Abstract
Polymyxins are currently used as the last-resort antibiotics against multidrug-resistant Acinetobacter baumannii. As resistance to polymyxins emerges in A. baumannii with monotherapy, combination therapy is often the only remaining treatment option. A novel approach is to employ the combination of polymyxin B with non-antibiotic drugs. In the present study, we employed metabolomics to investigate the synergistic mechanism of polymyxin B in combination with the antineoplastic drug mitotane against polymyxin-susceptible and -resistant A. baumannii. The metabolomes of four A. baumannii strains were analyzed following treatment with polymyxin B, mitotane and the combination. Polymyxin B monotherapy induced significant perturbation in glycerophospholipid (GPL) metabolism and histidine degradation pathways in polymyxin-susceptible strains, and minimal perturbation in polymyxin-resistant strains. Mitotane monotherapy induced minimal perturbation in the polymyxin-susceptible strains, but caused significant perturbation in GPL metabolism, pentose phosphate pathway and histidine degradation in the LPS-deficient polymyxin-resistant strain (FADDI-AB065). The polymyxin B – mitotane combination induced significant perturbation in all strains except the lipid A modified polymyxin-resistant FADDI-AB225 strain. For the polymyxin-susceptible strains, the combination therapy significantly perturbed GPL metabolism, pentose phosphate pathway, citric acid cycle, pyrimidine ribonucleotide biogenesis, guanine ribonucleotide biogenesis, and histidine degradation. Against FADDI-AB065, the combination significantly perturbed GPL metabolism, pentose phosphate pathway, citric acid cycle, and pyrimidine ribonucleotide biogenesis. Overall, these novel findings demonstrate that the disruption of the citric acid cycle and inhibition of nucleotide biogenesis are the key metabolic features associated with synergistic bacterial killing by the combination against polymyxin-susceptible and -resistant A. baumannii.
Keywords: polymyxin, mitotane, combination therapy, multidrug-resistance, metabolomics
Introduction
Multidrug-resistant (MDR) Acinetobacter baumannii has become a major global health threat (Talbot et al., 2006; Peleg et al., 2008; Boucher et al., 2009, 2013; Michalopoulos and Falagas, 2010). In 2013, the Centers for Disease Control and Prevention (CDC) classified A. baumannii as a “Serious threat” as approximately 63% of healthcare-associated Acinetobacter infections occurring in the United States were MDR (i.e., non-susceptible to ≥1 treating agent in ≥3 antimicrobial categories) (Magiorakos et al., 2012; Centers for Disease Control and Prevention [CDC], 2013). In 2017, carbapenem-resistant A. baumannii has been classified by the World Health Organization as one of the top priorities for research and development of new antibiotics, due to the current lack of novel antibiotic candidates in the drug development pipeline (Tacconelli and Magrini, 2017; WHO/EMP/IAU, 2017). Clearly, the development of novel therapeutic strategies to combat this deadly pathogen are urgently needed.
Polymyxin B and colistin are considered the last resort against MDR A. baumannii (Li et al., 2006a; Nation et al., 2015). Although polymyxins are believed to cause cell death primarily by disorganizing the Gram-negative outer membrane via binding to lipopolysaccharide (LPS), the precise antibacterial killing mechanism is not completely understood (Velkov et al., 2010; Trimble et al., 2016). Worryingly, we and others have demonstrated that polymyxin resistance rapidly emerges in polymyxin-susceptible A. baumannii following polymyxin monotherapy (Li et al., 2006b; Dudhani et al., 2010; Moffatt et al., 2010; Arroyo et al., 2011; Garonzik et al., 2011; Pelletier et al., 2013). A. baumannii becomes resistant to polymyxins by a reduction of the negative charge on the outer membrane (Velkov et al., 2010), which is achieved either through lipid A modification [with phosphoethanolamine (pEtN) and galactosamine (GalN) (Arroyo et al., 2005; Adams et al., 2009; Pelletier et al., 2013)] or loss of LPS (Moffatt et al., 2010).
Recently, the combination of an antibiotic (including polymyxins) and non-antibiotic drug has emerged as a potentially valuable and cost-effective approach to improve the clinical efficacy of currently available antibiotics against problematic MDR bacterial pathogens (Ejim et al., 2011; Hussein et al., 2016; Schneider et al., 2016, 2017). In a recent study, we demonstrated that the combination of clinically achievable concentrations of polymyxin B (2 mg/L) and the antineoplastic agent mitotane (4 mg/L) provided enhanced antimicrobial activity against MDR as well as polymyxin-resistant A. baumannii (Tran et al., 2018). Furthermore, the combination also prevented bacterial regrowth in polymyxin-susceptible strains. Mitotane is used for the treatment of adrenocortical carcinoma and Cushing’s syndrome (Lalli, 2015). Unlike other anticancer drugs that damage DNA and inhibit DNA replication, mitotane inhibits steroidogenesis in adrenocortical carcinoma cells (Lalli, 2015). The precise antimicrobial mechanism of mitotane, however, is presently unclear. Given the potential repositioning of mitotane to treat MDR A. baumannii, it is essential to understand the mechanisms by which the polymyxin/mitotane combination achieves this enhanced bacterial killing and suppression of bacterial regrowth.
Metabolomics has emerged as a valuable tool for elucidating the mechanisms of drug action in bacterial physiology and drug discovery (Mastrangelo et al., 2014; Vincent et al., 2016). Notably, metabolomics provides snapshots of cellular biochemical networks and helps explain how bacteria respond to antibiotic treatment at the systems level (Chen et al., 2007; Jia et al., 2009; Kaddurah-Daouk et al., 2014). Moreover, understanding how bacteria respond to antibiotic treatment at the metabolic level is valuable for the discovery of novel antibiotic targets (Jia et al., 2009). Accordingly, the primary aim of this study was to use untargeted metabolomics to elucidate the mechanism(s) of the enhanced antimicrobial activity of polymyxins against A. baumannii by mitotane.
Materials and Methods
Drugs and Bacterial Isolates
Polymyxin B (Beta Pharma, China, Batch number 20120204) solutions were prepared in Milli-QTM water (Millipore, Australia) and filtered through 0.22-μm syringe filters (Sartorius, Australia). Mitotane (Sigma-Aldrich, Australia, Lot number BCBG9480V) solutions were prepared in dimethyl sulfoxide (Sigma-Aldrich, Australia). All other reagents were purchased from Sigma-Aldrich (Australia) and were of the highest commercial grade available. Polymyxin-susceptible A. baumannii ATCC 17978 (polymyxin B MIC = 0.25 mg/L) and ATCC 19606 (polymyxin B MIC = 0.5 mg/L) were obtained from the American Type Culture Collection (Manassas, VA, United States). A. baumannii FADDI-AB225 (formally designated ATCC 17978-R2) is polymyxin-resistant (polymyxin B MIC = 16 mg/L) with phosphoethanolamine-modified lipid A and pmrB mutation derived from A. baumannii ATCC 17978 (Arroyo et al., 2011). A. baumannii FADDI-AB065 (formally designated ATCC 19606R) is a polymyxin-resistant (polymyxin B MIC = 64 mg/L), LPS-deficient, lpxA mutant derived from the ATCC 19606 strain (Moffatt et al., 2010). Isolates were stored in tryptone soy broth (Oxoid) with 20% glycerol (Ajax Finechem, Seven Hills, NSW, Australia) in cryovials at -80°C. Before use, A. baumannii ATCC 17978 and ATCC 19606 were subcultured onto nutrient agar plates (Media Preparation Unit, University of Melbourne, Melbourne, VIC, Australia) and A. baumannii FADDI-AB225 and FADDI-AB065 were subcultured onto Mueller–Hinton plates supplemented with 10 mg/L of polymyxin B (Media Preparation Unit) to maintain the selection pressure.
Bacterial Culture Preparation for Metabolomics Experiments
To investigate the possible molecular mechanisms of polymyxin B and mitotane combination, we employed untargeted metabolomics to determine the changes in different metabolite levels in all A. baumannii strains following 2-h treatment with 2 mg/L polymyxin B, 4 mg/L mitotane, or the combination. To ensure the clinical relevance of these findings, the concentrations of polymyxin B (2 mg/L) and mitotane (4 mg/L) employed were within the clinically achievable range of concentrations of each agent (Hermsen et al., 2011; Sandri et al., 2013). A 2 h exposure to the antibiotics was selected for investigation, as extensive bacterial killing normally occurs with polymyxins across this time (Li et al., 2006b; Owen et al., 2007; Percin et al., 2014). For each A. baumannii strain, single colonies grown on nutrient or Mueller–Hinton agar were selected and grown overnight (16–18 h) in 20 mL cation-adjusted Mueller–Hinton broth (CAMHB; Oxoid, England; 20–25 mg/L Ca2+ and 10–12.5 mg/L Mg2+) in 50 mL Falcon tubes (Thermo Fisher, Australia) incubated in a shaking water bath at 37°C (shaking speed, 180 rpm). Following overnight incubation, each culture was transferred to a 1,000 mL conical flask with 250 mL of fresh CAMHB at ∼50- to 100-fold dilutions. Flasks were incubated at 37°C with shaking at 180 rpm for ∼3–4 h to log-phase (OD600 ∼0.5). Cultures (50 mL) were transferred to four 500 mL conical flasks and solutions of polymyxin B, mitotane, or both added to three of four flasks to give a final concentration of 2 mg/L for polymyxin B and 4 mg/L for mitotane; the remaining flask acted as a drug-free control. To prevent excessive bacterial killing, the starting bacterial inoculum used was ∼108 cfu/mL. The flasks were further incubated at 37°C with shaking at 180 rpm. After 2 h, the OD600 reading for each flask was measured and normalized to ∼0.5 with fresh CAMHB and 10 mL samples transferred to 15 mL Falcon tubes (Thermo Fisher, Australia) for metabolite extraction. To minimize inherent random variation, for each strain four biological samples were prepared for each treatment condition.
Metabolite Extraction for Metabolomic Studies
Following bacterial culture preparation, extraction of metabolites was immediately performed to minimize further drug effects on metabolite levels. Samples were initially centrifuged at 3,220 × g at 4°C for 10 min. Supernatants were then removed and bacterial pellets washed twice in 2 mL of cold normal saline followed by centrifugation at 3,220 × g at 4°C for 5 min to remove residual extracellular metabolites and medium components. The washed pellets were then resuspended with cold chloroform:methanol:water (CMW; 1:3:1, v/v) extraction solvent containing 1 μM each of the internal standards (CHAPS, CAPS, PIPES, and TRIS). The selected internal standards are physicochemically diverse small molecules not naturally occurring in any microorganism. Samples were then thrice frozen in liquid nitrogen, thawed on ice and vortexed to release the intracellular metabolites. After the third cycle samples were centrifuged for 10 min at 3,220 × g at 4°C, whereby 300 μL of the supernatants was transferred to 1.5 mL Eppendorf tubes for immediate storage at -80°C. Prior to analysis samples were thawed and centrifuged at 14,000 × g at 4°C for 10 min to remove the presence of any particles, and 200 μL transferred into the injection vial for LC-MS analysis (described below). An equal volume of each sample was combined and used as a pooled quality control sample (QC); namely, a sample that contains all the analytes that will be encountered during the analysis (Gika et al., 2007).
LC-MS Analysis
Metabolites were detected with hydrophilic interaction liquid chromatography (HILIC) – high-resolution mass spectrometry (HRMS) using a Dionex high-performance liquid chromatography (HPLC) system (RSLCU3000, Thermo Fisher) with a ZIC-pHILIC column (5 μm, polymeric, 150 mm × 4.6 mm; SeQuant, Merck). The system was coupled to a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher) operated in both positive and negative electro-spray ionization (ESI) mode at 35,000 resolution with a detection range of 85 to 1,275 m/z. The LC solvents were (A) 20 mM ammonium carbonate and (B) acetonitrile, operated via a multi-step gradient system. The gradient system at 80% B and was reduced to 50% B over 15 min, then reduced from 50% B to 5% B over 3 min, followed by wash with 5% B for another 3 min, and finally 8 min re-equilibration with 80% B at a flow rate of 0.3 mL/min (Zhang et al., 2012). The total run time was 32 min with an injection sample volume of 10 μL. All samples were analyzed as a single LC-MS batch to reduce the batch-to-batch variation. Mixtures of pure standards containing over 250 metabolites were also included in the analysis to aid metabolite identification.
Data Processing, Bioinformatics, and Statistical Analyses
Conversion of LC-MS raw data to metabolites was conducted using IDEOM1 free software (Creek et al., 2012), which initially employed ProteoWizard to convert raw LC-MS data to mzXML format and XCMS to pick peaks to convert to peakML files (Smith et al., 2006; Scheltema et al., 2011). Mzmatch.R was subsequently used for the alignment of samples and the filtering of peaks using minimum detectable intensity of 100,000, relative standard deviation (RSD) of <0.5 (reproducibility), and peak shape (codadw) of >0.8. Mzmatch was also used to retrieve missing peaks and annotation of related peaks. Default IDEOM parameters were used to eliminate unwanted noise and artifact peaks. Loss or gain of a proton was corrected in negative and positive ESI mode, respectively, followed by putative identification of metabolites by the exact mass within 2 ppm. Retention times of authentic standards were used to confirm the identification of each metabolite (Level 1 identification based on MSI standards). Other metabolites were putatively identified (Level 2 identification based on MSI standards) using exact mass and predicted retention time based on the Kyoto Encyclopedia of Genes and Genomes (KEGG), MetaCyc, and LIPIDMAPS databases, using preference to bacterial metabolites annotated in EcoCyc. Raw peak intensity was used to quantify each metabolite. The free online tool MetaboAnalyst 3.0 was used for the statistical analysis. Briefly, putative metabolites with median RSD ≤ 0.2 (20%) within the QC group and IDEOM confidence level of ≥5 were incorporated into a table and uploaded to MetaboAnalyst. Data with >50% missing values were removed and remaining missing values replaced with half the minimum positive value in the original data. Data were filtered using interquartile range (IQR), normalized by the median, log2 transformed and auto scaled. Principal component analysis (PCA) was performed to identify and remove outliers. PCA is normally used to reduce the dimension of variables from a large data set (Ma and Dai, 2011). Outliers were defined as samples outside of ±2 standard deviations (SD) along the principal component 1 axis. One-way ANOVA was used to identify metabolites with significant level changes between all samples and Fisher’s least square difference (LSD) to determine the metabolites with significant level changes between treatment and control groups. Statistically significant metabolites were selected using a false discovery rate of ≤0.05 for one-way ANOVA and p ≤ 0.05 for Fisher’s LSD. KEGG mapper was used to determine the pathway modules by statistically significant metabolites containing the KEGG compound numbers.
Results
Multivariate and Univariate Analyses of the Metabolites Affected by Polymyxin B and Mitotane in A. baumannii
Untargeted metabolomics analysis using HILIC-based high resolution accurate mass LC-MS allowed detection of 1,769 putative metabolites in polymyxin-sensitive and -resistant strains of A. baumannii treated with polymyxin B and mitotane. The reproducibility of metabolite semi-quantitation was within acceptable limits based on the median RSD from independent biological replicates across the four A. baumannii strains, where the median RSD was 16% for all control groups and <20% for most treatment groups (Table 1) (Kirwan et al., 2014).
Table 1.
Data precision of different treatment groups represented as the median relative standard deviation (RSD) for all assessed metabolites.
| Median RSD (%) | |
|---|---|
| A. baumannii ATCC 17978 | |
| Control | 16 |
| Polymyxin B | 17 |
| Mitotane | 21 |
| Combination | 16 |
| A. baumannii FADDI-AB225 | |
| Control | 16 |
| Polymyxin B | 18 |
| Mitotane | 16 |
| Combination | 23 |
| A. baumannii ATCC 19606 | |
| Control | 16 |
| Polymyxin B | 23 |
| Mitotane | 14 |
| Combination | 14 |
| A. baumannii FADDI-AB065 | |
| Control | 16 |
| Polymyxin B | 15 |
| Mitotane | 16 |
| Combination | 14 |
| PBQCs | 14 |
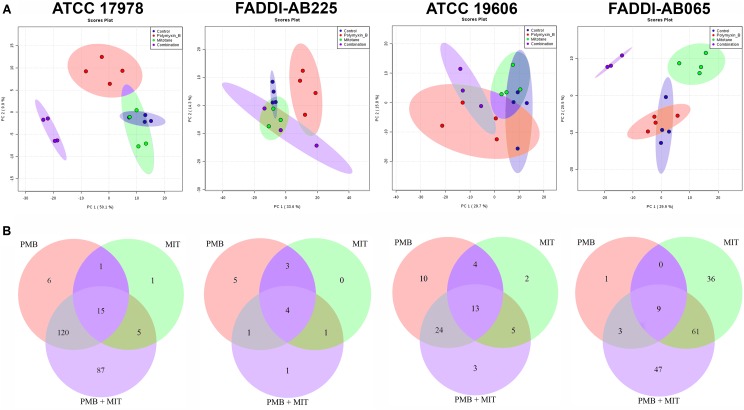
According to PCA, mitotane monotherapy clearly impacted the metabolome of FADDI-AB065 based on the first two principal components, but did not differentiate from controls for the other three tested strains (Figure 1A). Compared to polymyxin B and mitotane therapies, the combination produced more significant perturbations in the metabolomes of ATCC 17978 and FADDI-AB065 (Figure 1A). A minimal impact on the metabolome of FADDI-AB225 was observed for the combination (Figure 1A).
FIGURE 1.
(A) PCA score plots showing metabolomic variance for untreated samples (blue) and samples treated with polymyxin B (red), mitotane (green) and combination (purple) for each A. baumannii strain along principal components 1 and 2. The degree of separation shows the level of variance between these groups. Principal component 1 axis is responsible for higher level of variance. (B) Venn diagrams showing the number of statistically significant metabolites affected by different treatments (one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05) in each A. baumannii strain (PMB, polymyxin B; MIT, mitotane).
Polymyxin B monotherapy caused significant perturbations in a total of 142 metabolites in ATCC 17978, 51 in ATCC 19606, 13 in FADDI-AB225, and 13 in FADDI-AB065 (Figure 1B). For mitotane monotherapy, a total of 22 metabolites were perturbed in ATCC 17978, 24 in ATCC 19606, 8 in FADDI-AB225, and 106 in FADDI-AB065 (Figure 1B). The combination caused perturbations in a total of 227 metabolites in ATCC 17978, 45 in ATCC 19606, 7 in FADDI-AB225, and 120 in FADDI-AB065 (Figure 1B). Compared to mitotane monotherapy, polymyxin B monotherapy caused perturbation in more than twice the number of metabolites in the polymyxin-susceptible strains (ATCC 17978 and ATCC 19606). For the combination, over 50% of the perturbed metabolites in the polymyxin-susceptible strains were in common with those perturbed by polymyxin B and mitotane monotherapy. The common perturbed metabolites between combination therapy and polymyxin B monotherapy were much higher than the common perturbed metabolites between the combination therapy and mitotane monotherapy (Figure 1B). Although mitotane monotherapy had little impacts on the polymyxin-susceptible strains, it caused extensive metabolic changes in the LPS-deficient polymyxin-resistant FADDI-AB065 (Figure 1B). Most of the perturbed metabolites caused by the combination in this strain, consequently, were in common with those perturbed by mitotane monotherapy (Figure 1B).
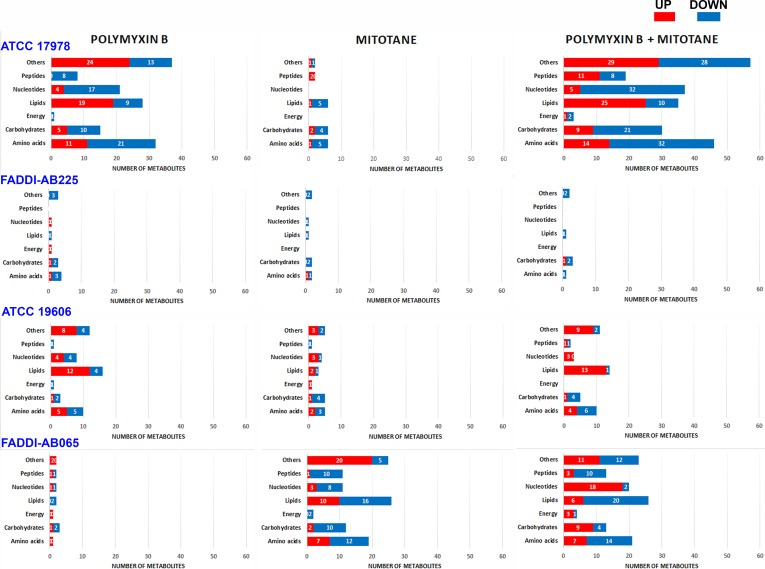
The statistically significant metabolites impacted (one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05) by different treatments in each A. baumannii strain were divided into seven different metabolite classes: amino acids, carbohydrates, energy, lipids, nucleotides, peptides, and others (the latter includes cofactors and vitamins, glycans, secondary metabolites and metabolites that could not be mapped into pathways). The number of metabolites impacted from each class that were higher or lower in abundance compared to the control group are shown in Figure 2. Details of all significantly impacted metabolites, including mass, retention time (RT), formula, putative identification, level of confidence (from IDEOM software), map, pathway, fold-change (FC; based on raw intensity), and FDR are shown in Supplementary Tables S1–S4 for all four strains.
FIGURE 2.
Bar graphs showing the number of significantly perturbed metabolites (ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05) in different metabolite classes following treatment with polymyxin B, mitotane, and the combination for polymyxin-susceptible A. baumannii ATCC 17978, polymyxin-resistant A. baumannii FADDI-AB225, polymyxin-susceptible A. baumannii ATCC 19606, and polymyxin-resistant A. baumannii FADDI-AB065. The class designated as ‘Others’ includes cofactors and vitamins, glycan, secondary metabolites and metabolites that could not be mapped into pathways based on existing bacterial metabolite databases.
Significantly Impacted Lipids and Lipid Metabolites
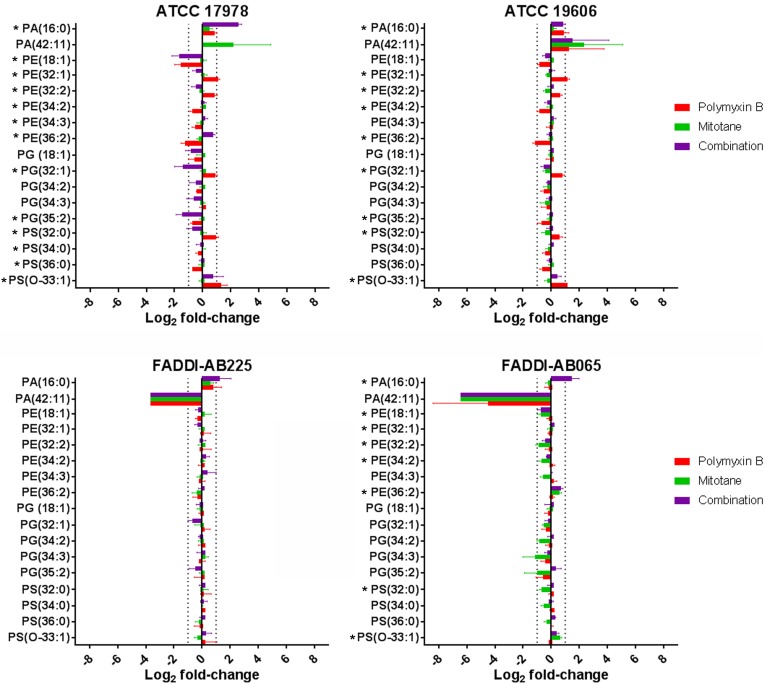
Glycerophospholipids (GPLs) across four A. baumannii strains detected by LC-MS and their relative abundance (based on raw peak intensity) compared to the control groups are shown in Figure 3. In the polymyxin-susceptible strains, polymyxin B monotherapy induced significant changes in a wide range of GPL while mitotane monotherapy induced minimal changes. Overall, polymyxin B monotherapy caused a higher level of GPL perturbation in ATCC 17978 than ATCC 19606. Compared to polymyxin B monotherapy, the combination substantially enhanced the perturbation of putative lysophospholipids PA(16:0) and PG(35:2) in ATCC 17978 to greater than two-fold change in both cases. Against ATCC 19606, where the perturbation caused by polymyxin B monotherapy was lower than two-fold change in most cases, the combination did not significantly affect the GPL. In both polymyxin-resistant strains, polymyxin B monotherapy had a minimal impact on GLP, while mitotane monotherapy significantly affected a wide range of GPL in FADDI-AB065. In FADDI-AB065, mitotane monotherapy caused over two-fold reduction in putative GPLs PG(34:3) and PG(35:2). Interestingly, the combination did not enhance the lipid perturbation caused by mitotane monotherapy in FADDI-AB065.
FIGURE 3.
Glycerophospholipids in A. baumannii ATCC 17978, ATCC 19606, FADDI-AB065, and FADDI-AB225 following treatment with polymyxin B, mitotane and the combination. (∗one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05, log2 fold-change ≥ |1| thresholds are indicated by vertical dotted lines).
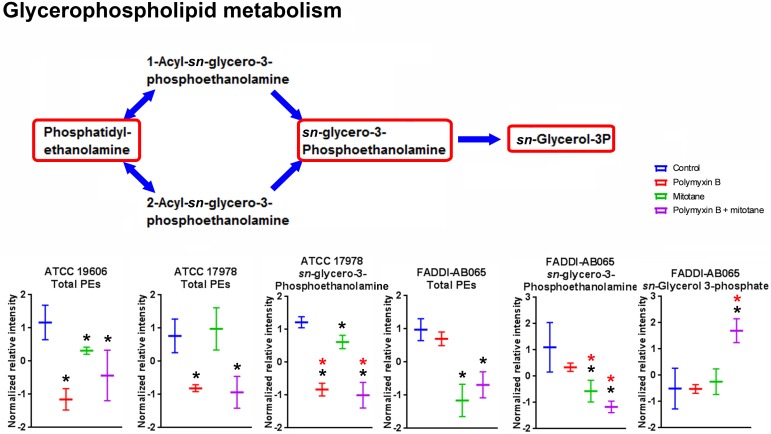
The impact of polymyxin B and mitotane on the metabolites involved in GPL metabolism in A. baumannii is shown in Figure 4. In ATCC 19606, polymyxin B monotherapy and combination therapy caused statistically significant perturbation in total putative PEs (the sum of all detected putative PE species), although the changes in relative abundance were less than two-fold. Mitotane monotherapy had minimal impacts on these metabolites in ATCC 19606. For ATCC 17978, in addition to total putative PEs, polymyxin B monotherapy and combination therapy also substantially reduced putative sn-glycero-3-phosphoethanolamine (Log2FC = -2.42 and -2.83, respectively). Mitotane monotherapy only caused minor reduction of putative sn-glycero-3-phosphoethanolamine (Log2FC = -0.47) in ATCC 17978. Against polymyxin-resistant FADDI-AB065, polymyxin B monotherapy did not impact any metabolites involved in GPL metabolism. However, mitotane monotherapy caused significant perturbations in total putative PEs (Log2FC = -0.45) and putative sn-glycero-3-phosphoethanolamine (Log2FC = -1.71). The combination caused significant perturbations in total putative PEs (Log2FC = -0.34), putative sn-glycero-3-phosphoethanolamine (Log2FC = -1.62), and sn-glycerol-3-phosphate (Log2FC = 1.55).
FIGURE 4.
Metabolites involved in glycerophospholipid metabolism in A. baumannii significantly impacted by polymyxin B, mitotane, and the combination. Red boxes indicate statistically significant metabolites (Mean ± SD; ∗one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05; ∗log2 fold-change ≥|1|).
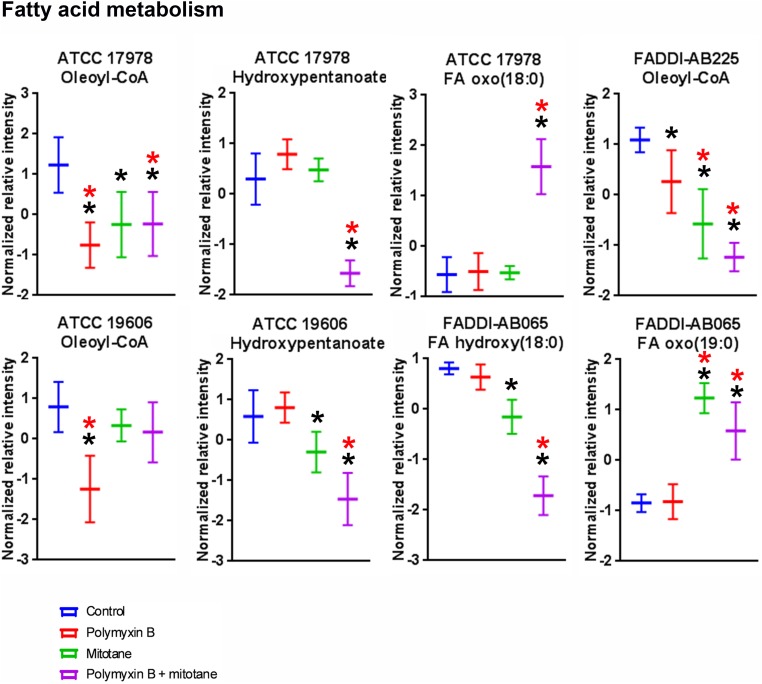
The statistically significant (ANOVA, FDR ≤ 0.05, Fisher’s LSD, p ≤ 0.05) fatty acyls impacted by polymyxin B monotherapy, mitotane monotherapy, or the combination are shown in Figure 5. In ATCC 17978, ATCC 19606, and FADDI-AB225, polymyxin B monotherapy induced significant perturbations in putative oleoyl-CoA, a metabolite involved in fatty acid metabolism; notably, the relative abundance of oleoyl-CoA was over two-fold lower in polymyxin-susceptible strains treated with polymyxin B monotherapy compared to the untreated group. For mitotane monotherapy, significant reduction of oleoyl-CoA was observed for FADDI-AB225 and its parent strain ATCC 17978. Compared to polymyxin B monotherapy, the combination caused a greater reduction of oleoyl-CoA in polymyxin-resistant FADDI-AB225. In addition, the combination also caused significant perturbation of oxidized fatty acids including putative hydroxypentanoate in both polymyxin-susceptible strains, putative FA oxo(18:0) in ATCC 17978, and putative FA hydroxy(18:0) and putative FA oxo(19:0) in FADDI-AB065.
FIGURE 5.
Putative fatty acyls from fatty acid metabolism in A. baumannii significantly impacted by polymyxin B, mitotane, and combination treatment (Mean ± SD; ∗one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05; ∗log2 fold-change ≥ |1|).
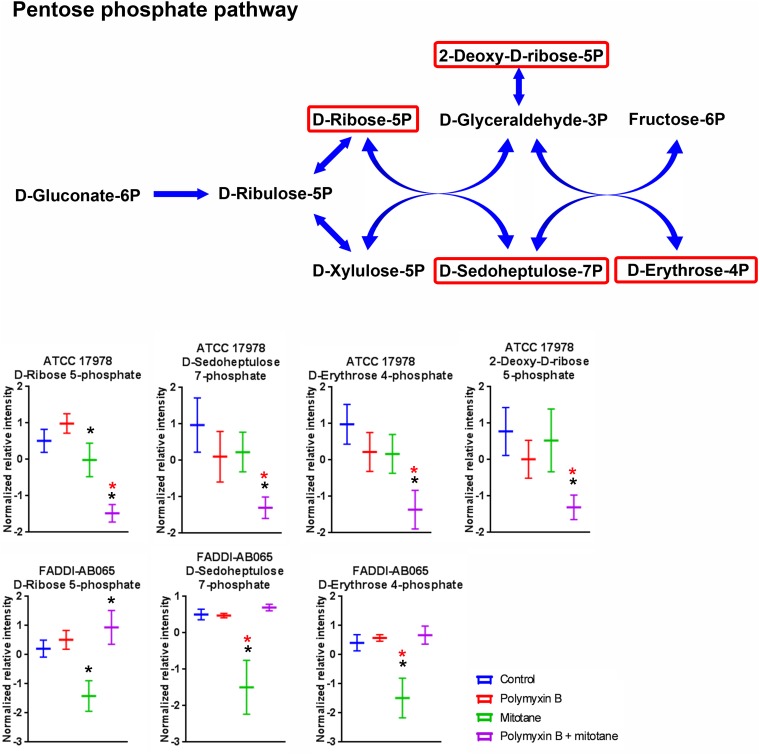
Significantly Impacted Metabolites in Pentose Phosphate Pathway
Metabolites involved in pentose phosphate pathway of A. baumannii were perturbed by polymyxin B and mitotane (Figure 6). For ATCC 17978, polymyxin B monotherapy had no impact on metabolites of the pentose phosphate pathway; however, the combination therapy caused a significant reduction in D-ribose-5-phosphate, putative D-sedoheptulose-7-phosphate, D-erythrose-4-phosphate, and 2-deoxy-D-ribose-5-phosphate (Log2FC = -1.82, -3.09, -3.07, and -1.79, respectively). For FADDI-AB065, mitotane monotherapy caused a statistically significant reduction of D-ribose-5-phosphate, putative D-sedoheptulose-7-phosphate and D-erythrose-4-phosphate (≥2-fold change) (Log2FC = -0.94, -1.93, and -2.74, respectively).
FIGURE 6.
Metabolites in pentose phosphate pathway in A. baumannii significantly impacted by polymyxin B, mitotane, and the combination. Red boxes indicate statistically significant metabolites (Mean ± SD; ∗one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05; ∗log2 fold-change ≥|1|).
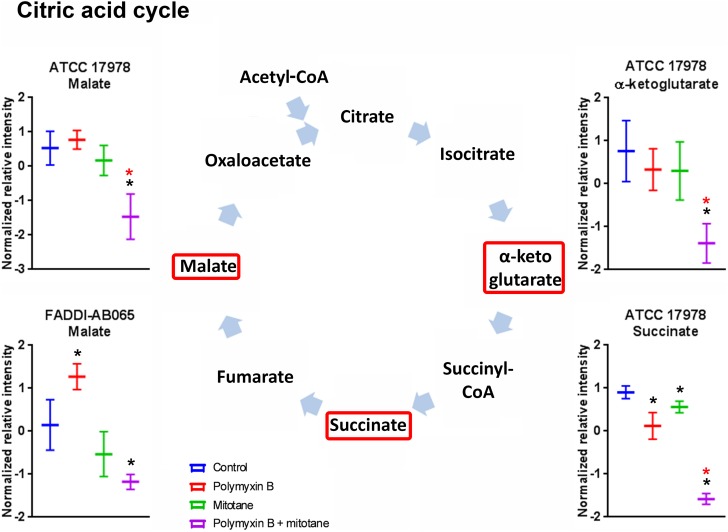
Significantly Impacted Metabolites in Citric Acid Cycle
The impact of polymyxin B and mitotane on citric acid cycle in A. baumannii is shown in Figure 7. In ATCC 17978, succinate was significantly reduced by both polymyxin B and mitotane monotherapies; however, the highest level of reduction was observed with combination treatment (Log2FC = -2.49). In addition, the combination also caused a significant reduction in abundance in α-ketoglutarate and malate (Log2FC = -2.01 and -1.47, respectively) in ATCC 17978. The abundance of malate was also reduced by the combination treatment in FADDI-AB065 (Log2FC = -0.82), but increased by polymyxin B monotherapy (Log2FC = 0.82).
FIGURE 7.
Metabolites in citric acid cycle in A. baumannii significantly impacted by polymyxin B, mitotane, and the combination. Red boxes indicate statistically significant metabolites (Mean ± SD; ∗one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05; ∗log2 fold-change ≥|1|).
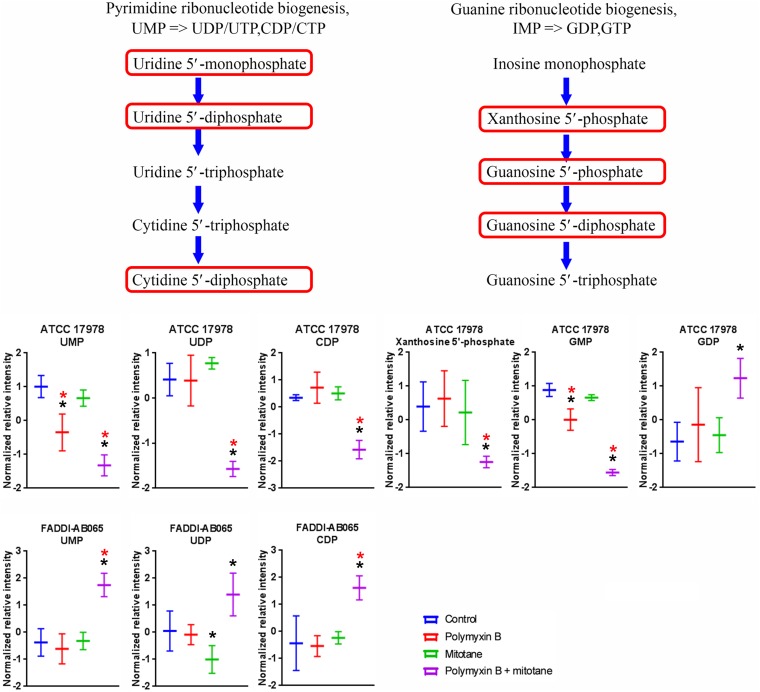
Significantly Impacted Metabolites in Nucleotide Metabolism
A high number of metabolites involved in nucleotide metabolism in A. baumannii were significantly impacted by polymyxin B and mitotane alone and in combination (Supplementary Tables S1–S4). In both ATCC 17978 and FADDI-AB065, the pyrimidine ribonucleotide biogenesis pathway was over represented (≥2 metabolites in the module affected) (Figure 8). In ATCC 17978, UMP was significantly reduced by both polymyxin B monotherapy and the combination (Log2FC = -1.19 and -2.07, respectively); and the combination also reduced UDP and putative CDP (Log2FC = -1.47 and -1.57, respectively). In FADDI-AB065, UDP was slightly reduced by mitotane monotherapy, while increased by the combination therapy (Log2FC = -0.04 and 0.87, respectively). Only the combination caused increases in UMP and putative CDP (Log2FC = 1.74 and 1.33, respectively). A related pathway, guanine ribonucleotide biogenesis was also over represented in ATCC 17978. In this pathway, GMP abundance was significantly reduced by polymyxin B monotherapy and the combination (Log2FC = -1.28 and -3.22, respectively) in ATCC 17978, with greater perturbation caused by the combination. Additionally, only the combination impacted putative xanthosine 5′-phosphate (XMP) and GDP (Log2FC = -1.40 and -0.05, respectively), with greater perturbation occurring for putative XMP.
FIGURE 8.
Metabolites in pyrimidine and guanine ribonucleotide biogenesis in A. baumannii significantly impacted by polymyxin B, mitotane, and the combination. Red boxes indicate statistically significant metabolites (Mean ± SD; ∗one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05; ∗log2 fold-change ≥|1|).
Significantly Impacted Metabolites in Amino Acid Metabolism
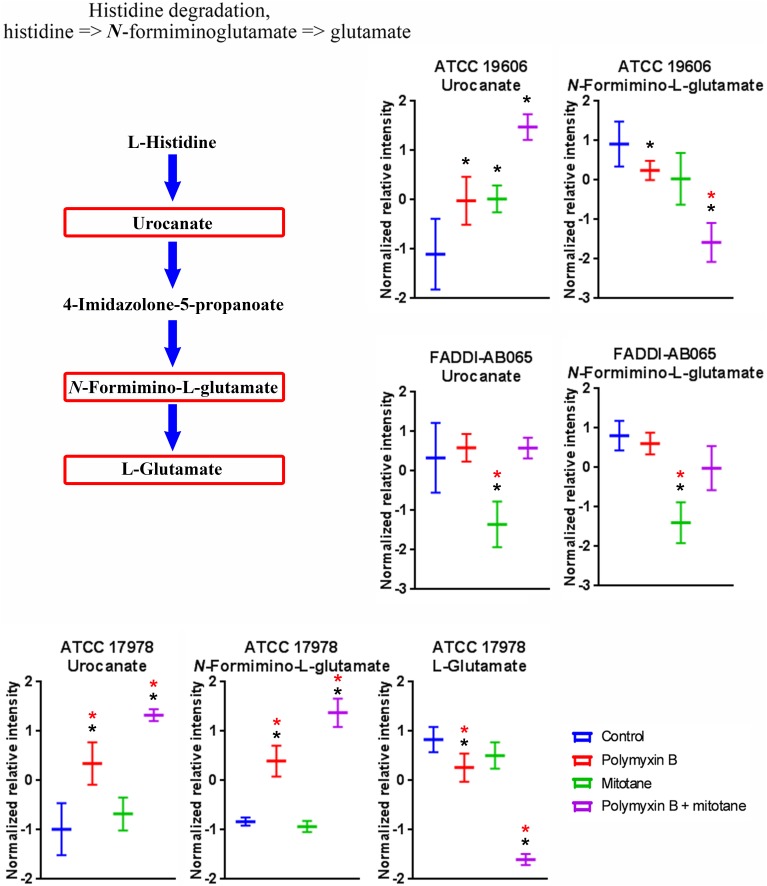
Treatments with polymyxin B and mitotane alone and in combination caused significant perturbations to a high number of metabolites involved in amino acid metabolism in A. baumannii (Supplementary Tables S1–S4). Across ATCC 17978, ATCC 19606 and FADDI-AB065, histidine degradation was over-represented (≥2 metabolites in the module affected) (Figure 9). In ATCC 17978, polymyxin B monotherapy and the combination treatment caused significant perturbations in putative urocanate (Log2FC = 1.46 and 2.49, respectively), putative N-formimino-L-glutamate (Log2FC = 1.27 and 2.29, respectively) and L-glutamate (Log2FC = -1.25 and -4.44, respectively). The combination treatment, however, produced the highest level of perturbation in all three metabolites. In ATCC 19606, the intracellular concentration of putative urocanate was significantly increased by polymyxin B, mitotane, and combination treatment (Log2FC = 0.11, 0.30, and 0.71, respectively), with the highest level of perturbation observed with the combination. Putative N-formimino-L-glutamate was significantly reduced by combination therapy (Log2FC = -1.56). Interestingly, only mitotane monotherapy caused significant reduction in putative urocanate and putative N-formimino-L-glutamate in FADDI-AB065 (Log2FC = -1.13 and -1.75, respectively).
FIGURE 9.
Metabolites in the histidine degradation pathway in A. baumannii significantly impacted by polymyxin B, mitotane, and the combination. Red boxes indicate statistically significant metabolites (Mean ± SD; ∗one-way ANOVA, FDR ≤ 0.05; Fisher’s LSD, p ≤ 0.05; ∗log2 fold-change ≥|1|).
Discussion
To improve the efficacy of polymyxins and prevent the emergence of resistance, novel polymyxin combination therapy against MDR A. baumannii is urgently needed (Pantopoulou et al., 2007; Pankuch et al., 2008, 2010; Lee et al., 2013; Ni et al., 2015). The combination of polymyxin B and the antineoplastic drug mitotane has been shown to produce synergistic bacterial killing and prevent regrowth in MDR A. baumannii (Tran et al., 2018). Mitotane is currently used for the treatment of adrenocortical carcinoma and Cushing’s syndrome where the mode of action is likely due to inhibition of sterol-O-acyl-transferase and inducing endoplasmic reticulum (ER) stress (Sbiera et al., 2015). However, the antibacterial mechanism of mitotane is unknown and has not been examined. Understanding the mechanism of the synergistic killing against A. baumannii by the combination of polymyxins and mitotane is essential for future repurposing of mitotane as an antimicrobial agent. This study is the first to investigate the potential antibacterial mechanism of this combination against A. baumannii using metabolomics. To ensure the observed responses were due to antibiotic treatments and not from extensive bacterial killing, a higher inoculum was employed for this metabolomics investigation.
It is well established that polymyxins exert their antimicrobial activity mainly through the disruption of the bacterial outer membrane (Hancock and Chapple, 1999; Hancock, 2001). Consequently, it was not unexpected that polymyxin B monotherapy impacted the membrane lipids of polymyxin-susceptible A. baumannii in the present study (Figures 4, 5). Similar to a previous metabolomics study (Maifiah et al., 2017), pathway analysis revealed the majority of the significantly perturbed metabolites caused by polymyxin B monotherapy were in fatty acid and GPL metabolism (Figures 4, 5). Our findings were also in agreement with a previous transcriptomic study that showed A. baumannii altered the expression of genes that are primarily associated with outer membrane biogenesis, fatty acid metabolism and phospholipid trafficking after 1-h exposure to colistin (Henry et al., 2015). Promisingly, the combination caused substantial reduction of sn-glycero-3-phosphoethanolamine in both polymyxin-susceptible and -resistant strains, while polymyxin B monotherapy only caused reduction of sn-glycero-3-phosphoethanolamine in the polymyxin-susceptible strain. This finding suggested that perturbation of sn-glycero-3-phosphoethanolamine in the polymyxin-resistant strains may be a synergistic killing mechanism of the combination.
In addition to the effect on the membrane lipids, pathway analysis also suggested that polymyxin B might affect the bacterial stress response through the degradation of L-histidine to L-glutamate. Since L-glutamate is an important metabolite involved in a wide range of bacterial metabolic processes including responses to acids and other stresses (Feehily and Karatzas, 2013), a reduced level of L-glutamate can stifle the stress response and result in cell death.
Remarkably, despite being a non-antibiotic, mitotane caused significant metabolic perturbation in the polymyxin-resistant strain lacking LPS (FADDI-AB065) (Figures 1, 2). LPS is a key component of the outer membrane, a permeability barrier in Gram-negative bacteria (Nikaido, 2003); hence, the loss of LPS likely enables hydrophobic mitotane to cross the outer membrane and access its intracellular target(s). Surprisingly, pathway analysis showed that mitotane monotherapy also affected GPL metabolism. It is, however, unclear if this effect is connected to mitotane role in steroidogenesis inhibition in ACC cells. Mitotane also affected the histidine degradation pathway in the LPS-deficient strain. Unlike that observed with polymyxin B, the levels of both urocanate and N-formimino-L-glutamate of the histidine degradation pathway were significantly reduced by mitotane monotherapy (Figure 9). It is possible that mitotane upregulated the histidine degradation pathway to produce additional essential L-glutamate for stress response (Feehily and Karatzas, 2013).
Apart from its potential impact on membrane structure and the bacterial stress response, mitotane monotherapy also affected pentose phosphate pathway in FADDI-AB065 (Figure 6). Pentose phosphate pathway is responsible for the production of NADPH during the oxidative phase and ribose during the non-oxidative phase, which are essential products for anabolic reactions and DNA/RNA synthesis, respectively (Wamelink et al., 2008; Barcia-Vieitez and Ramos-Martinez, 2014). As the metabolites affected by mitotane in FADDI-AB065 are involved in the non-oxidative phase of pentose phosphate pathway, it is likely that mitotane also affects DNA/RNA synthesis in A. baumannii.
In combination, our findings showed polymyxin B and mitotane additionally affected citric acid cycle and nucleotide metabolism. Given the significant reduction of the majority of the metabolites involved in citric acid cycle, it is highly likely that the combination compromised energy production in A. baumannii. In relation to nucleotide metabolism, our findings were in line with the proposed effect of mitotane on RNA/DNA synthesis through perturbation of pentose phosphate pathway.
For antibiotic combination therapies, several models have been proposed to describe the mechanism of synergism, notably subpopulation synergy and mechanistic synergy (Bulitta et al., 2009; Landersdorfer et al., 2013). Subpopulation synergy refers to the killing of the resistant subpopulations of one drug by the second drug, while mechanistic synergy refers to the targeting of different cellular pathways of each drug (Landersdorfer et al., 2013). From the present study, it is likely that the synergism of polymyxin B and mitotane is a result of both mechanisms. Since FADDI-AB065 is a polymyxin-resistant derivative of ATCC 19606 following colistin monotherapy (Moffatt et al., 2010), FADDI-AB065 might represent a polymyxin-resistant subpopulation of ATCC 19606. Consequently, the susceptibility of FADDI-AB065 to mitotane monotherapy supports the subpopulation synergy model. Mechanistic synergy is suggested since much higher metabolic perturbation was observed with the combination therapy compared to polymyxin B and mitotane monotherapies. Additionally, given the lack of activity of mitotane alone compared to the enhanced activity in combination with polymyxin B, it is likely that bioavailability synergy, which refers to the increased intracellular availability of one drug due to the action of a second drug (Cokol et al., 2011), contributed to the enhanced activity observed.
The current findings suggest that non-antibiotic drugs that affect DNA synthesis, protein synthesis, and lipid synthesis may potentially be used in combination with a polymyxin in order to enhance bacterial killing.
Conclusion
The present study is the first to investigate the synergistic killing mechanism of polymyxin B and mitotane in combination against A. baumannii. In addition to effects on lipid metabolism pathways identified in our previous metabolomic studies with colistin, the histidine degradation pathway has been shown to be impacted by polymyxin B monotherapy. As monotherapy, mitotane impacted lipid metabolism, histidine degradation and pentose phosphate pathway in a LPS-deficient polymyxin-resistant A. baumannii strain. Citric acid cycle and nucleotide metabolism were impacted by the combination in all strains. The novel finding from this study is that polymyxin B treatment per se causes significant perturbations in cellular lipids and amino acid metabolism, specifically histidine degradation, all of which were further enhanced by mitotane leading ultimately to the depletion of nucleotides. This study provides valuable mechanistic insights into the synergistic antibacterial killing of polymyxin and mitotane combinations against MDR A. baumannii, and is important for the potential repositioning of mitotane for an antimicrobial indication in combination with polymyxins.
Data Availability
The raw data supporting the conclusion of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
TT carried out the main experiments, data analysis, and wrote the manuscript draft. PB participated in experimental design. DC and TV participated in data analysis. JL designed the project and guided all experimental designs and data analysis. All authors participated in manuscript revision. All authors read and approved the final manuscript.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JL is an Australian National Health and Medical Research Council (NHMRC) Senior Research Fellow. TV is an Australian NHMRC Career Development Research Fellow. TT was supported by MBio Postgraduate Discovery Scholarship (MPDS), Monash University.
Funding. This study was supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI111965, JL, TV, and DC).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2018.00359/full#supplementary-material
References
- Adams M. D., Nickel G. C., Bajaksouzian S., Lavender H., Murthy A. R., Jacobs M. R., et al. (2009). Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53 3628–3634. 10.1128/AAC.00284-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo L. A., Garcia-Curiel A., Pachon-Ibanez M. E., Llanos A. C., Ruiz M., Pachon J., et al. (2005). Reliability of the E-test method for detection of colistin resistance in clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43 903–905. 10.1128/JCM.43.2.903-905.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo L. A., Herrera C. M., Fernandez L., Hankins J. V., Trent M. S., Hancock R. E. (2011). The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 55 3743–3751. 10.1128/AAC.00256-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcia-Vieitez R., Ramos-Martinez J. I. (2014). The regulation of the oxidative phase of the pentose phosphate pathway: new answers to old problems. IUBMB Life 66 775–779. 10.1002/iub.1329 [DOI] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Benjamin DK, Jr, Bradley J., Guidos R. J., Jones R. N., et al. (2013). 10 x ′20 Progress–development of new drugs active against gram-negative bacilli: an update from the infectious diseases society of America. Clin. Infect. Dis. 56 1685–1694. 10.1093/cid/cit152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Bulitta J. B., Li J., Poudyal A., Yu H. H., Owen R. J., Tsuji B. T., et al. (2009). “Quantifying synergy of colistin combinations against MDR Gram-negatives by mechanism-based models (abstract A1-573, p41),” in Proceedings of the Abstracts of the 49th Annual Interscience Conference of Antimicrobial Agents and Chemotherapy (ICAAC) (San Francisco, CA: American Society for Microbiology; ). [Google Scholar]
- Centers for Disease Control and Prevention [CDC] (2013). Antibiotic/Antimicrobial Resistance: Antibiotic Resistance Threats in the United States. Atlanta: National Center for Emerging and Zoonotic Infectious Diseases, CDC. [Google Scholar]
- Chen C., Gonzalez F. J., Idle J. R. (2007). LC-MS-based metabolomics in drug metabolism. Drug Metab. Rev. 39 581–597. 10.1080/03602530701497804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M., Chua H. N., Tasan M., Mutlu B., Weinstein Z. B., Suzuki Y., et al. (2011). Systematic exploration of synergistic drug pairs. Mol. Syst. Biol. 7:544. 10.1038/msb.2011.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek D. J., Jankevics A., Burgess K. E., Breitling R., Barrett M. P. (2012). IDEOM: an Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics 28 1048–1049. 10.1093/bioinformatics/bts069 [DOI] [PubMed] [Google Scholar]
- Dudhani R. V., Turnidge J. D., Nation R. L., Li J. (2010). fAUC/MIC is the most predictive pharmacokinetic/pharmacodynamic index of colistin against Acinetobacter baumannii in murine thigh and lung infection models. J. Antimicrob. Chemother. 65 1984–1990. 10.1093/jac/dkq226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejim L., Farha M. A., Falconer S. B., Wildenhain J., Coombes B. K., Tyers M., et al. (2011). Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 7 348–350. 10.1038/nchembio.559 [DOI] [PubMed] [Google Scholar]
- Feehily C., Karatzas K. A. (2013). Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 114 11–24. 10.1111/j.1365-2672.2012.05434.x [DOI] [PubMed] [Google Scholar]
- Garonzik S. M., Li J., Thamlikitkul V., Paterson D. L., Shoham S., Jacob J., et al. (2011). Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically-ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob. Agents Chemother. 55 3284–3294. 10.1128/AAC.01733-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gika H. G., Theodoridis G. A., Wingate J. E., Wilson I. D. (2007). Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine. J. Proteome Res. 6 3291–3303. 10.1021/pr070183p [DOI] [PubMed] [Google Scholar]
- Hancock R. E. (2001). Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 1 156–164. 10.1016/S1473-3099(01)00092-5 [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Chapple D. S. (1999). Peptide antibiotics. Antimicrob. Agents Chemother. 43 1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R., Crane B., Powell D., Deveson Lucas D., Li Z., Aranda J., et al. (2015). The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J. Antimicrob. Chemother. 70 1303–1313. 10.1093/jac/dku536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsen I. G., Fassnacht M., Terzolo M., Houterman S., Den Hartigh J., Leboulleux S., et al. (2011). Plasma concentrations of o,p’DDD, o,p’DDA, and o,p’DDE as predictors of tumor response to mitotane in adrenocortical carcinoma: results of a retrospective ENS@T multicenter study. J. Clin. Endocrinol. Metab. 96 1844–1851. 10.1210/jc.2010-2676 [DOI] [PubMed] [Google Scholar]
- Hussein M. H., Schneider E. K., Elliott A. G., Han M., Reyes-Ortega F., Morris F., et al. (2016). From breast cancer to antimicrobial: combating extremely resistant gram-negative “Superbugs” using novel combinations of polymyxin B with selective estrogen receptor modulators. Microb. Drug Resist. 23 640–650. 10.1089/mdr.2016.0196 [DOI] [PubMed] [Google Scholar]
- Jia J., Zhu F., Ma X., Cao Z., Cao Z. W., Li Y., et al. (2009). Mechanisms of drug combinations: interaction and network perspectives. Nat. Rev. Drug Discov. 8 111–128. 10.1038/nrd2683 [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R., Weinshilboum R. M. Pharmacometabolomics Research Network (2014). Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clin. Pharmacol. Ther. 95 154–167. 10.1038/clpt.2013.217 [DOI] [PubMed] [Google Scholar]
- Kirwan J. A., Weber R. J., Broadhurst D. I., Viant M. R. (2014). Direct infusion mass spectrometry metabolomics dataset: a benchmark for data processing and quality control. Sci. Data 1:140012. 10.1038/sdata.2014.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli E. (2015). Mitotane revisited: a new target for an old drug. Endocrinology 156 3873–3875. 10.1210/en.2015-1796 [DOI] [PubMed] [Google Scholar]
- Landersdorfer C. B., Ly N. S., Xu H., Tsuji B. T., Bulitta J. B. (2013). Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob. Agents Chemother. 57 2343–2351. 10.1128/AAC.00092-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Bergen P. J., Bulitta J. B., Tsuji B., Forrest A., Nation R. L., et al. (2013). Synergistic activity of colistin and rifampin combination against multidrug-resistant Acinetobacter baumannii in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 57 3738–3745. 10.1128/AAC.00703-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nation R. L., Turnidge J. D., Milne R. W., Coulthard K., Rayner C. R., et al. (2006a). Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6 589–601. 10.1016/S1473-3099(06)70580-1 [DOI] [PubMed] [Google Scholar]
- Li J., Rayner C. R., Nation R. L., Owen R. J., Spelman D., Tan K. E., et al. (2006b). Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50 2946–2950. 10.1128/AAC.00103-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Dai Y. (2011). Principal component analysis based methods in bioinformatics studies. Brief. Bioinform. 12 714–722. 10.1093/bib/bbq090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Maifiah M. H., Creek D. J., Nation R. L., Forrest A., Tsuji B. T., Velkov T., et al. (2017). Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against Acinetobacter baumannii. Sci. Rep. 7:45527. 10.1038/srep45527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo A., Armitage E. G., Garcia A., Barbas C. (2014). Metabolomics as a tool for drug discovery and personalised medicine, A review. Curr. Top. Med. Chem. 14 2627–2636. 10.2174/1568026614666141215124956 [DOI] [PubMed] [Google Scholar]
- Michalopoulos A., Falagas M. E. (2010). Treatment of Acinetobacter infections. Expert Opin. Pharmacother. 11 779–788. 10.1517/14656561003596350 [DOI] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Harrison P., Hale J. D., Vinogradov E., Seemann T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54 4971–4977. 10.1128/AAC.00834-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nation R. L., Li J., Cars O., Couet W., Dudley M. N., Kaye K. S., et al. (2015). Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect. Dis. 15 225–234. 10.1016/S1473-3099(14)70850-3 [DOI] [PubMed] [Google Scholar]
- Ni W., Shao X., Di X., Cui J., Wang R., Liu Y. (2015). In vitro synergy of polymyxins with other antibiotics for Acinetobacter baumannii: a systematic review and meta-analysis. Int. J. Antimicrob. Agents 45 8–18. 10.1016/j.ijantimicag.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Nikaido H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67 593–656. 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R. J., Li J., Nation R. L., Spelman D. (2007). In vitro pharmacodynamics of colistin against Acinetobacter baumannii clinical isolates. J. Antimicrob. Chemother. 59 473–477. 10.1093/jac/dkl512 [DOI] [PubMed] [Google Scholar]
- Pankuch G. A., Lin G., Seifert H., Appelbaum P. C. (2008). Activity of meropenem with and without ciprofloxacin and colistin against Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother. 52 333–336. 10.1128/AAC.00689-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankuch G. A., Seifert H., Appelbaum P. C. (2010). Activity of doripenem with and without levofloxacin, amikacin, and colistin against Pseudomonas aeruginosa and Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 67 191–197. 10.1016/j.diagmicrobio.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Pantopoulou A., Giamarellos-Bourboulis E. J., Raftogannis M., Tsaganos T., Dontas I., Koutoukas P., et al. (2007). Colistin offers prolonged survival in experimental infection by multidrug-resistant Acinetobacter baumannii: the significance of co-administration of rifampicin. Int. J. Antimicrob. Agents 29 51–55. 10.1016/j.ijantimicag.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21 538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M. R., Casella L. G., Jones J. W., Adams M. D., Zurawski D. V., Hazlett K. R., et al. (2013). Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob. Agents Chemother. 57 4831–4840. 10.1128/AAC.00865-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percin D., Akyol S., Kalin G. (2014). In vitro synergism of combinations of colistin with selected antibiotics against colistin-resistant Acinetobacter baumannii. GMS Hyg. Infect. Control 9:Doc14. 10.3205/dgkh000234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri A. M., Landersdorfer C. B., Jacob J., Boniatti M. M., Dalarosa M. G., Falci D. R., et al. (2013). Population pharmacokinetics of intravenous polymyxin B in critically-ill patients: implications for selection of dosage regimens. Clin. Infect. Dis. 57 524–531. 10.1093/cid/cit334 [DOI] [PubMed] [Google Scholar]
- Sbiera S., Leich E., Liebisch G., Sbiera I., Schirbel A., Wiemer L., et al. (2015). Mitotane inhibits sterol-O-Acyl transferase 1 triggering lipid-mediated endoplasmic reticulum stress and apoptosis in adrenocortical carcinoma cells. Endocrinology 156 3895–3908. 10.1210/en.2015-1367 [DOI] [PubMed] [Google Scholar]
- Scheltema R. A., Jankevics A., Jansen R. C., Swertz M. A., Breitling R. (2011). PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 83 2786–2793. 10.1021/ac2000994 [DOI] [PubMed] [Google Scholar]
- Schneider E. K., Azad M. A., Han M. L., Tony Zhou Q., Wang J., Huang J. X., et al. (2016). An “Unlikely” pair: the antimicrobial synergy of polymyxin B in combination with the cystic fibrosis transmembrane conductance regulator drugs KALYDECO and ORKAMBI. ACS Infect. Dis. 2 478–488. 10.1021/acsinfecdis.6b00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E. K., Reyes-Ortega F., Velkov T., Li J. (2017). Antibiotic-non-antibiotic combinations for combating extremely drug-resistant Gram-negative ’superbugs’. Essays Biochem. 61 115–125. 10.1042/EBC20160058 [DOI] [PubMed] [Google Scholar]
- Smith C. A., Want E. J., O’maille G., Abagyan R., Siuzdak G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using Nonlinear peak alignment, matching, and identification. Anal. Chem. 78 779–787. 10.1021/ac051437y [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Magrini N. (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization. [Google Scholar]
- Talbot G. H., Bradley J., Edwards JE, Jr, Gilbert D., Scheld M., Bartlett J. G., et al. (2006). Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the infectious diseases society of America. Clin. Infect. Dis. 42 657–668. 10.1086/499819 [DOI] [PubMed] [Google Scholar]
- Tran T. B., Wang J., Doi Y., Velkov T., Bergen P. J., Li J. (2018). Novel polymyxin combination with antineoplastic mitotane improved the bacterial killing against polymyxin-resistant multidrug-resistant gram-negative pathogens. Front. Microbiol. 9:721 10.3389/fmicb.2018.00721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble M. J., Mlynarcik P., Kolar M., Hancock R. E. (2016). Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 6:a025288. 10.1101/cshperspect.a025288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T., Thompson P. E., Nation R. L., Li J. (2010). Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 53 1898–1916. 10.1021/jm900999h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent I. M., Ehmann D. E., Mills S. D., Perros M., Barrett M. P. (2016). Untargeted metabolomics to ascertain antibiotic modes of action. Antimicrob. Agents Chemother. 60 2281–2291. 10.1128/AAC.02109-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamelink M. M., Struys E. A., Jakobs C. (2008). The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: a review. J. Inherit. Metab. Dis. 31 703–717. 10.1007/s10545-008-1015-6 [DOI] [PubMed] [Google Scholar]
- WHO/EMP/IAU (2017). Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline, Including Tuberculosis. Geneva: World Health Organization. [Google Scholar]
- Zhang T., Creek D. J., Barrett M. P., Blackburn G., Watson D. G. (2012). Evaluation of coupling reversed phase, aqueous normal phase, and hydrophilic interaction liquid chromatography with Orbitrap mass spectrometry for metabolomic studies of human urine. Anal. Chem. 84 1994–2001. 10.1021/ac2030738 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusion of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.