Abstract
Purpose
The pathophysiology of retinal ischemia involves mechanisms including inflammation and apoptosis. Ischemic post-conditioning (Post-C), a brief non-lethal ischemia, induces a long-term ischemic tolerance, but the mechanisms of ischemic post-conditioning in the retina have only been described on a limited basis. Accordingly, we conducted this study to determine the molecular events in retinal ischemic postconditioning and to identify targets for therapeutic strategies for retinal ischemia.
Methods
To determine global molecular events in ischemic Post-Conditioning, a comprehensive study of the transcriptome of whole retina was performed. We utilized RNA-Sequencing (RNA-Seq), a recently developed, deep sequencing technique enabling quantitative gene expression, with low background noise, dynamic detection range, and discovery of novel genes. Rat retina was subjected to ischemia in vivo by elevation of intraocular pressure above systolic blood pressure. At 24 h after ischemia, Post-C or sham Post-C was performed by another, briefer period of ischemia, and 24 h later, retinas were collected and RNA processed.
Results
There were 71 significantly affected pathways in post-conditioned/ischemic vs normals and 43 in sham post conditioned/ischemic vs normal. Of these, 28 were unique to Post C and ischemia. Seven biological pathways relevant to ischemic injury, in Post-C as opposed to sham Post C, were examined in detail. Apoptosis, p53, cell cycle, Jak-Stat, HIF-1, MAPK and PI3K-Akt pathways significantly differed in the number as well as degree of fold change in genes between conditions.
Conclusion
Post-C is a complex molecular signaling process with a multitude of altered molecular pathways. We identified potential gene candidates in Post-C. Studying the impact of altering expression of these factors may yield insight into new methods for treating or preventing damage from retinal ischemic disorders.
Keywords: ischemia, post-conditioning, retina, RNA-Seq
INTRODUCTION
Central retinal artery occlusion (CRAO) due to thrombus or embolism, occurring at an incidence of 1–15/10,000 yearly, develops into retinal ischemia, often impairing visual acuity to < 20/400. Severe visual loss increases healthcare costs, mortality and depression; and decreases productivity, independence, and quality of life. Retinal ischemia is also, significantly, a final common pathway in major, chronic vision-threatening diseases including retinal vein occlusion and diabetic retinopathy (DR), and may also be related to the development of age-related maculopathy (ARM). Rodent models of ischemia-reperfusion produce an injury resembling DR. [1] In chronic disease such as DR, there are few available effective treatments. While we, and, subsequently, others, have shown that the retina possesses a potent, endogenous neuroprotective capacity [2, 3], potentially accessible for clinical therapy, application to patients with DR at risk to develop DR has not yet been realized [4]. An obstacle to progress has been the incomplete understanding of retinal ischemia mechanisms and how to safely, efficiently, and precisely access therapeutic molecular targets. We previously demonstrated some of the novel mechanisms of the retina’s endogenous neuroprotective capacity. Ischemic tolerance is amongst the most promising modulators of ischemic injury shown to date because it can harness endogenous mechanisms to improve recovery even as late as 24-h after an ischemic event, a mechanism known as post-conditioning (or Post-C) [2].
Post conditioning is a brief period of ischemia applied to the retina after the damaging ischemia. It has also been shown to provide neuroprotection in stroke models in brain and in cardioprotection after myocardial ischemia. [5, 6] Some of the underlying mechanisms in Post-C have been described. Post-C attenuated apoptotic cell death [2]. Also, Post-C required activation of Akt, tied to numerous downstream cell survival mediators [7]. It is likely that multiple signaling pathways and altered gene expression are involved in the neuroprotective mechanism of Post-C. In order to identify changes in global gene expression a comprehensive approach is necessary. Accordingly, the goal of the present study was an in-depth analysis of the global molecular mechanisms by which Post-C produces its neuroprotective function.
Comprehensive analysis of the transcriptome is one of the most useful approaches in understanding global molecular events, with two main techniques based largely on hybridization or sequencing [8]. Hybridization, i.e. microarrays, is well-established and remains popular mostly due to its low-cost. Nevertheless, microarrays suffer from many limitations, e.g., their reliance upon existing knowledge about genome sequence; high background levels owing to cross-hybridization; and a limited dynamic range of detection owing to both background and saturation of signals. Moreover, comparing expression levels across different experiments is difficult and can require complicated normalization methods [9]. RNA sequencing (RNA-Seq) is a relatively new technique using a sequencing–based approach to enable global and quantitative examination of the entire transcriptome. It offers two key benefits over the microarray method: it is not limited to detecting transcripts corresponding to existing genomic sequences, and it has very low background signal because DNA sequences are unambiguously mapped to unique regions of the genome [9]. Accordingly, in this study, we used RNA-Seq to study the global gene expression changes in rat retina in vivo subjected to ischemia and Post-C. The goals were to define the changes in gene expression resulting from Post-C and to thus comprehensively describe molecular mechanisms of Post-C in the retina.
MATERIALS & METHODS
Animals
Procedures were approved by our institutional Animal Care Committee. Male Wistar rats (200–250 gm, Harlan, Indianapolis, IN, USA) were maintained on a 12 h on/12 h off light cycle. Experiments were performed during daylight hours. Eyes were treated with topical vigamox and cyclomydril, and for local anesthesia for needle placement, 0.5% proparacaine. For retinal ischemia, rats were anesthetized with ketamine 100 mg/kg and xylazine 7 mg/kg i.p. After sterile preparation, and working under an operating microscope, a 30-gauge, 5/8-inch metal needle (BD Precision Glide; Becton-Dickinson, Franklin Lakes, NJ, USA) was placed with its tip directed away from the lens, just inside the anterior chamber of the right eye, the experimental eye, while the left eye was control eye. To enable pressurization for increasing intraocular pressure (IOP), and measuring IOP, the needle was connected by a length of plastic tubing via a three-way stopcock to an electronic pressure transducer (Transpac 42661-04-27; Abbott, North Chicago, IL, USA) and to an elevated bag of balanced salt solution (BSS; by sterile technique, BSS was transferred from its bottle to an empty 1000-mL 0.9% saline plastic bag [Baxter, Deerfield, IL, USA]). IOP, continually displayed on a monitor (Hewlett-Packard HP78534C; Palo Alto, CA, USA), was increased to 130–135 mmHg and maintained constantly for 55 minutes by pressurizing the bag (Smiths Medical Clear-Cuff, Dublin, OH, USA). The temperature was maintained at 36–37 °C with a servo–controlled heating blanket (Harvard Apparatus, Natick, MA, USA). Oxygen saturation was measured by pulse oximetry (Ohmeda; Louisville, CO, USA) with a Band-Aid type probe on the tail. Supplemental oxygen, when necessary to maintain O2 saturation > 93%, was administered using a cannula in front of the nares and mouth. For the Post-C stimulus, the same procedure was used to increase IOP for 5 min; Post-C or sham Post-C was done 24 h after ischemia, and eyes were collected 24 h after Post-C or sham Post-C.
After euthanasia, eyes were removed and whole retinal homogenates were prepared. Thus a mixture of retinal cell types was analyzed and results must be interpreted in light of this limitation vs single cell analysis. Four biological groups, each with 4 replicates were used in the analysis: retina control (ShamN), retina + postconditioning (PCN), retina + ischemia + sham Post-C (ShamI), and retina + ischemia + Post-C (PCI). Four pairwise comparisons between groups were generated by the Fisher’s exact test: PCI vs ShamI, ShamI vs ShamN, PCI vs ShamN and PCN vs ShamN each yielding a separate table of fold changes, log2(counts per million), P and Q values. In order to examine the Post-C key regulator gene functions and locations within the molecular pathways, as well as the impact of Post-C on gene expression after ischemia, we performed a downstream functional and pathway analysis separately for each of our biological groups, i.e., the effects of Post-C on ischemia were derived by contrasting the individual differences between PCI vs ShamN and ShamI vs ShamN.
RNA Isolation and Library Preparation
Total RNA was extracted from frozen retinal tissue, homogenized using TRIzol (Invitrogen, Carlsbad, CA, USA) and Direct-Zol RNA microPrep (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocols. To purify the RNA, the homogenate was passed through Zymo-Spin columns (Zymo Research, Irvine, CA, USA). RNA quality and purity was assessed using the Agilent 2200 TapeStation (Wilmette, IL), Qubit 3.0 Fluorometer (Life Technologies, Grand Island, NY, USA), and Nanodrop. 260/280 ratios were measured for all RNA samples, with a range of 1.8–2.4. Most samples had a 260/280 ratio of 2–2.1. RNA-Seq libraries were prepared using the QuantSeq 3′ mRNA sequencing library preparation kit for Illumina sequencing platform-compatible libraries (Lexogen, Greenland, NH), according to the manufacturer’s instructions. The QuantSeq protocol (seq: https://www.lexogen.com/quantseq-3mrna-sequencing/) only requires single direction sequencing. This is because only the region adjacent to the poly(A) tail of mRNAs is sequencing, and the sequencing reaction heads towards the poly(A) tail. If the reverse read were implemented, only a poly(A) sequence would be recovered. Non-coding sequences are not detected using these methods.
50 ng of total RNA were used as input template, and 18 amplification cycles were performed. The amplification step is an essential part of the RNAseq library preparation protocol. This step has three purposes, including: (a) increase of total concentration of molecules via PCR to generate sufficient material for the sequencer, (b) incorporation of sample-specific barcode sequences into the final molecules, and (c) incorporation of Illumina sequencing adapters into the final molecules. The PCR step is part of the manufacturer’s instructions. Final libraries for each sample were quantified using quantitative PCR (Real-time PCR library amplification Kit; Kapa Biosystems, Boston, MA, USA). During preparation of the messenger RNAs for sequencing, a unique artificial sequence (barcode) is incorporated during the PCR amplification step. Since every molecule in a sample is labeled with the same barcode, samples from multiple samples can be pooled before sequencing. This approach is used because there is no easy way to physically separate samples on a sequencing flow cell, and because a single flow cell produces much more data than is needed for each sample. The combined library pool was sequenced using an Illumina Nextseq500 sequencer (Illumina, San Diego, CA, USA), employing a high-output kit with 80 base single-end reads. Libraries were prepared in the UIC Core Genomics Facility (CGF) and sequencing was performed in the DNA Services Facility (DNASF).
RNA-Seq Analysis
A) Count Matrix Preparation
First, we assessed the sequencing quality of raw reads with FastQC, which in contrast to quality control (QC) reports generated by the sequencer itself, detects problems which originate either in the sequencer or in the starting library material [10]. The raw sequence reads were aligned to the reference (rat) genome using the Burrows-Wheeler Aligner (BWA-MEM), an algorithm aligning long sequences < 1 Mb size against a large sequence database (e.g., human genome). This process efficiently maps reads with read-through into polyA tails and adapter sequences, as is common with 3′ RNA-Seq [11]. To quantitate the expression levels, we used featureCounts, a program that uses a highly efficient, general-purpose read summarization to approximate counts for genomic features including genes, exons, promoter, gene bodies, genomic bins and chromosomal locations [12]. The output, i.e., the table of raw counts, was used for the following analyses.
B) Data Exploration and Quality Assessment
Raw counts were generated for 17,311 genes across 16 samples (4 biological replicates in 4 groups). To demonstrate the structure of the data, Supp. Table 1 displays the first 20 rows of raw count data. For complete raw count data, see Supplemental Table 1, or the weblink: https://uofi.box.com/s/7cpuceq42fz0tk6jpgp8e9cvl9fiv2in. For preliminary quality assessment, genes with very low counts across all samples were first filtered using the cpm (counts per million) function in edgeR for cpm> 1 in at least 4 samples [13, 14]. Subsequently, to account for the RNA composition effect, i.e., highly expressed genes consuming a substantial proportion of the total library size, resulting in under-sampling of the remaining genes, Trimmed Mean Values (TMM) normalization was performed [13]. Results were then normalized with respect to library size (reads/million) using edgeR’s cpm function [13, 14]. A prior count of 2 was added to avoid logarithm evaluation errors and values were log2-transformed in order to make the counts approximately homoskedastic i.e. similar error variances across samples.
C) Differential Expression Analysis
The differential expression statistics of the data were also compiled using edgeR [14]. With the filtered raw count values as input, a negative binomial model was fitted, common dispersion estimates for each gene were obtained and Fisher’s exact test was performed for each of our pre-defined pairwise comparisons [14]. Genes were considered significantly up-regulated if log2(fold change) ≥ 1and False Discovery Rate-adjusted p value (FDR) ≤ 0.05 with Benjamin and Hochberg correction for multiple tests [15] (FDR will also sometimes be referred here as a Q value). Genes were considered significantly down-regulated with log2(fold change) ≤ −1 and Q ≤ 0.05.
D) Functional Enrichment Analysis in Ischemia and Post-conditioning
Functional enrichment analysis was performed on differentially expressed genes (absolute value of fold change ≥ 1 and Q ≤ 0.05) in each group using FunRich, a stand-alone software tool for functional enrichment and interaction network analysis [16] using the Gene Ontology rat database (Taxon ID: 10116) [17, 18]. FunRich was used due to its intuitive layout, built-in graphical tools that were publication quality as well as the ability to completely customize the background databases against which the gene sets were run. The functional enrichment tool DAVID is severely limited both in graphical outputs and the annotation databases are frequently outdated. Regulation of biological processes and molecular functions was considered significant when P ≤ 0.05, and “unique” if a given function’s P value was ≤ 0.05 in one group and > 0.05 in the comparison group. To illustrate, “Apoptotic Process” had P < 0.001 associated with PCI vs ShamN and P=0.12 associated with ShamI vs ShamN; hence “Apoptotic Process” was considered a uniquely affected process in PCI vs ShamN. The results of the functional enrichment analysis were used for inference of the gene pathways affected.
E) Pathway/Impact Analysis of Ischemia and Post-conditioning
To identify key regulator genes and to visualize the affected pathways in ischemic plus post-conditioned compared to ischemic plus sham post-conditioned retinae, iPathwayGuide [19] (Advaita, Plymouth, MI, USA) – a web-based impact analysis [20] tool was used. It utilizes the Kyoto Encyclopedia of Genes and Genomes (KEGG) [21] pathway database for molecular pathway representation, with results from edgeR pairwise comparisons (fold changes and Q values) as input. As opposed to functional analysis tools, iPathwayGuide takes into account the position of each gene on each pathway, and the type and directions of the interactions with the other genes. iPathwayGuide deems genes significantly altered if Q ≤ 0.05 and absolute value of log2(fold change) ≥ 2. Significantly affected pathways with adjusted P values, or FDR (false discovery rate) ≤ 0.05 were obtained from two of our comparison groups, i.e., PCI vs ShamN and ShamI vs ShamN, and were analyzed for uniqueness.
F) Downstream/Upstream Target Identification in Ischemia and Post-conditioning
To determine the overall changes between specific genes in our unique pathways, each pathway was analyzed with respect to the number, location and fold change difference of significantly up-regulated/down-regulated genes vs the other group.
RESULTS
1) Data Quality
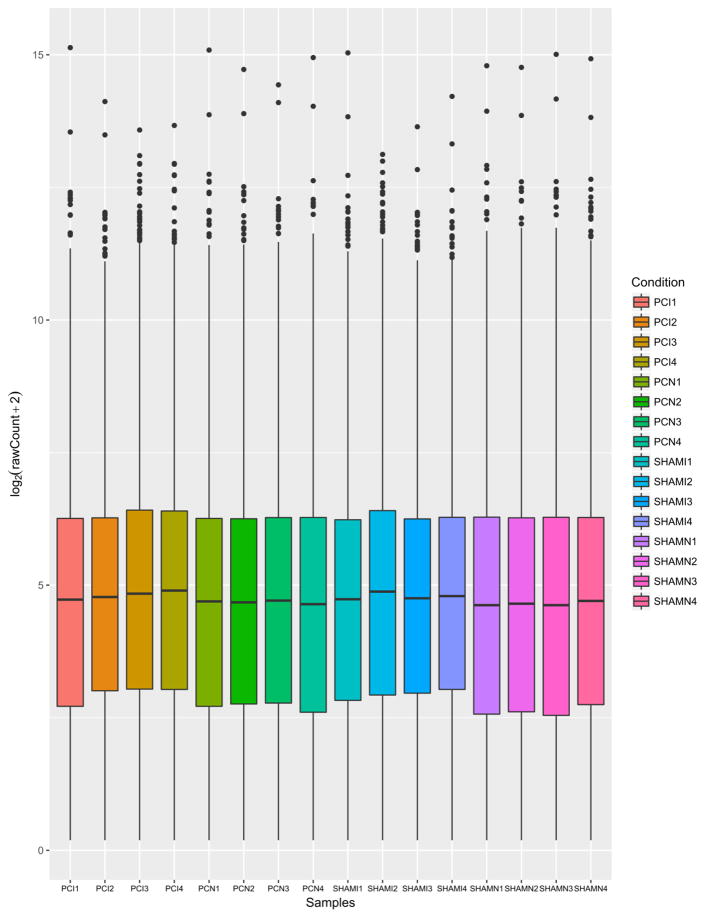
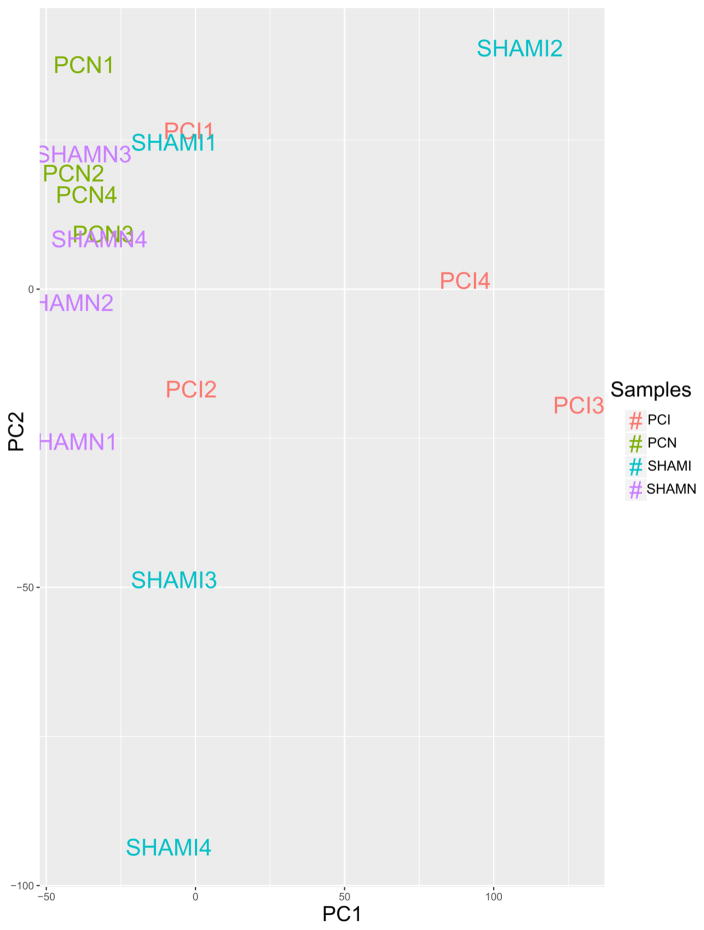
Low count-filtered, TMM and cpm-normalized, log2-transformed table of counts was used to generate boxplots (Fig 1) [22] visualizing the relative abundance of gene expression between samples. Results confirmed that the distribution of expression of individual genes was similar across the samples. Principal Component Analysis (PCA) [23] of the first two principal components and clustering image map of distances between samples demonstrated the variance between sample groups and sample replicates (Fig 2) [22]. PCA plots of additional components are available in the Data Supplement (Supp. Fig 1, Supp. Fig 2). The results indicated a relative homogeneity between samples within their biological groups with minimal variation.
Fig. 1.
Gene expression in each sample, contrasting their respective distributions; generated using R v. 3.2.2 and ggplot2_2.2.1. Individual samples (x-axis) and their log2-transformed expression count (y-axis) are shown. Prior count of 2 was added in order to avoid log evaluation errors. Results confirm that the distribution of expression of individual genes is quite similar across the samples after normalization was performed, as well as that treatment did not have a profound effect at the genome-wide expression level.
Fig. 2.
Principal Component Analysis (PCA) plot of the filtered, normalized and log2-transformed count matrix, assessing the overall effect of experimental covariates and batch effects - generated with R v. 3.2.2, ggplot2_2.2.1 andggfortify_0.4.1. The first two components (x-and y-axis) were used and next order components are available in the Data Supplement. We noticed a higher variation in PCI3 and PCI4 and ShamI2, most likely due to natural variation in biological samples.
2) Differential Gene Expression in Ischemia with Post-C or sham Post-C
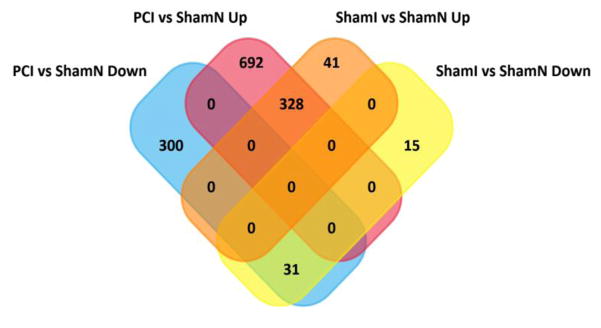
The overall results of the differential gene expression of PCI vs ShamN and ShamI vs ShamN are illustrated by their respective volcano plots (Supp. Fig 4, Supp. Fig 5) [24]. There were 1352 differentially expressed genes (DEGs), of which 332 were down-regulated and 1020 up-regulated in PCI vs ShamN, and 416 DEGs, with 46 down-regulated and 370 up-regulated in ShamI vs ShamN. The 4-way Venn diagram (Fig 3) summarizes the raw number of DEGs in each of the groups. The Supplemental Tables 8–11, and website link https://uofi.box.com/s/7cpuceq42fz0tk6jpgp8e9cvl9fiv2in contain the full set of data from which Fig 3 was derived. From Fig 3, 692 genes were uniquely up-regulated in PCI vs ShamN in contrast to 41 in ShamI vs ShamN. The vast majority of up-regulated genes in ShamI vs ShamN, were also up-regulated in PCI vs ShamN (328 genes). Similarly, 300 genes were significantly down-regulated in PCI vs ShamN and 15 in ShamI vs ShamN. The overall number of DEGs was much higher in PCI vs ShamN in contrast to ShamI vs ShamN. The top 20 DEGs arranged in ascending order of Q values, for PCI vs ShamN and ShamI vs ShamN are shown in Table 1 with their respective gene IDs and gene names.
Fig. 3.
Four-way quantitative summary of the differential expression analysis results generated with FunRich 3.0. Overall, of 1352 DEGs present in PCI vs ShamN and 416 DEGs present in ShamI vs ShamN, 328 were up-regulated and 31 were down-regulated in both groups, while 692 genes were uniquely up-regulated in PCI vs ShamN in contrast to 41 in ShamI vs ShamN. The vast majority of up-regulated genes in ShamI vs ShamN, were also up-regulated in PCI vs ShamN (328 genes). Similarly, 300 genes were found to be significantly down-regulated in PCI vs ShamN to 15 in ShamI vs ShamN. See Supplemental Tables 8–11 for the complete data set used to construct Fig 3. https://uofi.box.com/s/7cpuceq42fz0tk6jpgp8e9cvl9fiv2in
Table 1.
Top 20 differentially expressed genes according to descending Q values for PCI vs. ShamN and ShamI vs. ShamN respectively. The values were obtained as a result of the differential expression pairwise tests in edgeR. Table was generated using by edgeR_3.12.1.
| Top 20 Genes for ShamI vs. ShamN | Top 20 Genes for PCI vs. ShamN | ||||||
|---|---|---|---|---|---|---|---|
| ID | Gene Name | logFC | Q value | ID | Gene Name | logFC | Q value |
| Pde6h | phosphodiesterase 6H | −2.279 | 1.31E-10 | Spp1 | secreted phosphoprotein 1 | 4.780 | 1.92E-16 |
| Nckap 1l | NCK associated protein 1 like | 3.563 | 1.84E-07 | Mrpl43 | mitochondrial ribosomal protein L43 | 7.822 | 2.32E-15 |
| Rassf4 | Ras association domain family member 4 | 1.817 | 1.02E-06 | Top2a | topoisomerase (DNA) II alpha | 4.359 | 1.03E-13 |
| Txnip | thioredoxin interacting protein | 2.312 | 1.40E-06 | Alcam | activated leukocyte cell adhesion molecule | 2.529 | 1.03E-13 |
| RT1-Da | RT1 class II, locus Da | 4.833 | 4.25E-06 | Gpnmb | glycoprotein nmb | 4.443 | 1.36E-12 |
| Mpeg1 | macrophage expressed 1 | 3.683 | 5.02E-06 | Mki67 | marker of proliferation Ki-67 | 4.818 | 3.92E-12 |
| Tifab | TIFA inhibitor | 4.009 | 7.02E-06 | RT1-DMb | RT1 class II, locus DMb | 4.524 | 4.68E-12 |
| Arr3 | arrestin 3 | −2.146 | 1.58E-05 | Cenpf | centromere protein F | 4.224 | 6.16E-11 |
| Fcgr2b | Fc fragment of IgG, low affinity IIb, receptor | 10.693 | 1.58E-05 | Rassf4 | Ras association domain family member 4 | 2.172 | 1.66E-10 |
| Bin2 | bridging integrator 2 | 4.106 | 2.62E-05 | Alox5ap | arachidonate 5-lipoxygenase activating protein | 3.042 | 1.09E-09 |
| Mlf1 | myeloid leukemia factor 1 | 2.064 | 2.62E-05 | Ctsz | cathepsin Z | 4.345 | 3.04E-09 |
| Ctss | cathepsin S | 2.042 | 2.95E-05 | Vim | vimentin | 2.490 | 3.28E-09 |
| Laptm5 | lysosomal protein transmembrane 5 | 4.315 | 3.81E-05 | Pdpn | podoplanin | 2.526 | 2.68E-08 |
| Csf1r | colony stimulating factor 1 receptor | 3.103 | 3.84E-05 | Ptafr | platelet-activating factor receptor | 5.885 | 2.69E-08 |
| Gadd45g | growth arrest and DNA-damage-inducible, gamma | 1.882 | 3.84E-05 | Lmna | lamin A/C | 2.529 | 3.31E-08 |
| Clec4a1 | C-type lectin domain family 4, member A1 | 8.195 | 5.45E-05 | Cd44 | CD44 molecule (Indian blood group) | 4.005 | 3.70E-08 |
| Aif1 | allograft inflammatory factor 1 | 3.743 | 5.45E-05 | Mlf1 | myeloid leukemia factor 1 | 2.165 | 3.70E-08 |
| Mt2A | metallothionein 2A | 2.500 | 6.77E-05 | Lipa | lipase A, lysosomal acid type | 1.702 | 3.70E-08 |
| Ncf1 | neutrophil cytosolic factor 1 | 6.501 | 7.23E-05 | Ncf1 | neutrophil cytosolic factor 1 | 7.056 | 5.80E-08 |
| Apobec1 | apolipoprotein B mRNA editing enzyme catalytic subunit 1 | 4.109 | 0.000115 | Arpc1b | actin related protein 2/3 complex, subunit 1B | 3.784 | 5.92E-08 |
3) Functional Analysis of Ischemia with Post-C or sham Post-C
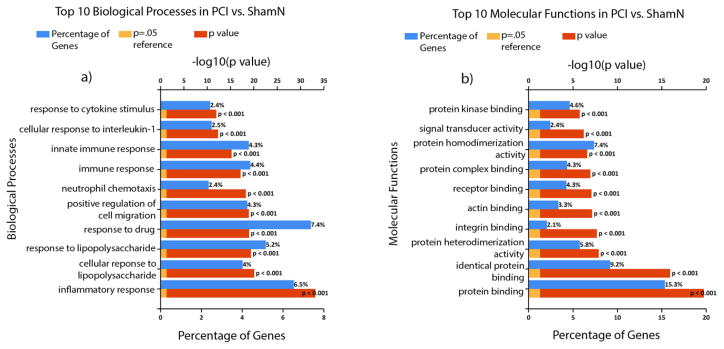
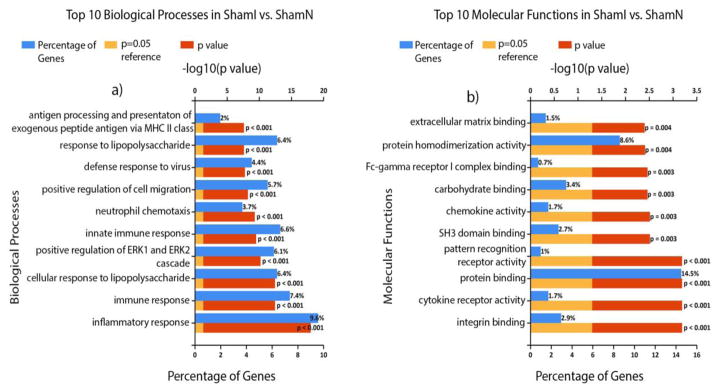
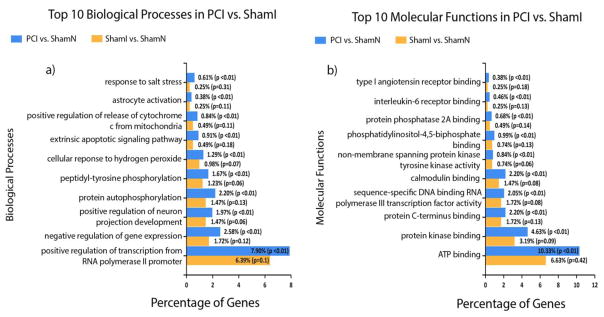
The top 10 (ordered by P value) biological processes and molecular functions for PCI vs ShamN as well as ShamI vs ShamN are displayed in Figs 4 & Fig 5, respectively. For PCI vs. ShamN, the highest % of genes making up the biological processes were: response to drug (7.4%; of course no drugs were used in this study, but common pathways may be detected), inflammatory response (6.5%), response to lipopolysaccharides (5.2%), and immune response (4.5%), while for molecular functions they included: protein binding (15.3%), identical protein binding (9.2%), protein homo-dimerization activity (7.4%) and protein kinase binding (4.6%). For ShamI vs ShamN, the biological processes with the highest % of differentially expressed genes included: inflammatory response (9.6%), immune response (7.4%), innate immune response (6.6%) and cellular response to lipopolysaccharide (6.4%), while for molecular functions they were: protein binding (14.5%), protein homo-dimerization activity (8.6%), carbohydrate binding (3.4%) and integrin binding (2.9%). In addition, the top 10 (ordered by P value) biological processes and molecular functions uniquely affected in PCI vs ShamN in comparison to ShamI vs ShamN are displayed in Fig 6. Biological processes involving the highest % of groups included: positive regulation of transcription from RNA polymerase II promoter (7.9%), negative regulation of gene expression (2.6%), protein auto-phosphorylation (2.2%) and positive regulation of neuron projection development (2%), while for molecular functions, we obtained: ATP binding (10.33%), protein kinase binding (4.6%), protein C-terminus binding (2.2%) and calmodulin binding (2.2%).
Fig. 4.
Functional analysis figures were generated with FunRich 3.0 [16] and Gene Ontology (GO) Norway rat (Taxon ID: 10116) database [17, 18]. Shown are the top 10 biological processes (a) and molecular functions (b) for PCI vs ShamN arranged according to increasing P (or decreasing log10(P value)). Percentage of differentially expressed genes belonging to each process is shown as blue bars. log10(P =0.05) is shown as a reference.
Fig. 5.
Functional analysis figures were generated with FunRich 3.0 [16] and Gene Ontology (GO) Norway rat (Taxon ID: 10116) database [17, 18]. Shown are the top 10 biological processes (a) and molecular functions (b) for ShamI vs ShamN, arranged according to increasing P (or decreasing −log10(P value)). Percentage of differentially expressed genes belonging to each process is shown as blue bars.−log10(P =0.05) is shown as a reference.
Fig. 6.
Top 10 biological processes (a) and molecular functions (b) “uniquely” affecting PCI vs ShamN, i.e. biological processes and functions with P ≤ 0.05 in PCI vs ShamN and P > 0.05 in ShamI vs ShamN. Figures were generated with FunRich 3.0[16] and Gene Ontology (GO) Norway rat (Taxon ID: 10116) database [17, 18].Percentage of DEGs belonging to a particular process/function is shown in blue bars (PCI) and yellow bars (ShamI).
4) Pathway/Impact Analysis in Ischemia with Post-C or sham Post-C
There were 71 significantly affected pathways in PCI vs ShamN and 43 in ShamI vs ShamN. Of these, 28 were unique to PCI vs ShamN. Table 2 summarizes the significantly affected pathways across our conditions. Of 28 uniquely affected pathways in PCI vs ShamN, 7 were chosen for further analysis due to their known or potential biological relevance to ischemic injury. These included: MAPK, apoptosis, Jak-STAT, PI3K-Akt, HIF-1, p53 and cell cycle.
Table 2.
KEGG pathways unique to PCI vs. ShamN group. The list was compiled by comparing significantly affected pathways in PCI vs. ShamN to ShamI vs. ShamN and keeping only the ones unique for the former. No pathways were unique for the latter. The list of affected pathways was generated by iPathwayGuide. Pathways with FDR value ≤ .05 in PCI vs. ShamN and Q value > .05 in ShamI vs. ShamN are shown. Biologically related pathways are bolded.
| PCI vs. ShamN Unique Pathways | |
|---|---|
| Pathway name | p-value |
| MAPK signaling pathway | 1.05E-06 |
| Regulation of actin cytoskeleton | 2.66E-05 |
| Apoptosis | 6.87E-05 |
| Measles | 7.73E-05 |
| Jak-STAT signaling pathway | 8.69E-05 |
| Focal adhesion | 1.75E-04 |
| Hepatitis B | 1.88E-04 |
| PI3K-Akt signaling pathway | 2.48E-04 |
| Non-alcoholic fatty liver disease (NAFLD) | 5.06E-04 |
| Insulin resistance | 8.77E-04 |
| Adipocytokine signaling pathway | 9.10E-04 |
| Pathways in cancer * | 9.84E-04 |
| Cytosolic DNA-sensing pathway | 0.002 |
| Amyotrophic lateral sclerosis (ALS) | 0.003 |
| HIF-1 signaling pathway | 0.005 |
| Viral carcinogenesis | 0.008 |
| Epstein-Barr virus infection | 0.008 |
| Rap1 signaling pathway | 0.008 |
| T cell receptor signaling pathway | 0.01 |
| p53 signaling pathway | 0.013 |
| Platelet activation | 0.017 |
| MicroRNAs in cancer * | 0.019 |
| Adherens junction * | 0.029 |
| Small cell lung cancer | 0.034 |
| RIG-I-like receptor signaling pathway | 0.047 |
| Arginine and proline metabolism * | 0.047 |
| Bladder cancer | 0.048 |
| Cell cycle | 0.049 |
5) Downstream/Upstream Potential Target Identification in Ischemia with Post-C or sham Post-C
The 7 unique pathway diagrams for PCI vs ShamN were juxtaposed to ShamI vs ShamN for comparison. They represent the overall positions and fold changes of the significantly affected genes in a chosen pathway for both groups. Genes were deemed significantly up-regulated if log2(fold change) ≥ 2 & Q ≤ 0.05, and significantly down-regulated if log2(fold change) ≤ −2 & Q ≤ 0.05. Diagrams present the up-regulated genes in red and down-regulated genes in blue.
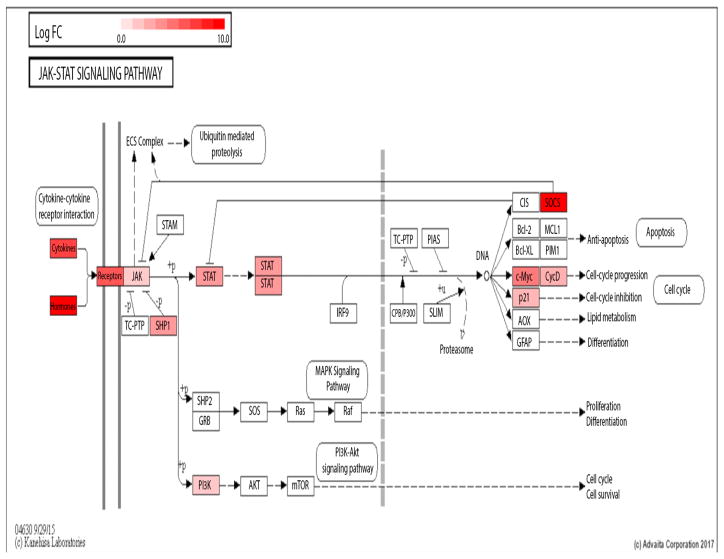
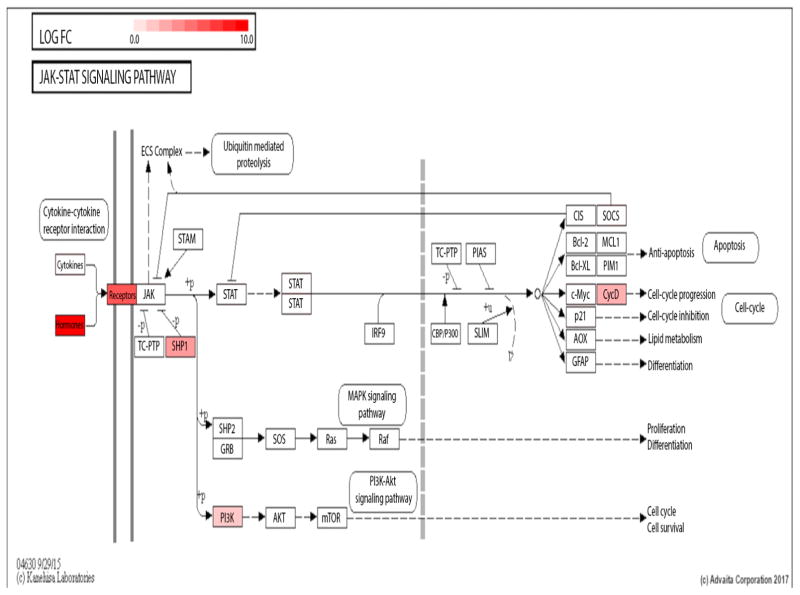
To illustrate, the Jak-Stat KEGG pathway diagrams for PCI vs ShamN (Fig 7) and ShamI vs ShamN (Fig 8) are presented. Genes uniquely up-regulated for PCI vs ShamN were: Socs3, Cdkn1a, Myc, Stat6, Jak2, Ifngr2, Il6r, Csf2ra, Il2rg, Il13ra2, Csf3r, Csf2rb, Il23a, Il11, Il6, and Csf2. (Table 3).
Fig. 7.
Jak-STAT KEGG pathway diagram for PCI vs ShamN and ShamI vs ShamN respectively. Diagrams were downloaded from iPathwayGuide’s pathways feature [19]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q ≤ 0.05 were considered significant.
Fig. 8.
Jak-STAT KEGG pathway diagram for PCI vs ShamN and ShamI vs ShamN respectively. Diagrams were downloaded from iPathwayGuide’s pathways feature [19]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q ≤ 0.05 were considered significant.
Table 3.
Uniquely affected genes in Jak-STAT KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
| Jak-Stat Pathway | |||||||
|---|---|---|---|---|---|---|---|
| ID | Gene Name | PCI vs. SHAMN: logFC | PCI vs. SHAMN: PValue | PCI vs. SHAMN: QValue | SHAMI vs. SHAMN: logFC | SHAMI vs. SHAMN: PValue | SHAMI vs. SHAMN: QValue |
| Cytokines | Csf2 | 7.415 | 0.001 | 0.014 | 4.590 | 0.147 | 0.658 |
| Il6 | 7.415 | 0.001 | 0.015 | 6.131 | 0.049 | 0.410 | |
| Il11 | 5.902 | 0.003 | 0.036 | 4.445 | 0.107 | 0.592 | |
| Il23a | 4.362 | 0.000 | 0.009 | 1.366 | 0.362 | 0.868 | |
| Receptors | Csf2rb | 6.545 | 0.002 | 0.026 | 4.496 | 0.092 | 0.560 |
| Csf3r | 5.245 | 0.000 | 0.002 | 4.638 | 0.004 | 0.097 | |
| Il13ra2 | 4.955 | 0.000 | 0.005 | 3.518 | 0.036 | 0.349 | |
| Il2rg | 4.643 | 0.001 | 0.017 | 4.282 | 0.004 | 0.090 | |
| Csf2ra | 2.544 | 0.002 | 0.022 | 2.225 | 0.009 | 0.164 | |
| Il6r | 2.475 | 0.000 | 0.000 | 1.830 | 0.013 | 0.196 | |
| Ifngr2 | 2.142 | 0.000 | 0.002 | 1.729 | 0.001 | 0.025 | |
| JAK | Jak2 | 2.079 | 0.000 | 0.008 | 1.140 | 0.071 | 0.501 |
| STAT | Stat6 | 3.999 | 0.003 | 0.034 | 1.881 | 0.323 | 0.851 |
| c-Myc | Myc | 5.167 | 0.000 | 0.001 | 3.766 | 0.007 | 0.142 |
| p21 | Cdkn1a | 2.876 | 0.000 | 0.008 | 1.663 | 0.034 | 0.339 |
| SOCS | Socs3 | 7.776 | 0.001 | 0.011 | 6.127 | 0.023 | 0.276 |
MAPK KEGG pathway diagrams for PCI vs ShamN (Supp. Fig 6) and ShamI vs ShamN (Supp. Fig 7) are presented. The MAPK KEGG pathway diagram yielded uniquely up-regulated genes in PCI vs ShamN: Dusp16, Dusp6, Dusp10, Dusp2, Gng12, Rps6ka1, Tgfb1, Rac2, Mapkapk3, Mknk1, Il1r2, Myc, Tnf, Nfkb1, Nfkb2, Fas, Map3k8, Map4k4, Gadd45b, and Fos. Fgf14 and Cacng2 were uniquely down-regulated in PCI vs ShamN. (Supp. Table 2).
Apoptosis KEGG pathway diagrams for PCI vs ShamN and ShamI vs ShamN (Supp. Fig 8 & Supp. Fig 9) respectively, revealed uniquely up-regulated genes in PCI vs ShamN: Traf1, Tnfrsf1a, Tnf, Parp3, Nfkbia, Nfkb1, Hrk, Gadd45b, Fos, Fas, Csf2rb, Cflar, Birc5, Birc3, Bak1 and Actb. (Supp. Table 3).
PI3K-Akt KEGG pathway diagrams (Supp. Fig 10 & Supp. Fig 11) showed uniquely up-regulated genes in PCI vs ShamN: Nfkb1, Creb5, Cdkn1a, Myc, Pik3cd, Gng12, Itgb7, Itga4, Itga5, Lamc1, Thbs3, Jak2, Il6r, Il2rg, Csf3r, Il6, Csf3, Pik3ap1, Syk, Met, Epha2, Csf1 and Angpt4. Fgf14 was down-regulated in PCI vs ShamN. Chrm2 was the only gene uniquely affected in ShamI vs ShamN (down-regulated, Supp. Table 4).
HIF-1 KEGG pathway diagrams (Supp. Fig 12& Supp. Fig 13) showed a set of uniquely up-regulated genes in PCI vs ShamN: Nos2, Nfkb1, Mknk1, Il6r, Il6, Hk3, Egln3, Cdkn1a, and Angpt4 (Supp. Table 5).
p53 KEGG pathway diagrams (Supp.Fig 14 & Supp. Fig 15), represent the overall positions and fold changes of the significantly up-regulated genes in the HIF-1 pathway for PCI vs ShamN and ShamI vs ShamN respectively. They revealed a set of uniquely up-regulated genes in PCI vs ShamN: Igfbp3, Cd82, Fas, Cdkn1a, Cdk1, Gadd45b, and Ccnb1. (Supp. Table 6).
Cell cycle KEGG pathway diagrams (Supp. Fig 16 & Supp. Fig 17) demonstrated genes that were uniquely up-regulated for PCI vs ShamN: Tgfb1, Myc, Mcm5, Cdkn1a, Cdk1, Gadd45b, Ccnb1, Bub1b, Pttg1, Mcm6, and Mcm3. (Supp. Table 7).
DISCUSSION
RNA-Seq profiling of the transcriptome of the whole retina yielded 17,300 genes. RNA-Seq is the optimal method for large-scale transcription analysis as it quantitates gene expression, enables novel gene discovery, and detects low abundance genes in a tissue such as the retina [25]. We observed significant differences in both the identities of, degree of changes in, and the pathways of differentially expressed genes in postconditioning following ischemia compared to sham post-conditioning following ischemia. Differentially expressed genes were then subsequently classified according to their biological processes and molecular functions. The results yield important clues regarding the molecular machinery of this remarkable endogenous protective mechanism in the retina. While we cannot state at this time with certainty which of these pathways will be therapeutically accessible, there are some promising candidates based upon existing studies.
Pathway analysis using iPathwayGuide™ [19] yielded 28 uniquely affected pathways in PCI vs ShamN. We chose seven specific pathways for further study of mechanisms of Post-C. These included MAPK, apoptosis, Jak-STAT, PI3K-Akt, HIF-1, p53 and cell cycle. Some of the genes overlap across the pathways, thus to simplify presentation, p53 and cell cycle pathways are combined in this Discussion.
These pathways were chosen based on previous studies suggesting biological relevance to ischemia, neuroprotection, or pre- or post-conditioning. Apoptosis is a common mechanism mediating cell death. We previously described that apoptosis peaks in the retina approximately 24–48 h after ischemia [26, 27]. Thus, the retinas were collected at 24 h after Post-C (48 h after ischemia), consistent with the previously described time course of apoptosis. We hypothesized that Post-C significantly attenuated ischemia-induced apoptosis-related gene expression. All of the genes present in Supp. Table 3 had significantly increased expression in PCI vs ShamN compared to ShamI vs Sham N. Some of these genes (e.g., Cflar, Gadd45b, Nfkbia, Birc3 and 5, and Nfkb1) are known anti-apoptosis, pro-survival genes, while others (e.g., Fos, Parp3, Fas, and Tnfrsf1a [28, 29]) are pro-apoptotic. Therefore, it is likely that survival or death from apoptosis is dependent upon a balance of these pro- and anti-apoptosis genes, and may also depend upon the timing of their expression, not specifically addressed in this study, where we only measured gene expression at one time point.
In the p53 pathway/cell cycle, eight genes were significantly increased in PCI vs ShamN, but not in ShamI vs ShamN: Ccnb1, Gadd45b and Gadd45g (overlapping with apoptosis gene expression), Cdk1, Cdkn1a, Fas (also overlapping with apoptosis gene expression), Igfbp3 and Cd82. The cell cycle pathway had: Myc, Ccnb1, Tgfb1, Cdkn1a, Bub1b, Pttg1, Cdk1, Gadd45b and g, and Mcm3, 5, and 6. Relatively little is known about cell cycle regulators and ischemic injury in the retina, rendering these results novel. Kuroiwa et al[30] showed increases in Ccnd1 at 24 h after retinal ischemia in rats, but the functional significance of the changes has not been described. Under normal conditions, neurons are post-mitotic and do not re-enter the cell cycle. However, following cerebral ischemia in rats, neurons resumed DNA synthesis as indicated by bromo-deoxyuridine (BrdU) incorporation and expression of G1/S-phase cell cycle transition markers, and were TUNEL positive, suggesting that they underwent apoptosis. BrdU+/TUNEL neurons had decreased G1-phase cyclin-dependent kinase (CDK) inhibitors Cdkn2a and Cdkn1b, induction of late G1/S-phase Cdk2 activity, and retinoblastoma protein phosphorylation [31]. It is difficult to explain why myc and Cdkn1a, with opposite effects on cell cycling, changed in the same direction, and further studies will be necessary to clarify the implications of these findings.
Notable in our results is that ischemic and post-conditioned retinas had increased Ccnb1, Cdk1, Cdkn1a, and Gadd45b. Ccnb1 regulates G2 to M transition, and Cdk1 favors cell cycle arrest in G2 phase, Gadd45 is involved in DNA repair and damage prevention, and Cdkn1a inhibits entrance into the cell cycle by blocking Ccnd1 and Cdk4/6 [32]. This suggests but does not prove, the possibility that Post-C prevents entrance or completion of the cell cycle in ischemia, and favors DNA repair, to prevent apoptosis and cell death.
With respect to the other cell cycle/p53 genes, Pttg1 de-regulation has recently been observed in several mouse models of retinal degeneration, [33, 34] suggesting a possible cell survival role in retina. Bub1b is a component of the mitotic spindle checkpoint, which prevents sister chromatid separation during cell division. Abnormal Bub1b signaling has been implicated in premature aging, although to date, no studies have addressed its role in retina [35, 36]. MCM proteins control DNA unwinding. [37] In retina, Mcm5 depletion resulted in apoptosis, likewise suggesting an essential survival function [38]. Because of the prominence of apoptosis in the damage from retinal ischemia, the p53 pathway may be a viable drug target. [39]
In the HIF-1 pathway, increases were found in Angpt4, Cdkn1a [40], Egln3, Hk3, Ifngr2, Il6, Il6r, Mknk1, Nfkb1, and Nos2. Cdkn1a has been associated with neuroprotection in cortical neurons subjected to oxidative stress as well as in a cerebral ischemia model in vivo [40]. Angpt4 has been identified as an angiogenic factor in diabetic retinopathy [41] and increases permeability while enhancing survival of endothelial cells [42]. With respect to Egln3 [43], HIF is regulated by prolyl hydroxylases, which induces hydroxylation on proline-402 and -564 on the HIFα subunit [44]. Inhibiting Egln3 prevented oxygen induced retinopathy [45]. However, blockade of Egln3 enhanced HIF1 and decreased myocardial infarct size [46], thus the significance of increased Egln3 in our study remains unclear.
JAK-STAT cytokines and their receptors, Jak2, Stat6, Myc, Cdkn1a, and Socs3 demonstrated increases. There is evidence implicating the JAK-STAT pathway in both neuroprotection from cerebral ischemia, and in myocardial preservation after ischemia. Specifically, the JAK-STAT pathway was involved in Post-C preventing damage from global cerebral ischemia and myocardial ischemia [47–50], suggesting a possible role in retinal ischemic Post-C as well. But there are novel genes in the pathway identified in this study which have not been previously studied in ischemic injury or neuroprotection, including Il23a and Csf2, which may be drug targets to examine in future studies.
For MAPK, previously, members of the pathway have been implicated in ischemic injury, pre- and post-ischemic conditioning [51]. But several novel genes were identified in this study. Fgf14 is a novel protein controlling channel gating, axonal targeting and phosphorylation in neurons, affecting synaptic transmission, plasticity and neurogenesis. To date, its role in retina or in ischemia has not been evaluated, although it may be involved in neurodegeneration [52, 53]. Cacng2 is an AMPA-receptor regulating protein [54]. Map4k4 and Map3k8 are inflammatory kinases; Map4k4 also inhibits Jnk. Their role in ischemia has not been previously studied [55–57].
Given our previously demonstrated essential role of Akt in retinal Post-C [7], it was not surprising that the PI3K-Akt pathway had a large number of differentially regulated genes in ischemia and Post-C. Many of these, such as the growth factors (e.g., Fgf14), the cytokines (e.g., Il6r, Il6), integrins (e.g., Itga4), receptor tyrosine kinase (e.g., Epha2), are involved in binding to cellular receptors and activation of the PI3K and Akt signaling pathways [58]. We also noted increases in downstream signals in PCI vs ShamN, including those involved in cell cycle regulation (e.g., Myc and Cdkn1a) [32], and cell survival mechanisms (e.g., Nfkb1, and Creb5) [59]. Excepting Nfkb1, these downstream factors have not been previously studied for a role in ischemic neuroprotection in the retina.
Complicating interpretation of our results is that we are unable to determine the role of specific retinal cells, since we studied the transcriptome in the whole retina. Importantly, neurons and Muller cells may behave differently after injury. Müller glial cells are the major support cell for neurons in the vertebrate retina. Following neuronal damage, Müller cells undergo reactive gliosis, characterized by proliferation and changes in gene expression. Downregulation of the tumor suppressor protein p27Kip1 and re-entry into the cell cycle occur within the first 24 hours after retinal injury. Müller glial cells upregulate genes for gliosis and then downregulate cyclin D3 [60]. Therefore, we could miss important effects specific to Muller cells. This has important implications for cell survival and regeneration of retina, because in mice, if neuronal death is coupled with the intraocular injection of growth factors, Müller glia dedifferentiate, enter the cell cycle, and support modest neuronal regeneration [61]. However, to date true neuronal regeneration from Muller cells has not been shown in mammalian retina. [62]
Comparing these results to descriptions of gene expression in human retina in diabetic retinopathy, ARM, and central retinal artery occlusion is complicated by the use of different measurement methods (e.g. microarray vs our use of RNA-seq), different sampling methods, and smaller sample sizes. However, it is of interest that in a study of fibrovascular membranes from patients with proliferative diabetic retinopathy, several of these pathways overlapped with ours including apoptosis, cell cycle, and immune response.[63]
The advantages and challenges of RNA-Seq have been reviewed elsewhere [9]. RNA-Seq also has drawbacks which may limit the implications of our results. Library construction remains a challenge [9], because it involves a number of discrete steps, each of which brings a set of unique complications. Library preparation may introduce noise, and skew read distribution. Long reads and shallow read depth are important trade-offs that could have affected our results [64]. Large or complex transcriptomes genome alignment poses significant computational and technical challenges. In complex transcriptomes, alternative splicing and splice junctions make reads difficult to map while for large transcriptomes a significant portion of reads map to the multiple locations of the genome [9]. For our experiment specifically, the main drawback is that RNA-Seq is a snapshot of the ever-shifting nature of the cell’s transcriptome and it might not be reflective of events over time. For this reason we chose a 24h post-ischemia period as our point of reference, which has been previously shown to be the peak time of apoptosis as well as the optimal time for treatment [2].
The results of this experiment have elucidated pathways as well as specific genes that may be involved in retinal ischemic post-conditioning. Studying the impact of increasing expression of these factors may yield insight into new methods for treating or preventing damage from retinal ischemic disorders.
Supplementary Material
Supp. Table 1: RNA-Seq raw count data. The values in the matrix contain the un-normalized counts of sequencing reads. The column names refer to samples names, while row names refer to gene names. The value in the i-th row and the j-th column of the matrix tells how many reads can be assigned to gene i in sample j. Sample labels’ refer to our experimental design: retina control (ShamN), retina + post-conditioning (PCN), retina + ischemia + sham post-c (ShamI), and lastly retina + ischemia + post-c (PCI). Each group had 4 biological replicates. The entirety (17338 rows) of Supp. Table 1 was used as a starting point for the differential expression analysis using edgeR (Double click within the text box to view all 17388 rows).
Supp. Table 2: Uniquely affected genes in MAPK KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 3: Uniquely affected genes in Apoptosis KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 4: Uniquely affected genes in PI3K-Akt KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 5: Uniquely affected genes in HIF-1 KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 6: Uniquely affected genes in p53 KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 7: Uniquely affected genes in Cell Cycle KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Tables 8–11: All genes used to construct Fig 3 are shown in Supp. Tables 8–11. From left to right are shown logFC of PCI vs ShamN, logCPM PCI vs ShamN, P value, and Q value. The rightmost column indicates change in expression UP or DOWN. Double click within in each text box to scroll up or down. Supp. Table 3 Supp. Table 2 Supp. Table 1
Supp. Fig 1, Supp. Fig 2: Principal Component Analysis (PCA) plots of the next order principal components (PC1-PC3 and PC2-PC3), generated using R v. 3.2.2, ggplot2_2.2.1 andggfortify_0.4.1. Markedly lower variation is apparent in these plots vs Fig 2 indicating that most inter-sample variation was contained within the first two principal components.
Supp. Fig 3: Clustering image map (CIM) of the filtered, normalized and log2-transformed count data in order to explore similarities/dissimilarities between samples; generated by R v. 3.2.2. and mixOmics v. 6.1.3 package. The rows and columns were independently rearranged using a hierarchical clustering method. Euclidean distances between samples were computed and most similar rows and columns were placed next to each other. The results indicate some variation in samples within groups – most likely due to natural variation in biological samples.
Supp. Fig 4, Fig 5: Volcano plots of PCI vs ShamN and ShamI vs ShamN respectively generated using R v. 3.2.2, ggplot2_2.2.1 and ggrepel_0.6.10. Log2- transformed fold change values (x-axis) and negative log10- transformed Q values (y-axis) summarize the overall distribution of the gene expression changes across our experimental conditions. Genes with significant fold changes (log (fold change) ≥ 2) and significant FDR-corrected p values were annotated with their gene names.
Supp. Fig 6: MAPK KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant
Supp. Fig 7: MAPK KEGG pathway diagram for ShamI vs ShamN respectively. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 8: Apoptosis KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 9: Apoptosis KEGG pathway diagram for ShamI vs ShamN respectively. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 10: PI3K-Akt KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 11: PI3K-Akt KEGG pathway diagram for ShamI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 12: HIF-1 pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 13: HIF-1 pathway diagram for ShamI vs ShamN respectively. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 14: p53 KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 15: p53 KEGG pathway diagram for ShamI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 16: Cell Cycle KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 17: Cell Cycle KEGG pathway diagram for ShamI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Acknowledgments
Funding:
This study was supported by National Institutes of Health (Rockville, MD, USA) grant EY10343 to Dr. Roth, UL1TR000050 to the Center for Clinical and Translational Sciences of the University of Illinois at Chicago, the Illinois Society for the Prevention of Blindness, Chicago, IL, USA (Ms. Stelman); the Craig Foundation (Chicago, IL, USA, Ms. Stelman), a medical student research fellowship grant from the Foundation for Anesthesia Education and Research (Schaumburg, IL, USA, Ms. Stelman), and Core Grant P30 EY001792 (to the Department of Ophthalmology, University of Illinois at Chicago from the National Institutes of Health, Rockville, MD, USA); There was no involvement of the funding bodies in the design of the study or in collection, analysis and interpretation of the data or the writing of the manuscript. None of the authors have any conflicts of interest.
Footnotes
Animal Experiments
Ethical approval: All procedures performed in studies involving animals were in accordance with the ethical standards of and approved by the Institutional Animal Care and Use Committee of the University of Illinois at Chicago.
References
- 1.Zheng L, Gong B, Hatala DA, Kern TS. Retinal ischemia and reperfusion causes capillary degeneration: similarities to diabetes. Invest Ophthalmol Vis Sci. 2007;48:361–367. doi: 10.1167/iovs.06-0510. [DOI] [PubMed] [Google Scholar]
- 2.Dreixler JC, Poston JN, Shaikh AR, Alexander M, Tupper KY, Marcet MM, Bernaudin M, Roth S. Delayed post-ischemic conditioning significantly improves the outcome after retinal ischemia. ExpEye Res. 2011;92:521–527. doi: 10.1016/j.exer.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez DC, Bordone MP, Chianelli MS, Rosenstein RE. Retinal neuroprotection against ischemia-reperfusion damage induced by postconditioning. Invest Ophthalmol Vis Sci. 2009;50:3922–3930. doi: 10.1167/iovs.08-3344. [DOI] [PubMed] [Google Scholar]
- 4.Osborne NN, Ugarte M, Chao M, Chidlow G, Bae JH, Wood JP, Nash MS. Neuroprotection in relation to retinal ischemia and relevance to glaucoma. Surv Ophthalmol. 1999;43(Suppl 1):S102–128. doi: 10.1016/s0039-6257(99)00044-2. [DOI] [PubMed] [Google Scholar]
- 5.Limalanathan S, Andersen GO, Hoffmann P, Klow N-E, Abdelnoor M, Eritsland J. Rationale and design of the POSTEMI (postconditioning in ST-elevation myocardial infarction) study. Cardiology. 2010;116:103–109. doi: 10.1159/000316965. [DOI] [PubMed] [Google Scholar]
- 6.Zhao H. Ischemic postconditioning as a novel avenue to protect against brain injury after stroke. J CerebBlood Flow Metabol. 2009;29:873–885. doi: 10.1038/jcbfm.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreixler JC, Sampat A, Shaikh AR, Alexander M, Marcet MM, Roth S. Protein kinase B (Akt) and mitogen-activated protein kinase p38alpha in retinal ischemic post-conditioning. J Mol Neurosci. 2011;45:309–320. doi: 10.1007/s12031-011-9523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda M, Tanaka Y, Omodaka K, Nishiguchi KM, Nakamura O, Tsuda S, Nakazawa T. Transcriptome profiling of the rat retina after optic nerve transection. Sci Rep. 2016;6:28736. doi: 10.1038/srep28736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown J, Pirrung M, McCue LA. FQC Dashboard: Integrates FastQC results into a web based interactive and extensible FastQ quality control tool. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx373. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. JRoyal StatSoc Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 16.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, Bacic A, Hill AF, Stroud DA, Ryan MT, Agbinya JI, Mariadason JM, Burgess AW, Mathivanan S. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 17.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collaborators. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahsan S, Draghici S. Identifying Significantly Impacted Pathways and Putative Mechanisms with iPathwayGuide. Curr Protoc Bioinformatics. 2017;57:7.15.1–7.15.30. doi: 10.1002/cpbi.24. [DOI] [PubMed] [Google Scholar]
- 20.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang HC, Niu Y, Qin LX. Differential Expression Analysis for RNA-Seq: An Overview of Statistical Methods and Computational Software. Cancer Inform. 2015;14(Suppl 1):57–67. doi: 10.4137/CIN.S21631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussein SMI, Puri MC, Tonge PD, Benevento M, Corso AJ, Clancy JL, Mosbergen R, Li M, Lee D-S, Cloonan N, Wood DLA, Munoz J, Middleton R, Korn O, Patel HR, White CA, Shin J-Y, Gauthier ME, Cao K-AL, Kim J-I, Mar JC, Shakiba N, Ritchie W, Rasko JEJ, Grimmond SM, Zandstra PW, Wells CA, Preiss T, Seo J-S, Heck AJR, Rogers IM, Nagy A. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- 24.Li W. Volcano plots in analyzing differential expressions with mRNA microarrays. J Bioinform Comput Biol. 2012;10:1231003. doi: 10.1142/S0219720012310038. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, Meehan J, Li X, Yang L, Li H, Labaj PP, Kreil DP, Megherbi D, Gaj S, Caiment F, van Delft J, Kleinjans J, Scherer A, Devanarayan V, Wang J, Yang Y, Qian H-R, Lancashire LJ, Bessarabova M, Nikolsky Y, Furlanello C, Chierici M, Albanese D, Jurman G, Riccadonna S, Filosi M, Visintainer R, Zhang KK, Li J, Hsieh J-H, Svoboda DL, Fuscoe JC, Deng Y, Shi L, Paules RS, Auerbach SS, Tong W. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotech. 2014;32:926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Rosenbaum DM, Shaikh AR, QL, Rosenbaum PS, Pelham DJ, Roth S. Ischemic preconditioning attenuates apoptosis following retinal ischemia in rats. Invest Ophthalmol Vis Sci. 2002;43:3059–3066. [PubMed] [Google Scholar]
- 27.Singh M, Savitz SI, Hoque R, Rosenbaum PS, Roth S, Rosenbaum DM. Cell-specific caspase expression by different neuronal phenotypes in transient retinal ischemia. J Neurochem. 2001;77:466–475. doi: 10.1046/j.1471-4159.2001.00258.x. [DOI] [PubMed] [Google Scholar]
- 28.Berger S, Savitz SI, Nijhawan S, Singh M, David J, Rosenbaum PS, Rosenbaum DM. Deleterious role of TNF-alpha in retinal ischemia-reperfusion injury. Invest Ophthalmol Vis Sci. 2008;49:3605–3610. doi: 10.1167/iovs.07-0817. [DOI] [PubMed] [Google Scholar]
- 29.Wan T, Xu Z, Zhou HJ, Zhang H, Luo Y, Li Y, Min W. Functional analyses of TNFR2 in physiological and pathological retina angiogenesis. Invest Ophthalmol Vis Sci. 2013;54:211–221. doi: 10.1167/iovs.12-10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroiwa S, Katai N, Shibuki H, Kurokawa T, Umihira J, Nikaido T, Kametani K, Yoshimura N. Expression of cell cycle-related genes in dying cells in retinal ischemic injury. Invest Ophthalmol Vis Sci. 1998;39:610–617. [PubMed] [Google Scholar]
- 31.Kuan CY, Schloemer AJ, Lu A, Burns KA, Weng WL, Williams MT, Strauss KI, Vorhees CV, Flavell RA, Davis RJ, Sharp FR, Rakic P. Hypoxia-ischemia induces DNA synthesis without cell proliferation in dying neurons in adult rodent brain. J Neurosci. 2004;24:10763–10772. doi: 10.1523/jneurosci.3883-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frade JM, Ovejero-Benito MC. Neuronal cell cycle: the neuron itself and its circumstances. Cell Cycle. 2015;14:712–720. doi: 10.1080/15384101.2015.1004937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keeley PW, Zhou C, Lu L, Williams RW, Melmed S, Reese BE. Pituitary tumor-transforming gene 1 regulates the patterning of retinal mosaics. Proc Natl AcadSciUSA. 2014;111:9295–9300. doi: 10.1073/pnas.1323543111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yetemian RM, Craft CM. Characterization of the pituitary tumor transforming gene 1 knockout mouse retina. Neurochem Res. 2011;36:636–644. doi: 10.1007/s11064-010-0334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuda D, Matsumoto T, Honma K, Ikawa-Yoshida A, Onimaru M, Furuyama T, Nakatsu Y, Tsuzuki T, Maehara Y. BUBR1 Insufficiency in Mice Increases their Sensitivity to Oxidative Stress. In Vivo. 2016;30:769–776. doi: 10.21873/invivo.10993. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Jun H, Choi CI, Yoo KH, Cho CH, Hussaini SMQ, Simmons AJ, Kim S, van Deursen JM, Baker DJ, Jang MH. Age-related decline in BubR1 impairs adult hippocampal neurogenesis. Aging Cell. 2017;16:598–601. doi: 10.1111/acel.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deegan TD, Diffley JF. MCM: one ring to rule them all. Curr Opin Struct Biol. 2016;37:145–151. doi: 10.1016/j.sbi.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Ryu S, Holzschuh J, Erhardt S, Ettl AK, Driever W. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. ProceNatl AcadSciUSA. 2005;102:18467–18472. doi: 10.1073/pnas.0506187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nivison-Smith L, Khoo P, Acosta ML, Kalloniatis M. Vinpocetine protects inner retinal neurons with functional NMDA glutamate receptors against retinal ischemia. Exp Eye Res. 2017;167:1–13. doi: 10.1016/j.exer.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Langley B, D’Annibale MA, Suh K, Ayoub I, Tolhurst A, Bastan B, Yang L, Ko B, Fisher M, Cho S, Beal MF, Ratan RR. Pulse inhibition of histone deacetylases induces complete resistance to oxidative death in cortical neurons without toxicity and reveals a role for cytoplasmic p21(waf1/cip1) in cell cycle-independent neuroprotection. J Neurosci. 2008;28:163–176. doi: 10.1523/jneurosci.3200-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babapoor-Farrokhran S, Jee K, Puchner B, Hassan SJ, Xin X, Rodrigues M, Kashiwabuchi F, Ma T, Hu K, Deshpande M, Daoud Y, Solomon S, Wenick A, Lutty GA, Semenza GL, Montaner S, Sodhi A. Angiopoietin-like 4 is a potent angiogenic factor and a novel therapeutic target for patients with proliferative diabetic retinopathy. Proc Natl Acad Sci USA. 2015;112:E3030–3039. doi: 10.1073/pnas.1423765112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kesler CT, Pereira ER, Cui CH, Nelson GM, Masuck DJ, Baish JW, Padera TP. Angiopoietin-4 increases permeability of blood vessels and promotes lymphatic dilation. Faseb J. 2015;29:3668–3677. doi: 10.1096/fj.14-268920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaakkola PM, Rantanen K. The regulation, localization, and functions of oxygen-sensing prolyl hydroxylase PHD3. Biol Chem. 2013;394:449–457. doi: 10.1515/hsz-2012-0330. [DOI] [PubMed] [Google Scholar]
- 44.Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trichonas G, Lee TJ, Hoppe G, Au J, Sears JE. Prolyl hydroxylase inhibition during hyperoxia prevents oxygen-induced retinopathy in the rat 50/10 model. Invest Ophthalmol Vis Sci. 2013;54:4919–4926. doi: 10.1167/iovs.13-12171. [DOI] [PubMed] [Google Scholar]
- 46.Vogler M, Zieseniss A, Hesse AR, Levent E, Tiburcy M, Heinze E, Burzlaff N, Schley G, Eckardt KU, Willam C, Katschinski DM. Pre- and post-conditional inhibition of prolyl-4-hydroxylase domain enzymes protects the heart from an ischemic insult. Pflugers Arch. 2015;467:2141–2149. doi: 10.1007/s00424-014-1667-z. [DOI] [PubMed] [Google Scholar]
- 47.Goodman MD, Koch SE, Fuller-Bicer GA, Butler KL. Regulating RISK: a role for JAK-STAT signaling in postconditioning? Am J Physiol. 2008;295:H1649–1656. doi: 10.1152/ajpheart.00692.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hausenloy DJ, Yellon DM. Survival kinases in ischemic preconditioning and postconditioning. Cardiovasc Res. 2006;70:240–253. doi: 10.1016/j.cardiores.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Kim HC, Kim E, Bae JI, Lee KH, Jeon YT, Hwang JW, Lim YJ, Min SW, Park HP. Sevoflurane Postconditioning Reduces Apoptosis by Activating the JAK-STAT Pathway After Transient Global Cerebral Ischemia in Rats. J Neurosurg Anesthesiol. 2017;29:37–45. doi: 10.1097/ana.0000000000000331. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Wang D, Zhang L, Ye F, Li M, Wen K. Role of JAK-STAT pathway in reducing cardiomyocytes hypoxia/reoxygenation injury induced by S1P postconditioning. Eur J Pharmacol. 2016;784:129–136. doi: 10.1016/j.ejphar.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 51.Cox-Limpens KE, Gavilanes AW, Zimmermann LJ, Vles JS. Endogenous brain protection: what the cerebral transcriptome teaches us. Brain Res. 2014;1564:85–100. doi: 10.1016/j.brainres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Di Re J, Wadsworth PA, Laezza F. Intracellular Fibroblast Growth Factor 14: Emerging Risk Factor for Brain Disorders. Front Cell Neurosci. 2017;11:103. doi: 10.3389/fncel.2017.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pablo JL, Pitt GS. FGF14 is a regulator of KCNQ2/3 channels. Proc Natl Acad Sci USA. 2017;114:154–159. doi: 10.1073/pnas.1610158114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziff EB. TARPs and the AMPA receptor trafficking paradox. Neuron. 2007;53:627–633. doi: 10.1016/j.neuron.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Machida N, Umikawa M, Takei K, Sakima N, Myagmar BE, Taira K, Uezato H, Ogawa Y, Kariya K. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J Biol Chem. 2004;279:15711–15714. doi: 10.1074/jbc.C300542200. [DOI] [PubMed] [Google Scholar]
- 56.Roth Flach RJ, Skoura A, Matevossian A, Danai LV, Zheng W, Cortes C, Bhattacharya SK, Aouadi M, Hagan N, Yawe JC, Vangala P, Menendez LG, Cooper MP, Fitzgibbons TP, Buckbinder L, Czech MP. Endothelial protein kinase MAP4K4 promotes vascular inflammation and atherosclerosis. Nat Commun. 2015;6:8995. doi: 10.1038/ncomms9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowley SM, Kuriakose T, Dockery LM, Tran-Ngyuen T, Gingerich AD, Wei L, Watford WT. Tumor progression locus 2 (Tpl2) kinase promotes chemokine receptor expression and macrophage migration during acute inflammation. J Biol Chem. 2014;289:15788–15797. doi: 10.1074/jbc.M114.559344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghigo A, Laffargue M, Li M, Hirsch E. PI3K and Calcium Signaling in Cardiovascular Disease. Circ Res. 2017;121:282–292. doi: 10.1161/circresaha.117.310183. [DOI] [PubMed] [Google Scholar]
- 59.Carloni S, Girelli S, Scopa C, Buonocore G, Longini M, Balduini W. Activation of autophagy and Akt/CREB signaling play an equivalent role in the neuroprotective effect of rapamycin in neonatal hypoxia-ischemia. Autophagy. 2015;6:366–377. doi: 10.4161/auto.6.3.11261. [DOI] [PubMed] [Google Scholar]
- 60.Dyer MA, Cepko CL. Control of Muller glial cell proliferation and activation following retinal injury. Nat Neurosci. 2000;3:873–880. doi: 10.1038/78774. [DOI] [PubMed] [Google Scholar]
- 61.Vetter ML, Hitchcock PF. Report on the National Eye Institute Audacious Goals Initiative: Replacement of Retinal Ganglion Cells from Endogenous Cell Sources. Transl Vis Sci Technol. 2017;6:5. doi: 10.1167/tvst.6.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilken MS, Reh TA. Retinal regeneration in birds and mice. Curr Opin Gen Devel. 2016;40:57–64. doi: 10.1016/j.gde.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 63.Ishikawa K, Yoshida S, Kobayashi Y, Zhou Y, Nakama T, Nakao S, Sassa Y, Oshima Y, Niiro H, Akashi K, Kono T, Ishibashi T. Microarray Analysis of Gene Expression in Fibrovascular Membranes Excised From Patients With Proliferative Diabetic Retinopathy. Invest Ophthalmol Vis Sci. 2015;56:932–946. doi: 10.1167/iovs.14-15589. [DOI] [PubMed] [Google Scholar]
- 64.Kratz A, Carninci P. The devil in the details of RNA-seq. Nat Biotech. 2014;32:882–884. doi: 10.1038/nbt.3015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Table 1: RNA-Seq raw count data. The values in the matrix contain the un-normalized counts of sequencing reads. The column names refer to samples names, while row names refer to gene names. The value in the i-th row and the j-th column of the matrix tells how many reads can be assigned to gene i in sample j. Sample labels’ refer to our experimental design: retina control (ShamN), retina + post-conditioning (PCN), retina + ischemia + sham post-c (ShamI), and lastly retina + ischemia + post-c (PCI). Each group had 4 biological replicates. The entirety (17338 rows) of Supp. Table 1 was used as a starting point for the differential expression analysis using edgeR (Double click within the text box to view all 17388 rows).
Supp. Table 2: Uniquely affected genes in MAPK KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 3: Uniquely affected genes in Apoptosis KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 4: Uniquely affected genes in PI3K-Akt KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 5: Uniquely affected genes in HIF-1 KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 6: Uniquely affected genes in p53 KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Table 7: Uniquely affected genes in Cell Cycle KEGG pathway for PCI vs. ShamN along with their associated FDR values and fold changes in both comparison groups. Genes are displayed along with their respective log2FC ratio, p value and Q value.
Supp. Tables 8–11: All genes used to construct Fig 3 are shown in Supp. Tables 8–11. From left to right are shown logFC of PCI vs ShamN, logCPM PCI vs ShamN, P value, and Q value. The rightmost column indicates change in expression UP or DOWN. Double click within in each text box to scroll up or down. Supp. Table 3 Supp. Table 2 Supp. Table 1
Supp. Fig 1, Supp. Fig 2: Principal Component Analysis (PCA) plots of the next order principal components (PC1-PC3 and PC2-PC3), generated using R v. 3.2.2, ggplot2_2.2.1 andggfortify_0.4.1. Markedly lower variation is apparent in these plots vs Fig 2 indicating that most inter-sample variation was contained within the first two principal components.
Supp. Fig 3: Clustering image map (CIM) of the filtered, normalized and log2-transformed count data in order to explore similarities/dissimilarities between samples; generated by R v. 3.2.2. and mixOmics v. 6.1.3 package. The rows and columns were independently rearranged using a hierarchical clustering method. Euclidean distances between samples were computed and most similar rows and columns were placed next to each other. The results indicate some variation in samples within groups – most likely due to natural variation in biological samples.
Supp. Fig 4, Fig 5: Volcano plots of PCI vs ShamN and ShamI vs ShamN respectively generated using R v. 3.2.2, ggplot2_2.2.1 and ggrepel_0.6.10. Log2- transformed fold change values (x-axis) and negative log10- transformed Q values (y-axis) summarize the overall distribution of the gene expression changes across our experimental conditions. Genes with significant fold changes (log (fold change) ≥ 2) and significant FDR-corrected p values were annotated with their gene names.
Supp. Fig 6: MAPK KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant
Supp. Fig 7: MAPK KEGG pathway diagram for ShamI vs ShamN respectively. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 8: Apoptosis KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 9: Apoptosis KEGG pathway diagram for ShamI vs ShamN respectively. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 10: PI3K-Akt KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 11: PI3K-Akt KEGG pathway diagram for ShamI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 12: HIF-1 pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 13: HIF-1 pathway diagram for ShamI vs ShamN respectively. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 14: p53 KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 15: p53 KEGG pathway diagram for ShamI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 16: Cell Cycle KEGG pathway diagram for PCI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.
Supp. Fig 17: Cell Cycle KEGG pathway diagram for ShamI vs ShamN. Diagrams were downloaded from iPathwayGuide’s pathways feature [10]. Significantly affected genes between conditions are colored and the intensity of the color reveals the magnitude of the fold change of a given gene (genes in red indicate up-regulation; genes in blue indicate down-regulation). Only the genes with an absolute value of log2(fold change) ≥ 2 and Q value ≤ 0.05 were considered significant.