Abstract
How DNA lesions in nucleosomes are recognized for global genome nucleotide excision repair (GG-NER) remains poorly understood, and the roles that histone tails may play remains to be established. Histone H3 and H4 N-terminal tails are of particular interest as their acetylation states are important in regulating nucleosomal functions in transcription, replication and repair. In particular the H3 tail has been the focus of recent attention as a site for the interaction with XPC, the GG-NER lesion recognition factor. Here we have investigated how the structure and dynamics of the DNA lesion cis-B[a]P-dG, derived from the environmental carcinogen benzo[a]pyrene (B[a]P), is impacted by the presence of flanking H3 and H4 tails. This lesion is well-repaired by GG-NER, and adopts a base-displaced/intercalated conformation in which the lesion partner C is displaced into the major groove. We used molecular dynamics simulations to obtain structural and dynamic characterizations for this lesion positioned in nucleosomal DNA so that it is bracketed by the H3 and H4 tails. The H4 tail was studied in unacetylated and acetylated states, while the H3 tail was unacetylated, its state when it binds XPC (Kakumu, Nakanishi et al. 2017). Our results reveal that upon acetylation, the H4 tail is released from the DNA surface; the H3 tail then forms a pocket that induces flipping and capture of the displaced lesion partner base C. This reveals synergistic effects of the behavior of the two tails. We hypothesize that the dual capability of the H3 tail to sense the displaced lesion partner base and to bind XPC could foster recognition of this lesion by XPC for initiation of GG-NER in nucleosomes.
Keywords: nucleosome core particle, histone tails, lysine acetylation, DNA adduct, MD simulations, nucleotide excision repair
Graphical abstract
Introduction
In nucleosomes [1, 2], there is considerable interest in the functions of the histone N-terminal tails [3]. The tails are important sites for posttranslational modifications (PTMs) [4–8]. Among the PTMs, lysine acetylation, which neutralizes the lysine positive charge, has been identified to play a critical role for opening chromatin structure for transcription, replication and repair [9–11]. It has been established for many years that hyperacetylation in nucleosomes is associated with DNA damage [12–14] to create access to DNA lesions for repair [15–18]. DNA becomes more accessible in nucleosomes upon acetylation [11], as well as in nucleosomal arrays when the H4 tail is neutralized with acetylation mimics [19]. The functions of the H3 and H4 tails (Figure 1, A and B) are under intensive investigation. The H4 tail is implicated in chromatin condensation through its interaction with the negative acidic patch of an adjacent nucleosome, because it contains a highly positively charged basic region [3, 20–22]. The H3 tail is involved in gaining access to DNA lesions for repair [23, 24]. Recently, it was found that chromatin assembly that is replication-dependent requires an H3/H4 complex containing the relevant N-terminal tail domains in vivo [25]. However, whether and how histone tails cooperate to carry out their functions is poorly understood. Nonetheless, there are indications that the H3 and H4 tails act synergistically to provide access to local DNA upon lysine acetylation [19, 26].
Figure 1.
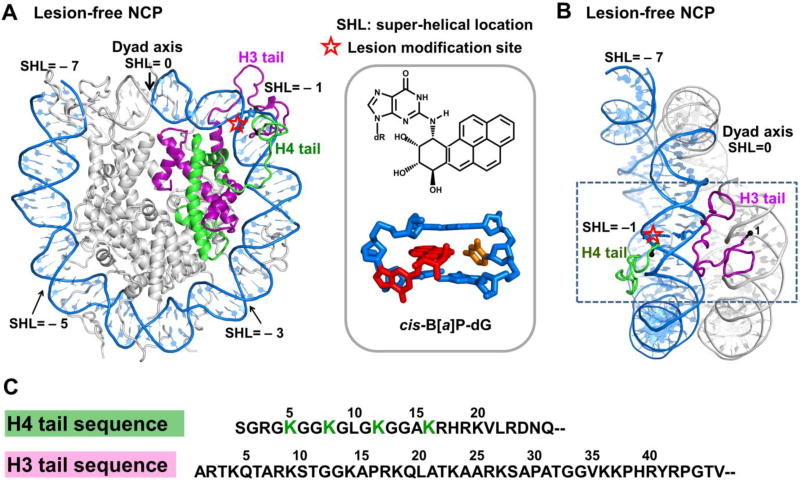
(A) Top view of the nucleosome core particle structure (NCP) in the investigated lesion-free model, indicating the positioning of histones H3 (purple) and H4 (green) and their tails on the DNA surface. The other histones are grey. The position of the dyad axis and super-helical locations (SHLs) are indicated. The model is based on the crystal structure with PDB [40] ID 1KX5 [45]. The H3 and H4 tails investigated are from Chain A and Chain B, respectively, in the crystal structure. The red star indicates the modification site for the lesion-containing cases. The chemical and NMR solution structures of the cis-B[a]P-dG adduct [36] are shown in the inset box. (B) Side view of the same NCP structure as in (A), with the core histones not displayed except for the H3 and H4 tails investigated here. The two gyres of the DNA are shown: the one from SHL = 0 to SHL = ~ − 7 is blue, and the other, from SHL = 0 to SHL = ~ 7, is grey. For clarity, the gyre with SHL = 1 to SHL = ~ 7 is not displayed in (A). In the window which is the region of interest, the DNA is flanked by the H4 tail on one side, and the H3 tail on the other. (C) The amino acid sequences of the H4 and H3 tails are given. The acetylation sites lysines 5, 8, 12, 16 on the H4 tail are designated in green.
An interesting recent study has shown that the H3 tail, when unacetylated, binds to XPC [27], the global genome nucleotide excision repair (GG-NER) lesion recognition protein [28]. GG-NER is responsible for the ultimate excision of bulky DNA lesions such as those derived from the ubiquitous environmental pre-carcinogen benzo[a]pyrene (B[a]P) [29, 30]. Metabolic activation of B[a]P through the well-studied diol-epoxide pathway [31] leads to a variety of DNA adducts [32], including the 10R (+)-cis-anti-B[a]P-N2-dG adduct (cis-B[a]P-dG) (Figure 1, box) that is well-repaired by GG-NER in both HeLa cell extracts [33, 34] and with purified proteins [35], and it is repaired in nucleosomes (Geacintov N.E., manuscript in preparation). The cis-B[a]P-dG adduct adopts a base-displaced/intercalated conformation (Figure 1, box); in this conformation, the modified base G is displaced from the double helix and resides in the minor groove, the B[a]P aromatic rings are inserted into the helix with the hydroxyl-containing benzylic ring in the minor groove, and the partner C to the damaged G is displaced into the major groove [36]. A recent computational study determined the binding free energy pathway of Rad4, the yeast orthologue of human XPC, with cis-B[a]P-dG, using umbrella sampling with restrained molecular dynamics (MD) simulations and free energy calculations [37]. A key finding was the initial capture of the extruded partner C by the Rad4 protein. Since lesion partner base flipping is required for productive lesion binding by Rad4 [38], the capture of the pre-flipped partner C should play a critical role in the initial recognition of this lesion for subsequent successful NER.
Our prior MD simulations investigated the cis-B[a]P-dG adduct placed at the dyad axis in a nucleosome core particle (NCP); this study was based on a nucleosome crystal structure [39] with PDB [40] ID: 2NZD, in which histone N-terminal tails were truncated [41]. The results showed that this base-displaced/intercalated adduct weakens local histone-DNA interactions and causes severe local DNA distortions [41]. Additionally, we have studied a stereoisomerically different B[a]P-derived DNA adduct, the 10S (+)-trans-B[a]P-N2-dG adduct which resides in the B-DNA minor groove [42]; it was placed at super-helical location (SHL) ~ 3 in the nucleosome environment [43, 44]. In this case, a single nearby histone H2B tail was stably engulfed by the B[a]P ring system [43], and upon lysine acetylation of the tail, tail-B[a]P interactions were destabilized [44]. To further investigate the interactions of histone tails with DNA lesions, here we investigated the well-repaired cis-B[a]P-dG DNA adduct [33–35] bracketed by the H3 and H4 tails in a nucleosome with all tails present (PDB [40] ID: 1KX5 [45]), and considered the H4 tail in unacetylated and acetylated states. We wished to determine whether the cis-B[a]P-dG lesion that is well-repaired by NER, when flanked by the H3 and H4 tails, would induce local structural and dynamic changes that might be sensed by XPC; furthermore, we wished to establish whether such changes would be modulated by the DNA accessibility provided by H4 tail acetylation.
Methods
We have carried out 3 µs MD simulations for a system that has the cis-B[a]P-dG DNA adduct (Figure 1, box) embedded in a tail-containing nucleosome core particle (PDB [40] ID: 1KX5 [45]). We have placed this lesion at SHL= − 1, where it is flanked by the H3 and the H4 tails (Figure 1, A and B). The amino acid sequences of the two tails are given in Figure 1C. We investigated the H3 tail in an unacetylated state, as required for loading XPC [27]; the H4 tail was studied in unacetylated and acetylated states at lysines 5, 8, 12, and 16, which are observed to be acetylated in vivo [46–51]. We modeled the cis-B[a]P-dG lesion with its displaced partner C (Figure 1, box) into the nucleosome based on an MD [34] equilibrated NMR solution structure [36]. Thus, we have here prepared two models: lesion-containing NCP with unacetylated H4 tail and with acetylated H4 tail. An unmodified control with unacetylated H4 tail (lesion-free NCP) was also investigated. Additional simulations were performed to further explore transitions observed in the 3 µs simulation and to investigate the role of the H3 tail, that are described below.
The AMBER14 simulation package [52] with force field ff14SB [53] and previously published additional parameters for the cis-B[a]P-dG lesion [35] was utilized for carrying out MD simulations. Table S1 gives box sizes, number of waters, and length of MD simulations. The CPPTRAJ module [54] of the AMBER14 package [52] was used for structural analyses. The best representative structures were obtained using the cluster analysis in the CPPTRAJ module with the average linkage hierarchical agglomerative method [55]. VMD [56] was used for molecular modeling and trajectory viewing. PyMOL (The PyMOL Molecular Graphic System, version 1.3×, Schrödinger, LLC) was used for structural viewing, images and movies. Full details of the force field, parameters, modeling, MD protocols, further analyses, and ensemble sampling are given in Supplementary Data.
Results
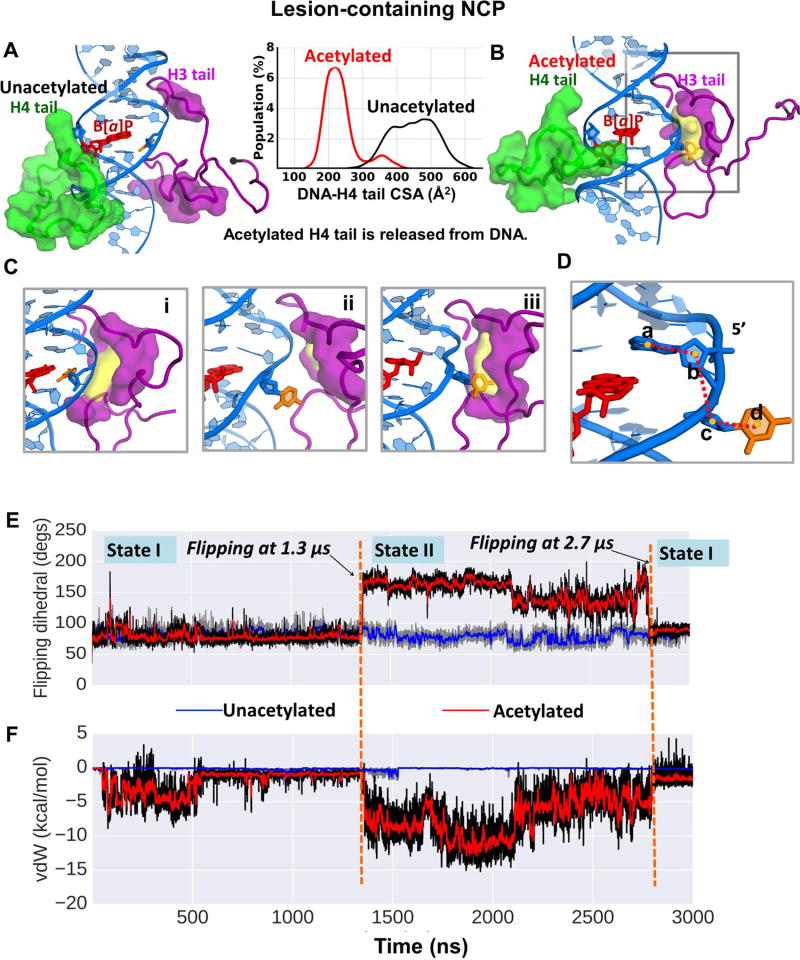
Our study reveals that interactions of the two tails with the lesion-containing DNA depend on the acetylation state of the H4 tail. Best representative structures from our MD simulations are shown in Figure 2 for the cis-B[a]P-dG lesion-containing NCPs, containing unacetylated (Figure 2A) and acetylated (Figure 2B) H4 tails. We observe that when the H4 tail is unacetylated (Figure 2A, and Movie S1), it condenses on the DNA minor and major grooves and interacts with the DNA and cis-B[a]P-dG lesion. The contact surface area (CSA) shows the interaction between the unacetylated H4 tail and the DNA grooves (Figure 2, between A and B). Hydrogen bonding interactions between the H4 tail and the lesion-containing DNA are given in Table S2, and van der Waals interactions are given in Figure S1, Supplementary Data. The bulky B[a]P aromatic ring system is intercalated, with the hydroxyl groups in the minor groove and the displaced partner C is on the major groove side, similar to the NMR solution structure [36]. The cis-B[a]P-dG lesion in its base-displaced/intercalated conformation (Figure 1, box) opens the minor groove both in free DNA [35, 36] and in the nucleosome (Figure S2, Supplementary Data).
Figure 2.
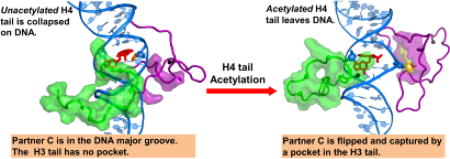
(A) When the H4 tail (green surface) is unacetylated, it is collapsed on the DNA minor and major grooves, and interacts with the cis-B[a]P-dG lesion. (B) When acetylated, the H4 tail is released from the DNA surface, as reflected in the inserted histogram which gives the contact surface area (CSA) between the H4 tail and the DNA surface. Also in (B), the partner base C to the cis-B[a]P-dG lesion is flipped out and can be captured by the nearby H3 tail, which is rearranged to form a pocket (yellow surface) for the C base. The pocket and its volume were computed using the POCASA web server [64]. The pocket is surrounded by amino acid residues Pro16, Arg17, Lys18, Ser28, Ala29, Pro30, Ala31, Pro38, His39, and Tyr41 (purple surface). The amino acids comprising the pocket were computed with the CASTp web server [65–67]. These residues are rendered in purple surface in both (A) and (B) to highlight the rearrangement of the H3 tail upon H4 tail acetylation. (A) and (B) show the same region of interest as in Figure 1B. The N-terminus of each tail is shown as a black sphere. The color code is the same as in Figure 1. See also Movies S1 and S2. (C) Transition of the C opposite the cis-B[a]P-dG lesion into the varying-sized pocket of the H3 tail when the H4 tail is acetylated. Only the window highlighted in the grey box in (B) is shown. (i) A snapshot at ~1.2 µs, where the partner C is on the major groove side and the pocket has formed. (ii) an intermediate snapshot between 1.2 µs and 1.3 µs, in which the partner C is flipped out but is not yet captured by the pocket. (iii) A snapshot at ~1.3 µs, in which the partner C has been captured by the pocket in the H3 tail. The dihedral angles (defined in (D)) for (i), (ii), and (iii) are respectively, 76 °, 140 °, and 157 °. The pocket volumes are respectively, 61, 39, and 59 Å2. See also Movie S3. (D) The flipping dihedral angle a-b-c-d is defined as the center of mass of 4 groups, a, b, c, and d; a is the base A that is the 5’- neighbor to the partner C; b is the sugar of the 5’- neighbor base A to the partner C; c is the sugar of the partner C; and d is the partner base C. (E) Time dependence of the flipping dihedral angle of the partner base C for the lesion-containing NCP with the H4 tail in unacetylated state (blue line) and in acetylated state (red line). (F) Time dependence of van der Waals interactions between the partner C base and the H3 tail residues that are components of the pocket that can capture the partner base C when the H4 tail is acetylated.
By contrast, when the H4 tail is acetylated, it is released from the DNA (Figure 2B, and Movie S2); this results from the reduction in positive charges on the tail, which weakens the interactions between the tail and the negatively charged DNA backbone. Hydrogen bonding is also reduced (Table S2, Supplementary Data). The acetylation of the H4 tail exposes the local DNA and provides greater accessibility to the vicinity of the lesion-containing DNA. The CSA between the H4 tail and the DNA is significantly lower in the acetylated case (Figure 2, between A and B).
In the presence of the lesion and upon the H4 tail acetylation, the H3 tail forms a pocket that is flexible due to rotation of the sidechains; this pocket captures the flipping, extruding partner C. Prior to the flipping, the partner C remains in the major groove (State I). The flipping and capture occur at ~ 1.3 µs in the MD simulation. The captured state (State II) persists to ~ 2.7 µs; then the C flips back to its original position in the major groove (State I) and remains there till the end of the ~ 3.0 µs MD simulation). Figure 2C gives three snapshots along the flipping transition with their varying flipping dihedral angles (defined in Figure 2D). Figure 2E shows time dependence of the flipping dihedral angle. Movie S3 shows the process of flipping the C into the pocket in the H3 tail. The pocket that captures the base C is composed of H3 tail residues Pro16, Arg17, Lys18, Ser28, Ala29, Pro30, Ala31, Pro38, His39, and Tyr41 (Figures 1C and 2B). Figure 2F shows time dependence of the van der Waals interaction energies between the partner C and the residues that surround the pocket, which reveals that the interaction is minimal until the C is captured. Furthermore, Pro30 plays an important role in interacting with the flipped out partner C base, anchoring it near the bottom of the pocket (Figure S3, Supplementary Data). The importance of CH/π interactions between aromatic rings and proline has been discussed by Zondlo et al. [57].
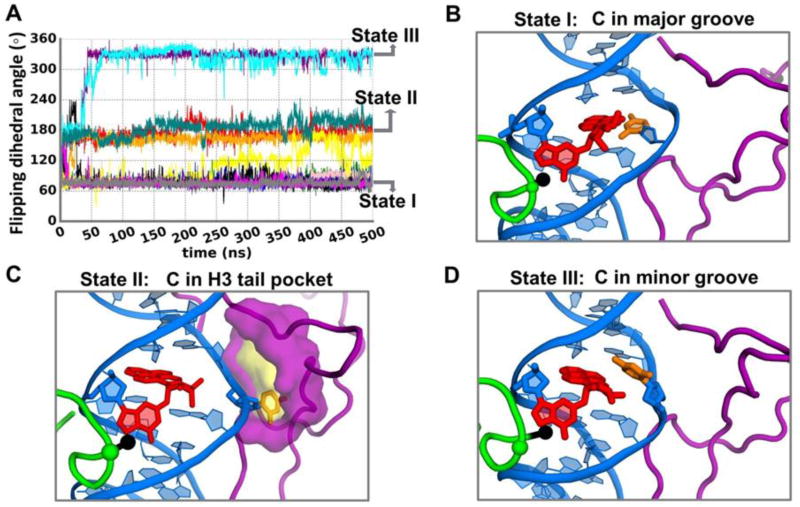
In order to further explore the transitions and states observed in the ~ 3 µs simulation, we carried out additional 12 independent 500 ns MD simulations. Each simulation started from the same structure (Figure S4) just prior to the flipping and capture at ~ 1.3 µs and had a flipping dihedral angle of ~ 90° (Figure 2E). In this structure, the partner C is in the major groove and a pocket had already formed in the H3 tail (Details are given in Supplementary Data). Figure 3A shows the time dependence of the flipping dihedral angles in these 12 simulations. In 5 of them there was no transition, and the partner base C stayed close to the initial state (State I, with partner C in the major groove, Figure 3B). In addition, one simulation made a brief transition to State II in the beginning of the simulation and then returned to State I for the remainder of the simulation. In 3 simulations, the transition from State I to II (in which partner C is captured) occurred within the first 50 ns and persisted throughout the simulation (Figure 3C). In one simulation, the partner C episodically transited between State I and State II, especially in the second half of the simulation. In two of the simulations we observed a transition from State I to II, and then rapidly (all within the first 65 ns) from State II to III, in which the partner C resides in the minor groove (Figure 3D); in this case, the transient capture of the C by the H3 tail facilitates the shift of the C from the major to the minor groove, which does not happen directly and was not observed in the 3 µs long MD simulation.
Figure 3.
Conformational transitions of the partner C in the presence of the H3 tail when H4 tail is acetylated. (A) Time-dependence of flipping dihedral angle (defined in Figure 2D) during the 12 additional MD simulations of 500 ns, showing the transitions between the distinct states. (B), (C) and (D) are the best representative structures of these states. Each color in Figure 3A provides the time dependence of the flipping dihedral angle from one MD run of the 12 simulations.
To further substantiate the role played by the H3 tail in the flipping of the displaced C, we also carried out 10 additional 200 ns MD simulations that had the full length H3 tail truncated when the H4 tail is acetylated (Details given in Supplementary Data). The results revealed that, without the H3 tail, the partner C remains stably on the major groove side. Thus, these studies show that the partner C explores more conformations in the presence of the nearby H3 tail and with the H4 tail acetylated; this suggests a synergistic effect of the H3 and H4 tails in bringing about an enhanced dynamic range of the partner C.
Conclusion
We used MD simulations to investigate a base-displaced/intercalated cis-B[a]P-dG lesion positioned in nucleosomal DNA placed at SHL = −1, where it is bracketed by the H3 and H4 tails. We found that the behavior of the H3 tail and the base C that is partner to the cis-B[a]P-dG lesion is greatly impacted by the state of acetylation of the H4 tail, indicating synergy between the behavior of the two tails. In particular, when the H4 tail is in the unacetylated state, it is collapsed on the lesion-containing DNA surface and the lesion partner C stays in the major groove, so that the H3 tail does not interact with it. However, upon charge-neutralizing acetylation, the H4 tail is released from the DNA surface. In this altered electrostatic environment of the DNA, the H3 tail forms a pocket near the partner base C. This pocket induces the flipping and capture of the partner C. The key role played by the H3 tail in this flipping process is substantiated by the observation that the absence of this tail abolishes the flipping transition.
Thus, our work reveals that a nucleosomal tail can capture the displaced partner base C of the cis-B[a]P-dG lesion. The generality of this phenomenon if the cis-B[a]P-dG lesion were placed at different rotational and translational positions in the nucleosome will require further investigation. We envision that the amino acid sequence and the length of the tail, its state of acetylation, and the location of the cis-B[a]P-dG lesion vis-à-vis one or more tails would interplay to govern the outcome, representing a complex set of variables whose combined impact is hard to predict. However, the capacity of the partner C to be captured is clearly revealed here and it is plausible that the phenomenon would be manifested at other nucleosomal settings. The SHL = + 1 position, flanked by the second set of H3/H4 tails could be such a location. More broadly, our work shows the capacity of the displaced lesion partner C to interact with nearby proteins in general; an example is its capture by Rad4 [37]. It will be interesting to learn if DNA adducts that adopt base-displaced/intercalated conformations generally utilize their displaced lesion partner bases to bind with interacting proteins.
The H3 tail/partner base complex could foster detection of the lesion by the GG-NER lesion recognition factor XPC. The recent work of Kakumu et al. [27] showed that XPC binds to the unacetylated H3 tail and these studies suggested that the H3 tail could act as a platform for recruiting XPC in chromatin. The importance of the H3 tail in XPC recruitment has recently been reinforced by a study of the cis-syn cyclobutane pyrimidine dimer (CPD); this lesion specifically requires DDB2 for its primary recognition and its handoff to XPC. It was found that a methylase is recruited by DDB2 that methylates lysine 4 of the histone H3 tail to aid in the docking of the XPC to nucleosomes [58].
A single flipped out deoxyribonucleotide present on the undamaged partner strand has been shown to trigger assembly of excision complexes in the case of the 10R (+)-cis-anti-B[a]P-N2-dG adduct (cis-B[a]P-dG), its (−) stereoisomeric adduct, as well as the adduct derived from the binding of N-acetylaminofluorene (AAF) to the C8-position of guanine [59]. The relevance of the displaced partner C in the well-repaired cis-B[a]P-dG lesion to its recogntion has been computationally elucidated; it was found [37] that Rad4, the yeast homolog of XPC, captures the displaced partner C of cis-B[a]P-dG as an initial recognition event in GG-NER.
Our present findings suggest the hypothesis that lesion recognition by XPC could be fostered by the dual capability of the H3 tail to interact with XPC and also to sense specific intrinsic lesion-induced DNA distortions, such as a flipped out base observed for the base-displaced/intercalated conformational theme [34, 60] reviewed in [61]. Future studies are needed to understand how the interactions of lesion-containing DNA with the histone H3 tail in nucleosomes and higher order structures would affect the tail’s biological roles, including in NER. Striking single molecule imaging studies are providing insights on the dynamic process of lesion-recognition by XPC [62]. In addition, recent work has provided evidence that UV-DDB2 opens chromatin around a site that is damaged by UVC, based on real-time tracking of parental H3 and H4 histones in human cells, and suggests that histone reorganization or eviction accompanies XPC recognition of the lesion [63]. Understanding these higher order dynamic processes in molecular detail is a future challenge.
Supplementary Material
Highlights.
Nucleosomal histone tails play key roles in chromatin unpacking for NER.
H3 and H4 tails impact the structure of a nearby benzo[a]pyrene derived DNA lesion.
H3 tail induces capture of the displaced partner C to the cis-B[a]P-dG lesion.
Acetylation state of the H4 tail modulates the H3 tail’s capture of partner C.
H3 tail’s dual ability to bind XPC and partner C could foster lesion recognition.
Acknowledgments
We gratefully acknowledge resources provided by the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation (NSF) Grant MCB060037 to S.B., and the NYU IT High Performance Computing Resources and Services. We thank Dr. Hong Mu for careful reading of the manuscript.
Funding Information
This work was supported by NIH, NIEHS Grant R01-ES025987 and NCI Grant R01-CA75449 (to S.B.) and R01-CA168469 (to N.E.G.), and NIGMS Grant R01-GM079223 (to Y.Z.).
ABBREVIATIONS
- B[a]P
benzo[a]pyrene
- PTM
posttranslational modification
- NER
nucleotide excision repair
- NCP
nucleosome core particle
- PDB
protein data bank
- MD
molecular dynamics
- cis-B[a]P-dG
10R (+)-cis-anti-B[a]P-N2-dG adduct
- SHL
super-helical location
- RMSF
root mean square fluctuations
- CSA
contact surface area
- HB
hydrogen bonds
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Associated Content
The Supplementary Data is available free of charge alongside the electronic version of the article in Elsevier, including ScienceDirect: http://www.sciencedirect.com.
Supplementary Methods: initial models; force field; protonation; MD simulation protocols; structural analyses including tools for structural analyses, molecular modeling, image and movies visualization and rendering; best representative structures; minor groove widths; contact surface area; hydrogen bonds; protein pockets calculation. Supplementary Tables: Table S1. Box sizes, number of waters, and length of MD simulations Table S2. Number of hydrogen bonds between the H4 tail and DNA. Supplementary Figures: Figure S1. Time dependence of van der Waals interactions between the unacetylated H4 tail and local DNA; Figure S2. Impact of lesion modification on the local DNA minor groove widths when the H4 tail is unacetylated; Figure S3, Time dependence of the van der Waals interactions between the partner C base and the H3 tail residue Pro30; Figure S4. The initial structure for the 12 independent 500 ns MD simulations. Supplementary Movies: Movie S1. Best representative structure of the cis-B[a]P-dG containing DNA flanked by the H3 and the unacetylated H4 tails; Movie S2. Best representative structure of the cis-B[a]P-dG containing DNA flanked by the H3 and the acetylated H4 tails after the partner C to the lesion is captured; Movie S3. Transition of the C opposite the cis-B[a]P-dG lesion into a pocket in the H3 tail when the H4 tail is acetylated.
The authors declare no competing financial interest.
References
- 1.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGinty RK, Tan S. Nucleosome structure and function. Chem. Rev. 2015;115:2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pepenella S, Murphy KJ, Hayes JJ. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2014;123:3–13. doi: 10.1007/s00412-013-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman GD, Poirier MG. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015;115:2274–2295. doi: 10.1021/cr500350x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Mao P, Wyrick JJ. Emerging roles for histone modifications in DNA excision repair. FEMS Yeast Res. 2016;16 doi: 10.1093/femsyr/fow090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 10.Koprinarova M, Schnekenburger M, Diederich M. Role of histone acetylation in cell cycle regulation. Curr. Top Med. Chem. 2016;16:732–744. doi: 10.2174/1568026615666150825140822. [DOI] [PubMed] [Google Scholar]
- 11.Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 12.Smerdon MJ, Lan SY, Calza RE, Reeves R. Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J. Biol. Chem. 1982;257:13441–13447. [PubMed] [Google Scholar]
- 13.Ramanathan B, Smerdon MJ. Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis. 1986;7:1087–1094. doi: 10.1093/carcin/7.7.1087. [DOI] [PubMed] [Google Scholar]
- 14.Ramanathan B, Smerdon MJ. Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J. Biol. Chem. 1989;264:11026–11034. [PubMed] [Google Scholar]
- 15.Li S. Implication of posttranslational histone modifications in nucleotide excision repair. Int. J. Mol. Sci. 2012;13:12461–12486. doi: 10.3390/ijms131012461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polo SE, Almouzni G. Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model. DNA Repair (Amst) 2015;36:114–121. doi: 10.1016/j.dnarep.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smerdon MJ. DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 18.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA-reapir synthesis. Proc. Natl. Acad. Sci. U. S. A. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra LN, Pepenella S, Rogge R, Hansen JC, Hayes JJ. Acetylation mimics within a single nucleosome alter local DNA accessibility in compacted nucleosome arrays. Sci. Rep. 2016;6:34808. doi: 10.1038/srep34808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 21.Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J. Mol. Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q, Yang R, Korolev N, Liu CF, Nordenskiold L. Regulation of nucleosome stacking and chromatin compaction by the histone H4 N-terminal tail-H2A acidic patch interaction. J. Mol. Bio. 2017;429:2075–2092. doi: 10.1016/j.jmb.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Gospodinov A, Herceg Z. Shaping chromatin for repair. Mutat. Res. 2013;752:45–60. doi: 10.1016/j.mrrev.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Meas R, Smerdon MJ, Wyrick JJ. The amino-terminal tails of histones H2A and H3 coordinate efficient base excision repair, DNA damage signaling and postreplication repair in Saccharomyces cerevisiae. Nucleic Acids Res. 2015;43:4990–5001. doi: 10.1093/nar/gkv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ejlassi A, Menil-Philippot V, Galvani A, Thiriet C. Histones H3 and H4 require their relevant amino-tails for efficient nuclear import and replication-coupled chromatin assembly in vivo. Sci. Rep. 2017;7:3050. doi: 10.1038/s41598-017-03218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gansen A, Toth K, Schwarz N, Langowski J. Opposing roles of H3- and H4-acetylation in the regulation of nucleosome structure--a FRET study. Nucleic Acids Res. 2015;43:1433–1443. doi: 10.1093/nar/gku1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kakumu E, Nakanishi S, Shiratori HM, Kato A, Kobayashi W, Machida S, Yasuda T, Adachi N, Saito N, Ikura T, Kurumizaka H, Kimura H, Yokoi M, Sakai W, Sugasawa K. Xeroderma pigmentosum group C protein interacts with histones: regulation by acetylated states of histone H3. Genes Cells. 2017;22:310–327. doi: 10.1111/gtc.12479. [DOI] [PubMed] [Google Scholar]
- 28.Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips DH. Fifty years of benzo(a)pyrene. Nature. 1983;303:468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- 30.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 31.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 32.Szeliga J, Dipple A. DNA adduct formation by polycyclic aromatic hydrocarbon dihydrodiol epoxides. Chem. Res. Toxicol. 1998;11:1–11. doi: 10.1021/tx970142f. [DOI] [PubMed] [Google Scholar]
- 33.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol. Cell Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeves DA, Mu H, Kropachev K, Cai Y, Ding S, Kolbanovskiy A, Kolbanovskiy M, Chen Y, Krzeminski J, Amin S, Patel DJ, Broyde S, Geacintov NE. Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion-base stacking interactions. Nucleic Acids Res. 2011;39:8752–8764. doi: 10.1093/nar/gkr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–4155. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 37.Mu H, Geacintov NE, Min JH, Zhang Y, Broyde S. Nucleotide excision repair lesion-recognition protein Rad4 captures a pre-flipped partner base in a benzo[a]pyrene-derived DNA lesion: How structure impacts the binding pathway. Chem. Res. Toxicol. 2017;30:1344–1354. doi: 10.1021/acs.chemrestox.7b00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 39.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J. Mol. Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 40.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai Y, Kropachev K, Terzidis MA, Masi A, Chatgilialoglu C, Shafirovich V, Geacintov NE, Broyde S. Differences in the access of lesions to the nucleotide excision repair machinery in nucleosomes. Biochemistry. 2015;54:4181–4185. doi: 10.1021/acs.biochem.5b00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu I, Cai Y, Zhang Y, Geacintov NE, Broyde S. Entrapment of a histone tail by a DNA lesion in a nucleosome suggests the lesion impacts epigenetic marking: A molecular dynamics study. Biochemistry. 2016;55:239–242. doi: 10.1021/acs.biochem.5b01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu I, Cai Y, Geacintov NE, Zhang Y, Broyde S. Nucleosome histone tail conformation and dynamics: Impacts of lysine acetylation and a nearby minor groove benzo[a]pyrene-derived lesion. Biochemistry. 2017;56:1963–1973. doi: 10.1021/acs.biochem.6b01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 46.Turner BM, O'Neill LP, Allan IM. Histone H4 acetylation in human cells. Frequency of acetylation at different sites defined by immunolabeling with site-specific antibodies. FEBS Lett. 1989;253:141–145. doi: 10.1016/0014-5793(89)80947-0. [DOI] [PubMed] [Google Scholar]
- 47.Thorne AW, Kmiciek D, Mitchelson K, Sautiere P, Crane-Robinson C. Patterns of histone acetylation. Eur. J. Biochem. 1990;193:701–713. doi: 10.1111/j.1432-1033.1990.tb19390.x. [DOI] [PubMed] [Google Scholar]
- 48.Munks RJ, Moore J, O'Neill LP, Turner BM. Histone H4 acetylation in Drosophila. Frequency of acetylation at different sites defined by immunolabelling with site-specific antibodies. FEBS Lett. 1991;284:245–248. doi: 10.1016/0014-5793(91)80695-y. [DOI] [PubMed] [Google Scholar]
- 49.Clarke DJ, O'Neill LP, Turner BM. Selective use of H4 acetylation sites in the yeast Saccharomyces cerevisiae. Biochem. J. 1993;294(Pt 2):557–561. doi: 10.1042/bj2940557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sobel RE, Cook RG, Allis CD. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J. Biol. Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 51.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 52.Case DA, Darden TA, Cheatham TE, 3rd, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Gusarov S, Kovalenko A, Kollman PA. AMBER 14. University of California; San Francisco: 2014. [Google Scholar]
- 53.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 55.Shao J, Tanner SW, Thompson N, Cheatham TE. Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 2007;3:2312–2334. doi: 10.1021/ct700119m. [DOI] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 57.Zondlo NJ. Aromatic-proline interactions: Electronically tunable CH/pi interactions. Acc. Chem. Res. 2013;46:1039–1049. doi: 10.1021/ar300087y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balbo Pogliano C, Gatti M, Ruthemann P, Garajova Z, Penengo L, Naegeli H. ASH1L histone methyltransferase regulates the handoff between damage recognition factors in global-genome nucleotide excision repair. Nat. Commun. 2017;8:1333. doi: 10.1038/s41467-017-01080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 60.Wang F, DeMuro NE, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for-2 bp deletions. J. Am. Chem. Soc. 2006;128:10085–10095. doi: 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geacintov NE, Broyde S. Repair-resistant DNA lesions. Chem. Res. Toxicol. 2017;30:1517–1548. doi: 10.1021/acs.chemrestox.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kong LLM, Chen X, Driscoll KI, Mao P, Böhm S, Kad NM, Watkins SC, Bernstein KA, Wyrick JJ, Min JH, Van Houten B. Single-molecule imaging reveals that Rad4 employs a dynamic DNA damage recognition process. Mol. Cell. 2016;64:376–387. doi: 10.1016/j.molcel.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adam S, Dabin J, Chevallier O, Leroy O, Baldeyron C, Corpet A, Lomonte P, Renaud O, Almouzni G, Polo SE. Real-Time tracking of parental histones reveals their contribution to chromatin integrity following DNA damage. Mol. Cell. 2016;64:65–78. doi: 10.1016/j.molcel.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu J, Zhou Y, Tanaka I, Yao M. Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics. 2010;26:46–52. doi: 10.1093/bioinformatics/btp599. [DOI] [PubMed] [Google Scholar]
- 65.Naghibzadeh S, Binkowski TA, Liang J. castP: Computed atlas of surface topography of proteins. Biophys. J. 2001;80:320a. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Binkowski TA, Naghibzadeh S, Liang J. CASTp: Computed atlas of surface topography of proteins. Nucleic Acids Res. 31(13):3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.