Abstract
Objective
To explore the expression level of FGF5 in the peripheral blood of primary hypertension patients and its clinical significance.
Methods
The 34 patients with primary hypertension treated in this hospital from June 2012 to June 2014 were selected as the observation group, while the 25 patients at this hospital who had physical exam with heathy results were selected as control group. Venous blood was drawn early in the morning after an overnight fast. FGF5, mRNA and protein level changes in the peripheral blood cells and peripheral blood serum were analyzed by real-time fluorescence based quantitative PCR (RT-PCR) and enzyme-linked immunosorbent assay (ELISA). FGF5 gene SNP (rs16998073) were amplified by PCR and inserted into T vector, and its genetic variation were analyzed by sequencing. The relationship of FGF5 protein levels and genetic variation with diastolic/systolic blood pressure was also analyzed.
Results
Comparing with the control group, the observation group’s FGF5 mRNA and protein levels significantly increased in the peripheral blood cells and peripheral blood. The difference was statistically significant (P < .05). Correlation analysis showed that FGF5 protein level and systolic/diastolic blood pressure were positively correlated (P < .05). T/A genetic variation of FGF5 gene SNP (rs16998073) and diastolic/systolic blood pressure were positively correlated (P < .05).
Conclusion
The FGF5 mRNA and protein expression levels of the patients with primary hypertension were abnormal and had genetic variation, which were associated with blood pressure of the patients with primary hypertension.
Keywords: FGF5, Primary hypertension, Peripheral blood, Genetic variation, Blood pressure
0. Introduction
Hypertension is a common clinical cardiovascular and cerebrovascular disease and is classified as either primary (essential) hypertension or secondary hypertension. The incidence of primary hypertension is more than 95% of all the hypertension cases. Hypertension has seriously threatened human health and influenced the quality of life (Milouk et al., 2014, Benjelloun et al., 2009). Hypertension is also the risk factor for multiple diseases, such as stroke, coronary heart disease, cardiac functional insufficiency and kidney diseases. Every year, there are tens of millions of people died of hypertension; therefore, hypertension has become the leading cause of human death (Kearney et al., 2005). However, currently, the pathogenesis of hypertension has not been fully understood. The general research believes that hypertension is the result of generic and environmental interactions. The relevant studies of exactly what role generics plays in hypertension are very rare. In recent years, with the continuous development of the whole gene sequencing and restriction fragment length polymorphism (RFLP) study, the genome-wide association studies (GWAS) based on single nucleotide polymorphism (SNP) chip technology provided a great progress for the study of pathogenesis of primary hypertension (Wellcome Trust Case Control Consortium, 2007, Levy et al., 2007). A large number of GWAS reported several hypertension susceptibility gene in primary hypertension patients from Europe, China, Japan and Korea, which laid the foundation for the study of the pathogenesis of hypertension (Hong et al., 2009, Ehret et al., 2008, Maruthamuthu and Kandasamy, 2016, Yang et al., 2016, Zhu et al., 2011). FGF5 is one of the fibroblast growth factors and the existing studies showed that it played an important role in the control of the animal hair growth process (Higgins et al., 2014, Kubelkova and Macela, 2015, Haitham, 2016, Chen et al., 2013). The GWAS found that inside or nearby area of FGF5 gene was the susceptible region of primary hypertension (Dierks et al., 2013, Gao et al., 2017a, Gao et al., 2017b, Lin et al., 2011, Liu et al., 2011b). No study was conducted on the expression level changes of FGF5 in the blood serum of primary hypertension or genetic variation of SNP (rs16998073) in the FGF5 gene susceptible region. Therefore, this study analyzed the FGF5 expression of the primary hypertension patients and the genetic variation in order to provide the theoretical basis for the genetic factor of primary hypertension.
1. Materials and methods
1.1. Clinical data
The 34 patients with primary hypertension treated in this hospital from June 2012 to June 2014 were selected as observation group. Screening criteria: (1) All the patients met the clinical diagnostic standards developed by “Chinese Hypertension Prevention Guide” (Higgins et al., 2014, Kubelkova and Macela, 2015, Gao et al., 2017a, Gao et al., 2017b, Haitham, 2016); (2) Age between 50 and 75; (3) All patients signed informed consent forms; (4) this study was granted permission and supervised by the Medical Ethics Committee of this hospital. Exclusion criteria: (1) Secondary hypertension patients; (2) Coronary heart disease patients diagnosed by coronary angiography; (3) Diabetes, high cholesterol or cerebral thrombus patients; (4) Patients with liver diseases, kidney diseases and autoimmune diseases/tumor; (5) Patients who had surgery recently or currently have trauma. Among the 34 selected patients in the observation group, there were 23 male and 11 female; Age was from 50 to 75 years old averaged at 60.4 ± 10.1. The 25 patients at this hospital who had physical exam with heathy results were selected as control group which had 17 male and 8 female; Age was from 52 to 76 years old averaged at 62.6 ± 10.8. The difference of the age and gender of the patients in both groups had no statistical significance (P > .05).
1.2. Materials
The erythrocyte lysate was purchased from Sigma. The reverse transcription kit was purchased from Nanjing Vazyme Biotech Co., Ltd. 2 × Taq DNA polymerase mix and T vector kit were purchased from TAKARA. Human FGF5 ELISA was purchased from Shanghai Co., Ltd. 2 × SYBR Green universal qPCR Master Mix was purchased from Roche. Real-time PCR machine was purchased from Bio-Rad Company. Trizol was purchased from Invitrigen. The rest of organic reagents were purchased from Sinopharm.
1.3. Blood sample pretreatment
Five (5) ml of venous blood samples were draw from both observation group and control group in the early morning with an overnight fast. The 2.5 ml of the blood samples were set at 4 °C. The blood serum was separated out and then centrifuged at 3000 rpm/min for 5 mins. The blood serum was collected. The other 2.5 ml blood samples were lysed using erythrocyte lysate. All the red blood cells were lysed and then centrifuged at 1500 rpm/min to collect the white blood cells which were used to extract the total RNA.
1.4. Experimental methods
1.4.1. FGF5 level in peripheral blood cells analyzed by real-time quantitative PCR
The 1 ml Trizol was added to the white blood cell sedimentation separated from the outer periphery. The 200 μl Chloroform was added after the solution was shake and mixed well. The mixture was placed on ice until stratification and was then centrifuged at 12,000 rpm/min for 15 mins. The 500 μl supernatant was transferred to equal volume of isopropanol and placed on ice for 30 mins after the solution was shake and mixed well. The mixture was then centrifuged at 15,000 rpm/min for 15 mins. The supernatant was discarded and the precipitate was washed with precooled 70% ethanol for one time. The precipitate was then centrifuged at 8000 rpm/min for 6 mins and was dissolved in double-distilled water treated by DEPC. RNA concentration was measured and its quality was determined. The extracted RNA was reverse transcribed to cDNA according to the instruction of the reverse transcription kit, which was used as the RT-PCR template.
The FGF5 primer, FGF5-F1: FGF5-R1, was designed according to the FGF5 mRNA sequence. First, the conventional PCR was used to optimize the specificity and annealing temperature of the primer. The following reaction systems were prepared: 2 × SbGreen RT-PCR Mix 25 μl, FGF5-F1 1 μl, FGF5-R1 1 μl and cDNA 1 μl, which were made up to 50 μl by adding water. A certain volume of reactants were prepared according to the number of samples and were added to the real time quantitative PCR 96 Well Plates which were centrifuged at 1500 rpm/min for 1 min to throw the reaction solution to the bottom of the wells. The following reaction conditions were set on the real time PCR instrument: 94 °C start PCR and read the values directly from the instrument after the reaction.
1.4.2. FGF protein level in the peripheral blood detected by ELISA
The FGF5 protein level in the peripheral blood was analyzed strictly in accordance with the operating instruction of the kit. The FGF5 level in the peripheral blood for both control group and observation group were averaged for comparison.
1.4.3. FGF5 gene SNP (rs16998073) amplification and mutation analysis
According to the FGF5 gene sequence, the following primers were designed: FGF5-F2: FGF5-R2. The following PCR reaction system was prepared: 2 × Taq DNAPolymerase mix 25 μl, FGF5-F2 1 μl, FGF5-R2 1 μl, and cDNA 1 μl, which were made up to 50 μl by adding water. PCR was conducted in accordance with the following reaction conditions: 94 °C pre-denaturation for 5 min, 94 °C denaturation for 30 s, 55 °C annealing for 30 s, 72 °C extension for 2 mins, 30 cycles and finally cool to 4 °C. The PCR product was ligated into T vector according to the instruction manual of T vector, transformed to E. coli DH5a, spread on the solid LB plates and incubated overnight at 37 °C. Monoclonal cells were picked for sequencing analysis after the colonies grew the next day.
1.5. Statistical analysis
All the data were analyzed using SPSS 17.0 statistical software. The measurement data were expressed as xs and compared using the t test. The enumeration data were compared using chi-square test. Correlation was analyzed using…; P < .05 indicated that the difference was statistically significant.
2. Results
2.1. Clinical data comparison
The difference between the control group and the observation group had no statistical significant (P > .05) from the aspects of age, gender, TC, HDL - C, LDC - C, fasting glucose and TG. The SBP and DBP levels in the observation group were significantly higher than those in the control group, which had statistically significant (P < .05) (see Table 1).
Table 1.
Clinical data comparison between observation group and control group.
| Group | Control group | Observation group | P value |
|---|---|---|---|
| Gender (Male/Female) | 23/11 | 17/8 | 0.88 |
| Age | 62.6 ± 10.8 | 60.4 ± 10.1 | 0.57 |
| TC (mmol/L) | 4.23 ± 0.60 | 4.33 ± 0.51 | 0.67 |
| TG (mmol/L) | 1.26 ± 0.34 | 1.21 ± 0.39 | 0.44 |
| HDL-C (mmol/L) | 1.68 ± 0.32 | 1.60 ± 0.37 | 0.53 |
| LDL-C (mmol/L) | 2.67 ± 0.76 | 2.55 ± 0.71 | 0.78 |
| Fasting glucose (mmol/L) | 5.15 ± 0.67 | 5.23 ± 0.61 | 0.45 |
| SBP | 125.3 ± 10.2 | 175.3 ± 12.9 | 0.01* |
| DBP | 75.2 ± 8.9 | 95.55 ± 10.2 | 0.03* |
Compare with the control group, P < .05
2.2. Comparison of FGF5 mRNA level in the peripheral blood cells between the two groups
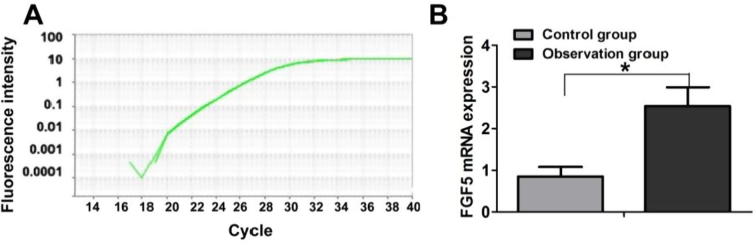
This experiment established a real time fluorescence based quantitative PCR to test the FGF5 mRNA level. The experiment designed RT-PCR primer had very high specificity. Amplification curve had very good linearity and there were no noise peaks (see Fig. 1A). Quantitative analysis showed that the FGF5 mRNA level in the peripheral blood cells in the observation group was significantly higher than that in the control group and the difference was statistically significant (P < .05).
Fig. 1.
Comparison of FGF5 mRNA level in the peripheral blood cells of control group and observation group.
2.3. Comparison of FGF5 protein level in the peripheral blood between the two groups
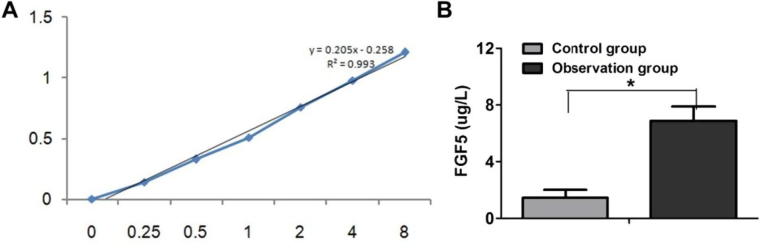
The standard curve for FGF5 protein ELISA test was established according to the instructions of the kit (see Fig. 2A). The calibration curve was linear and r = 0.993. The FGF5 protein level in the peripheral blood of the control group and observation group was calculated using the standard curve. The results were shown in Fig. 2B. The FGF5 protein level in the peripheral blood in the observation group were significantly higher than that in the control group and the difference was statistically significant (P < .05).
Fig. 2.
Comparison of FGF5 protein level in peripheral blood of control group and observation group.
2.4. FG5 gene SNP genetic variation analysis
FGF5 Gene SNP (rs16998073) fragments were amplified using PCR and were ligated into T vectors. The clones were picked for sequencing analysis. The FGF5 Gene SNP (rs16998073) genetic variation in the peripheral blood was analyzed for both observation group and control group and the results were shown in Table 2. The FGF5 Gene SNP (rs16998073) genetic variation in the observation group was significantly higher than that in the control group and the difference was statistically significant (P < .05).
Table 2.
Analysis of FGF5 gene SNP genetic variation in the control group and observation group.
| Variation type | Control group | Observation group | P value |
|---|---|---|---|
| T/A | 1 (4.00%) | 24 (53.33) | 0.000a |
| T/C | 0 (0.00%) | 1 (2.22%) | 0453 |
| T/G | 2 (8.00%) | 5 (11.11%) | 0.687 |
Compare with the control group, P < .05.
2.5. Relationship study of FGF5 protein level, genetic variation and blood pressure
Linear correlation analysis showed that FGF5 protein level in the peripheral blood was positively correlated with systolic and diastolic blood pressure (r = 0.590, P < .05; r = 0.391, P < .05). FGF5 gene SNP (rs16998073) T/C gene was positively correlated with systolic and diastolic blood pressure (r = 1.015, P < .05; r = 0.754, P < .05).
3. Discussion
Primary hypertension is a type of complex disease, which is the result of generic and environmental interactions. The environmental factors result in different clinical phenotypes in different genetic backgrounds. The genetic factors play pivotal role in the occurrence and progression of primary hypertension (Chen et al., 2013, Dierks et al., 2013, Liu et al., 2011b, Takeuchi et al., 2010, Zhang and Liuxing, 2008, Lackland and Weber, 2015). In recent years, with the development of molecular genetics and related technologies, gene research on the patients with primary hypertension is carried out gradually and achieved gratifying results (Mohammed et al., 2016, Peng et al., 2017, Ge et al., 2016). The most commonly used method of studying hypertension related genes include candidate gene study and genome-wide association study. The genome-wide association study discovered a number of primary hypertension related genes, such as ATP2B1, CYP17A1, PLEKHA7, SH2B3, MTHFR, CASZ1, ULK4, HFE, EBF1 and FGF5 (Hong et al., 2010, Lin et al., 2011, Liu et al., 2011b, Haddad et al., 2016, Kathirvel and Subban, 2016, Xi et al., 2013, Weili, 2008, Fox et al., 2011). Numerous studies have shown that FGF5 is the hypertension susceptibility gene and its genetic variability is positively correlated with patients’ blood pressure. Currently, the studies on FGF5 gene are very few. As one of the fibroblast growth factor, FGF5 plays a regulating role in the hair growth of mice, goats and other species. The expression level of FGF5 in hypertension patients is still unknown.
This study used control study to analyze the FGF5 expression level in the peripheral blood of healthy individuals and patients with primary hypertension. From the aspect of the mRNA level, the FGF5 mRNA level in the peripheral blood cells in the patients with primary hypertension were significantly higher than that in the healthy individuals, which was further verified though ELISA on the protein level. It was found that FGF5 protein levels in the peripheral blood of patients with hypertension were significantly higher than that in the healthy subjects. This result suggested that the expression levels of FGF5 mRNA and protein in patients with hypertension were abnormal, which may be involved in the occurrence and development of hypertension disease.
Reports also showed that hypertensive patients had genetic variation at or near the FGF5 gene. The patients’ genetic variation was related with the diastolic and systolic blood pressure. This study amplified FGF5 gene SNP (rs16998073) fragments, analyzed what genetic variations were in this gene section and explored the relationship between these variants and blood pressure among the hypertensive patients. The results showed that FGF5 gene SNP (rs16998073) had T/C and T/G mutations and correlation analysis showed T/C mutation in the patients was positively correlated with diastolic and systolic blood pressure. This result confirmed the above findings, which provided a theoretical basis for the etiology of hypertension from the aspect of the genetic factors.
It is worth noting that this study had small sample size; therefore, the results may require more samples, multiple research centers and multiple ethnic population to re-verify. In conclusion, this study provided a basis for the etiology of hypertension from the aspect of the genetic factors. With the development of molecular genetics and molecular biology, the incidence of primary hypertension can be well controlled in the near future.
Footnotes
Peer review under responsibility of King Saud University.
References
- Benjelloun H., Aboudrar S., Jroundi I. Sympathetic response in primary hypertension. Ann. Cardiol. Angeiol. (Paris). 2009;58(3):139–143. doi: 10.1016/j.ancard.2008.05.023. [DOI] [PubMed] [Google Scholar]
- Chen Z., Wang Z., Xu S. Characterization of hairless (Hr) and FGF5 genes provides insights into the molecular basis of hair loss in cetaceans. BMC Evol. Biol. 2013;13:34. doi: 10.1186/1471-2148-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks C., Mömke S., Philipp U., Distl O. Allelic heterogeneity of FGF5 mutations causes the long-hair phenotype in dogs. Anim. Genet. 2013;44(4):425–431. doi: 10.1111/age.12010. [DOI] [PubMed] [Google Scholar]
- Ehret G.B., Morrison A.C., O'Connor A.A. Replication of the Wellcome Trust genome-wide association study of essential hypertension: the Family Blood Pressure Program. Eur. J. Hum. Genet. 2008;16(12):1507–1511. doi: 10.1038/ejhg.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox E.R., Young J.H., Li Y. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum. Mol. Genet. 2011;20(11):2273–2284. doi: 10.1093/hmg/ddr092. Jun 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Wang Y., Basavanagoud B., Jamil M.K. Characteristics studies of molecular structures in drugs. Saudi Pharm. J. 2017;25(4):580–586. doi: 10.1016/j.jsps.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad M., Soukkarieh C., Khalaf H., Eddin A., Abdul Q. Purification of polyclonal IgG specific for Camelid’s antibodies and their recombinant nanobodies. Open Life Sci. 2016;11(1):1–9. [Google Scholar]
- Gao W., Wang Y., Wang W., Shi L. The first multiplication atom-bond connectivity index of molecular structures in drugs. Saudi Pharm. J. 2017;25(4):548–555. doi: 10.1016/j.jsps.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S., Liu Z., Furuta Y., Peng W. Characteristics of activated carbon remove sulfur particles against smog. Saudi J. Biol. Sci. 2016 doi: 10.1016/j.sjbs.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitham Q. Chemical and bioactive diversities of marine sponge Neopetrosia. Bangladesh J. Pharmacol. 2016;11(2):433–452. [Google Scholar]
- Higgins C.A., Petukhova L., Harel S. FGF5 is a crucial regulator of hair length in humans. Proc. Natl. Acad. Sci. USA. 2014;111(29):10648–10653. doi: 10.1073/pnas.1402862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K.W., Go M.J., Jin H.S. Genetic variations in ATP2B1, CSK, ARSG and CSMD1 loci are related to blood pressure and/or hypertension in two Korean cohorts. J. Hum. Hypertens. 2010;24(6):367–372. doi: 10.1038/jhh.2009.86. [DOI] [PubMed] [Google Scholar]
- Hong K.W., Jin H.S., Cho Y.S. Replication of the Wellcome Trust genome-wide association study on essential hypertension in a Korean population. Hypertens Res. 2009;32(7):570–574. doi: 10.1038/hr.2009.68. [DOI] [PubMed] [Google Scholar]
- Kathirvel P., Subban R. Phytochemical investigation and in vitro antimicrobial activity of Richardia scabra. Bangladesh J. Pharmacology. 2016;11(2):348–352. [Google Scholar]
- Kearney P.M., Whelton M., Reynolds K. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365(9455):217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- Kubelkova K., Macela A. Putting the jigsaw together – a brief insight into the tularemia. Open Life Sci. 2015;10(1):195–216. [Google Scholar]
- Lackland D.T., Weber M.A. Global burden of cardiovascular disease and stroke: hypertension at the core. Can. J. Cardiol. 2015 doi: 10.1016/j.cjca.2015.01.009. pii: S0828-282X(15)00034-3. [DOI] [PubMed] [Google Scholar]
- Levy D.1., Larson M.G., Benjamin E.J. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med. Genet. 2007;8(Suppl. 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Lai X., Chen B. Genetic variations in CYP17A1, CACNB2 and PLEKHA7 are associated with blood pressure and/or hypertension in She ethnic minority of China. Atherosclerosis. 2011;219(2):709–714. doi: 10.1016/j.atherosclerosis.2011.09.006. Dec. [DOI] [PubMed] [Google Scholar]
- Liu C.1., Li H., Qi Q. Common variants in or near FGF5, CYP17A1 and MTHFR genes are associated with blood pressure and hypertension in Chinese Hans. J. Hypertens. 2011;29(1):70–75. doi: 10.1097/HJH.0b013e32833f60ab. [DOI] [PubMed] [Google Scholar]
- Maruthamuthu V., Kandasamy R. Ferric reducing anti-oxidant power assay in plant extract. Bangladesh J. Pharma. 2016;11(3):570–572. [Google Scholar]
- Milouk F.Z., El Bakkali M., Coghlan L. Kinetics of orthostatic blood pressure in primary hypertension. Int. Cardiovasc. Res. J. 2014;8(3):83–88. [PMC free article] [PubMed] [Google Scholar]
- Mohammed N., Messiha B., Abo-Saif A.A. Effect of amlodipine, lisinopril and allopurinol on acetaminophen-induced hepatotoxicity in rats. Saudi Pharma. J. 2016;24(6):635–644. doi: 10.1016/j.jsps.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W., Ge S., Liu Z., Furuta Y. Adsorption characteristics of sulfur powder by bamboo charcoal to restrain sulfur allergies. Saudi J. Biol. Sci. 2017;24(1):103–107. doi: 10.1016/j.sjbs.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F.1., Isono M., Katsuya T., Yamamoto K. Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation. 2010;121(21):2302–2309. doi: 10.1161/CIRCULATIONAHA.109.904664. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi B., Shen Y., Reilly K.H. Recapitulation of four hypertension susceptibility genes (CSK, CYP17A1, MTHFR, and FGF5) in East Asians. Metabolism. 2013;62(2):196–203. doi: 10.1016/j.metabol.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Yan Weili, 2008. Genome-wide association study on complex diseases – statistical analysis of genetic inheritance, 30 (5): 545-549. [DOI] [PubMed]
- Yang L., Li R., Xiang S.X., Xiao W.H. MafB. A target of MicroRNA-155, regulates dendritic cell maturation. open. Life Sci. 2016;11(1):46–54. [Google Scholar]
- Zhang Lingling, Liuxing De, 2008. Expression in peripheral blood of patients with essential hypertension and significance of interleukin-6 gene Guiyang Medical College 33 (6): 665–667.
- Zhu X., Young J.H., Fox E. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum. Mol. Genet. 2011;20(11):2285–2295. doi: 10.1093/hmg/ddr113. [DOI] [PMC free article] [PubMed] [Google Scholar]