys Eng Sci. 2009 Nov 13;367:4313–37. doi: 10.1098/rsta.2009.0164. [DOI] [PubMed] [Google Scholar]
- 3.Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–44. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009 Feb;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 5.Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007 Mar;25:309–16. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 6.Kim ES, Ahn EH, Chung E, Kim DH. Recent advances in nanobiotechnology and high-throughput molecular techniques for systems biomedicine. Mol Cells. 2013 Dec;36:477–84. doi: 10.1007/s10059-013-0304-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertram L, Tanzi RE. Genome-wide association studies in Alzheimer’s disease. Hum Mol Genet. 2009 Oct 15;18:R137–45. doi: 10.1093/hmg/ddp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010 May;6:265–73. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005 Nov;15:869–77. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010 Sep;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015 Oct;10:1556–66. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002 Jun;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015 Aug;33:831–8. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 14.Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015 Jun;47:569–76. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, Guan Q, Nie ZY, Jin LJ. Gene expression profile and functional analysis of Alzheimer’s disease. Am J Alzheimers Dis Other Demen. 2013 Nov;28:693–701. doi: 10.1177/1533317513500838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong W, Mou X, Zhang N, Zeng W, Li S, Yang Y. The construction of common and specific significance subnetworks of Alzheimer’s disease from multiple brain regions. Biomed Res Int. 2015;2015:394260. doi: 10.1155/2015/394260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubio-Perez JM, Morillas-Ruiz JM. A review: inflammatory process in Alzheimer’s disease, role of cytokines. Scientific World Journal. 2012;2012:756357. doi: 10.1100/2012/756357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamburov A, Wierling C, Lehrach H, Herwig R. Consensus Path DB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009 Jan;37:D623–8. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000 Jan;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 21.Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008 Mar 18;105:4441–6. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010 Sep 15;26:2347–8. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Chiang HC, Wu W, Liang B, Xie Z, Yao X, et al. Epidermal growth factor receptor is a preferred target for treating amyloid-beta-induced memory loss. Proc Natl Acad Sci U S A. 2012 Oct 9;109:16743–8. doi: 10.1073/pnas.1208011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali-Rahmani F, Schengrund CL, Connor JR. HFE gene variants, iron, and lipids: a novel connection in Alzheimer’s disease. Front Pharmacol. 2014;5:165. doi: 10.3389/fphar.2014.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spear PG. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 2004 May;6:401–10. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 26.Piacentini R, De Chiara G, Li Puma DD, Ripoli C, Marcocci ME, Garaci E, et al. HSV-1 and Alzheimer’s disease: more than a hypothesis. Front Pharmacol. 2014;5:97. doi: 10.3389/fphar.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam KT, Chan PK, Zhang W, Law PP, Tian Z, Fung Chan GC, et al. Nucleic Acids Res. Sep 19, 2016. Identification of a novel distal regulatory element of the human Neuroglobin gene by the chromosome conformation capture approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jun G, Vardarajan BN, Buros J, Yu CE, Hawk MV, Dombroski BA, et al. Comprehensive search for Alzheimer disease susceptibility loci in the APOE region. Arch Neurol. 2012 Oct;69:1270–9. doi: 10.1001/archneurol.2012.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruchaga C, Nowotny P, Kauwe JS, Ridge PG, Mayo K, Bertelsen S, et al. Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch Neurol. 2011 Aug;68:1013–9. doi: 10.1001/archneurol.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyall DM, Harris SE, Bastin ME, Munoz Maniega S, Murray C, Lutz MW, et al. Alzheimer’s disease susceptibility genes APOE and TOMM40, and brain white matter integrity in the Lothian Birth Cohort 1936. Neurobiol Aging. 2014 Jun;35:1513e25–33. doi: 10.1016/j.neurobiolaging.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayasena CS, Bronner ME. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-beta signaling. J Cell Biol. 2012 Oct 29;199:453–66. doi: 10.1083/jcb.201204138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Town T, Laouar Y, Pittenger C, Mori T, Szekely CA, Tan J, et al. Blocking TGF-beta-Smad2/3 innate immune signaling mitigates Alzheimer-like pathology. Nat Med. 2008 Jun;14:681–7. doi: 10.1038/nm1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogunshola OO, Antoniou X. Contribution of hypoxia to Alzheimer’s disease: is HIF-1alpha a mediator of neurodegeneration? Cell Mol Life Sci. 2009 Nov;66:3555–63. doi: 10.1007/s00018-009-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cermakian N, Lamont EW, Boudreau P, Boivin DB. Circadian clock gene expression in brain regions of Alzheimer ‘s disease patients and control subjects. J Biol Rhythms. 2011 Apr;26:160–70. doi: 10.1177/0748730410395732. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Le W. Pathological role of hypoxia in Alzheimer’s disease. Exp Neurol. 2010 Jun;223:299–303. doi: 10.1016/j.expneurol.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 36.Li JS, Yao ZX. Modulation of FGF receptor signaling as an intervention and potential therapy for myelin breakdown in Alzheimer’s disease. Med Hypotheses. 2013 Apr;80:341–4. doi: 10.1016/j.mehy.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Kitano H. A robustness-based approach to systems-oriented drug design. Nat Rev Drug Discov. 2007 Mar;6:202–10. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- 38.Hart T, Dider S, Han W, Xu H, Zhao Z, Xie L. Toward Repurposing Metformin as a Precision Anti-Cancer Therapy Using Structural Systems Pharmacology. Sci Rep. 2016 Feb 04;6:20441. doi: 10.1038/srep20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan H, Akoglu L, Tong H. Make it or break it: Manipulating robustness in large networks. SIAM International Conference on Data Mining 2014 SDM 2014.1 ed: Society for Industrial and Applied Mathematics Publications; 2014; pp. 325–333. [Google Scholar]
- 40.Braskie MN, Kohannim O, Jahanshad N, Chiang MC, Barysheva M, Toga AW, et al. Relation between variants in the neurotrophin receptor gene, NTRK3, and white matter integrity in healthy young adults. Neuroimage. 2013 Nov 15;82:146–53. doi: 10.1016/j.neuroimage.2013.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh A, Giese KP. Calcium/calmodulin-dependent kinase II and Alzheimer’s disease. Mol Brain. 2015 Nov 24;8:78. doi: 10.1186/s13041-015-0166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W, Yang Y, Yuan G, Zhu W, Ma D, Hu S. Exendin-4, a glucagon-like peptide-1 receptor agonist, reduces Alzheimer disease-associated tau hyperphosphorylation in the hippocampus of rats with type 2 diabetes. J Investig Med. 2015 Feb;63:267–72. doi: 10.1097/JIM.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 43.Xie L, Xie L, Kinnings SL, Bourne PE. Novel computational approaches to polypharmacology as a means to define responses to individual drugs. Annu Rev Pharmacol Toxicol. 2012;52:361–79. doi: 10.1146/annurev-pharmtox-010611-134630. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Z, Xie L, Xie L, Bourne PE. Delineation of Polypharmacology across the Human Structural Kinome Using a Functional Site Interaction Fingerprint Approach. J Med Chem. 2016 May 12;59:4326–41. doi: 10.1021/acs.jmedchem.5b02041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.