Abstract
Cytochrome P2C (CYP2C) subfamily members (CYP2C8,CYP2C9,CYP2C18, and CYP2C19) are known to participate in clinical drug metabolism. However, the association between CYP2C subfamily members and hepatocellular carcinoma (HCC) remains unclear. This study investigated the prognostic value of CYP2C subfamily gene expression levels with HCC prognosis. Data of 360 HCC patients in The Cancer Genome Atlas database and 231 in the Gene Expression Omnibus database were analyzed. Kaplan–Meier analysis and a Cox regression model were used to ascertain overall survival and recurrence‐free survival, and to calculate median survival time using hazard ratios (HR) and 95% confidence intervals (CI). In TCGA database, low expression of CYP2C8,CYP2C9, and CYP2C19 in tumor tissue was associated with a short median survival time (all crude P = 0.001, adjusted P = 0.004, P = 0.047, and P = 0.020, respectively). In TCGA database, joint effects analysis of the combinations of CYP2C8 and CYP2C9,CYP2C8 and CYP2C19, and CYP2C9 and CYP2C19 revealed that high expression of two genes (group 4; group IV, group d) was associated with a reduced risk of death as compared to low expression (group 1, group I, and group a) (adjusted P = 0.005, P = 0.013, and P = 0.016, respectively). In TCGA database, joint effects analysis of CYP2C8,CYP2C9, and CYP2C19 showed that the risk of death from HCC was lower for groups C and D than for group A (adjusted P = 0.012 and P = 0.008, respectively). CYP2C8,CYP2C9, and CYP2C19 gene expression levels are potential prognostic markers of HCC following hepatectomy.
Keywords: CYP2C subfamily, gene expression, hepatocellular carcinoma, prognosis, serum biomarker
Introduction
Liver cancer is the fifth most commonly diagnosed cancer and the second most frequent cause of cancer‐related deaths in men and the seventh most frequently diagnosed cancer and the sixth leading cause of cancer‐related deaths in women worldwide 1. There were about 4,292,000 newly diagnosed cases and 2,814,000 deaths from cancer in China in 2015 2. Hepatocellular carcinoma (HCC), the major histological type, accounts for most (70–85%) cases of primary liver cancer worldwide 3. Etiologically, infection of hepatitis C or B virus (HBV), aflatoxin exposure, obesity, diabetes, nonalcoholic steatohepatitis, alcohol ingestion, hemochromatosis, and other metabolic diseases are the primary risk factors for HCC 4. Despite advances in several treatment strategies, such as liver resection, liver transplantation, percutaneous ethanol injection, transcatheter arterial chemoembolization, transarterial radiation, microwave ablation, and systemic therapy, the prognosis of HCC remains unsatisfactory because of late‐stage diagnosis 5, which has resulted in a reported 5‐year survival rate of only 7% 6. Thus, the identification of molecular biomarkers for the early diagnosis of HCC is crucial to provide more effective therapies and improve patient prognosis.
Cytochrome P2 (CYP2) family members of the CYP superfamily include many subfamilies, such as CYP2A, CYP2B, CYP2C, CYP2D, CYP2E, and CYP2F. The human CYP2C subfamily consists of four members (CYP2C8, CYP2C9, CYP2C18, and CYP2C19) that are localized in a single gene locus on chromosome 10 7, 8. Members of the CYP2C subfamily are known to be involved in the metabolism of roughly 20% of clinically used drugs, such as the anticancer drug paclitaxel 9, the antidiabetic agent tolbutamide 8, proton pump inhibitors 10, as well as various endogenous and exogenous substances 11. In addition, CYP2C8 is reportedly related with an increased risk of essential hypertension and coronary artery disease in Bulgarians 12 and has also been associated with anemia 13, breast cancer 14, and vascular inflammatory diseases 15. Moreover, CYP2C9 is reportedly associated with the risk of colorectal cancer 16, while CYP2C18 was found to have no contribution to cancer risk 11 and CYP2C19 has been associated with peptic ulcer disease 17, colorectal adenoma recurrence 18, breast cancer 19, and cardiovascular diseases 20. However, little is known about the associations of the expression levels of these four genes with the risk of HCC. Thus, the aim of this study was to identify relationships between CYP2C expression levels and HCC prognosis.
Material and Methods
Patient data
First, the Metabolic gEne RApid Visualizer database (http://merav.wi.mit.edu/) was accessed on September 10, 2017 to determine whether any of the four members of the CYP2C subfamily are differentially expressed between normal liver tissues and primary liver tumors. Then, the GTExPortal (https://gtexportal.org/home/) was accessed on September 10, 2017 to obtain gene expression levels of CYP2C subfamily in different tissues 21. Moreover, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database was accessed on September 10, 2017 to construct protein–protein interaction networks between CYP2C subfamily members and other proteins.
The OncoLnc (http://www.oncolnc.org/) and The Cancer Genome Atlas (TCGA), (http://tcga-data.nci.nih.gov/tcga) databases were accessed on September 10, 2017 to acquire data regarding the gene expression levels of CYP2C8, CYP2C9, CYP2C18, and CYP2C19, as well as the corresponding 50% cutoff values. The results presented here, in part, are based on TCGA studies 22. Data of 360 HCC patients, including sex, race, age, body mass index (BMI), tumor, node, metastasis (TNM) stage, survival time, and survival status, were collected. Gene expression data were downloaded from the GSE14520 dataset of the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520) on September 12, 2017 23. The GSE14520 dataset included gene expression levels originated from [HT_HG–U133A] Affymetrix HT Human Genome U133A 23 and [HT_HG–U133A_2] Affymetrix HT Human Genome U133A_2.0 24 arrays. In order to prevent batch effects, the former array of 231 HCC patients (more patients than the latter, 445 samples) was chosen.
Functional enrichment analysis of the CYP2C subfamily
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) v.6.7 (https://david-d.ncifcrf.gov/) was accessed on September 15, 2017 25, 26 for enrichment analysis, gene ontology (GO) functional analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. GO analysis is composed of terms of biological processes (BP), cellular components (CC), and molecular functions (MF); in the latter, KEGG pathways were drawn between CYP2C and other subfamilies.
Survival analysis
From the TCGA database, 360 HCC patients were divided into two groups of 180 patients each at a 50% cutoff value. The median survival time (MST) was applied to estimate patient prognosis and TNM stage in a Cox regression model adjusted for patient age and sex. In order to assure a rational comparison between the above two databases, the 50% cutoff was used for the GEO database. In the GEO database, overall survival (OS) and recurrence‐free survival (RFS) were applied to evaluate patient prognosis. In addition, the Cox regression model was adjusted for age, sex, alanine aminotransferase level, nodal status, HBV status, primary tumor size, alpha‐fetoprotein (AFP) level, cirrhosis status, and Barcelona Clinic Liver Cancer (BCLC) stage.
Joint effects analysis of CYP2C8, CYP2C9, and CYP2C19
In the TCGA database, only CYP2C8, CYP2C9, and CYP2C19 were statistically significant. Joint effects analysis was conducted with the following combinations: (1) CYP2C8 and CYP2C9; (2) CYP2C8 and CYP2C19; (3) CYP2C9 and CYP2C19; and (4) CYP2C8, CYP2C9, and CYP2C19.
Combinations of CYP2C8 and CYP2C9 were composed of four groups: group 1 (low CYP2C8 and low CYP2C9 expression), group 2 (low CYP2C8 and high CYP2C9 expression), group 3 (high CYP2C8 and low CYP2C9 expression), and group 4 (high CYP2C8 and high CYP2C9 expression).
Combinations of CYP2C8 and CYP2C19 were composed of four groups: group I (low CYP2C8 and low CYP2C19 expression), group II (low CYP2C8 and high CYP2C19 expression), group III (high CYP2C8 and low CYP2C19 expression), and group IV (high CYP2C8 and high CYP2C19 expression).
Combinations of CYP2C9 and CYP2C19 were composed of four groups: group a (low CYP2C9 and low CYP2C19 expression), group b (low CYP2C9 and high CYP2C19 expression), group c (high CYP2C9 and low CYP2C19 expressions), and group d (high CYP2C9 and high CYP2C19 expression).
Combinations of CYP2C8, CYP2C9, and CYP2C19 were composed of four groups: group A (low CYP2C8, low CYP2C9, and low CYP2C19 expression); group B (high CYP2C8, low CYP2C9, and low CYP2C19 expression; low CYP2C8, high CYP2C9, and low CYP2C19 expression; and low CYP2C8, low CYP2C9, and high CYP2C19 expression); group C (high CYP2C8, high CYP2C9, and low CYP2C19 expression; high CYP2C8, low CYP2C9, and high CYP2C19 expression; and low CYP2C8, high CYP2C9, and high CYP2C19 expression); and group D (high CYP2C8, high CYP2C9, and high CYP2C19 expression). The Cox regression model was adjusted for TNM stage, age, and sex in keeping with the above combinations.
Statistical analysis
The Pearson correlation coefficient was used to assess correlations among the CYP2C8, CYP2C9, CYP2C18, and CYP2C19 genes. Correlation plots were depicted by R v.3.2.0 (https://www.r-project.org/). Interactions among these four genes and others as well as the four proteins encoded by these with others were drawn with the Cytoscape v.3.5.1 open source software platform for visualizing complex networks (http://www.cytoscape.org/). MST and probability (P) values were calculated by Kaplan–Meier survival analysis and the log‐rank test. Univariate and multivariate survival analysis were performed using the Cox hazards regression model. Scatter diagrams and survival curves were constructed using GraphPad Prism v.7 software (GraphPad Software, Inc., La Jolla, CA). All statistical analyses were performed using SPSS v.16 software (SPSS, Inc., Chicago, IL, USA). A P < 0.05 was considered statistically significant.
Results
Basic patient data
Detailed characteristics of the 360 patients in the TCGA database are shown in Table 1. TNM stage was significantly associated with MST (P < 0.001), but not sex, age, BMI, or race (all P > 0.05).
Table 1.
Basic characteristics of 360 HCC patients
| Variables | Patients (n = 360) | No. of events (%) | MST (days) | HR (95% CI) | Log‐rank P value |
|---|---|---|---|---|---|
| Race | |||||
| Asian | 155 | 44 (28.4%) | NA | Ref. | 0.185 |
| White + others | 196 | 78 (39.8%) | 1397 | 1.29 (0.89–1.87) | |
| MissingĐ | 9 | ||||
| Sex | |||||
| Male | 244 | 78 (32.0%) | 2486 | Ref. | 0.309 |
| Female | 116 | 48 (41.4%) | 1560 | 1.21 (0.84–1.73) | |
| Age(year) | |||||
| <60 | 168 | 54 (32.1%) | 2532 | Ref. | 0.363 |
| ≥60 | 189 | 70 (37.0%) | 1685 | 1.18 (0.83–1.68) | |
| Missing† | 3 | ||||
| BMI | |||||
| ≤25 | 193 | 66 (34.2%) | 2456 | Ref. | 0.478 |
| >25 | 137 | 45 (32.8%) | 2116 | 0.87 (0.60–1.27) | |
| Missingý | 30 | ||||
| TNM stage | |||||
| A + B | 252 | 66 (26.2%) | 2532 | Ref. | <0.001 |
| C + D | 87 | 48 (55.2%) | 770 | 2.50 (1.72–3.63) | |
| MissingĹ | 21 | ||||
BMI, body mass index; TNM stage, tumor, node and metastasis stage; MST, median survival time; HR, hazard ratio; 95% CI, 95% confidence interval; Ref, reference; MissingĐ, information of race was unavailable in 9 patients; Missing†, information of age was unavailable in 3 patients; Missingý, information of BMI was unavailable in 30 patients; MissingĹ, information of TNM stage was unavailable in 21 patients. The significance is that all the values are statistically significant.
The data of the 231 patients from the GEO database are presented in Table 2. Sex, nodal status, primary tumor size, BCLC stage, cirrhosis status, and AFP level were related to OS (all P = 0.048, 0.003, <0.001, <0.001, 0.004, and 0.001, respectively), while sex, cirrhosis status, primary tumor size, and BCLC stage were related to RFS (P = 0.001, 0.019, 0.020, and <0.001, respectively).
Table 2.
Basic characteristics of 231 HCC patients
| Variables | Patients (n = 231) | Overall survival | Recurrence‐free survival | ||||
|---|---|---|---|---|---|---|---|
| MST (months) | HR (95% CI) | Log‐rank P | MST (months) | HR (95% CI) | Log‐rank P | ||
| Sex | |||||||
| Male | 191 | NA | Ref. | 0.048 | 40 | Ref. | 0.001 |
| Female | 30 | NA | 0.59 (0.34–1.00) | NA | 0.47 (0.29–0.75) | ||
| MissingƷ | 10 | ||||||
| Age | |||||||
| ≤60 | 181 | NA | Ref. | 0.852 | 46 | Ref. | 0.937 |
| >60 | 40 | NA | 0.96 (0.65–1.44) | 37 | 1.01 (0.73–1.41) | ||
| MissingƷ | 10 | ||||||
| HBV–virus status | |||||||
| AVR–CC | 56 | NA | Ref. | 0.149 | 30 | Ref. | 0.092 |
| CC + NO | 162 | NA | 0.78 (0.56–1.09) | 48 | 0.78 (0.59–1.04) | ||
| Missingƛ | 13 | ||||||
| ALT | |||||||
| ≤50 U/L | 130 | NA | Ref. | 0.710 | 53 | Ref. | 0.090 |
| >50 U/L | 91 | NA | 1.06 (0.78–1.44) | 40 | 1.25 (0.97–1.61) | ||
| MissingƷ | 10 | ||||||
| Main tumor size | |||||||
| ≤5 cm | 140 | NA | Ref. | <0.001 | 51 | Ref. | 0.020 |
| >5 cm | 80 | 53 | 1.87 (1.38–2.55) | 30 | 1.37 (1.05–1.78) | ||
| Missingƥ | 11 | ||||||
| Multinodular | |||||||
| Yes | 45 | 48 | Ref. | 0.003 | 27 | Ref. | 0.136 |
| No | 176 | NA | 0.59 (0.42–0.84) | 49 | 0.79 (0.58–1.08) | ||
| MissingƷ | 10 | ||||||
| Cirrhosis | |||||||
| Yes | 203 | NA | Ref. | 0.004 | 38 | Ref. | 0.019 |
| No | 18 | NA | 0.23 (0.09–0.63) | NA | 0.50 (0.28–0.89) | ||
| MissingƷ | 10 | ||||||
| BCLC stage | |||||||
| 0+A | 168 | NA | Ref. | <0.001 | 58 | Ref. | <0.001 |
| B+C | 51 | 20 | 3.63 (2.64–5.00) | 18 | 2.84 (2.14–3.75) | ||
| MissingƜ | 12 | ||||||
| AFP | |||||||
| ≤300 ng/ml | 100 | NA | Ref. | 0.001 | 49 | Ref. | 0.094 |
| >300 ng/ml | 118 | NA | 1.67 (1.23–2.27) | 31 | 1.24 (0.96–1.61) | ||
| Missingƛ | 13 | ||||||
AVR–CC, active viral replication chronic carrier; CC, chronic carrier; ALT, alanine aminotransferase; AFP, alpha fetoprotein; BCLC stage, Barcelona Clinic Liver Cancer; MissingƷ, information of sex, age, ALT, multinodular, cirrhosis was unavailable in 10 patients; Missingƥ, information of main tumor size was unavailable in 11 patients; MissingƜ, information of BCLC stage was unavailable in 12 patients; Missingƛ, information of HBV–virus status and AFP was unavailable in 13 patients. The significance is that all the values are statistically significant.
Analysis of CYP2C subfamily gene expression levels in tumor and nontumor tissues
Expression levels of CYP2C8, CYP2C9, CYP2C18, and CYP2C19 in different organs are exhibited in the supplementary material. Box diagrams of the gene expression levels of CYP2C8, CYP2C9, CYP2C18, and CYP2C19 were downloaded from an online website (Fig. 1A–D, respectively). The expression levels of these genes were high in normal liver tissues, but low in primary liver tumors. Scatter diagrams of these four genes from the GEO database showed that all generated statistically significant results between tumor and nontumor tissues (all P < 0.0001, Fig. 1E).
Figure 1.
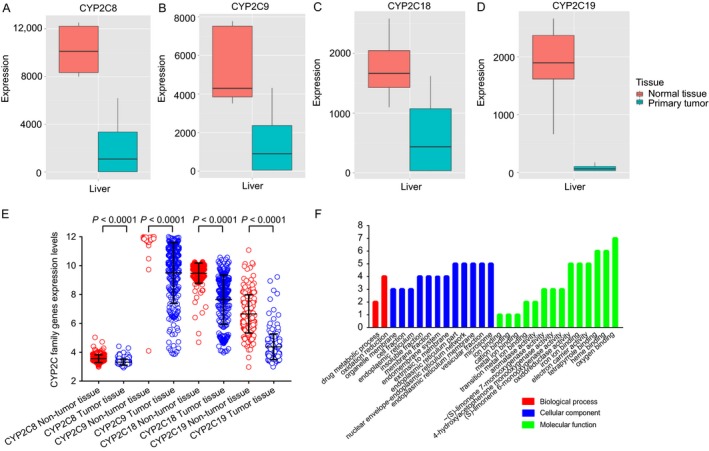
Gene expression levels of CYP2C8 (A), CYP2C9 (B), CYP2C18 (C), and CYP2C19 (D) in normal liver tissue and primary liver tumors. Expression levels in the GEO database (E) and GO analysis (F) of the four genes.
Analysis of the GO and KEGG pathways of the CYP2C subfamily
The biological functions (BP, CC, and MF) of CYP2C8, CYP2C9, CYP2C18, and CYP2C19 were evaluated using GO analysis, which showed that each were involved in drug metabolism and oxidation–reduction reactions. Detailed outcomes are shown in Figure 1F. In the KEGG pathway analysis, DAVID determined associations between CYP2C subfamily members and other genes. Benzo[a]pyrene can be metabolized by CYP2C subfamily members and finally transformed into DNA adducts, including (+)‐trans‐benzo[a]pyrene‐7, 8‐dihydrodiol‐9, and 10‐oxide (BPDE)‐N2‐dG, which are known to induce cancers of the skin, lung, and stomach (Fig. 2).
Figure 2.
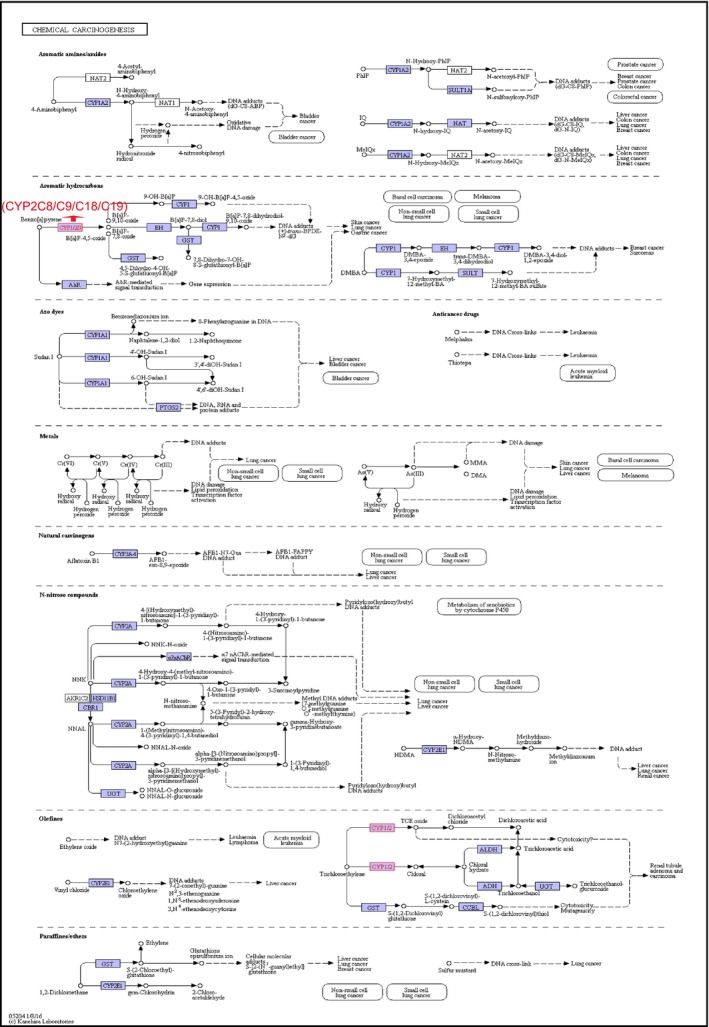
Metabolic pathways of the CYP2C8,CYP2C9,CYP2C18, and CYP2C19 genes in chemical carcinogenesis.
Correlation analysis of the expression levels among CYP2C subfamily members
The Pearson correlation coefficients of the four CYP2C members were calculated. In the TCGA database, each of these four genes was positively and significantly correlated with the other three members (all P < 0.05) (Fig. 3A). In the GEO database, all four genes were positively and statistically significantly correlated with the other three genes as well (all P < 0.05) (Fig. 3B).
Figure 3.
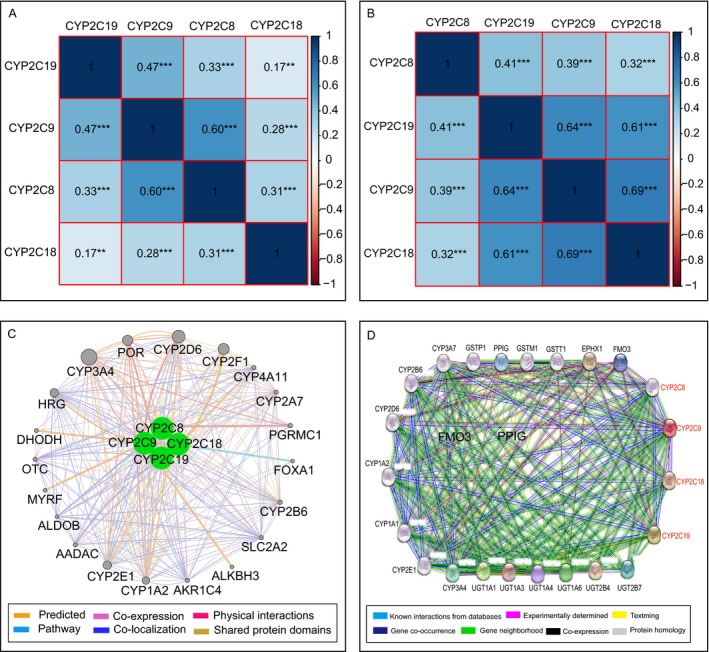
Matrix graphs of Pearson correlations of CYP2C8,CYP2C9,CYP2C18, and CYP2C19 gene expression levels in the TCGA database (A) and GEO database (B). Gene–gene interaction networks among the four genes of interest with other genes (C) and protein–protein interaction networks among the four proteins of interest with other proteins (D).
Analysis of gene–gene interactions between CYP2C subfamily and other genes showed that these four genes were associated with other CYP subfamily members (CYP1A2, CYP2A7, CYP2B6, CYP2D6, CYP2E1, CYP2F1, CYP3A4, and CYP4A11) and other genes (ALDOB, OTC, SLC2A2, PGRMC1, FOXC1, etc.) (Fig. 3C). Moreover, protein–protein interaction networks were constructed using STRING database, which showed that the CYP family member proteins CYP1A1, CYP1A2, CYP2B6, CYP2D6, CYP2E1, CYP3A4, and CYP3A7 were also associated with CYP2C8, CYP2C9, CYP2C18, and CYP2C19 (Fig. 3D).
Survival analysis of CYP2C subfamily members
The prognostic‐related characteristics in the TCGA database of age, TNM stage, and sex were analyzed using a multivariate Cox regression model, which showed that CYP2C8, CYP2C9, and CYP2C19 exhibited significant relationships with MST (adjusted P = 0.004, hazard ratio (HR) = 0.57, 95% confidence interval (CI) = 0.39–0.84; adjusted P = 0.047, HR = 0.67, 95% CI = 0.46–1.00; and adjusted P = 0.020, HR = 0.63, 95% CI = 0.43–0.93, respectively, Table 3). In the GEO database, sex, age, HBV status, alanine aminotransferase level, primary tumor size, nodal status, BCLC stage, AFP level, and cirrhosis status were analyzed using a multivariate Cox regression model, which showed that CYP2C8, CYP2C9, CYP2C18, and CYP2C19 were not statistically associated with OS or RFS (all P > 0.05, Table 4).
Table 3.
Prognostic survival analysis of CYP2C8, CYP2C9, CYP2C18 and CYP2C19 genes in TCGA databases
| Gene | Patients (n = 360) | MST (days) | Crude HR (95% CI) | Crude P value | Adjusted HR (95% CI)a | Adjusted P valuea |
|---|---|---|---|---|---|---|
| CYP2C8 | ||||||
| Low | 180 | 1229 | Ref. | 0.001 | Ref. | 0.004 |
| High | 180 | 2456 | 0.56 (0.39–0.79) | 0.57 (0.39–0.84) | ||
| CYP2C9 | ||||||
| Low | 180 | 1271 | Ref. | 0.001 | Ref. | 0.047 |
| High | 180 | 2456 | 0.56 (0.39–0.80) | 0.67 (0.46–1.00) | ||
| CYP2C18 | ||||||
| Low | 180 | 2456 | Ref. | 0.794 | Ref. | 0.845 |
| High | 180 | 1560 | 0.95(0.67–1.35) | 0.96(0.66–1.40) | ||
| CYP2C19 | ||||||
| Low | 180 | 1229 | Ref. | Ref. | ||
| High | 180 | 2456 | 0.55 (0.38–0.78) | 0.001 | 0.63 (0.43–0.93) | 0.020 |
Adjusted P, adjustment for sex, age, TNM stage; CYP2C8, cytochrome P450 family 2 subfamily C member 8; CYP2C9, cytochrome P450 family 2 subfamily C member 9; CYP2C18, cytochrome P450 family 2 subfamily C member 18; CYP2C19, cytochrome P450 family 2 subfamily C member 19. The significance is that all the values are statistically significant.
Table 4.
Prognostic survival analysis of CYP2C8, CYP2C9, CYP2C18 and CYP2C19 genes in GEO databases
| Gene | Samples (n = 445) | Overall survival | Recurrence‐free survival | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Crude HR (95% CI) | Crude P value | Adjusted HR(95% CI) | Adjusted P value | Crude HR (95% CI) | Crude P value | Adjusted HR (95% CI)a | Adjusted P valuea | ||
| CYP2C8 | |||||||||
| Low | 223 | Ref. | 0.415 | Ref. | 0.721 | Ref. | 0.2 | Ref. | 0.198 |
| High | 222 | 0.88 (0.65–1.20) | 0.94(0.69–1.29) | 0.85(0.66–1.10) | 19 | 0.84(0.65–1.10) | |||
| CYP2C9 | |||||||||
| Low | 223 | Ref. | Ref. | Ref. | Ref. | ||||
| High | 222 | 0.81 (0.59–1.09) | 0.165 | 0.81 (0.60–1.11) | 0.194 | 0.92 (0.71–1.19) | 0.523 | 0.96 (0.75–1.25) | 0.774 |
| CYP2C18 | |||||||||
| Low | 223 | Ref. | 0.502 | Ref. | 0.561 | Ref. | 0.954 | Ref. | 0.945 |
| High | 222 | 0.90 (0.66–1.22) | 0.91 (0.67–1.24) | 0.99 (0.77–1.28) | 1.01 (0.78–1.31) | ||||
| CYP2C19 | |||||||||
| Low | 223 | Ref. | 0.605 | Ref. | 0.460 | Ref. | 0.826 | Ref. | 0.850 |
| High | 222 | 0.92 (0.68–1.25) | 0.89 (0.65–1.21) | 0.97 (0.75–1.25) | 0.98 (0.75–1.26) | ||||
Adjusted P, adjustment of sex, age, HBV–virus status, ALT, main tumor size, multinodular, cirrhosis, AFP and BCLC stage.
As shown by the survival curves of CYP2C8, CYP2C9, CYP2C18, and CYP2C19, based on data retrieved from the TCGA database, which are presented in Figure 4A–D, CYP2C8, CYP2C9, and CYP2C19 were significantly associated with survival (P = 0.001, <0.001, and <0.001, respectively). However, survival curves of these genes, based on data retrieved from the GEO database, as presented in Figure 4A–H, showed that none were significantly associated with OS or RFS (all P > 0.05). In addition, scatter diagrams of the expression levels of these genes, based on data retrieved from both databases, are presented in Figure 4E and F.
Figure 4.
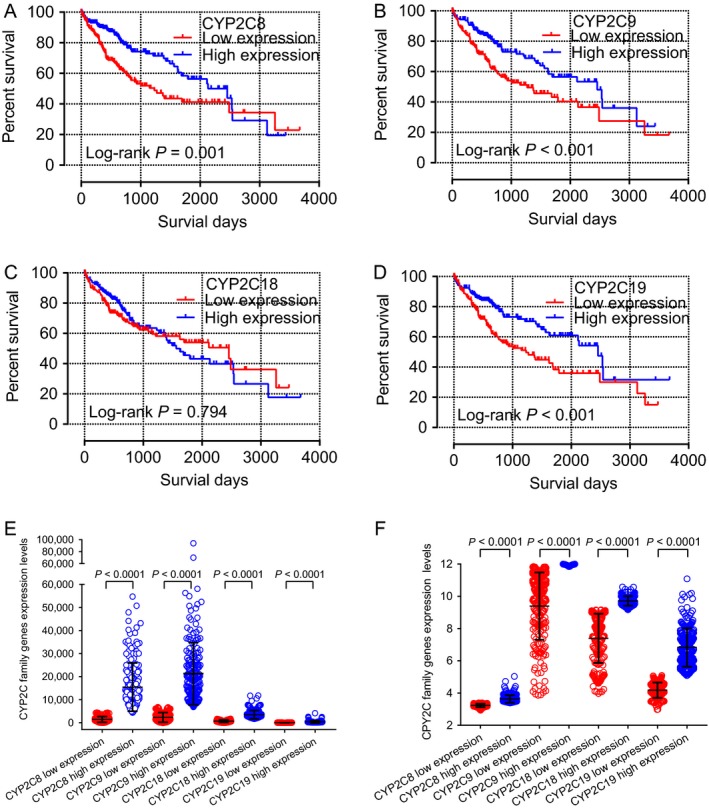
Kaplan–Meier survival curves of the CYP2C8 (A), CYP2C9 (B), CYP2C18 (C), and CYP2C19 (D) genes in the TCGA database. Scatter plots of CYP2C8,CYP2C9,CYP2C18, and CYP2C19 genes expression levels in the TCGA database (E) and GEO database (F).
Joint effects analysis of CYP2C subfamily members
Joint effects analysis of the CYP2C8 and CYP2C9 combination showed that MST was poorest in group 1 (931 days; adjusted P = 0.031) and best in group 4 (2456 days; adjusted P = 0.005). Meanwhile, analysis of the CYP2C8 and CYP2C19 combination showed that MST was poorest in group I (899 days; adjusted P = 0.005) and best in group IV (2456 days; adjusted P = 0.013), and that of the CYP2C9 and CYP2C19 combination showed the poorest MST in group a (1005 days; adjusted b = 0.097) and the best in group d (2456 days; adjusted P = 0.016). Detailed joint effects analysis results are shown in Table 5 and associated survival curves are shown in Figure 5A–C.
Table 5.
Joint effects analysis of the combinations of CYP2C8 and CYP2C9; CYP2C8 and CYP2C19; CYP2C9 and CYP2C19 genes
| Group | CYP2C8 expression | CYP2C9 expression | CYP2C19 expression | Patients (n = 360) | MST (days) | Crude HR (95% CI) | Crude P value | Adjusted HR (95% CI)a | Adjusted P valuea |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Low | Low | 126 | 931 | Ref. | 0.002 | Ref. | 0.031 | |
| 2 | Low | High | 54 | 1694 | 0.61 (0.36–1.04) | 0.071 | 0.82 (0.46–1.44) | 0.483 | |
| 3 | High | Low | 54 | 1791 | 0.60 (0.35–1.03) | 0.064 | 0.61 (0.33–1.10) | 0.102 | |
| 4 | High | High | 126 | 2456 | 0.44 (0.29–0.67) | <0.001 | 0.51 (0.32–0.81) | 0.005 | |
| I | Low | Low | 123 | 899 | Ref. | <0.001 | Ref. | 0.005 | |
| II | Low | High | 57 | NA | 0.52 (0.30–0.90) | 0.020 | 0.80 (0.50–1.29) | 0.356 | |
| III | High | Low | 57 | 1685 | 0.54 (0.32–0.92) | 0.023 | 0.24 (0.10–0.61) | 0.003 | |
| IV | High | High | 123 | 2456 | 0.43 (0.28–0.66) | <0.001 | 0.54 (0.34–0.88) | 0.013 | |
| a | Low | Low | 144 | 1005 | Ref. | 0.003 | Ref. | 0.097 | |
| b | Low | High | 36 | NA | 0.54 (0.27–1.08) | 0.082 | 0.63 (0.31–1.28) | 0.200 | |
| c | High | Low | 36 | 1694 | 0.60 (0.32–1.12) | 0.109 | 0.75 (0.39–1.42) | 0.374 | |
| d | High | High | 144 | 2456 | 0.49 (0.33–0.72) | <0.001 | 0.58 (0.37–0.90) | 0.016 |
Adjusted P, adjustment for sex, age, TNM stage. The significance is that all the values are statistically significant.
Figure 5.
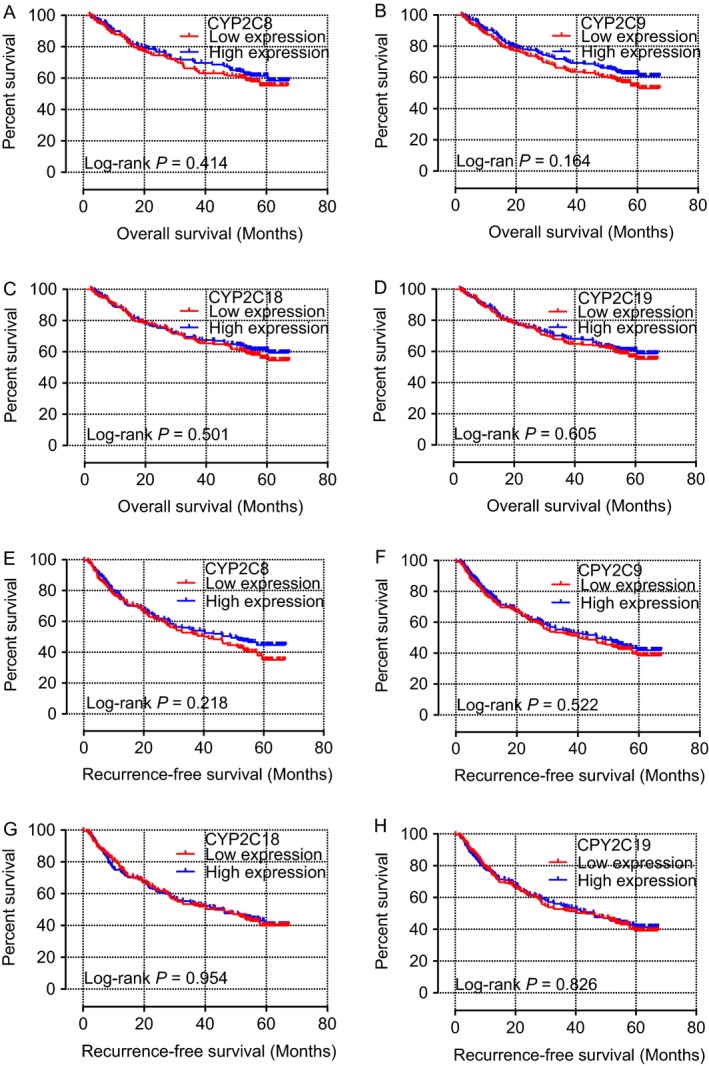
Kaplan–Meier overall survival curves of CYP2C8 (A), CYP2C9 (B), CYP2C18 (C), and CYP2C19 (D), as well as recurrence‐free survival of CYP2C8 (E), CYP2C9 (F), CYP2C18 (G), and CYP2C19 (H) in the GEO database.
Finally, joint effects analysis of the CYP2C8, CYP2C9, and CYP2C19 combinations showed that MST was poorest in group A (827 days; adjusted P = 0.017) and best in group C (3125 days; adjusted P = 0.012). Surprisingly, MST could not be determined for group D, which contained the best factors for patients, possibly due to the influence of other potential elements (Table 6). Survival curves of the above analysis are presented in Figure 6D.
Table 6.
Joint effects analysis of the combination of CYP2C8, CYP2C9, and CYP2C19 genes
| Group | CYP2C8 expression | CYP2C9 expression | CYP2C19 expression | Patients (n = 360) | MST (days) | Crude HR (95% CI) | Crude P value | Adjusted HR (95% CI)a | Adjusted P valuea |
|---|---|---|---|---|---|---|---|---|---|
| A | Low | Low | Low | 103 | 827 | Ref. | <0.001 | Ref. | 0.017 |
| B | Low | Low | High | 84 | 1694 | 0.66 (0.42–1.05) | 0.080 | 0.77 (0.47–1.26) | 0.298 |
| Low | High | Low | |||||||
| High | Low | Low | |||||||
| C | High | High | Low | 63 | 3125 | 0.40 (0.23–0.69) | 0.001 | 0.47 (0.27–0.85) | 0.012 |
| Low | High | High | |||||||
| High | Low | High | |||||||
| D | High | High | High | 110 | 2456 | 0.42(0.27–0.66) | <0.001 | 0.51(0.31–0.84) | 0.008 |
Adjusted P, adjustment for sex, age, TNM stage. The significance is that all the values are statistically significant.
Figure 6.
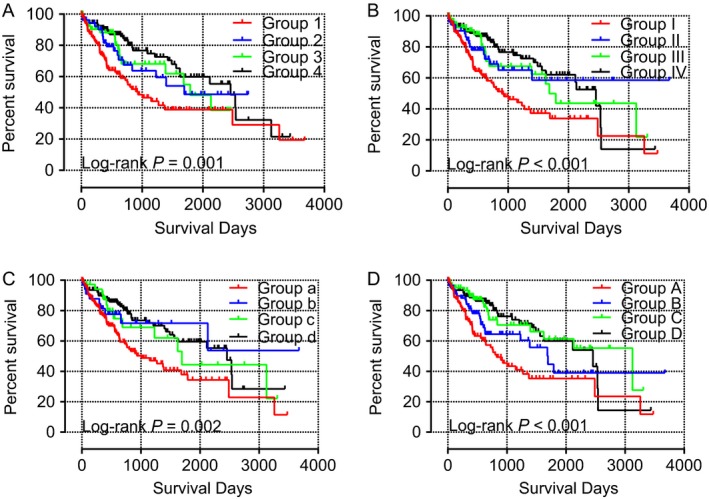
Survival curves of the joint effects analysis of the combination of CYP2C8 and CYP2C9 (A), CYP2C8 and CYP2C19 (B), CYP2C9 and CYP2C19 (C), and CYP2C8,CYP2C9, and CYP2C19 (D) in the TCGA database.
Discussion
In this study, the associations between CYP2C subfamily genes with HCC were investigated in both TCGA and GEO databases. The results showed that low gene expression levels of CYP2C8, CYP2C9, and CYP2C19 in TCGA database were associated with poor prognosis of HCC. Moreover, the groups, in TCGA database analysis, with the most poor prognostic factors had the poorest prognosis in the combination analysis of the above three genes. Thus, gene expression levels of CYP2C8, CYP2C9, and CYP2C19—in TCGA database— both alone and in combination, may serve as potential biomarkers of HCC.
CYP2C subfamily members participate in the metabolism of many endogenous and exogenous substances. It is estimated that approximately 30% of all drugs are metabolized by CYP2C8, CYP2C9, CYP2C18, and CYP2C19 27. Moreover, CYP2C9, CYP2C19, and CYP2C8 contribute to 17%, 10%, and 6% of drug biotransformations, respectively 28. Specifically, CYP2C8 is reported to metabolize analgesics 29 as well as antidiabetics and cholesterol‐lowering drugs 30, while CYP2C9 was found to metabolize analgesics 31 and neurological drugs 32, and CYP2C19 has been linked to the metabolism of antidepressants and antipsychotics 33, as well as drugs for treatment of respiratory diseases and allergies 34. Among them, CYP2C18 has been less well studied. Furthermore, members of the CYP2C subfamily have been implicated in drug metabolism and have also been explored in many diseases, including several cancers. Specifically, genetic variants of CYP2C8 have been associated with an increased risk of myocardial infarction 35, paclitaxel‐induced neuropathy 36, and bisphosphonate‐related osteonecrosis of the jaw in multiple myeloma 37 and esophageal squamous cell carcinoma 38. A CYP2C9 gene polymorphism has been associated with increased susceptibility to colorectal cancer and adenoma 39, increased progression of nonalcoholic fatty liver disease 40, and excessive anticoagulation and bleeding risk in patients taking warfarin 41. Also, mutant alleles of CYP2C18 have been linked to CYP2C19 in a Japanese population 42. Genetic polymorphisms of CYP2C19 were found to be associated with a greater risk of HCC in Japanese cirrhotic patients with HCV infection 43, as well as a significant risk of triple‐negative breast cancer 44 and lung cancer in combination analysis with smoking in a Chinese population 45.
CYP2C subfamily members are highly expressed in normal liver tissue and mainly metabolize endogenous and exogenous substances as well as clinical drugs. A previous study reported that CYP2C subfamily members in human hepatocytes were affected by different inflammatory cytokines, including bacterial lipopolysaccharide, interleukin 6, tumor necrosis factor–α, interferon γ, transforming growth factor β, and interleukin 1β. Meanwhile, with regard to the four members, CYP2C8 was downregulated by each of the above elements, CYP2C9 and CYP2C19, which had almost identical response patterns, gave rise to cytokine‐specific outcomes. However, CYP2C18 was not affected by any treatment 46. Moreover, CYP2C subfamily members are involved in the metabolic pathways of arachidonic acid, linoleic acid, retinol, as well as drug metabolism of cytochrome P450, serotonergic synapses, and chemical carcinogenesis.
In chemical carcinogenesis metabolism, benzo[a]pyrene can be metabolized by CYP2C8, CYP2C9, CYP2C18, and CYP2C19, and then finally transformed into the DNA adduct (+)‐trans‐BPDE‐N2‐dG, which has been shown to promote cancers of the skin, lung, and stomach. In addition, the CYP2C8, CYP2C9, CYP2C18, and CYP2C19 genes are linked to CYP1A2 in physical interactions, co‐expression, shared protein domains, co‐localization, other various pathways, and even predicted relationships. At the protein–protein interaction level, CYP2C8, CYP2C9, CYP2C18, and CYP2C19 were related to CYP1A1 and CYP1A2 in coexpression, protein homology, text mining, predicted gene neighborhood interactions, predicted gene fusions interactions, predicted gene co‐expression interactions, and other known interactions, as noted in curated databases and as determined experimentally.
These results further confirmed that CYP2C subfamily members exhibit many interactions with CYP1A1 and CYP1A2. CYP1A1 is known to participate in the metabolism of Sudan I to 8‐(phenylazo)guanine in DNA, 1, 2‐naphthoquinone, 3′,4′–diOH–Sudan I, and 4′,6′ –diOH–Sudan I, as well as DNA, RNA, and protein adducts. Among them, 8‐(phenylazo)guanine in DNA and DNA, RNA, and protein adducts can result in cancers of the liver and bladder. Meanwhile, CYP1A2 can metabolize IQ and MeIQx and finally into DNA adducts (dG‐C8‐MeIQx, dG‐N‐MeIQx). The above DNA adducts can lead to tumorigenesis in cancers of the liver, lung, colon, and breast. In view of these results, CYP2C8, CYP2C9, CYP2C18, and CYP2C19 may be associated with the occurrence of HCC. Therefore, CYP2C8, CYP2C9, CYP2C18, and CYP2C19 may serve as potential diagnostic and prognostic serum biomarkers for HCC diagnosis.
It is well‐known that serum AFP is the most widely used biomarker for early diagnosis and monitoring of HCC recurrence 47. However, the prognostic value of AFP remains controversial. Several studies refuted the prognostic value of AFP in single, small HCC, and even for the prediction of HCC recurrence 48, 49. Several literatures reported its sensitivity of less than 70% at a cutoff value of 20 ng/mL 50, 51.
Many novel serum biomarkers of HCC have been identified in recent years, including osteopontin 52, UQCRH 53, CXCL1 54, integrator complex subunit 6 55, PIVKA–II 56, TIP 30 57, cavin–2 58, and annexin A2 59, among others. Although a variety of potential serum biomarkers were put forward by different research centers, clinical applications have been limited because of the highly heterogeneous nature of HCC. In the present population, CYP2C subfamily gene expression levels were associated with HCC prognosis. Thus, we postulate that the CYP2C subfamily members may serve as potential serum biomarkers for the early diagnosis of HCC.
However, there were some limitations in this study. First, larger population studies are required to increase the credibility of these conclusions. Second, other potential influencing factors regarding tumor evolution and prognosis, such as drinking status, smoking status, cirrhosis status, Child–Pugh score, tumor number, primary tumor size, pathological differentiation diagnosis, tumor capsule status, and vascular invasion should be included for analysis to better evaluate the relationships between CYP2C subfamily members and HCC prognosis. Third, more commonly used indicators, such as disease‐free survival, should be considered to estimate HCC prognosis. Fourth, further well‐designed studies concentrating on functional validation are warranted with a greater number of research centers and more racially diverse countries. Fifth, other significant drug‐metabolizing CYPs, including CYP1A2, CYP2A6, CYP2B6, CYP2D6, CYP2E1, and CYP3A4/5, will be explored for HCC in our future studies. To summarize, the results of this study indicate that CYP2C8, CYP2C9, and CYP2C19 present potential serum biomarkers for the early diagnosis of HCC and combination analysis showed significant interactions that were better prognostic indicators of HCC. However, because of the incomplete clinical data and small sample size in this study, further research is necessary to validate these findings.
Conflict of Interest
No conflicts of interest were disclosed in this study.
Supporting information
Figure S1. Expression levels of CYP2C8, CYP2C9, CYP2C18, and CYP2C19 genes in different tissues.
Acknowledgment
This work was supported in part by the National Nature Science Foundation of China (No: 81560535, 81072321, 30760243, 30460143 and 30560133), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Nature Sciences Foundation (No: GuiKeGong 1104003A‐7), and Guangxi Health Ministry Medicine Grant (Key‐Scientific Research‐Grant Z201018). Self‐raised Scientific Research Fund of the Health and Family Planning Commission of Guangxi Zhuang Autonomous Region (Z2016318). The authors thank the contributors of The Cancer Genome Atlas and Gene Expression Omnibus databases for their contribution to share the sequencing dataset on open access. The authors thank the contributors of STRING database for their contribution to share open access authority.
Cancer Medicine 2018; 7(4):966–980
Reference
- 1. Jemal, A. , Bray F., Center M. M., Ferlay J., Ward E., and Forman D.. 2011. Global cancer statistics. CA Cancer J. Clin. 61:69–90. [DOI] [PubMed] [Google Scholar]
- 2. Chen, W. , Zheng R., Baade P. D., Zhang S., Zeng H., Bray F., et al. 2016. Cancer statistics in China, 2015. CA Cancer J. Clin. 66:115–132. [DOI] [PubMed] [Google Scholar]
- 3. Perz, J. F. , Armstrong G. L., Farrington L. A., Hutin Y. J., and Bell B. P.. 2006. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 45:529–538. [DOI] [PubMed] [Google Scholar]
- 4. Kgatle, M. M. , Setshedi M., and Hairwadzi H. N.. 2016. Hepatoepigenetic Alterations in Viral and Nonviral‐Induced Hepatocellular Carcinoma. Biomed. Res. Int. 2016:3956485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balogh, J. , Victor D. III, Asham E. H., Burroughs S. G., Boktour M., Saharia A., et al. 2016. Hepatocellular carcinoma: A review. J. Hepatocell. Carcinoma. 3:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tien, A. J. , Chien C. Y., Chen Y. H., Lin L. C., and Chien C. T.. 2017. Fruiting Bodies of Antrodia cinnamomea and Its Active Triterpenoid, Antcin K, Ameliorates N‐Nitrosodiethylamine‐Induced Hepatic Inflammation, Fibrosis and Carcinogenesis in Rats. Am. J. Chin. Med. 45:173–198. [DOI] [PubMed] [Google Scholar]
- 7. Gelboin, H. V. , and Krausz K.. 2006. Monoclonal antibodies and multifunctional cytochrome P450: drug metabolism as paradigm. J. Clin. Pharmacol. 46:353–372. [DOI] [PubMed] [Google Scholar]
- 8. Goldstein, J. A. , and de Morais S. M.. 1994. Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics 4:285–299. [DOI] [PubMed] [Google Scholar]
- 9. Rahman, A. , Korzekwa K., Grogan J., Gonzalez F., and Harris J.. 1994. Selective biotransformation of taxol to 6 alpha‐hydroxytaxol by human cytochrome P450 2C8. Cancer Res. 54:5543–5546. [PubMed] [Google Scholar]
- 10. Andersson, T. , Regårdh C., Lou Y., Zhang Y., Dahl M., and Bertilsson L.. 1992. Polymorphic hydroxylation of S‐mephenytoin and omeprazole metabolism in Caucasian and Chinese subjects. Pharmacogenetics 2:25–31. [DOI] [PubMed] [Google Scholar]
- 11. Agundez, J. 2004. Cytochrome P450 gene polymorphism and cancer. Curr. Drug Metab. 5:211–224. [DOI] [PubMed] [Google Scholar]
- 12. Tzveova, R. , Naydenova G., Yaneva T., Dimitrov G., Vandeva S., Matrozova Y., et al. 2015. Gender‐Specific Effect of CYP2C8*3 on the Risk of Essential Hypertension in Bulgarian Patients. Biochem Genet. 53:319–333. [DOI] [PubMed] [Google Scholar]
- 13. Bosó, V. , Herrero M. J., Santaballa A., Palomar L., Megias J. E., de la Cueva H., et al. 2014. SNPs and taxane toxicity in breast cancer patients. Pharmacogenomics 15:1845–1858. [DOI] [PubMed] [Google Scholar]
- 14. Wei, X. , Zhang D., Dou X., Niu N., Huang W., Bai J., et al. 2014. Elevated 14,15‐ epoxyeicosatrienoic acid by increasing of cytochrome P450 2C8, 2C9 and 2J2 and decreasing of soluble epoxide hydrolase associated with aggressiveness of human breast cancer. BMC Cancer 14:841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu, W. , Wang B., Ding H., Wang D. W., and Zeng H.. 2014. A potential therapeutic effect of CYP2C8 overexpression on anti‐TNF‐α activity. Int. J. Mol. Med. 34:725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martínez, C. , García‐Martín E., Ladero J., Sastre J., Garcia‐Gamito F., Diaz‐Rubio M., et al. 2001. Association of CYP2C9 genotypes leading to high enzyme activity and colorectal cancer risk. Carcinogenesis 22:1323–1326. [DOI] [PubMed] [Google Scholar]
- 17. Musumba, C. O. , Jorgensen A., Sutton L., Van Eker D., Zhang E., O'Hara N., et al. 2013. CYP2C19*17 gain‐of‐function polymorphism is associated with peptic ulcer disease. Clin. Pharmacol. Ther. 93:195–203. [DOI] [PubMed] [Google Scholar]
- 18. Barry, E. L. , Poole E. M., Baron J. A., Makar K. W., Mott L. A., Sandler R. S., et al. 2013. CYP2C9 variants increase risk of colorectal adenoma recurrence and modify associations with smoking but not aspirin treatment. Cancer Causes Control 24:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jernström, H. , Bågeman E., Rose C., Jönsson P. E., and Ingvar C.. 2009. CYP2C8 and CYP2C9 polymorphisms in relation to tumour characteristics and early breast cancer related events among 652 breast cancer patients. Br. J. Cancer 101:1817–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shuldiner, A. R. , O'Connell J. R., Bliden K. P., Gandhi A., Ryan K., Horenstein R. B., et al. 2009. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 302:849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carithers, L. , Ardlie K., Barcus M., Branton P., Britton A., Buia S., et al. 2015. A novel approach to high‐quality postmortem tissue procurement: the GTEx project. Biopreserv. Biobank. 13:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anaya, J. 2016. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ. Comput. Sci. 2:e67. [Google Scholar]
- 23. Roessler, S. , Jia H. L., Budhu A., Forgues M., Ye Q. H., Lee J. S., et al. 2010. A unique metastasis gene signature enables prediction of tumor relapse in early‐stage hepatocellular carcinoma patients. Cancer Res. 70:10202–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roessler, S. , Long E. L., Budhu A., Chen Y., Zhao X., Ji J., et al. 2012. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 142(957–66):e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang, D. W. , Sherman B. T., and Lempicki R. A.. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang, D. W. , Sherman B. T., and Lempicki R. A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- 27. Vormfelde, S. V. , Schirmer M., Toliat M. R., Meineke I., Kirchheiner J., Nurnberg P., et al. 2007. Genetic variation at the CYP2C locus and its association with torsemide biotransformation. Pharmacogenomics J. 7:200–211. [DOI] [PubMed] [Google Scholar]
- 28. Zanger, U. M. , Turpeinen M., Klein K., and Schwab M.. 2008. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 392:1093–1108. [DOI] [PubMed] [Google Scholar]
- 29. Hamman, M. A. , Thompson G. A., and Hall S. D.. 1997. Regioselective and stereoselective metabolism of ibuprofen by human cytochrome P450 2C. Biochem. Pharmacol. 54:33–41. [DOI] [PubMed] [Google Scholar]
- 30. Jaakkola, T. , Laitila J., Neuvonen P. J., and Backman J. T.. 2006. Pioglitazone is metabolised by CYP2C8 and CYP3A4 in vitro: potential for interactions with CYP2C8 inhibitors. Basic Clin. Pharmacol. Toxicol. 99:44–51. [DOI] [PubMed] [Google Scholar]
- 31. Tougou, K. , Gotou H., Ohno Y., and Nakamura A.. 2004. Stereoselective glucuronidation and hydroxylation of etodolac by UGT1A9 and CYP2C9 in man. Xenobiotica 34:449–461. [DOI] [PubMed] [Google Scholar]
- 32. Kiang, T. K. , Ho P. C., Anari M. R., Tong V., Abbott F. S., and Chang T. K.. 2006. Contribution of CYP2C9, CYP2A6, and CYP2B6 to valproic acid metabolism in hepatic microsomes from individuals with the CYP2C9*1/*1 genotype. Toxicol. Sci. 94:261–271. [DOI] [PubMed] [Google Scholar]
- 33. Hiemke, C. , and Hartter S.. 2000. Pharmacokinetics of selective serotonin reuptake inhibitors. Pharmacol. Ther. 85:11–28. [DOI] [PubMed] [Google Scholar]
- 34. Akutsu, T. , Kobayashi K., Sakurada K., Ikegaya H., Furihata T., and Chiba K.. 2007. Identification of human cytochrome p450 isozymes involved in diphenhydramine N‐demethylation. Drug Metab. Dispos. 35:72–78. [DOI] [PubMed] [Google Scholar]
- 35. Rodenburg, E. M. , Visser L. E., Danser A. H., Hofman A., van Noord C., Witteman J. C., et al. 2010. Genetic variance in CYP2C8 and increased risk of myocardial infarction. Pharmacogenet. Genomics 20:426–434. [DOI] [PubMed] [Google Scholar]
- 36. Hertz, D. L. , Roy S., Motsinger‐Reif A. A., Drobish A., Clark L. S., McLeod H. L., et al. 2013. CYP2C8*3 increases risk of neuropathy in breast cancer patients treated with paclitaxel. Ann. Oncol. 24:1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhong, D. N. , Wu J. Z., and Li G. J.. 2013. Association between CYP2C8 (rs1934951) polymorphism and bisphosphonate‐related osteonecrosis of the jaws in patients on bisphosphonate therapy: a meta‐analysis. Acta Haematol. 129:90–95. [DOI] [PubMed] [Google Scholar]
- 38. Bergheim, I. , Wolfgarten E., Bollschweiler E., Hölscher A. H., Bode C., and Parlesak A.. 2007. Cytochrome P450 levels are altered in patients with esophageal squamous‐cell carcinoma. World J. Gastroenterol. 13:997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang, H. , Ren L., He Y., Wei Y., Chen Z., Yang W., et al. 2014. Association between cytochrome P450 2C9 gene polymorphisms and colorectal cancer susceptibility: evidence from 16 case–control studies. Tumor Biol. 35:4317–4322. [DOI] [PubMed] [Google Scholar]
- 40. Fisher, C. D. , Lickteig A. J., Augustine L. M., Ranger‐Moore J., Jackson J. P., Ferguson S. S., et al. 2009. Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab. Dispos. 37:2087–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ucar, M. , Alagozlu H., Sahin S., and Ozdemir O.. 2013. The relationship between CYP2C9 gene polymorphisms and upper gastrointestinal bleeding in patients who used warfarin. Med. Glas (Zenica). 10:50–54. [PubMed] [Google Scholar]
- 42. Kubota, T. , Hibi N., and Chiba K.. 1998. Linkage of mutant alleles of CYP2C18 and CYP2C19 in a Japanese population. Biochem. Pharmacol. 55:2039–2042. [DOI] [PubMed] [Google Scholar]
- 43. Chau, T. K. , Murakami S., Kawai B., Nasu K., Kubota T., and Ohnishi A.. 2000. Genotype analysis of the CYP2C19 gene in HCV‐seropositive patients with cirrhosis and hepatocellular carcinoma. Life Sci. 67:1719–1724. [DOI] [PubMed] [Google Scholar]
- 44. Tervasmäki, A. , Winqvist R., Jukkola‐Vuorinen A., and Pylkäs K.. 2014. Recurrent CYP2C19 deletion allele is associated with triple‐negative breast cancer. BMC Cancer 14:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yan, F. , Xu J. F., Liu X. F., and Li X. H.. 2014. Interaction between smoking and CYP2C19*3 polymorphism increased risk of lung cancer in a Chinese population. Tumour Biol. 35:5295–5298. [DOI] [PubMed] [Google Scholar]
- 46. Aitken, A. E. , and Morgan E. T.. 2007. Gene‐specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab. Dispos. 35:1687–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chaiteerakij, R. , Addissie B. D., and Roberts L. R.. 2015. Update on biomarkers of hepatocellular carcinoma. Clin. Gastroenterol. Hepatol. 13:237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giannini, E. G. , Marenco S., Borgonovo G., Savarino V., Farinati F., Del Poggio P., et al. 2012. Alpha‐fetoprotein has no prognostic role in small hepatocellular carcinoma identified during surveillance in compensated cirrhosis. Hepatology 56:1371–1379. [DOI] [PubMed] [Google Scholar]
- 49. Shim, J. H. , Yoon D.‐L., Han S., Lee Y.‐J., Lee S.‐G., Kim K. M., et al. 2012. Is serum alpha‐fetoprotein useful for predicting recurrence and mortality specific to hepatocellular carcinoma after hepatectomy? A test based on propensity scores and competing risks analysis. Ann. Surg. Oncol. 19:3687–3696. [DOI] [PubMed] [Google Scholar]
- 50. Sherman, M. , Peltekian K. M., and Lee C.. 1995. Screening for hepatocellular carcinoma in chronic carriers of hepatitis B virus: incidence and prevalence of hepatocellular carcinoma in a North American urban population. Hepatology 22:432–438. [PubMed] [Google Scholar]
- 51. Gambarin‐Gelwan, M. , Wolf D. C., Shapiro R., Schwartz M. E., and Min A. D.. 2000. Sensitivity of commonly available screening tests in detecting hepatocellular carcinoma in cirrhotic patients undergoing liver transplantation. Am. J. Gastroenterol. Suppl. 95:1535–1538. [DOI] [PubMed] [Google Scholar]
- 52. Al‐Zoubi, S. , Wassouf A., and Zetoune A. B.. 2017. Measuring levels of osteopontin as potential biomarker for hepatocellular carcinoma in Syrian patients. Gastroenterol. Hepatol. Bed. Bench. 10:97–101. [PMC free article] [PubMed] [Google Scholar]
- 53. Park, E. R. , Kim S. B., Lee J. S., Kim Y. H., Lee D. H., Cho E. H., et al. 2017. The mitochondrial hinge protein, UQCRH, is a novel prognostic factor for hepatocellular carcinoma. Cancer Med. 6:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Han, K. Q. , Han H., He X. Q., Wang L., Guo X. D., Zhang X. M., et al. 2016. Chemokine CXCL1 may serve as a potential molecular target for hepatocellular carcinoma. Cancer Med. 5:2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lui, K. Y. , Zhao H., Qiu C., Li C., Zhang Z., Peng H., et al. 2017. Integrator complex subunit 6 (INTS6) inhibits hepatocellular carcinoma growth by Wnt pathway and serve as a prognostic marker. BMC Cancer 17:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu, R. , Tan Z., Xiang X., Dan Y., and Deng G.. 2017. Effectiveness of PIVKA‐II in the detection of hepatocellular carcinoma based on real‐world clinical data. BMC Cancer 17:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang, X. , Lv L., Ouyang X., Zhang S., Fang J., Cai L., et al. 2016. Association of TIP30 expression and prognosis of hepatocellular carcinoma in patients with HBV infection. Cancer Med. 5:2180–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jing, W. , Luo P., Zhu M., Ai Q., Chai H., and Tu J.. 2016. Prognostic and Diagnostic Significance of SDPR‐Cavin‐2 in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 39:950–960. [DOI] [PubMed] [Google Scholar]
- 59. Shaker, M. K. , Abdel Fattah H. I., Sabbour G. S., Montasser I. F., Abdelhakam S. M., El Hadidy E., et al. 2017. Annexin A2 as a biomarker for hepatocellular carcinoma in Egyptian patients. World J. Hepatol. 9:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression levels of CYP2C8, CYP2C9, CYP2C18, and CYP2C19 genes in different tissues.


