Abstract
Posidonia oceanica is a marine phanerogam, largely widespread in the Mediterranean sea, representing an important food substrate for many marine organisms. A progressive reduction of P. oceanica meadows has been reported, due to anthropogenic coastal activity. Studying mechanisms by which this species responds to environmental stresses, three DNA sequences putatively encoding metallothioneins (MTs) have been isolated, by PCR. Two sequences, Pomt2a (accession no. AJ249603) and Pomt2b (accession no. AJ249602), show high similarities with genes encoding type-II MTs and are interrupted by two and one intron, respectively. The third sequence, Pomt2c (accession no. AJ249604), is supposed to be a pseudogene, originated by retrotranscription of the Pomt2b mRNA. These sequences belong to a multigene family with at least five members. Northern hybridizations indicated that MT transcripts accumulation is constitutive and seasonally regulated. MT encoding RNAs increase after rhyzome harvesting and (at a lesser extent) after 15 d of cultivation in an aquarium. As for animal MTs, transcripts accumulation is observed also after exposure to trace metals such as copper and cadmium. In the case of copper, the effect depends on concentration. Finally, taking into consideration the great interest in studying the biogeochemical cycle of mercury in the Mediterranean basin and since P. oceanica is commonly considered a bioindicator of this metal, the effect of mercury treatments on the accumulation of MT transcripts has been analyzed: in only a few experiments a small increase in the level of transcripts was recorded, suggesting that MTs are not key elements in the mercury accumulation by this species.
Seagrasses of the genus Posidonia are marine phanerogams of the Potamogetonaceae family. They grow along coastal waters of the Mediterranean basin and Australia and form dense infralittoral populations that frame the so-called Posidonia meadow ecosystem (Boudouresque and Meinesz, 1982). In particular, Posidonia oceanica (L.) Delile is largely spread in the Mediterranean basin, where large prairies of this plant represent a dynamic substrate that exceeds the area of the sediment surface several times over and allows settlement of epiphyte organisms. P. oceanica prairies extend from a few meters up to 30 to 40 m in depth and occupy an area of approximately 50,000 km2 (Maserti et al., 1991).
Because of its high productivity, P. oceanica represents an important food substrate for many marine organisms (Novak, 1982; Pirc, 1985). The productivity is different in the various Mediterranean areas due to the sensitivity of the plants to changing local and temporal factors of its environment (Giaccone et al., 1981). During recent years in many areas of the Mediterranean sea, the progressive reduction of P. oceanica meadows due to coastal activity has been reported (Peres, 1984; Pergent-Martini and Pergent, 1990). This reduction was also related to the widespread vegetative propagation in Posidonia prairies (Loques et al., 1990; Buia and Mazzella, 1991; Capiomont et al., 1996; Procaccini and Mazzella, 1998). The resulting genetic uniformity should not allow plants to face environmental changes.
Like other seagrasses, i.e. Zostera and Cymodocea, Posidonia plays a major role in maintaining marine environments. However, basic understanding of seagrass molecular physiology is still limited (Cavallini et al., 1995; Fukuhara et al., 1996), in particular with regards to molecular mechanisms involved in environmental adaptation that could appear peculiar to these plant species compared to terrestrial phanerogams.
It is known that P. oceanica may absorb and accumulate metals from sediments in its organs and tissues (Maserti et al., 1988; Ferrara et al., 1989; Rainbow and Phillips, 1993; Warnau et al., 1996; Schlacher-Hoenlinger and Schlacher, 1998) determining metal bioavailability in the marine ecosystem. In particular, since P. oceanica can live on mercury-rich sites, it has been proposed to be a bioindicator of this metal (Maserti et al., 1988; Pergent-Martini, 1998).
Trace metal accumulation in plants may induce the production of small peptides, the phytochelatins, and/or 7,000- to 10,000-kD peptides, metallothioneins (MTs), whose involvements in the response of plants to trace metals is still controversial (Robinson et al., 1993; Zenk, 1996).
MTs are defined as low-Mr Cys-rich proteins that bind heavy metals. They are widely distributed in eukaryotic and prokaryotic species (Robinson et al., 1993). In higher plants, MT proteins have been purified and sequenced only in Arabidopsis (Murphy et al., 1997). In most cases, however, the presence of proteins that are enriched in Cys and bind metals has been largely inferred from DNA sequences, so that the term MT-like is preferred if protein-metal characterization is not established (Hsieh et al., 1996). Plants may carry three major classes of MT proteins, classes I, II, and III, which are distinguishable on the basis of their sequences (Robinson et al., 1993; Murphy et al., 1997). Class-I MT-like proteins are characterized by two Cys-rich domains separated by a central Cys-free spacer; class-II MTs are represented by the wheat Ec protein, lacking the internal spacer (Kawashima et al., 1992); class-III MTs include diverse glutathione-derived peptides as phytochelatins (Zenk, 1996). Class-I MT-like proteins are further distinguished as type I or type II, based on their Cys spacing; in both types there are six Cys residues in each of the two domains, arranged as CXC motifs (C stands for Cys and X for variable amino acids), but in the amino terminus of type-II MT-like proteins, two additional Cys are present in tandem and one of the CXC motifs exists as CXXC (Robinson et al., 1993). Moreover, in recent years other MT-like coding sequences have been isolated showing different Cys distribution, mainly at the amino terminus, for which the term type-III MT-like proteins has been proposed (Reid and Ross, 1997).
In animals and fungi, MTs have been shown to play a role in the detoxification of heavy metals (Robinson et al., 1993), although their exact function is not completely understood. In plants a correlation has been reported between MT RNA levels and naturally observed differences in the tolerance to heavy metals in Arabidopsis ecotypes, suggesting a role in metal homeostasis (Murphy and Taiz, 1995; Murphy et al., 1997).
The effect of metals on the expression of MTs varies with the plant species, tissue, and MT type. For example, in Arabidopsis (Zhou and Goldsbrough, 1994; Murphy and Taiz, 1995), wheat (Snowden et al., 1995), pea (Fordham-Skelton et al., 1997) and rice (Hsieh et al., 1995), transcription of MTs was enhanced by certain metals. By contrast, in certain species (De Miranda et al., 1990; Kawashima et al., 1991; Okumura et al., 1991), MT mRNA levels were decreased by copper treatment, whereas in other (Lane et al., 1987; Choi et al., 1996; Foley et al., 1997), MT expression was not affected by metals. As reported for animals (Robinson et al., 1993), a number of other stimuli, including abscisic acid, heat shock, cold shock, wounding, viral infection, senescence, fruit ripening, salinity, and Suc starvation, have been shown to affect the accumulation of MT transcripts in plants (Foley and Singh, 1994; Hsieh et al., 1995; Snowden et al., 1995; Choi et al., 1996; Buchanan-Wollaston and Ainsworth, 1997; Foley et al., 1997; Butt et al., 1998; Woodhead et al., 1998; Nam et al., 1999).
In the terrestrial environment, plants uptake trace metals mainly by the root system, whereas leaves can absorb metals only if associated to the particulate matter in the atmosphere or in volatile forms. By contrast, marine phanerogams may uptake trace metals by both roots and leaves, thus providing a valuable tool to study cellular mechanisms of response to the presence of trace metals in the environment. During a study on the mechanisms by which P. oceanica responds to trace metals, three DNA sequences putatively coding MTs were isolated by PCR using primers homologous to genes coding other plant MTs. In this paper the genetic and functional characterization of these sequences are reported.
RESULTS
Isolation of DNA Sequences Putatively Encoding Type-II MTs
Three genomic sequences putatively encoding MTs were isolated by PCR amplification, using primers homologous to genes encoding other plant MTs. These sequences are named Pomt2a, Pomt2b, and Pomt2c and are 747, 364, and 226 bp long, respectively. The deduced proteins of the three sequences were analyzed by the BLAST program at National Center for Biotechnology Information (Bethesda, MD) (Altschul et al., 1990) and showed high similarities to putative plant MTs.
After comparison to other MT sequences and using the program FEX at Baylor College of Medicine (Houston), it was inferred that Pomt2a should contain two introns, from bases 65 to 499 and from bases 576 to 660. Pomt2b should contain only one intron, from bases 143 to 279. The second intron of Pomt2a is located at the same site of the unique Pomt2b intron, but it is shorter than the unique Pomt2b intron (84 versus 136 bp).
The deduced amino acid sequences of Pomt2a and Pomt2b contain the Cys-rich regions typical of plant class-I type-II MTs, including the presence of a CXXC motif at the amino terminus (see the introduction). The alignment of Pomt2a and Pomt2b deduced proteins to other eight plant type-II MTs is reported in Figure 1. Only minor differences are observed between Pomt2a and Pomt2b with regard to their deduced amino acid sequence in the Cys domains, in particular three conservative substitutions in the 5′ domain and one conservative substitution in the 3′ domain, respectively. More differences are found in the internal spacer: 16 amino acids were substituted, nine of which were conservative and seven non-conservative.
Figure 1.
Sequence alignment of Pomt2a and Pomt2b deduced proteins to other type-II MTs (for which GenBank code and species name is reported). Sequences were aligned by the CLUSTAL program. Conserved amino acids are reported in bold. Dashes are included for optimal alignment. Target sequences of Pomt2a and Pomt2b are not included in the comparison.
The third sequence, Pomt2c, varies from the others for one base deletion at base 62 and some base substitutions and for the absence of the DNA sequences corresponding to the introns of Pomt2a and Pomt2b. The deletion at base 62 determines a frame shift and the presence of a stop codon in the RNA. For this reason, and because of the absence of introns, Pomt2c can probably be considered as a pseudogene that originated by reverse transcription of the mRNA produced by other MT sequences. In particular it is conceivable that Pomt2c derived from reverse transcription of a mRNA encoded by Pomt2b, because only five base substitutions are observed between Pomt2c and the coding regions of Pomt2b, versus the 41 base substitutions between Pomt2c and those of Pomt2a.
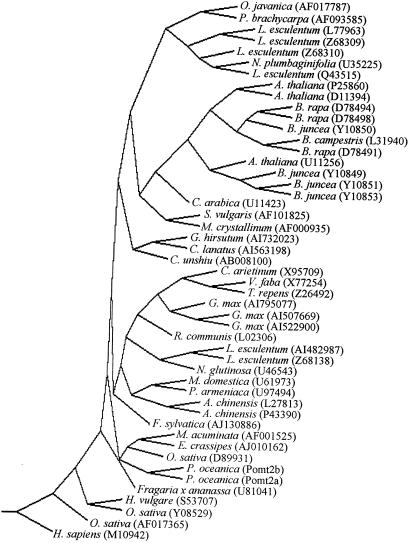
The sequence comparison between Pomt2a-b and other 46 plant type-II MTs produced a single most parsimonious tree, reported in Figure 2.
Figure 2.
Strict consensus of 100 most parsimonious trees of plant type-II MTs, obtained using PROTPARS software (PHYLIP package version 3.572). An MT coding sequence of Homo sapiens (M10942) was used as the outgroup. Bootstrap support values (%) are shown at the forks. For each sequence the GenBank code is reported.
Genomic Organization of the Isolated MT Coding Sequences
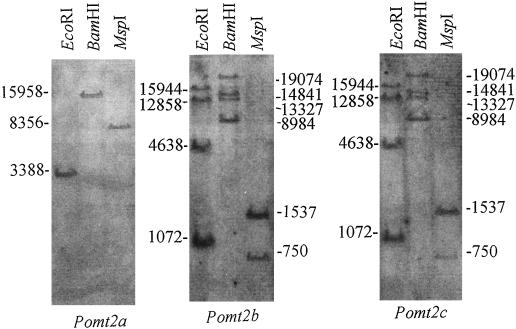
Southern hybridizations were performed to estimate the copy number of each gene. Southern-blot experiments were then made using DNAs restricted with enzymes that do not recognize sites within any of the three MT-like coding genomic sequences (Fig. 3).
Figure 3.
Southern blots of P. oceanica DNA, digested with enzymes that do not recognize sites within isolated putative MT coding sequences, hybridized with digoxygenin-labeled Pomt2a, Pomt2b, or Pomt2c. Molecular masses of bands are reported in bp.
It may be observed that Pomt2b and Pomt2c show the same restriction pattern (four EcoRI, four BamHI, and two MspI bands), indicating the presence of at least four members of these two sequences in the P. oceanica genome, altogether. By contrast, Pomt2a shows only one band, suggesting that this may be a single-copy gene. On the whole, at least five MT genes should be present in P. oceanica genome.
Steady-State Level of MT mRNAs in Leaf Tissues of P. oceanica in Vivo
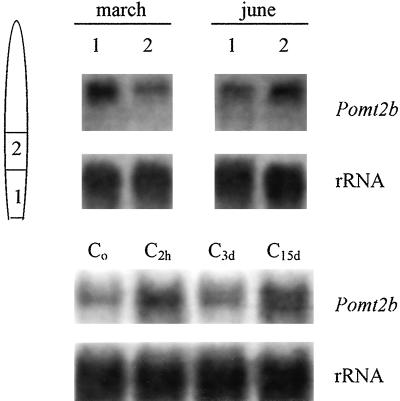
The accumulation of MT-like transcripts was analyzed in vivo by northern-blot experiments and hybridization with Pomt2b probe. The minor differences in the coding regions between Pomt2a and Pomt2b did not distinguish Pomt2a or Pomt2b mRNA sequences in northern blots. In Figure 4, the steady-state levels of MT transcripts in basal and central portions of leaves collected in March or June of 1999 and frozen in liquid nitrogen immediately after explantations are reported.
Figure 4.
Northern blots of RNAs isolated from: different regions of leaves of P. oceanica (1, basal; 2, central; a schematic representation of leaf is reported) collected from Antignano meadows in different months and immediately frozen in liquid nitrogen (top) or isolated from the basal region of leaves of P. oceanica collected in March, immediately frozen in liquid nitrogen (Co) or frozen after 2 h (C2h), 3 d (C3d), or 15 d (C15d) in an aquarium, hybridized with digoxygenin-labeled Pomt2b probe (bottom). Ten micrograms of total RNA per sample was loaded. A Phaseolus coccineus 25S rDNA fragment was used as a control probe (rRNA).
Very low metal concentrations were measured in the seawater where seagrasses were collected at least for copper, cadmium, and mercury (Table I). Total (i.e. including both bioavailable and chelated) metal concentrations were about 10−2 μm for copper, 10−3 μm for cadmium, and 10−4 μm for mercury; these concentrations are much lower than those commonly used in experiments involving metal treatments for MT induction in plants. In this sense, the accumulation of MT transcripts in vivo should be considered as constitutive.
Table I.
Copper, cadmium, and mercury concentrations in seawater and in plant tissues in vivo
| Copper (Seawater 10−2) | Cadmium (Seawater 10−3) | Mercury (Seawater 10−4) | ||||
|---|---|---|---|---|---|---|
| Sampling time | 1 | 2 | 1 | 2 | 1 | 2 |
| March | 20.7 | 17.5 | 2.3 | 3.5 | 0.3 | 0.7 |
| June | 21.6 | 13.4 | 2.8 | 2.3 | 0.2 | 0.2 |
Cu2+, Cd2+, and Hg2+ concentrations measured in the seawater where plants were collected (μm) and in young (1) or differentiated (2) portion of P. oceanica leaves (μg/g dry wt; see Fig. 4) of plants collected in March and June of 1999 at the same sampling site. For each metal determination in plant tissues, values are not significantly different at the 5% level with Tukey's ranking test.
However, differences were observed between leaf portions: younger leaf portions accumulated more MT transcripts than differentiated ones in March, whereas in June, more transcripts were found in the differentiated portions than in the basal ones (Fig. 4, top).
The copper, cadmium, and mercury content of leaf tissues collected in March and June of 1999 is reported in Table I. No relationship may be found between metal concentrations and MT transcripts accumulation in the different leaf portions and time periods.
Steady-State Level of MT mRNAs in Leaf Tissues of P. oceanica in the Aquarium
The steady-state level of MT transcripts in the basal leaf portions of plants acclimated in the aquarium was studied by northern blot and hybridization with labeled Pomt2b as probe (Fig. 4, bottom). An increase in the amount of MT transcripts was observed after 2 h from shoot explantation, with a subsequent decrease in shoots acclimated for 3 d in the aquarium and a slight increase after 15 d in the aquarium.
Trace-Metal-Induced Accumulation of MT Transcripts in Leaf Tissues of Plants Cultivated in the Aquarium
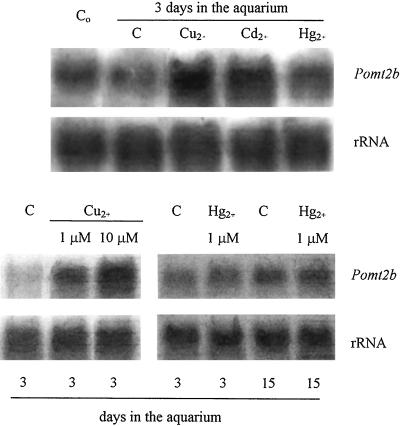
To study metal induction of MT transcripts accumulation, experiments were performed on shoots of P. oceanica after 1 d of acclimation in aquarium. After this time, seawater was substituted and CuCl2, Cd(NO3)2, or Hg(NO3)2 was added at final concentrations of 10 μm, 10 μm, and 10−2 μm, respectively. After 2 d, total RNA was isolated from basal leaf portions, of which the copper, cadmium, and mercury concentrations are reported in Table II. Experiments of northern blot and hybridization with labeled Pomt2b probe (Fig. 5) indicate that MT transcripts accumulation is increased by copper and cadmium, whereas no apparent effect is observed after mercury treatment.
Table II.
Copper, cadmium, and mercury concentrations in metal-treated plants
| Treatment Time | Copper
|
Cadmium
|
Mercury
|
|||
|---|---|---|---|---|---|---|
| 1 μm | 10 μm | 10 μm | 10−2 μm | 1 μm | 5 μm | |
| 2 d (Control) | 18.0a | 15.7a | 3.3a | 0.4a | 0.2a | 0.3a |
| 2 d (Treated) | 27.9b | 80.2c | 302.9b | 40.8b | 252.0c | 400.8d |
| 15 d (Control) | – | – | – | 0.4a | 0.5a | 0.2a |
| 15 d (Treated) | – | – | – | 50.3b | 290.0c | 635.8e |
Cu2+, Cd2+ and Hg2+ levels (μg/g dry wt) measured in P. oceanica leaves from plants experienced, at laboratory conditions, with different metals and metals concentrations, independently. For each metal, means followed by the same letter are not significantly different at the 5% level with Tukey's ranking test.
Figure 5.
Northern blots of RNAs isolated from the basal region of leaves of P. oceanica collected in June from Antignano meadows. Top, Immediately frozen in liquid nitrogen (Co) or frozen after 3 d in an aquarium supplemented (for the last 2 d) with 10 μm CuCl2 (Cu2+), 10 μm Cd(NO3)2 (Cd2+), 1 μm Hg(NO3)2 (Hg2+) or with no trace metal added (C); bottom left, frozen after 3 d in an aquarium supplemented with no trace metal (C) or treated (for the last 2 d) with 1 μm or 10 μm CuCl2; bottom right, frozen after 3 or 15 d in an aquarium supplemented with no trace metal (C) or treated (for the last 2 or 14 d, respectively) with 1 μm Hg(NO3)2. Digoxygenin-labeled Pomt2b was used as probe. Ten micrograms of total RNA per sample was loaded. A P. coccineus 25S rDNA fragment was used as a control probe (rRNA).
In other experiments, plants were treated with different (1 and 10 μm) CuCl2 concentrations. Northern-blot experiments (Fig. 5) show that the increase in steady-state level of MT transcripts depends on the copper concentration, at least in the range of concentrations used in the experiments undertaken here. The increase of copper concentration in leaves treated with 1 μm CuCl2 is very small, hence CuCl2 concentrations lower than 1 μm were not used. On the other hand, treatments with CuCl2 concentrations higher than 10 μm was fatal to the plants in the aquarium after 1 d.
Finally, since P. oceanica is known to be mercury tolerant and a bioindicator of environmental mercury contamination, shoots have been treated with different mercury concentrations [10−2, 1, and 5 μm Hg(NO3)2] for 2 and 14 d. Only a small (and not regularly observed) increase of MT transcripts was found in treated plants for every mercury concentration and treatment duration tested. In Figure 5, northern blot of RNA of leaves of plants treated with 5 μm Hg(NO3)2 hybridized with labeled Pomt2b probe is reported. The results obtained in the other mercury treatments are similar and are not shown.
DISCUSSION
Physiological studies on marine phanerogams are very rare, probably due to difficulties on collecting and cultivating plants, though fundamental for the great importance in maintaining the marine ecosystem. In particular, studies on the response of these plants to environmental stimuli can be useful to establish adequate strategies of protection of marine environment.
P. oceanica is commonly studied as a trace-metal bioaccumulator (see the introduction), but no information on molecular mechanisms of metal tolerance is currently available. On the other hand, such molecular mechanisms are yet to be completely established for every plant species. It is known, for example, that genes coding for putative MTs are present in plant genomes, but their role in stress response is still controversial (Zenk, 1996).
In our experiments, three DNA sequences had been isolated whose deduced proteins show strong similarities with deduced class-I type-II MTs of other plant species (Fig. 1). Two of them, Pomt2a and Pomt2b, are probably different genes, as shown by the different hybridization-labeling pattern in Southern-blot experiments (Fig. 3). Both Pomt2a and Pomt2b are related to type-II MTs of other monocotyledons, as shown by phylogenetic analysis (Fig. 2).
It is interesting that the third sequence, Pomt2c, is very similar to Pomt2b except for the absence of the intron and some point mutations, of which one (in position 62) determines the presence of a stop codon. These characteristics support the hypothesis that this sequence is a pseudogene derived from retrotranscription of a Pomt2b mRNA. A similar case was described in human genome (Hamer, 1986), where five MT pseudogenes were isolated in addition to six functional genes. The origin of Pomt2c from a transcribed Pomt2b sequence is also indicated by the results of Southern-blot and hybridization experiments: the same pattern is obtained using the two sequences as probes (Fig. 3). However, only analysis of sequences flanking Pomt2c in the genome of P. oceanica may definitely clarify the nature of this sequence.
Other considerations may derive from the results of Southern-blot experiments. For example, the single band observed for Pomt2a indicates that this is a single-copy gene; moreover, unless two different allele hybridize on fragments of the same length, the single band should indicate that these plants are probably homozygous, supporting the theory that, besides through vegetative propagation, these plants reproduce (even rarely) autogamously, as shown by microsatellite analyses on different populations (Procaccini and Mazzella, 1998).
According to this hypothesis, since Pomt2b or Pomt2c show four hybridization bands, it should be conceivable that at least four of these genes are present in Posidonia genome, with other pseudogenes than Pomt2c possibly contributing to this number. Obviously, it is also possible that these plants are homozygous only for Pomt2a gene, and heterozygous for the other MT genes, that consequently would be in number lower than four.
The expression of the isolated sequences was studied by northern-blot and hybridization experiments. No specificity could be established in expression pattern between Pomt2a and Pomt2b in northern blots, because of the high homology between coding regions of these sequences. We assume that hybridization signals using labeled Pomt2b as probe are due to Pomt2b mRNAs; however, it is also possible that those signals were partly due to Pomt2a mRNAs. Experiments of quantitative RT-PCR (using specific primers) are being planned to establish if different genes are expressed in response to different stimuli or metals, as observed for MT genes of Arabidopsis (Murphy and Taiz, 1995).
MT mRNAs were found in leaves collected from plants in the sea and immediately frozen (Fig. 4, above). Actually, the concentration of copper, cadmium, and mercury in the seawater where seagrasses were collected was very low (Table I), i.e. much lower than metal concentrations commonly used to induce gene expression in other plants. On the other hand, total metal concentrations in the water is similar to those measured in other sites of the Tyrrhenian Sea: hence, it is conceivable that MT genes are expressed constitutively, as reported for Arabidopsis roots (Murphy et al., 1997). Alternatively, it is possible that metals induce MT gene expression in P. oceanica at thresholds lower than those established for other plant species. Moreover, it is also possible that MT mRNA accumulation can be induced by metals different from those studied in the experiments described here.
The steady-state level of MT mRNA in young and differentiated portions of the leaf and in different time periods was also analyzed (Fig. 4, top). Differences were observed between leaves collected in March or in June, in particular related to the leaf portion analyzed. Younger tissues showed higher MT mRNA levels than older ones in March; whereas in June, a somehow opposite result was obtained. Copper, cadmium, and mercury concentrations in the water did not vary between March and June. Concerning the leaf portions from which RNAs were isolated, no significant differences were observed in copper, cadmium, and mercury concentrations (Table I), indicating that variations found in the steady-state MT mRNA level between young and differentiated leaf portions are not related to metal translocations in the plant.
Different stimuli are known to determine high steady-state levels of MT mRNAs in other plant species (see the introduction). For example, tissue wounding is known to induce MT mRNA increase in Nicotiana spp. (Choi et al., 1996). MT mRNA levels were studied in leaves from plants after 2 h from harvesting, after 3 and 15 d in the aquarium without adding metals (Fig. 4, bottom). Leaves of plants 2 h after sampling show an increase of MT mRNA levels, probably induced by wounding. After 3 d, MT mRNA signal is similar to those recorded at time 0, i.e. MT mRNA accumulation reduces with acclimation to aquarium conditions. A further increase in MT mRNA levels is found in leaves from plants cultivated for 15 d in the aquarium. This is probably because of the stress imposed on plants by prolonged time periods in the aquarium, which indicates the necessity of studying P. oceanica physiology only in the first days after harvesting.
Among the trace metals that can induce the expression of MT genes, the effects of copper, cadmium and mercury were studied. Concerning copper and cadmium (Fig. 5), an increase of MT mRNA levels after 2-d treatments with 10 μm copper or cadmium was noted, confirming that these metals can induce MT gene expression, as already reported for other plant species (for example, see Murphy and Taiz, 1995). It is worth noting that an MT gene product (with similarities to plant MT sequences) has recently been isolated from a marine microalga Fucus vesicolosus; this gene is induced by copper and its encoded protein binds cadmium and copper (Morris et al., 1999).
In particular the effect of copper was studied using different CuCl2 concentrations (1 and 10 μm). Even at a concentration of 1 μm, copper induces a strong increase of MT mRNA levels in leaves of P. oceanica (Fig. 5); even relatively small increases of copper uptake (Table II) may determine an increase in MT mRNAs. It is possible that the stimulus inducing mRNA accumulation is not (or it is not only) the increase of total metal concentration, but possibly a variation in copper cellular compartmentalization following copper treatment. Such a variation has been assumed to result also after stresses as senescence and Suc deficiency (Fordham-Skelton et al., 1997).
In other experiments, the effect of mercury on the level of MT transcripts was analyzed (Fig. 5). Although in human tissues mercury is known to induce MT synthesis (Palmiter, 1994), studies on MT induction by this metal in plants do not exist.
Different mercury concentrations for 2 or 14 d in the aquarium were used. A slight increase in the steady-state level of MT transcripts was recorded in only a few experiments (independently of concentration and treatment time) indicating that MTs are not (or are only slightly) involved in the high-mercury tolerance shown by P. oceanica.
In conclusion, the experiments reported in this paper show that putative MT genes are present in P. oceanica genome, where they form a multigene family. Our data show that MT transcripts are present constitutively in the plant and are increased by different stimuli, supporting the hypothesis that MTs participate to processes of metal homeostasis and possibly tolerance. Moreover, MT induction seems to be metal-specific. MT mRNAs are increased by copper and cadmium, but not by mercury.
To further elucidate these aspects, we plan to isolate flanking sequences of these genes to verify the presence of metal-responsive elements in the promoters, as described for other MT genes, both in animals (Culotta and Hamer, 1989) and in plants (Whitelaw et al., 1997). Moreover, other experiments are programmed to isolate type-I MT genes from Posidonia genome.
Such studies will help to clarify the role of these genes in metal homeostasis and to isolate P. oceanica genotypes particularly tolerant to stress, to be used in experiments of recovery of P. oceanica populations in the Mediterranean Sea.
MATERIALS AND METHODS
Plant Materials
Posidonia oceanica (L.) Delile shoots were collected in different periods by scuba diving at between 2 and 5 m depth in the P. oceanica meadow of Antignano (Leighorn, Italy). Excess sediment was removed by washing the plants gently in seawater, before placing them in plastic bags filled with seawater. Some plants were frozen in liquid nitrogen immediately after explantation for RNA analyses and metals determinations.
Shoots were then acclimated to laboratory conditions (closed circuit aquaria, constantly aerated; 16.5°C ± 0.5°C; light/dark cycle, 16 h/8 h; six shoots per aquarium containing 12 L of natural seawater, collected in the same place) for 24 h before starting metal treatments.
Metal Treatments
Seawater culture medium was spiked with CuCl2, Cd(NO3)2, or Hg(NO3)2 solutions to reach total concentrations of 1 or 10 μm copper; 10 μm cadmium; and 10−2, 1, or 5 μm mercury. Metal treatment duration was 2 d for plants coping with CuCl2 and Cd(NO3)2 and 2 and 14 d for those experiencing Hg(NO3)2. During mercury prolonged treatments, seawater in the aquarium was substituted every 2 d.
Analyses were performed on two leaf portions: the basal region (undifferentiated and light-green) and the intermediate region (differentiated and green). The leaf tip was not used because of the presence of epiphytes.
Metal Content Determination
Samples Preparation
Leaf portions were rinsed carefully in seawater and dried until constant weight in an electric oven at 40°C. One hundred milligrams of each dried sample was mineralized by a 7-mL solution (5:2) of H(NO3):H2O2 in a microwave oven (Milestone MSL 1200, Bergamo, Italy) for 25 min. The mineralized samples were diluted 1:100 with Milli-Q (Millipore, Bedford, MA) and then analyzed for metal determination.
Copper and Cadmium Determination in Plants
The copper and cadmium content of each leaf sample was independently determined by an atomic absorption spectrometer (1100B, Perkin Elmer, Foster City, CA) equipped with a graphite furnace (HGA 700, Perkin Elmer) and fitted with autosampler. Pro-analysis grade reagents (Merck) were used in every case. Standards were prepared by serial dilution of commercially available stock solutions within the linear range of respective metals.
Copper and Cadmium Determination in Water
The copper and cadmium content in seawater was measured by differential pulse anodic stripping voltammetry using a MetroHom (Zurich) 646 VA processor and 647 electrode system with hanging mercury drop electrode. Due to the very low background levels in seawater the metals concentrations were estimated about 10−2 μm for copper and 10−3 μm for cadmium.
Mercury Determination in Plants
The mercury content of each leaf sample was determined independently by cold vapor atomic absorption spectrometry with a detection limit of 8 ng, and was the mean of three replicates. The typical contribution of the blank was 1 to 2 ng of mercury. Standards were prepared with serial dilution of commercially available stock solutions within the linear range of the metal.
Mercury Determination in Water
A 400-mL seawater sample was acidified with 400 μL of a solution of mercury-free potassium dichromate (0.4% [w/v] K2Cr207 in 9 m H2SO4) and then photooxidized for 15 min using a UV immersion lamp (90 W). The mercury was reduced with 10 mL of mercury-free tin chloride solution (10% SnCl2 in 4 m H2SO4) as described elsewhere (Seritti et al., 1980). Mercury was measured by cold vapor atomic absorption spectometry after preconcentration on a gold absorber (Seritti et al., 1980).
Isolation of Genomic DNA
DNA was purified according to the method devised by Doyle and Doyle (1989) with modifications. Leaf portions were homogenized in liquid nitrogen in a mortar and lysed in hexadecyl-trimethyl ammonium bromide (CTAB) isolation buffer (3% [w/v] CTAB [Sigma], 1.4 m NaCl, 0.2% [v/v] 2-mercaptoethanol, 20 mm EDTA, and 100 mm Tris-HCl, pH 8.0), and 1 mL/g tissue, at 60°C. Samples were incubated at 60°C for 30 min with occasional gentle swirling and then extracted once with chloroform:isoamyl alcohol (24:1, v/v). After centrifugation (5,000 rpm) at room temperature, nucleic acids were precipitated from the aqueous phase by adding 0.67 volume of cold isopropanol and then spooled with a glass hook, washed in 76% (v/v) ethanol and 10 mm ammonium acetate for 1 to 2 h, allowed to dry briefly, and resuspended in water.
For further purification, solid CsCl and ethidium bromide were added to the nucleic acids solution up to final concentrations of about 0.8 mg/mL (refraction index of 1.3890–1.3895) and 200 μg/mL, respectively. The solution was centrifuged at 44,000 rpm in an ultracentrifuge (L5–65, Beckman Instruments, Fullerton, CA) for 30 h at 15°C using the model 65 Ti rotor, and the DNA band, which was visualized under long-wave UV illumination, was collected. Ethidium bromide was then removed by gentle inversion of the solution with n-butanol and dialyzed against water at 4°C for 3 h. Finally, DNA was ethanol-precipitated and resuspended in the appropriate buffer.
Isolation of Genomic Sequences Coding Putative Type-II MTs
DNA sequences homologous to MT coding genes were isolated by PCR on P. oceanica genomic DNA. PCR was performed using two degenerated oligonucleotides based on the published DNA sequences encoding MTs in other plant species: 5′-ATGTCTTGCTGYGGAGGAARCTGT-3′ (sense) and 5′-ACAAKYGCARGGGTYACASK TGCA-3′ (antisense). Sequences were amplified using 100 ng of genomic DNA as a template; thermocycling was performed at 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s, for 30 cycles, using Taq-DNA polymerase (Promega, Madison, WI).
The amplified fragments were cloned into a pGEM-T Easy plasmid vector (Promega). The cloned fragments were sequenced by the dideoxy-chain termination method (Sanger et al., 1977) using the PRISM dye terminator cycle sequencing kit (Perkin-Elmer) according to the manufacturer's instructions; sequences were analyzed using the Sequencing Analysis 2.1.2 and the Sequencer 3.0 analysis programs (Perkin-Elmer Applied Biosystems, Foster City, CA).
Southern Analysis
Ten micrograms of genomic DNA was digested with the restriction endonucleases EcoRI, BamHI, and MspI in a 5-fold excess according to the instructions of the suppliers (Boehringer, Mannheim, Germany). Complete digestion was checked by including unmethylated bacteriophage λ-DNA, which, when digested with EcoRI plus HindIII (DNA molecular size marker III, Boehringer), was also used as a fragment size marker. Southern blotting of digested DNAs was performed according to standard protocols (Sambrook et al., 1989) and hybridizations were made with digoxigenin-labeled riboprobes transcribed from Pomt2a, Pomt2b, or Pomt2c clones using the DIG RNA labeling kit SP6/T7 (Boehringer).
Hybridizations were performed using the labeled RNA probes under high-stringency conditions, at 50°C in 50% (v/v) formamide, 5× SSC (1× SSC is 0.15 m NaCl and 0.015 m trisodium citrate, pH 7.0), 2% (w/v) blocking reagent (Boehringer), 0.02% (w/v) SDS, 0.1% (w/v) sodium lauroyl sarcosinate. Filters were washed twice in 2× SSC, 0.1% (w/v) SDS for 15 min at room temperature, once in 1× SSC, 0.1% (w/v) SDS for 30 min at 68°C, and once in 0.3× SSC, 0.1% (w/v) SDS for 30 min at 68°C.
RNA Isolation and Northern-Blot Analysis
Total RNA extraction was performed by the same CTAB method used for DNA isolation, except that higher volumes extraction buffer were used in this case to increase the RNA yield. RNA was solubilized in diethylpirocarbonate-treated water. RNA integrity was assayed on agarose gels, before northern blotting.
Total RNAs (10 μg) were separated by 1% (w/v) formaldehyde agarose gel electrophoresis and blotted onto positively charged nylon membranes (Boehringer). The integrity and the equal amount of RNA loading were confirmed by ethidium bromide staining and subsequent densitometric image analysis and hybridization with a Phaseolus coccineus rDNA probe. The ribosomal probe was the EcoRI-Sau3A 25S-rDNA fragment of clone pPH1 (Maggini et al., 1992), subcloned in pBluescript II SK+ (Stratagene, La Jolla, CA).
The Pomt2b DNA sequence (or the P. coccineus rDNA) was digoxygenin-labeled using the DIG RNA labeling kit SP6/T7 (Boehringer). Hybridizations and subsequent washes were performed under high-stringency conditions, as described for Southern analysis, except for hybridization temperature (68°C). Northern blots were repeated three times per experiment. When hybridization signals seemed over saturation, experiments were repeated loading reduced amounts of RNA.
DNA Sequence Analysis
Intron delimitation within genomic sequences was made by comparing other putative MT-like genomic sequences and confirmed by the use of the program FEX (Baylor College of Medicine).
Database searches were conducted using the program BLAST (National Center for Biotechnology Information). Alignment of deduced amino acid sequences of Pomt2a and Pomt2b to other 46 type-II MT sequences was obtained using the program CLUSTALW (GenomeNet, Kyoto).
Maximum parsimony analysis was performed using the PHYLIP program package version 3.572 (Felsenstein, 1989): using the SEQBOOT program, 100 versions of the original alignment were generated. Trees were constructed using the PROTPARS and NEIGHBOR programs. A strict consensus tree was generated from the available trees using the CONSENSE program.
ACKNOWLEDGMENTS
We thank Prof. Fabio Maggini (Università della Tuscia, Viterbo, Italy) who gave us the rDNA probe, Dr. Elisabetta Morelli and Luciano Nannicini (Consiglio Nazionale delle Ricerche, Institute of Biophysics, Pisa, Italy) for their help in copper and cadmium determinations, and students Ylenia Chiari and Pierluigi Maestrini for plant harvesting.
Footnotes
This research was supported by Consiglio Nazionale delle Ricerche, Target Project on Biotechnology.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Boudouresque CF, Meinesz A. Decouverte de l'herbier de Posidonie. Marseille, France: Parc National Port-Cros/Parc National Region Corse, GIS Posidonie; 1982. [Google Scholar]
- Buchanan-Wollaston V, Ainsworth C. Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridization. Plant Mol Biol. 1997;33:821–834. doi: 10.1023/a:1005774212410. [DOI] [PubMed] [Google Scholar]
- Buia MC, Mazzella L. Reproductive phenology of the Mediterranean seagrasses Posidonia oceanica (L.) Delile, Cymodocea nodosa (Ucria) Aschers and Zostera noltii Hornem. Aquat Bot. 1991;40:343–362. [Google Scholar]
- Butt A, Mousley C, Morris K, Beynon J, Can C, Holub E, Greenberg JT, Buchanan-Wollaston V. Differential expression of a senescence-enhanced metallothionein gene in Arabidopsis in response to isolates of Peronospora parasitica and Pseudomonas syringae. Plant J. 1998;16:209–221. doi: 10.1046/j.1365-313x.1998.00286.x. [DOI] [PubMed] [Google Scholar]
- Capiomont A, Sandmeier M, Caye G, Meinesz A. Enzyme polymorphism in Posidonia oceanica, a seagrass endemic to the Mediterranean. Aquat Bot. 1996;54:265–277. [Google Scholar]
- Cavallini A, Natali L, Giordani T, Polizzi E, Balestri E, Cinelli F, Maserti BE, Ferrara R. Cytophotometric and biochemical characterization of Posidonia oceanica L. (Potamogetonaceae) genome. Caryologia. 1995;48:201–209. [Google Scholar]
- Choi D, Kim HM, Yun HK, Park JA, Kim WT, Bok SH. Molecular cloning of a metallothionein-like gene from Nicotiana glutinosa L. and its induction by wounding and tobacco mosaic virus infection. Plant Physiol. 1996;112:353–359. doi: 10.1104/pp.112.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta VC, Hamer DH. Fine mapping of a mouse metallothionein gene metal response element. Mol Cell Biol. 1989;9:1376–1380. doi: 10.1128/mcb.9.3.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda JR, Thomas MA, Thurman DA, Tomsett AB. Metallothionein-like gene from the flowering plant Mimulus guttatus. FEBS Lett. 1990;262:29–32. doi: 10.1016/0014-5793(90)80122-y. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1989;12:13–15. [Google Scholar]
- Felsenstein J. PHYLIP-phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Ferrara R, Maserti BE, Paterno P. Mercury distribution in maritime sediment and its correlation with the Posidonia oceanica prairie in a coastal area affected by a chlor-alkali complex. Toxicol Environ Chem. 1989;22:131–134. [Google Scholar]
- Foley RC, Liang ZM, Singh KB. Analysis of type 1 metallothionein cDNAs in Vicia faba. Plant Mol Biol. 1997;33:583–591. doi: 10.1023/a:1005790927581. [DOI] [PubMed] [Google Scholar]
- Foley RC, Singh KB. Isolation of a Vicia faba metallothionein-like gene: expression in foliar trichomes. Plant Mol Biol. 1994;26:435–444. doi: 10.1007/BF00039552. [DOI] [PubMed] [Google Scholar]
- Fordham-Skelton AP, Wilson JR, Groom Q, Robinson NJ. Accumulation of metallothionein transcripts in response to iron, copper and zinc: metallothionein and metal-chelate reductase. Acta Physiol Plant. 1997;19:451–457. [Google Scholar]
- Fukuhara T, Pak J-Y, Ohwaki Y, Tsujimura H, Nitta T. Tissue-specific expression of the gene for a putative plasma membrane H+-ATPase in a seagrass. Plant Physiol. 1996;110:35–42. doi: 10.1104/pp.110.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccone G, Barone R, Muscetta PN, De Lorenzo R. Effet de polluants petrochimiques sur les vegetaux marins de Porto Torres (Sardaigne, Italie): methodologies d'analyze et d'interpretation de données. J Etud Pollut CIESM. 1981;5:87–93. [Google Scholar]
- Hamer DH. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- Hsieh HM, Liu WK, Chang A, Huang PC. RNA expression pattern of a type 2 metallothionein-like gene from rice. Plant Mol Biol. 1996;32:525–529. doi: 10.1007/BF00019104. [DOI] [PubMed] [Google Scholar]
- Hsieh HM, Liu WK, Huang PC. A novel stress-inducible metallothionein-like gene from rice. Plant Mol Biol. 1995;28:381–389. doi: 10.1007/BF00020388. [DOI] [PubMed] [Google Scholar]
- Kawashima I, Inokuchi Y, Chino M, Kimura M, Shimizu N. Isolation of a gene for a metallothionein-like protein from soybean. Plant Cell Physiol. 1991;32:913–916. [Google Scholar]
- Kawashima I, Kennedy TD, Chino M, Lane BG. Wheat Ec metallothionein genes: like mammalian Zn++ metallothionein genes, wheat Zn++ metallothionein genes are conspicuously expressed during embryogenesis. Eur J Biochem. 1992;209:971–976. doi: 10.1111/j.1432-1033.1992.tb17370.x. [DOI] [PubMed] [Google Scholar]
- Lane B, Kajioka R, Kennedy T. The wheat germ Ec protein is a zink-containing metallothionein. Biochem Cell Biol. 1987;65:1001–1005. [Google Scholar]
- Loques F, Caye G, Meinesz A. Axenic culture of selected tissue of Posidonia oceanica. Aquat Bot. 1990;37:171–188. [Google Scholar]
- Maggini F, Tucci G, Demartis A, Gelati MT, Avanzi S. Ribosomal RNA genes of Phaseolus coccineus: I. Plant Mol Biol. 1992;18:1073–1082. doi: 10.1007/BF00047710. [DOI] [PubMed] [Google Scholar]
- Maserti BE, Ferrara R, Morelli M. Posidonia oceanica: uptake and mobilization of mercury in the mediterranean basin. In: Gabrielides GP, editor. Proceedings of the FAO/UNEP/IAEA Meeting on the Accumulation and Transformation of Chemical Contaminants by Biotic and Abiotic Processes in the Marine Environment. MAP Techn Rep 59: 243–249. 1991. [Google Scholar]
- Maserti BE, Ferrara R, Paterno P. Posidonia as an indicator of mercury contamination. Mar Pollut Bull. 1988;19:381–382. [Google Scholar]
- Morris CA, Nicolaus B, Sampson V, Harwood JL, Kille P. Identification and characterization of a recombinant metallothionein protein from a marine alga, Fucus vesicolosus. Biochem J. 1999;338:553–560. [PMC free article] [PubMed] [Google Scholar]
- Murphy A, Taiz L. Comparison of metallothionein gene expression and nonprotein thiols in ten Arabidopsis ecotypes. Plant Physiol. 1995;109:945–954. doi: 10.1104/pp.109.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A, Zhou JM, Goldsbrough PB, Taiz L. Purification and immunological identification of metallothioneins 1 and 2 from Arabidopsis thaliana. Plant Physiol. 1997;113:1293–1301. doi: 10.1104/pp.113.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YW, Tichit L, Leperlier M, Cuerq B, Marty I, Lelievre JM. Isolation and characterization of mRNAs differentially expressed during ripening of wild strawberry (Fragaria vesca L.) fruits. Plant Mol Biol. 1999;39:629–636. doi: 10.1023/a:1006179928312. [DOI] [PubMed] [Google Scholar]
- Novak R. Spatial and seasonal distribution of the meiofauna in the seagrass Posidonia oceanica. Neth J Sea Res. 1982;16:380–388. [Google Scholar]
- Okumura N, Neshizawa NK, Umehara Y, Mori S. An iron deficiency-specific cDNA from barley roots having two homologous cysteine-rich MT domains. Plant Mol Biol. 1991;17:531–533. doi: 10.1007/BF00040651. [DOI] [PubMed] [Google Scholar]
- Palmiter RD. Regulation of metallothionein genes by heavy metals appears to be mediated by a zinc-sensitive inhibitor that interacts with a constitutively active transcription factor, MTF-1. Proc Natl Acad Sci USA. 1994;91:1219–1223. doi: 10.1073/pnas.91.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres JM. La regression des herbiers àPosidonia oceanica. In: Boudouresque CF, Jeudy de Grissac A, Olivier J, editors. Proceedings International Workshop Posidonia oceanica Beds. Marseille, France: GIS Posidonie; 1984. pp. 445–454. [Google Scholar]
- Pergent-Martini C. Posidonia oceanica: a biological indicator of past and present mercury contamination in the Mediterranean sea. Mar Environ Res. 1998;45:101–111. [Google Scholar]
- Pergent-Martini C, Pergent G. Suivi de l'evolution de l'herbier àPosidonia oceanica après la mise en service de la station d'épuration de Marseille, Contrat Ville de Marseille/Université Aix-Marseille II No. 9261, GIS Posidonie, Marseille, France. 1990. p. 58. [Google Scholar]
- Pirc H. Growth dynamics in Posidonia oceanica (L.) Delile. Mar Ecol. 1985;6:141–145. [Google Scholar]
- Procaccini G, Mazzella L. Population genetic structure and gene flow in the seagrass Posidonia oceanica assessed using microsatellite analysis. Mar Ecol Prog Ser. 1998;169:133–141. [Google Scholar]
- Rainbow PS, Phillips DJH. Cosmopolitan biomonitors of trace metals. Mar Pollut Bull. 1993;26:593–601. [Google Scholar]
- Reid SJ, Ross GS. Up-regulation of two cDNA clones encoding metallothionein-like proteins in apple fruit during cool storage. Physiol Plant. 1997;100:183–189. [Google Scholar]
- Robinson NJ, Tommey AM, Kuske C, Jackson PJ. Plant metallothioneins. Biochem J. 1993;295:1–10. doi: 10.1042/bj2950001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher-Hoenlinger MA, Schlacher TA. Accumulation, contamination, and seasonal variability of trace metals in the coastal zone-patterns in a seagrass meadow from the Mediterranean. Mar Biol. 1998;131:401–410. [Google Scholar]
- Seritti A, Petrosino A, Ferrara R, Barghigiani C. A contribution to the determination of “reactive ” and “total” mercury in sea water. Environ Technol Lett. 1980;1:50–57. [Google Scholar]
- Snowden KC, Richards KD, Gardner RC. Aluminum-induced genes: induction by toxic metals, low calcium, and wounding and pattern of expression in root tips. Plant Physiol. 1995;107:341–348. doi: 10.1104/pp.107.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnau M, Fowler SW, Teyssie JL. Biokinetics of selected heavy metals and radionuclides in two marine macrophytes: the seagrass Posidonia oceanica and the alga Caulerpa taxifolia. Mar Environ Res. 1996;41:343–362. [Google Scholar]
- Whitelaw CA, Le Huquet JA, Thurman DA, Tomsett AB. The isolation and characterization of type II metallothionein-like genes from tomato (Lycopersicum esculentum L.) Plant Mol Biol. 1997;33:503–511. doi: 10.1023/a:1005769121822. [DOI] [PubMed] [Google Scholar]
- Woodhead M, Taylor MA, Brennan R, McNicol RJ, Davies HV. Cloning and characterization of the cDNA clones of five genes that are differentially expressed during ripening in the fruit of blackcurrant (Ribes nigrum L.) J Plant Physiol. 1998;153:381–393. [Google Scholar]
- Zenk MH. Heavy metal detoxification in higher plants. Gene. 1996;179:21–30. doi: 10.1016/s0378-1119(96)00422-2. [DOI] [PubMed] [Google Scholar]
- Zhou JM, Goldsbrough PB. Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell. 1994;6:875–884. doi: 10.1105/tpc.6.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]