Abstract
The 16.5 kb mitochondrial genome is subjected to damage from reactive oxygen species (ROS) generated in the cell during normal cellular metabolism and external sources such as ionizing radiation and ultraviolet light. ROS cause harmful damage to DNA bases that could result in mutagenesis and various diseases, if not properly repaired. The base excision repair (BER) pathway is the primary pathway involved in maintaining the integrity of mtDNA. Several enzymes that partake in BER within the nucleus have also been identified in the mitochondria. The nei-like (NEIL) DNA glycosylases initiate BER by excising oxidized pyrimidine bases and others such as the ring-opened formamidopyrimidine and the hydantoin lesions. During BER, the NEIL enzymes interact with proteins that are involved with DNA replication and transcription. In the current manuscript, we detected NEIL1 in purified mitochondrial extracts from human cells and showed that NEIL1 interacts with the human mitochondrial single-stranded DNA binding protein (mtSSB) via its C-terminal tail using protein painting, far-western analysis, and gel-filtration chromatography. Finally, we scrutinized the NEIL1-mtSSB interaction in the presence and absence of a partial-duplex DNA substrate using a combination of multi-angle light scattering (MALS) and small-angle X-ray scattering (SAXS). The data indicate that NEIL1 and homotetrameric mtSSB form a larger ternary complex in presence of DNA, however, the tetrameric form of mtSSB gets disrupted by NEIL1 in the absence of DNA as revealed by the formation of a smaller NEIL1-mtSSBmonomer complex.
Keywords: Base Excision Repair, NEIL1 DNA glycosylase, mitochondrial single-stranded DNA binding protein, protein painting, size exclusion chromatography, small angle X-ray scattering
1. Introduction
Reactive oxygen species (ROS) are generated in a cell as a result of normal cellular respiration as well as from external sources that include ultraviolet light, ionizing radiation, and chemicals found in air pollutants such as cigarette smoke [1, 2]. ROS elicit stress responses in the cell by causing lipid peroxidation, damage to DNA bases, and oxidization of proteins [3]. Oxidative stress if not mitigated can result in several disease states including mutagenesis that leads to malignancies and aging-related neurodegenerative disorders [4–6]. Both the nuclear and mitochondrial genomes are susceptible to the harmful effects of ROS [7, 8]. The ~16.5 kb mitochondrial genome encodes for 13 polypeptides, 22 tRNAs, and 2 rRNAs involved with oxidative phosphorylation, while all other proteins required for mitochondrial DNA (mtDNA) maintenance (replication, repair, signaling etc.) are encoded by nuclear genes and transported to the mitochondria [9–11]. MtDNA is not packaged into nucleosomes, but instead is organized within large multi-protein-DNA complexes called nucleoids [12]. The most abundant protein involved in the compact packing of mtDNA is the mitochondrial transcription factor A (TFAM) [13]. The other nucleoid-associated proteins include polymerase gamma (Polγ), the ATPase family AAA-domain-containing protein 3 (ATAD3), the mitochondrial AAA proteases Lon peptidase 1 (LONP1), and single-stranded DNA binding protein (mtSSB) [12, 14–16].
To combat the harmful effects of ROS, akin to the nucleus, mitochondria also possess DNA repair mechanisms that are involved in the repair of mtDNA [17, 18]. The base excision repair (BER) pathway that is involved with the repair of DNA base damage caused by deamination, alkylation, oxidation, or single-strand breaks, participates in the repair of both nuclear and mtDNA [19–22]. BER is a step-wise, highly conserved pathway, and is carried out in 5 basic steps: damaged base recognition and excision, removal of the resulting abasic site, end processing, gap synthesis, and ligation [20, 21, 23, 24]. The first step of BER is catalyzed by enzymes called DNA glycosylases and of the 11 mammalian DNA glycosylases, 7 have been identified within the mitochondria [19, 25]. DNA glycosylases that only catalyze excision of the damaged base are monofunctional, whereas those that are able to excise the damaged base and further cleave the DNA backbone 3’ to the lesion (i.e. possess lyase activity) are termed bifunctional glycosylases. There are 6 known oxidized-base specific glycosylases belonging to 2 structurally distinct families: the endonuclease III (or Nth) family and the formamidopyrimidine DNA glycosylase/ endonuclease VIII (or Fpg/Nei) family, both named after their bacterial prototypes [24]. The Nei-like (NEIL) NEIL1, 2, and 3 are the human orthologs of bacterial Fpg and Nei and thus far, NEIL1 and NEIL2 have been reported to be involved with mtDNA maintenance [26–28].
The human DNA glycosylases are structurally similar to their prokaryotic counterparts; however, the mammalian enzymes possess regions of disorder that are not present in the bacterial enzymes [29]. In NEIL1 and NEIL2, the disordered regions participate in protein-protein interactions that mediate the remainder of the repair process as well as enable multiple interactions with proteins involved in other cellular processes including replication, and transcription [29]. The current prevailing hypothesis is that NEIL1 participates in repair during replication and serves as a “cowcatcher” ahead of the replication fork where it binds to base lesions generated [30]. This is typically followed by stalling of the replication fork and binding of downstream BER factors [30]. The disordered C-terminal tail of NEIL1 spans 100 amino acid residues (290–390) that are involved in interactions with proteins including heterotrimeric replication protein A (RPA), homotrimeric proliferating cellular nuclear antigen (PCNA), flap endonuclease 1 (FEN-1), the Werner Syndrome RecQ like helicase (WRN helicase), and others, thereby reinforcing its role during replication [30–34].
MtSSB plays an indispensable role in mtDNA replication where it protects single-stranded DNA (ssDNA) from damage caused by ROS, and stimulates Polγ and TWINKLE activities [35, 36]. It also plays a role in mtDNA repair as evidenced by inhibition of AAG and UNG1-mediated base excision from ssDNA as well as inhibits APE1-mediated DNA strand nicking [37, 38]. While binding partners of NEIL1 in nuclear DNA repair have been previously documented [31–34, 39], thus far, there is no direct evidence for the role of NEIL1 in mtDNA repair or its association with proteins involved with mtDNA replication and transcription. Studying protein-protein interactions that occur during mtDNA repair will help us understand the role of repair factors during mtDNA replication and transcription. In this work, we probed the interaction between NEIL1 and mtSSB using 9 truncated constructs of NEIL1, in vitro. Our data indicate that amino acid residues 289–312 in the C-terminal tail of NEIL1 are required for minimal binding to mtSSB. We isolated a complex of NEIL1 with mtSSB in the absence and presence of a partial duplex DNA substrate and scrutinized the resulting complexes formed using small angle X-ray scattering (SAXS) and multi-angle light scattering (MALS).
2. Materials and methods
2.1. Plasmids and DNA oligonucleotides
The bacterial expression plasmid containing C-terminal His-tagged NEIL1 (pET30a) was obtained from the laboratories of Dr. Sylvie Doublié and Dr. Susan Wallace (University of Vermont). The expression plasmid for mtSSB (pET21a) was obtained from Dr. William C. Copeland (NIEHS, North Carolina). Construction of E. coli expression plasmids for the full-length (FL) and truncated forms of NEIL1 (Δ40, Δ56, Δ78, and Δ100) were described previously [32, 39]. A C-terminal GST fused NEIL1 construct comprising residues 289–389 was synthesized by Genscript (Piscataway, NJ) and subcloned into the pGEX-4T2 vector. The pGEX-4T2 GST fused NEIL1 C-terminal constructs 289–349, 312–389, and 312–349 were further created using PCR techniques.
The partial duplex oligonucleotide substrate used in these studies consists of a short 13-nt oligomer 5’-CGTCCAXGTCTAC-3’ (X = tetrahydrofuran, an abasic site analog) and a long oligomer 5’-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGTAGACCTGGACG-3’. For optimal annealing, equimolar mixtures of the oligomers were heated at 94 °C for 2 min, and then slowly cooled to room temperature. For mtSSB-DNA interactions, a poly dT60 substrate was also used.
2.2. Overexpression and purification of NEIL1- constructs and mtSSB
MtSSB was overexpressed and purified as described previously [40]. Full length NEIL1 (NEIL1-FL) and C-terminal truncation constructs were transformed into Rosetta2 (DE3) pLysS Escherichia coli cells (Novagen) and protein expression was induced with 0.4 mM isopropyl-D-thiogalactopyranoside (IPTG) overnight at 12–16 °C. The cell pellets were resuspended in a buffer containing 50 mM sodium phosphate (pH 7.5), 300 mM NaCl, 10 mM imidazole, 10% (v/v) glycerol, 2 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and sonicated. Clarified cell lysates were loaded onto pre-equilibrated TALON cobalt resin (Clontech). Protein elution was performed in the above buffer containing 250–500 mM imidazole. The proteins were further purified using a HiTrap SP-FF column (GE healthcare) using a linear NaCl gradient (from 150 mM – 1 M). The purified fractions were pooled, concentrated and applied over a Superdex 200 size exclusion column (GE Healthcare) in a buffer containing 20 mM HEPES (pH 7.5), 300 mM NaCl, 10% (v/v) glycerol, and 1 mM dithiothreitol (DTT). Fractions were pooled, concentrated, and flash-frozen aliquots were stored at −80°C. GST-fused NEIL1 C-terminal constructs were transformed similarly as above and protein expression was initiated with the addition of 0.4 mM IPTG for 4 hrs at 25 °C. The recombinant proteins were purified using glutathione sepharose beads (GE healthcare) following the manufacturer’s instructions. The proteins were further purified using Hi-Trap SP-FF and Q-FF columns (GE healthcare).
2.3. Complex formation and size exclusion chromatography (SEC)
The complex of NEIL1-mtSSB was prepared by mixing NEIL1 and mtSSB tetramer in a 1:1 molar ratio in a buffer containing 20 mM HEPES (pH 7.5), 300 mM NaCl, 10% (v/v) glycerol, and 1 mM DTT. The complex was incubated on ice for 30 min prior to SEC analysis. The ternary complex of NEIL1-DNA-mtSSB was prepared by mixing NEIL1 and DNA in a 1:1 molar ratio followed by incubation on ice for 30 min. The mtSSB tetramer was then added to this complex such that the final ratio of NEIL1:DNA:mtSSB tetramer was 1:1:1. The ternary complex was further incubated for 30 min on ice prior to SEC performed with a Superdex 200 column using a buffer containing 20 mM HEPES (pH 7.5), 300 mM NaCl, 10% (v/v) glycerol, and 1 mM DTT. The NEIL1-DNA and mtSSB-DNA complexes were prepared by mixing the proteins and the DNA in a 1:1 molar ratio and incubated for 30 min on ice prior to loading on the SEC column. The column was calibrated using blue dextran (to determine the void volume) and four standards of known molecular weights (MW) and Stokes radii; Aprotinin (19.68 Å), Ribonuclease A (18.24 Å), Ovalbumin (15.36 Å) and Conalbumin (14.64 Å).
2.4. Cell culture and whole cell extract preparation
HCT-116 and HEK-293T cells were cultured in McCoy's 5A medium and Dulbecco's Modified Eagle's Medium, respectively, supplemented with 10% fetal bovine serum. To prepare whole cell extracts for far-western analysis, cells were washed with cold PBS and scraped into Cell Lytic M reagent (Sigma) containing Halt protease inhibitor cocktail (Thermo Fisher Scientific). The scraped cells were incubated on ice for 15 min and centrifuged at 10,000 × g for 10 min to remove cell debris. The supernatant was collected and stored at −80 °C.
2.5. Far-Western Analysis
Far-western analysis was performed as described previously [32]. Briefly, proteins (50 pmol of all NEIL1 constructs, bovine serum albumin, and glutathione S-transferase; 10 pmol mtSSB) were separated by 12% SDS-PAGE, transferred to a PVDF membrane (BioRad), treated with 6M guanidine HCl in PBS containing 1 mM DTT, and then gradually refolded with successive dilutions of guanidine HCl in PBS containing 1 mM DTT. The membranes were then blocked for 3 hr with 3% BSA in PBS, 0.05% Tween 20 at 4 °C and incubated with pre-prepared HCT-116/HEK-293T whole cell extracts (1 mg/ml) or purified protein (10 pmol/ml), overnight. Immunoblot analysis was performed using an anti-SSB antibody (Abcam- ab74710).
2.6. Small-angle X-ray scattering (SAXS) and multi-angle light scattering (MALS)
SEC-MALS-SAXS/ SEC-SAXS data were collected at beamline 18-ID (BioCAT) of the advanced photon source (APS) at Argonne National Laboratory. All measurements were performed in a buffer containing 25 mM HEPES pH 7.4, 5% glycerol, 300 mM NaCl, and 1 mM DTT. Samples include NEIL1-FL, mtSSB tetramer, the mtSSB-DNA (dT60/partial duplex) complex, the NEIL1-mtSSB complex, and the NEIL1-DNA-mtSSB complex at concentrations of 7–11 mg/ml. The samples were loaded on the SEC column with an exclusion limit of 2.5 MDa (Wyatt Technologies), which was followed by a UV detector, MALS detector (DAWN Helios II, Wyatt Technologies) and a SAXS flow cell. At a flow rate of 0.75 ml/min, 0.5s exposures were acquired every 3 seconds. The exposures flanking the elution peak were averaged to generate a buffer file that was subtracted from the exposures corresponding to the elution peak to get the I(q) vs q SAXS curves. Data analysis was performed using the ATSAS package [41]. Buffer blank subtraction, Guinier approximation based forward scattering (I0) analysis, and radius of gyration (Rg) determination were performed using PRIMUS [42]. The normalized Kratky plots were calculated using Scatter 3.0 [43]. Scattering curves were further analyzed using GNOM [44] for the calculation of I0, Rg, distance distribution P(r), maximum dimension (Dmax), Porod volume (Vρ), and excluded volume (Ve). I0 and Rg obtained from GNOM were compared with those obtained from PRIMUS. Molecular weights were estimated using Porod volume [45] and volume of correlation (BioXTAS RAW) [46, 47]. Three-dimensional shapes were reconstructed by 10 iterations of ab initio modeling using DAMMIF [48] followed by averaging of models using DAMAVER [49]. The crystal structures of NEIL1 and mtSSB were fitted into the envelopes of NEIL1, mtSSB and mtSSB-DNA complexes using SUPCOMB [50]. For NEIL1, owing to the presence of the large disordered region, the structure had to be manually adjusted after SUPCOMB was initially used to place the model in the envelope. The figures were prepared using PyMOL (Schrödinger LLC). Molecular weights were also calculated from the MALS data for comparison with the SAXS estimates using the ASTRA software (Wyatt Technologies). The resulting absolute molar masses obtained are reported in Table 2.
Table 2.
SAXS data collection parameters
| (A) Sample details | |||||
|
| |||||
| mtSSB (tetramer) | NEIL1 | mtSSB-DNA complex | NEIL1-mtSSB complex | NEIL1-DNA-mtSSB complex | |
|
| |||||
| Organism | Homo sapiens | Homo sapiens | Homo sapiens | Homo sapiens | Homo sapiens |
| Source | E. coli | E. coli | E. coli | E. coli | E. coli |
|
| |||||
| UniProt sequence ID (residues in constructs) | Q04837 (17–148) | Q96FI4 (1–390) | |||
| Extinction coefficients [A280, 0.1%(w/v)] | 1.313 | 0.725 | |||
|
| |||||
| SEC–SAXS column, 10/300 mm Superdex S200 | |||||
| Loading concentration (mg ml−1) | 7 | 11 | 8 | 10 | 10 |
| Injection volume (µl) | 300 | 300 | 300 | 300 | 300 |
| Flow rate (ml min−1) | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 |
| Solvent (blanks taken from SEC flow through prior to elution of protein) | 25 mM HEPES pH 7.4, 5% glycerol, 300 mM NaCl, and 1 mM DTT | ||||
|
| |||||
| (B) Data collection parameters | |||||
|
| |||||
| Beamline | APS | APS | APS | APS | APS |
| Wavelength (Å) | 1.03 | 1.03 | 1.03 | 1.03 | 1.03 |
| Q Range (Å−1) | 0.0059–0.3892 | 0.0059–0.3898 | 0.0058–0.3587 | 0.0058–0.3746 | 0.0052–0.3606 |
| Temperature (°C) | 25 | 25 | 25 | 25 | 25 |
|
| |||||
| (C) Softwares used for data reduction, analysis and interpretation | |||||
|
| |||||
| SAXS data reduction | PRIMUS (ATSAS 2.8.1) | ||||
| Extinction coefficient estimate | ProtParam (Expasy) [83] | ||||
| Basic analyses: Guinier, P(r), MW (Vp, Vc) | PRIMUS (ATSAS 2.8.1), Scatter [43], BioXTAS RAW [47] | ||||
| Shape/bead modelling | DAMMIF and DAMAVER (ATSAS 2.8.1) | ||||
|
| |||||
| (C) Structural parameters | |||||
|
| |||||
| I(0) from P(r) | 44.88 | 51.49 | 22.74 | 102.8 | 116 |
| Rg (Å) from P(r) | 27.52 | 37.15 | 28.18 | 28.12 | 46.86 |
| I(0) from Guinier | 44.83±0.10 | 49.20±0.24 | 22.77±0.03 | 104.00±0.27 | 114.52±0.46 |
| Rg from Guinier | 27.44±1.21 | 33.04±2.46 | 28.40±0.86 | 28.48±0.80 | 44.83±1.80 |
| Dmax (Å) | 103.21 | 149.96 | 98.74 | 94.32 | 165.72 |
|
| |||||
| (D) Molecular weight determination (kDa) | |||||
|
| |||||
| Expected Theoretical (Expasy) | 60.78 | 44.72 | 78 | 105.5 | 123 |
| MW(Vp) | 61 | 46 | 79 | 60.3 | 146.6 |
| MW(Vc) | 59 | 36 | 71 | 55.4 | 134.3 |
| MW (MALS) | 59.8±1.67% | 48.5±2.23% | 69±1.46% | 58±1.54% | 129.6±1.54% |
|
| |||||
| (E) Modeling parameters | |||||
|
| |||||
| Symmetry | P4 | P1 | P4 | P1 | P1 |
| Particle anisometry | Oblate | Unknown | Oblate | Oblate, Prolate, Unknown | Oblate, Prolate, Unknown |
| # of modeling iterations | 10 | 10 | 10 | 10 | 10 |
| X2 of the model | 1.072 | 1.041 | 0.945 | 1.311(O) | 1.072(O) |
| 1.311(P) | 1.066(P) | ||||
| 1.312(U) | 1.069(U) | ||||
3. Results
3.1. Mapping the interaction between NEIL1 and mtSSB
The importance of NEIL1 in the maintenance of the mitochondrial genome is highlighted by studies conducted in NEIL1 knockout mice where a percentage of the mice developed a metabolic phenotype [27, 28]. NEIL1 was also previously observed in mitochondrial extracts derived from mouse liver and served as a backup glycosylase in extracts lacking Ogg1 and Nth1 [51]. In this current study, we identified NEIL1 in purified mitochondria isolated from HCT-116 cells (Supplemental Fig. 1). Given that NEIL1 associates with multiple proteins involved with DNA replication in the nucleus including RPA, PCNA, and others [30–34], we sought to study whether NEIL1 associates with the mitochondrial single-stranded DNA binding protein (mtSSB). Previous reports indicate that the C-terminal residues (290–390) of NEIL1 are disordered and responsible for the binding of the enzyme to its protein partners [29]. We used protein painting to identify the residues that mediate the interaction between NEIL1 and mtSSB (Supplemental Fig. 2A) [52]. Briefly, with this technique, the complexes of the interacting partners are coated with molecular paints that bind to solvent accessible surfaces and prevent cleavage by proteases such as trypsin. When the protein partners are dissociated the only residues that remain uncoated by the paints are the interaction interfaces, which are susceptible to proteolytic digestion. The tryptic peptides can then be identified via mass spectrometry (MS). For this purpose, we tested five molecular paints that were used previously to successfully map the interaction of interleukin 1b with the interleukin 1 receptor type I protein [52] (Supplemental Fig. 2B). In our hands, the disodium;1-amino-9,10-dioxo-4-[3-(2-sulphonatooxyethylsulphonyl)anilino]anthracene-2-sulphonate (RBB) molecule optimally coated the surfaces of both NEIL1 and mtSSB yielding the least number of peptides after mass-spectrometry analysis (Supplemental Fig. 2C, D). With mtSSB alone, the only peptide that was revealed in our analysis encompassed residues 107–132 which is present at the homotetramer interface (Supplemental Fig. 2D). When the NEIL1-mtSSB complex was coated with RBB, the peptides observed for the individual proteins subtracted from the peptides obtained after complex dissociation, yielded the interaction interface residues (Supplemental Fig. 2E). Our data confirm that the residues within the C-terminal 100 amino acids of NEIL1 mediate its interaction with mtSSB (Supplemental Fig. 2E, F). Two additional NEIL1 peptides were also obtained as part of the interaction with mtSSB: one peptide (residues 30–52) forms a surface-exposed β-strand in the N-terminal domain and other comprising residues 186–195 is disordered in the crystal structure and is located proximal to the C-terminal interaction domain (Supplemental Fig. 2F).
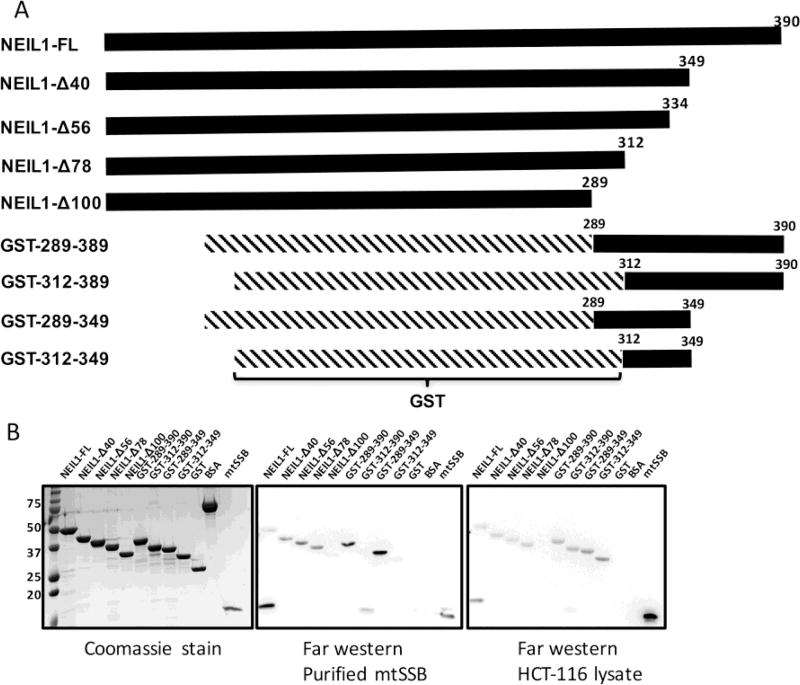
To identify the minimum interacting region of NEIL1 that binds to mtSSB, we purified histidine tagged full-length NEIL1 (NEIL1-FL) and C-terminal truncation constructs NEIL1-Δ40, NEIL1-Δ56, NEIL1-Δ78, and NEIL1-Δ100 (Fig. 1A, 1B left panel). We also expressed and purified GST-tagged NEIL1 C-terminal constructs 289–390, 289–349, 312–390, and 312–349 (Fig. 1A, 1B left panel) and performed far-western analysis as previously described [32, 53]. BSA and GST alone were used as negative controls. The membrane with the refolded NEIL constructs was incubated with either purified mtSSB or HCT-116/HEK-293T whole cell extracts and then probed with an antibody against mtSSB. Our results showed that purified mtSSB interacts with NEIL1-FL, NEIL1-Δ40, NEIL1-Δ56, NEIL1-Δ78 but not with NEIL1-Δ100 (Fig. 1B, middle panel). We also observed an interaction of mtSSB with residues 289–390 and 289–349 but not with 312–349 and 312–390, allowing us to conclude that the minimum interaction domain of NEIL1 required for interactions with mtSSB is present between residues 289–312 (Fig. 1B middle panel). Similar results were also obtained using the whole cell extracts, however, we also observed binding with residues 312–349 and 312–390 (Fig. 1B right panel, Supplemental Fig. 3). We attribute the differences observed in the far-western analysis between the whole cell extracts and purified protein to the possibility that third-party protein interactions, the presence of DNA, or post-translational modifications mediate the interaction between NEIL1 and mtSSB in the whole cell extracts.
Figure 1.
Residues within the C-terminal tail of NEIL1 interact with mtSSB. (A) Domain organization of full-length (FL) His-tagged NEIL1, C-terminal deletion constructs, and GST-tagged C-terminal constructs of the NEIL1 DNA glycosylase. (B) Far-western analysis to determine the minimal construct of NEIL1 required for an interaction with mtSSB. All constructs used in this study were expressed in E. coli, purified to homogeneity and verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis stained with Coomassie blue (left panel). 50 pmol of all NEIL1 constructs, bovine serum albumin (negative control), glutathione S-transferase (negative control), and 10 pmol of mtSSB were loaded onto the gel. Far-western analysis was performed where proteins were transferred to a PVDF membrane, denatured, slowly renatured on the membrane, and incubated with either 10 pmol/ml purified mtSSB (middle panel) or 1 mg/ml HCT-116 whole cell extract (right panel), and probed with an anti-mtSSB antibody to detect an interaction.
3.2. Interaction of NEIL1 and mtSSB in vitro
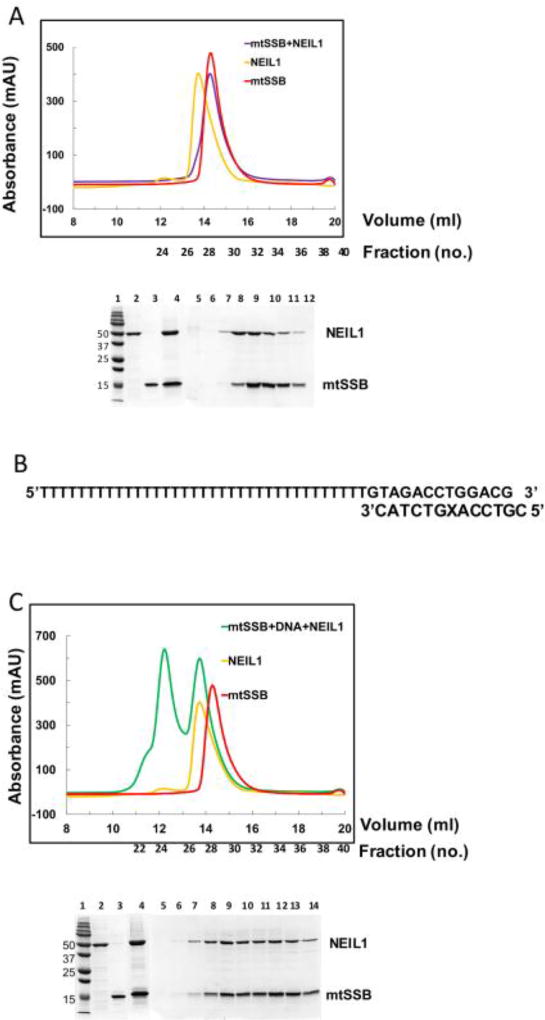
To isolate a binary complex of NEIL1 and mtSSB, we employed size exclusion chromatography (SEC) using a calibrated Superdex 200 column and documented the Stokes radii of the eluting proteins and complexes (Table 1). SEC separates proteins based on their Stokes radius which depends on MW as well as shape of the molecule (globular vs. elongated) [54–56]. First, we evaluated the proteins alone to determine their individual Stokes radii. NEIL1-FL (44.75 kDa) elutes at a volume of 13.73 ml and a Stokes radius of 42.2 Å similar to values reported previously [32]. The homotetrameric mtSSB (60.78 kDa) forms a compact globular complex and elutes at a volume 14.29 ml with a Stokes radius of 39.2 Å. The higher Stokes radius observed with NEIL1 corresponds to its elongated shape that results from the presence of a disordered C-terminal tail, which is absent in the globular mtSSB tetramer. NEIL1-Δ100 (34 kDa) that lacks the C-terminal tail elutes at 15.79 ml and a Stokes radius of 31.2 Å (Table 1) [32]. The NEIL1-mtSSB complex elutes as a single peak at a volume of 14.26 ml and a Stokes radius of 39.4 Å. Both NEIL1 and mtSSB were present in the fractions obtained from the eluted peak as observed by SDS-PAGE analysis (Fig. 2A). As predicted, NEIL1-Δ100, which lacks the putative protein interaction domain did not form a complex with mtSSB and the two proteins eluted separately (confirmed by SDS-PAGE analysis, Supplemental Fig. 4A). Thereafter, we performed SEC analysis of the NEIL1-DNA-mtSSB complex combined in a 1:1:1 molar ratio where the DNA used in these studies was a partial duplex substrate containing tetrahydrofuran, an abasic site analog (Fig. 2B, C). NEIL1-DNA-mtSSB elutes as a large complex at an elution volume of 12.22 ml with a Stokes radius 50.3 Å and a second peak at an elution volume of 13.7 ml (Fig. 2C). The samples from the eluted peaks were analyzed by SDS-PAGE, which confirmed the presence of both mtSSB and NEIL1. As a control, we performed SEC analysis of the mtSSB-DNA and NEIL1-DNA complexes (1:1 molar ratio). The NEIL1-DNA complex elutes at a volume of 13.44 ml and Stokes radius 43.7 Å (Table 1, Supplemental Fig. 4B). The mtSSB-DNA complex elutes at a volume of 13.14 ml and Stokes radius of 45.6 Å (Supplemental Fig. 4B). We observed a second peak at an elution volume of 13.7 ml, similar to the peak observed with the NEIL1-DNA-mtSSB complex (Supplemental Fig. 4B). This peak could be the result of a different binding mode of mtSSB to DNA [57–59] or could reflect that the theoretical 1:1 calculated molar ratios do not translate to 1:1 stoichiometric complexes. Overall, the results from our SEC analysis indicate that NEIL1 and mtSSB form a complex either in the absence or presence of DNA in vitro. These data are further corroborated by SAXS and MALS analyses as described in the following section.
Table 1.
Stokes radius calculated by size exclusion chromatography
| Protein/complex | Molecular weight (kDa) | Elution Volume (ml) | Stokes radius (Å) |
|---|---|---|---|
| mtSSB | 60.78 | 14.29 | 39.2 |
| NEIL1 | 44.75 | 13.73 | 42.2 |
| NEIL1Δ-100 | 34 | 15.79 | 31.2 |
| mtSSB-NEIL1 | 105 | 14.26 | 39.4 |
| mtSSB-NEIL1-DNA | 128 | 12.22 | 50.3 |
| mtSSB-DNA | 78 | 13.14 | 45.6 |
| NEIL1-DNA | 62.75 | 13.44 | 43.7 |
Figure 2.
Size exclusion chromatography (SEC) analysis of the NEIL1-mtSSB complexes in the presence and absence of a partial-duplex DNA substrate. (A) Complex of mtSSB and NEIL1 in the absence of DNA. Top panel, elution profiles from a pre-calibrated Superdex 200 column. Purple line, complex of NEIL1 and mtSSB (1:1 molar ratio); red line, mtSSB alone; and yellow line, NEIL1 alone. Corresponding fraction numbers are shown for clarity. Bottom panel, SDS-PAGE analysis of fractions collected after SEC. Lane 1, marker; lane 2, NEIL1-FL; lane 3, mtSSB; lane 4, NEIL1-mtSSB complex input prior to SEC; lane 5 – 12, fractions 26 – 33 obtained after SEC corresponding to elution volume 13 – 16 ml. 0.5 ml fractions were collected for all samples. (B) Sequence of partial duplex oligonucleotide used to study the NEIL1-DNA-mtSSB complex. The X represents tetrahydrofuran, an abasic site analog. (C) Complex of mtSSB and NEIL1 in the presence of partial DNA duplex. Top, SEC elution profile from the Superdex 200 column, green line, NEIL1-DNA-mtSSB (1:1:1) complex; red line, mtSSB alone; and yellow line, NEIL1 alone. Corresponding fraction numbers are indicated for clarity since 0.5 ml fractions were collected. Bottom, SDS-PAGE analysis of fractions collected after SEC. Lane 1, marker; lane 2, NEIL1-FL; lane 3, mtSSB; lane 4, NEIL1-DNA-mtSSB complex input prior to SEC analysis; lane 5 – 14, fractions 22 – 31 obtained after SEC corresponding to elution volume 11 – 15.5 ml. 0.5 ml fractions were collected for all samples.
3.3. Characterization of NEIl1:mtSSB complexes via SEC-MALS-SAXS and SEC-SAXS
Previously, SAXS analysis of NEIL1 provided evidence for the flexible conformation of the C-terminal region of the enzyme [32, 60]. In order to extend our knowledge of the structural assembly of the NEIL1-mtSSB and the NEIL1-DNA-mtSSB complexes, we employed in-line SEC-SAXS methods where SEC was performed on the samples prior to X-ray exposure. This method is advantageous for studying multi-component complexes over a concentration-series as it is more effective at removing aggregates that are frequently generated during sample concentration, reduces interference from inter-particle interactions, and allows for accurate buffer/background subtraction [61, 62]. SEC-SAXS and SEC-MALS-SAXS data were collected for the mtSSB-DNA (poly dT60 single-stranded/partial duplex DNA), NEIL1-mtSSB, and NEIL1-DNA-mtSSB (using the partial duplex DNA substrate) complexes along with the NEIL1 and mtSSB proteins individually. A distinct pattern was observed for each scatterer when a relative curve of the scattering intensity as function of momentum transfer was plotted for each individual protein or complex (Fig. 3A). The linearity of the Guinier region [63] indicated lack of aggregation for each sample (Supplemental Fig. 5).
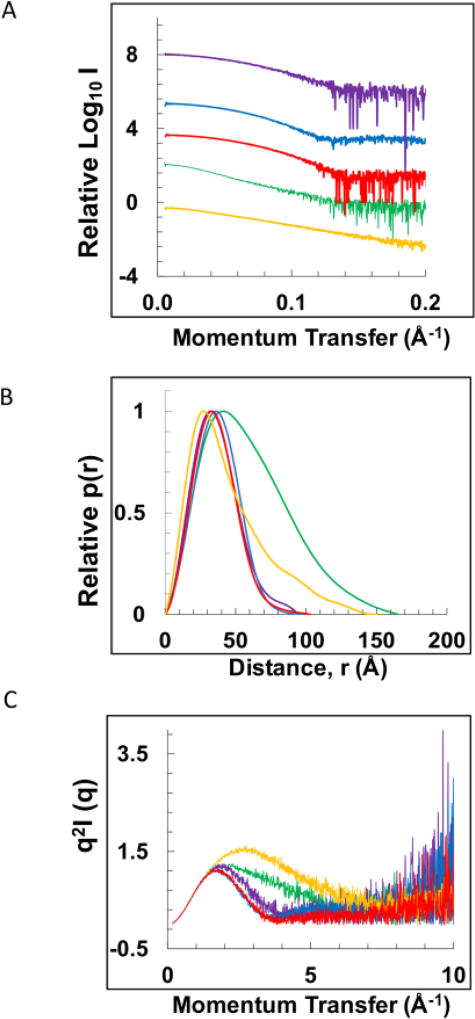
Figure 3.
Small angle X-ray scattering (SAXS) analysis of NEIL1, mtSSB, mtSSB-DNA, NEIL1-mtSSB, and NEIL1-DNA-mtSSB. (A) Scattering intensity curves for NEIL1 (yellow), mtSSB (red), mtSSB-DNA (blue), NEIL1-mtSSB (purple), and NEIL1-DNA-mtSSB (green). The same color scheme is used throughout the figure. For the mtSSB-DNA complex, a dT60 single-stranded DNA substrate was used and for the NEIL1-DNA-mtSSB complex, the partial duplex DNA substrate (Fig. 2B) was used in the SAXS analysis. (B) Normalized pairwise interatomic distance distribution function P(r) is shown for all proteins and complexes. (C) Normalized Kratky analysis representing the degree of disorder of all proteins and complexes.
The pairwise distance distribution function P(r) analysis provides a better understanding of the overall shape and size of the molecules/ complexes scrutinized here by SAXS (Fig. 3B). GNOM-based calculations suggested a maximum particle dimension (Dmax) for NEIL1 of 149.96 Å (Table 2). The shape of the curve with long tail at higher Dmax values (Fig. 3B, yellow) corresponds to an elongated shape of the molecule similar to that reported previously [32, 60]. The Dmax values of mtSSB and mtSSB-DNA are 103.21 Å, 98.74 Å, respectively. The bell-shaped curves for mtSSB and the mtSSB-DNA complex are indicative of their globular nature in solution (Fig. 3B, red, blue). The marginally larger particle size of unliganded mtSSB compared with the mtSSB-DNA complex indicates tighter packing of mtSSB when it forms a complex with DNA. The Dmax values for the NEIL1-mtSSB complex and the NEIL1-DNA-mtSSB complex are 94.32 Å and 165.72 Å, respectively (Fig. 3B, purple and green respectively). The curves at higher Dmax values indicate a partially elongated shape for the complexes. MW values were estimated from the volume of correlation and the Porod volume for NEIL1, mtSSB, mtSSB-DNA, NEIL1-mtSSB, and NEIL1-DNA-mtSSB (Table 2) [45, 46]. Furthermore, each protein and complex were subjected to SEC-MALS analysis to obtain the absolute molar masses for each species. For SEC-MALS measurement of the mtSSB-DNA complex, the partial duplex DNA substrate was used. The values obtained by MALS are within the experimental error of the theoretical masses for all the proteins and complexes and indicate the formation of a 1:1:1 NEIL1-DNA-mtSSB complex (123 kDa theoretical and 129.6 kDa from MALS, Table 2). Interestingly, in the absence of the partial duplex DNA substrate, the molar mass obtained by MALS (58 kDa, Table 2) for the NEIL1-mtSSB complex corresponds to NEIL1 bound to a monomer of mtSSB. This destabilization of a multimeric protein by NEIL1 was observed previously with the interaction between NEIL1 and PCNA [32].
As observed previously [32, 60], the Kratky plot of NEIL1 showed an initial parabolic curve with an upward trend at higher angles that is explained by the flexibility of the C-terminal region (Fig. 3C, yellow). The Kratky plots for mtSSB and the mtSSB-DNA complex displayed Gaussian curves characteristic of globular proteins confirming the compact nature of mtSSB in its apo and DNA bound states (Fig. 3C, red, blue). The Kratky curve of the NEIL1-mtSSB and the NEIL1-DNA-mtSSB complexes displayed characteristics of both mtSSB and NEIL1 with a relatively less elevated baseline at higher angles compared to NEIL1 alone (Fig. 3C, purple and green, respectively). Taken together, Kratky analysis of all proteins/complexes indicates that the disordered tail of NEIL1 adopts a more ordered conformation when present in a complex with mtSSB and DNA.
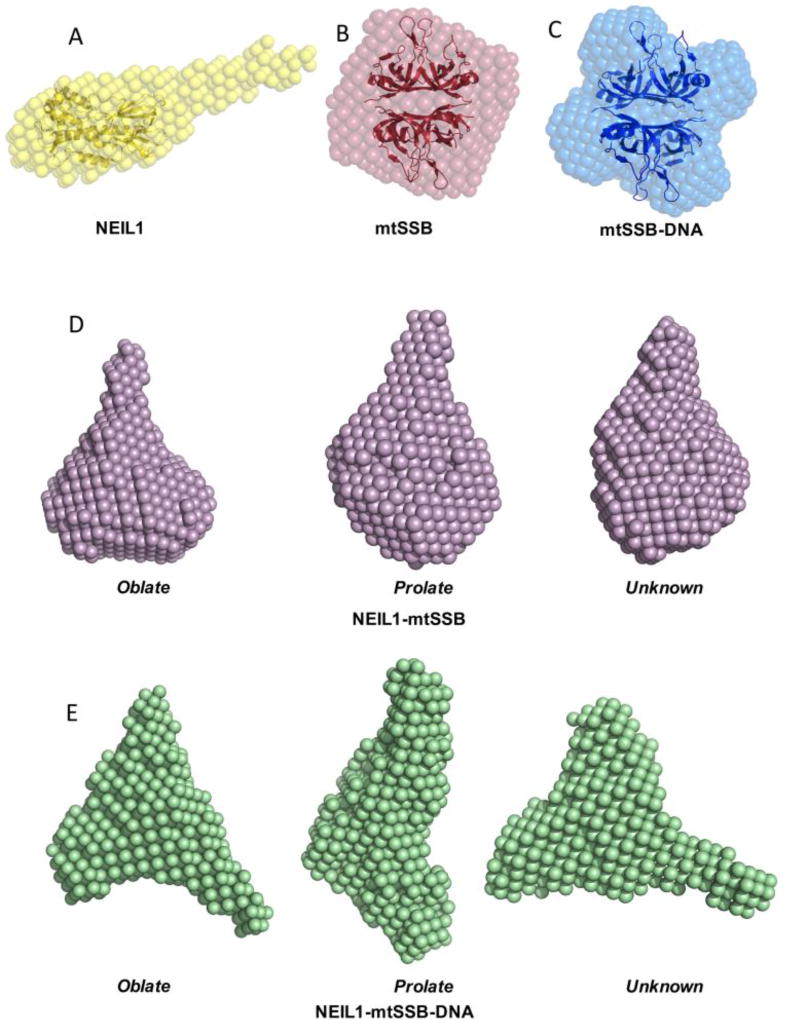
The program DAMMIF was used to generate ab initio molecular envelopes for all proteins and complexes scrutinized here. Shape reconstruction for NEIL1 produced an envelope with a globular region and a long tubular structure consistent with that reported previously [32, 60] (Fig. 4A). The envelope generated for mtSSB and mtSSB-DNA displayed a globular fold with a distinct tetrameric shape (Fig. 4B, C) very similar to the electron micrographs obtained for mtSSB previously [64]. The models of NEIL1 (PDB ID: 1TDH) [65] and the mtSSB tetramer (PDB ID: 3ULL) [66] were fitted into the SAXS envelopes using SUPCOMB (Fig. 4A–C). Ab initio modeling for the NEIL1-mtSSB complex and the NEIL1-DNA-mtSSB assembly produced an ensemble of three potential shapes when different particle anisometry parameters (oblate, prolate, and unknown) were used (Fig. 4D, E, Table 2). Caution is recommended while interpreting the larger envelope sizes that represent greater scattering produced by multiple components of the complex. The ab initio models together with the scattering data, MW assessments, and distance distribution plots confirm complex formation of NEIL1-mtSSB and NEIL1-DNA-mtSSB in solution.
Figure 4.
Ab initio shape reconstruction using DAMMIF. (A) NEIL1, (B) mtSSB, (C) mtSSB-DNA, (D) NEIL1-mtSSB, and (E) NEIL1-mtSSB-DNA complex. The envelopes obtained for NEIL1 and mtSSB agree well with the corresponding crystal structures (PDB IDs: 1TDH [65] and 3ULL [66], respectively).
4. Discussion and Concluding Remarks
The discovery of the three NEIL enzymes over a decade ago led to a deeper understanding of glycosylase biology in human cells [25, 67, 68]. As DNA glycosylases, the NEIL enzymes have overlapping functions in that they can excise oxidized pyrimidine lesions such as thymine glycol, 5-hydroxyuracil, dihyrouracil, as well as spiroiminodihydantoin and guanidinohydantoin, the further oxidation products of 7,8-dihydro-8-oxoguanine [69–71]. The NEIL enzymes vary in their ability to excise lesions from different DNA substrates [70]. For instance, NEIL2 has a strong preference for lesions present in single-stranded and bubble DNA whereas the NEIL1 enzyme can efficiently excise lesions from double-stranded DNA substrates [70, 72]. In addition, NEIL3 is able to excise lesions from G-quadruplex DNA structures lending diversity to the requirement of the NEIL enzymes in maintaining genomic integrity [73, 74]. Owing to the substrate overlap, it was proposed that the NEIL enzymes can serve as back up enzymes for each other, however, emerging data suggests that the NEIL enzymes may have distinct functions beyond excision of oxidized bases [28, 30].
In the current manuscript, we evaluated the in vitro interaction between NEIL1 and mtSSB, a highly stable homotetramer that is essential for mtDNA replication [66, 75]. The transient nature of the protein-protein interactions involved in DNA replication and repair makes it challenging to evaluate the three-dimensional crystal structures of the full-length native complexes (i.e. without chemical crosslinking or other protein modifications). However, structural features such as overall shape and conformation can be probed using solution scattering techniques such as SAXS [76, 77]. Here, we isolated NEIL1-mtSSB complexes in the presence and absence of a partial-duplex DNA substrate using size exclusion chromatography. This substrate provides a binding platform for both proteins and comprises a 13-mer duplex DNA with an abasic site analog to facilitate NEIL1 binding and a poly dT35 overhang that is suitable for the binding of mtSSB [57]. We chose this substrate because mtSSB has a much greater affinity for single-stranded (ss) DNA over duplex DNA and inhibits the activity of NEIL1 on ssDNA substrates (Supplemental Fig. 6). SEC-SAXS and MALS data reveal that the two proteins form a complex in the presence of DNA and the MW information estimated from the data acquired indicates formation of a 1:1:1 NEIL1-DNA-mtSSB complex. Previously, we studied the complex formed between NEIL1 and PCNA using techniques similar to those employed here. Our data indicated that the homotrimeric ring of PCNA is disrupted upon interaction with NEIL1 [32]. Based on our previous work and the data presented in the current manuscript we surmise that the flexible C-terminal tail of NEIL1 is capable of forming complexes with its binding partners in distinct ways. Both PCNA and mtSSB are multimeric proteins and their interactions with NEIL1 appear to be dependent upon their quaternary architecture. In vitro, mtSSB forms a stable homotetramer where the individual subunits begin to dissociate only at elevated temperatures (ranging from ~60 °C – 100 °C), whereas human PCNA forms a donut-shaped homotrimeric ring with a central cavity that is stable up to ~50 °C [78–80]. In the presence of NEIL1, both proteins appear to be monomeric. This destabilization of multimeric proteins involved with DNA replication suggests that NEIL1 may serve as a sentinel ahead of the replication fork and prevents replication from proceeding when a lesion is encountered.
Thus far, of the three NEIL enzymes, NEIL1 and NEIL2 have been implicated in mitochondrial genome maintenance [26, 27]. Yet, several questions regarding their role within the mitochondria remain to be answered. In the nucleus, some evidence exists for the cell cycle regulation of NEIL1 where the levels of the enzyme are upregulated during S-phase, while the levels of NEIL2 remain unchanged [81, 82]. Given that mitochondrial replication is not regulated in a manner similar to the cell cycle, it is still not clear whether the two enzymes have overlapping or distinct functions within this organelle.
Supplementary Material
Highlights.
NEIL1 glycosylase interacts with mitochondrial single-stranded DNA binding protein.
Protein painting reveals that the disordered tail of NEIL1 binds to mtSSB.
The minimum binding domain of NEIL1 with mtSSB lies within residues 289–312.
SEC-MALS-SAXS data revealed NEIL1-mtSSB complex formation in absence and presence of DNA.
Acknowledgments
We would like to thank Dr. Sylvie Doublié and Dr. Brian Eckenroth for advice as this project was initiated while AP was a post-doctoral fellow at the University of Vermont. We would like to thank Dr. Mohammad Saki for assistance with tissue culture experiments, and Dr. Lewis Pannell and Lindsay Schambeau for conducting the mass-spectrometry analyses (MCI, core facility). NS and AP were supported by a grant from the National Institutes of Health (NIEHS grant 5R00ES024417-04 to AP). Funds were also provided by the University of South Alabama Cancer Center Research Fund to NS and AP. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This project was supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health. ML and WC were supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ES 065078). The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None
References
- 1.Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature reviews. Molecular cell biology. 2014;15:411. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiological reviews. 2014;94:329–354. doi: 10.1152/physrev.00040.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harbor perspectives in biology. 2013;5:a012559. doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, Zhou T, Ziegler AC, Dimitrion P, Zuo L. Oxidative Stress in Neurodegenerative Diseases: From Molecular Mechanisms to Clinical Applications. Oxidative Medicine and Cellular Longevity. 2017;2017 doi: 10.1155/2017/2525967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasad S, Gupta SC, Tyagi AK. Reactive oxygen species (ROS) and cancer: Role of antioxidative nutraceuticals. Cancer letters. 2017;387:95–105. doi: 10.1016/j.canlet.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 6.Fang EF, Scheibye-Knudsen M, Chua KF, Mattson MP, Croteau DL, Bohr VA. Nuclear DNA damage signalling to mitochondria in ageing. Nature reviews. Molecular cell biology. 2016;17:308. doi: 10.1038/nrm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cline SD. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2012;1819:979–991. doi: 10.1016/j.bbagrm.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasiak J, Glowacki S, Kauppinen A, Kaarniranta K. Mitochondrial and nuclear DNA damage and repair in age-related macular degeneration. International journal of molecular sciences. 2013;14:2996–3010. doi: 10.3390/ijms14022996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson S, Bankier AT, Barrell BG, De Bruijn M, Coulson AR, Drouin J, Eperon I, Nierlich D, Roe BA, Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nature reviews. Molecular cell biology. 2010;11:655. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 11.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2013;1833:274–285. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Gilkerson R, Bravo L, Garcia I, Gaytan N, Herrera A, Maldonado A, Quintanilla B. The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harbor perspectives in biology. 2013;5:a011080. doi: 10.1101/cshperspect.a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kukat C, Larsson N-G. mtDNA makes a U-turn for the mitochondrial nucleoid. Trends in cell biology. 2013;23:457–463. doi: 10.1016/j.tcb.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Kolesnikov A. The mitochondrial genome. The nucleoid. Biochemistry (Moscow) 2016;81:1057–1065. doi: 10.1134/S0006297916100047. [DOI] [PubMed] [Google Scholar]
- 15.Young MJ, Copeland WC. Human mitochondrial DNA replication machinery and disease. Current opinion in genetics & development. 2016;38:52–62. doi: 10.1016/j.gde.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SR, Han J. Mitochondrial Nucleoid: Shield and Switch of the Mitochondrial Genome. Oxidative Medicine and Cellular Longevity. 2017;2017 doi: 10.1155/2017/8060949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harbor perspectives in biology. 2013;5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saki M, Prakash A. DNA damage related crosstalk between the nucleus and mitochondria. Free Radical Biology and Medicine. 2017;107:216–227. doi: 10.1016/j.freeradbiomed.2016.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prakash A, Doublié S. Base excision repair in the mitochondria. Journal of cellular biochemistry. 2015;116:1490–1499. doi: 10.1002/jcb.25103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fromme JC, Verdine GL. Base excision repair. Advances in protein chemistry. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 21.Krokan HE, Bjørås M. Base excision repair. Cold Spring Harbor perspectives in biology. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. Journal of Biological Chemistry. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer letters. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David SS, O’Shea VL, Kundu S. Base excision repair of oxidative DNA damage. Nature. 2007;447:941. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace SS. Base excision repair: a critical player in many games. DNA repair. 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3′-phosphatase in maintenance of mitochondrial genome. Journal of Biological Chemistry. 2012;287:2819–2829. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolseth V, Luna L, Olsen AK, Suganthan R, Scheffler K, Neurauter CG, Esbensen Y, Kuśnierczyk A, Hildrestrand GA, Graupner A. No cancer predisposition or increased spontaneous mutation frequencies in NEIL DNA glycosylases-deficient mice. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-04472-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegde ML, Hazra TK, Mitra S. Functions of disordered regions in mammalian early base excision repair proteins. Cellular and molecular life sciences. 2010;67:3573–3587. doi: 10.1007/s00018-010-0485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangaswamy S, Pandey A, Mitra S, Hegde ML. Pre-Replicative Repair of Oxidized Bases Maintains Fidelity in Mammalian Genomes: The Cowcatcher Role of NEIL1 DNA Glycosylase. Genes. 2017;8:175. doi: 10.3390/genes8070175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theriot CA, Hegde ML, Hazra TK, Mitra S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA repair. 2010;9:643–652. doi: 10.1016/j.dnarep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prakash A, Moharana K, Wallace SS, Doublié S. Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase. Nucleic acids research. 2017;45:2897–2909. doi: 10.1093/nar/gkw1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. Journal of Biological Chemistry. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the Human DNA Glycosylase NEIL1 with Proliferating Cell Nuclear Antigen THE POTENTIAL FOR REPLICATION-ASSOCIATED REPAIR OF OXIDIZED BASES IN MAMMALIAN GENOMES. Journal of Biological Chemistry. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 35.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE has 5′→ 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. Journal of Biological Chemistry. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 36.Genuario R, Wong TW. Stimulation of DNA polymerase gamma by a mitochondrial single-strand DNA binding protein. Cellular & molecular biology research. 1992;39:625–634. [PubMed] [Google Scholar]
- 37.Van Loon B, Samson LD. Alkyladenine DNA glycosylase (AAG) localizes to mitochondria and interacts with mitochondrial single-stranded binding protein (mtSSB) DNA repair. 2013;12:177–187. doi: 10.1016/j.dnarep.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steen KW, Doseth B, Westbye MP, Akbari M, Kang D, Falkenberg M, Slupphaug G. mtSSB may sequester UNG1 at mitochondrial ssDNA and delay uracil processing until the dsDNA conformation is restored. DNA repair. 2012;11:82–91. doi: 10.1016/j.dnarep.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 39.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. Journal of Biological Chemistry. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 40.Longley MJ, Smith LA, Copeland WC. Preparation of human mitochondrial single-stranded DNA-binding protein. Mitochondrial DNA: Methods and Protocols. 2009:73–85. doi: 10.1007/978-1-59745-521-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franke D, Petoukhov M, Konarev P, Panjkovich A, Tuukkanen A, Mertens H, Kikhney A, Hajizadeh N, Franklin J, Jeffries C. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. Journal of Applied Crystallography. 2017;50 doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konarev PV, Volkov VV, Sokolova AV, Koch MH, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of applied crystallography. 2003;36:1277–1282. [Google Scholar]
- 43.Förster S, Apostol L, Bras W. Scatter: software for the analysis of nano-and mesoscale small-angle scattering. Journal of Applied Crystallography. 2010;43:639–646. [Google Scholar]
- 44.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. Journal of applied crystallography. 1992;25:495–503. [Google Scholar]
- 45.Porod G. General theory. In: Glatter O, Kratky O, editors. Small angle X-ray scattering. Academic Press; London: 1982. pp. 17–51. [Google Scholar]
- 46.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hopkins JB, Gillilan RE, Skou S. BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. Journal of Applied Crystallography. 2017;50 doi: 10.1107/S1600576717011438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of applied crystallography. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of applied crystallography. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozin MB, Svergun DI. Automated matching of high-and low-resolution structural models. Journal of applied crystallography. 2001;34:33–41. [Google Scholar]
- 51.Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of Formamidopyrimidines in DNA Involves Different Glycosylases ROLE OF THE OGG1, NTH1, AND NEIL1 ENZYMES. Journal of Biological Chemistry. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 52.Luchini A, Espina V, Liotta LA. Protein painting reveals solvent-excluded drug targets hidden within native protein–protein interfaces. Nature communications. 2014;5 doi: 10.1038/ncomms5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hegde ML, Hegde PM, Arijit D, Boldogh I, Mitra S. Human DNA glycosylase NEIL1’s interactions with downstream repair proteins is critical for efficient repair of oxidized DNA base damage and enhanced cell survival. Biomolecules. 2012;2:564–578. doi: 10.3390/biom2040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.La Verde V, Dominici P, Astegno A. Determination of Hydrodynamic Radius of Proteins by Size Exclusion Chromatography. Protein Science. 2016 doi: 10.21769/BioProtoc.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong P, Koza S, Bouvier ES. A review size-exclusion chromatography for the analysis of protein biotherapeutics and their aggregates. Journal of liquid chromatography & related technologies. 2012;35:2923–2950. doi: 10.1080/10826076.2012.743724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biological procedures online. 2009;11:32. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian Y, Johnson KA. The human mitochondrial single strand DNA binding protein displays distinct kinetics and thermodynamics of DNA binding and exchange. Journal of Biological Chemistry. 2017 doi: 10.1074/jbc.M117.791392. jbc. M117. 791392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morin JA, Cerrón F, Jarillo J, Beltran-Heredia E, Ciesielski GL, Arias-Gonzalez JR, Kaguni LS, Cao FJ, Ibarra B. DNA synthesis determines the binding mode of the human mitochondrial single-stranded DNA-binding protein. Nucleic Acids Research. 2017 doi: 10.1093/nar/gkx395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. Journal of Biological Chemistry. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 60.Hegde ML, Tsutakawa SE, Hegde PM, Holthauzen LMF, Li J, Oezguen N, Hilser VJ, Tainer JA, Mitra S. The disordered C-terminal domain of human DNA glycosylase NEIL1 contributes to its stability via intramolecular interactions. Journal of molecular biology. 2013;425:2359–2371. doi: 10.1016/j.jmb.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeffries CM, Graewert MA, Blanchet CE, Langley DB, Whitten AE, Svergun DI. Preparing monodisperse macromolecular samples for successful biological small-angle X-ray and neutron-scattering experiments. Nature protocols. 2016;11:2122–2153. doi: 10.1038/nprot.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathew E, Mirza A, Menhart N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. Journal of synchrotron radiation. 2004;11:314–318. doi: 10.1107/S0909049504014086. [DOI] [PubMed] [Google Scholar]
- 63.Guinier A. La diffraction des rayons X aux tres petits angles: applications a l'etude de phenomenes ultramicroscopiques. 1939 [Google Scholar]
- 64.Curth U, Urbanke C, Greipel J, Gerberding H, Tiranti V, Zeviani M. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. The FEBS Journal. 1994;221:435–443. doi: 10.1111/j.1432-1033.1994.tb18756.x. [DOI] [PubMed] [Google Scholar]
- 65.Doublié S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang C, Curth U, Urbanke C, Kang C. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 Å resolution. Nature Structural & Molecular Biology. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 67.Hazra TK, Izumi T, Kow YW, Mitra S. The discovery of a new family of mammalian enzymes for repair of oxidatively damaged DNA, and its physiological implications. Carcinogenesis. 2003;24:155–157. doi: 10.1093/carcin/24.2.155. [DOI] [PubMed] [Google Scholar]
- 68.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA repair. 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 69.Prakash A, Doublié S, Wallace SS. The Fpg/Nei family of DNA glycosylases: substrates, structures and search for damage. Progress in molecular biology and translational science. 2012;110:71. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. Journal of Biological Chemistry. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 73.Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. Journal of Biological Chemistry. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic acids research. 2015;43:4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruhanen H, Borrie S, Szabadkai G, Tyynismaa H, Jones AW, Kang D, Taanman J-W, Yasukawa T. Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organisation. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2010;1803:931–939. doi: 10.1016/j.bbamcr.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 76.Kikhney AG, Svergun DI. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS letters. 2015;589:2570–2577. doi: 10.1016/j.febslet.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 77.Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH. 2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update. Acta Crystallographica Section D: Structural Biology. 2017;73 doi: 10.1107/S2059798317011597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li K, Williams RS. Tetramerization and single-stranded DNA binding properties of native and mutated forms of murine mitochondrial single-stranded DNA-binding proteins. Journal of Biological Chemistry. 1997;272:8686–8694. doi: 10.1074/jbc.272.13.8686. [DOI] [PubMed] [Google Scholar]
- 79.Schurtenberger P, Egelhaaf SU, Hindges R, Maga G, Jónsson ZaO, May RP, Glatter O, HuÈbscher U. The solution structure of functionally active human proliferating cell nuclear antigen determined by small-angle neutron scattering. Journal of molecular biology. 1998;275:123–132. doi: 10.1006/jmbi.1997.1435. [DOI] [PubMed] [Google Scholar]
- 80.De Biasio A, Sánchez R, Prieto J, Villate M, Campos-Olivas R, Blanco FJ. Reduced stability and increased dynamics in the human proliferating cell nuclear antigen (PCNA) relative to the yeast homolog. PloS one. 2011;6:e16600. doi: 10.1371/journal.pone.0016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proceedings of the National Academy of Sciences. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. Journal of Biological Chemistry. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 83.Gasteiger E, Hoogland C, Gattiker A, Duvaud Se, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. Springer; 2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.