Abstract
Current vaccines against Streptococcus pneumoniae, a bacterial species that afflicts people by causing a wide spectrum of diseases, do not protect against all pneumococcal serotypes. Thus, alternative vaccines to fight pneumococcal infections that target common proteins are under investigation. One promising strategy is to take advantage of immune cross-reactivity between commensal and pathogenic microbes for cross-protection. In this study, we examined the antibody-mediated cross-reactivity between S. pneumoniae and Streptococcus mitis, a commensal species closely related to S. pneumoniae. Western blot analysis showed that rabbit antisera raised against S. mitis reacted with multiple proteins of virulent S. pneumoniae strains (6B, TIGR4, and D39). Rabbit anti-S. pneumoniae IgG antibodies also showed binding to S. mitis antigens. Incubation of rabbit antisera raised against S. mitis with heterologous or homologous bacterial lysates resulted in marked inhibition of the developments of bands in the Western blots. Furthermore, plasma IgG antibodies from adult human volunteers intranasally inoculated with S. pneumoniae 6B revealed enhanced S. mitis-specific IgG titers compared with the pre-inoculation samples. Using an on-chip protein microarray representing a number of selected membrane and extracellular S. pneumoniae proteins, we identified choline-binding protein D (CbpD), cell division protein (FtsH), and manganese ABC transporter or manganese-binding adhesion lipoprotein (PsaA) as common targets of the rabbit IgG antibodies raised against S. mitis or S. pneumoniae. Cumulatively, these findings provide evidence on the antibody-mediated cross-reactivity of proteins from S. mitis and S. pneumoniae, which may have implications for development of effective and wide-range pneumococcal vaccines.
Keywords: Streptococcus pneumoniae, Streptococcus mitis, antibody, antigens, cross-reactivity
Introduction
Streptococcus pneumoniae is a Gram-positive bacterial species that resides in the nasopharynx and oral cavity of humans and causes a wide spectrum of diseases, including pneumonia, meningitis, and septicemia (1). On the other hand, Streptococcus mitis, a commensal that exhibits genetic and ecological similarity with S. pneumoniae, is one of the most abundant members of the oral microbiota and rarely causes disease (2). Clinic-epidemiological studies indicate that S. pneumoniae is the fourth most frequent cause of lethal infections and inflicts death on more than a million children under the age of 5 years worldwide (3, 4). In addition, immunocompromised and elderly individuals are highly susceptible to S. pneumoniae infections (3, 5). Although current pneumococcal vaccines, both conjugated (PCV7, 10, and 13) and unconjugated (PPSV23), have reduced the occurrence of diseases, they are unable to confer protective immunity against all S. pneumoniae serotypes (more than 90) (6, 7). Recent reports showing replacement by non-vaccine serotypes pose grave concern (7). Therefore, novel prophylactic strategies are needed to develop better vaccines that could provide long-term protection against all pneumococcal serotypes.
Understanding host immunity to pneumococcal infections is crucial for identifying immune targets for vaccine development. Several studies involving mouse models of S. pneumoniae indicate that both arms of the adaptive immunity (antibody and T cell) play an important role in mounting protective immunity to infection (8–14). In a mouse model of nasopharyngeal colonization by S. pneumoniae, it was shown that mice deficient in CD4+ T cells or IL-17 had a reduced ability to clear streptococcal carriage compared with control mice (12). By contrast, protective immunity to septicemia after previous colonization is conferred by antibody-mediated (IgG and IgA) immunity (14). This suggests that the type of protective immune response against pneumococcal infections is largely influenced by the site of infection. Using a murine model of nasopharyngeal colonization by S. pneumoniae EF3030 followed by lung infection, Wilson et al. recently showed that colonization-induced protection in mice deficient in B cells, CD4+ T cells or IL-17 is impaired, suggesting that naturally acquired immunity to S. pneumoniae lung infection requires both humoral and cellular immunity (13). This has also been confirmed in controlled human infection studies with pneumococcus. Following experimental colonization, volunteers protected against subsequent re-colonization with the same strain presented increased pneumococci-specific antibodies as well as CD4+ T cells (8). On the other hand, how the host responds to S. mitis is poorly understood. Salivary IgA antibodies reactive to S. mitis antigens have been reported in human infants (15–17). Our recent study has shown that human CD4+ T cells, which express IL-17 (Th17 cells) and are reactive with S. mitis, also respond to pneumococcal proteins when cocultured with antigen-presenting cells pulsed with S. pneumoniae (18).
Currently available pneumococcal vaccines provide immunity that is mediated by serotype-specific antibody responses, which has limitations as this protection is restricted to only 7–23 serotypes. In addition, capsular polysaccharide vaccines are more protective against invasive pneumococcal diseases than non-invasive diseases, such as otitis media (5, 6). A novel approach to deal with these limitations and provide broader serotype coverage could be to exploit the immune cross-reactivity between commensal and pathogenic microbes. The antibodies generated against commensal microbes have been reported to cross-react with pathogens (18–20). Although a few studies have examined the serological cross-reactivity of polysaccharide antigens between S. mitis and S. pneumoniae, it remains unclear whether antibodies specific to S. mitis cross-react with pneumococcal proteins (21–23). In this study, we hypothesized that antibodies elicited against S. mitis and S. pneumoniae show cross-reactivity to S. pneumoniae and S. mitis proteins, respectively. To test this hypothesis, we examined the ability of antibodies raised against S. mitis or S. pneumoniae to cross-react with S. pneumoniae and S. mitis and attempted to identify pneumococcal protein antigens that react with the antibodies. Our findings shed light on the antibody-mediated cross-reactivity between S. mitis and S. pneumoniae. This may have implications for designing effective prophylactic strategies against pneumococcal infections. S. mitis-based vaccines can induce the production of antibodies reactive to S. pneumoniae proteins, resulting in serotype-independent protection against pneumococci.
Materials and Methods
Bacterial Strains, Culture, and Lysis
Streptococcus mitis strain used was CCUG31611 (type strain, equivalent to NCTC12261), whereas S. pneumoniae strains included TIGR4, D39, and BHN418 (6B). The BHN418 6B strain was used in Experimental Human Pneumococcal Carriage (EHPC) studies (8). All strains were stored at −80°C in trypticase soy broth (Becton Dickinson, Franklin Lakes, NJ, USA), and 15% glycerol. Before use, stock cultures were diluted 1:10, followed by growth at 37°C to an optical density (OD) of 0.5 at 600 nm in a 5% CO2 incubator. The bacterial cells were harvested by centrifugation at 5,000 g for 10 min at 4°C and washed in endotoxin free Dulbecco’s-PBS (Sigma-Aldrich, St. Louis, MO, USA) for further processing. To lyse the bacterial cells, a Precellys Lysing Kit and Homogenizer (Precellys 24, Bertin Instruments) was used as per the manufacturer’s instruction. In brief, the bacterial cells dissolved in cold PBS were poured into the Precellys Lysing tube prefilled with high quality glass beads (0.5 mm) and homogenized for disruption of bacterial cells (Precellys 24 homogenizer; program 2). After homogenization, the bacterial cell lysate was centrifuged at 1,000 g for 5 min at 4°C and the supernatant was collected and stored at −80°C for further use. The total protein in the cell lysate was quantified using BCA Protein Assay Kit (ThermoFischer Scientific).
Raising Antisera
Streptococcus mitis antisera were obtained from specific pathogen free New Zealand White rabbits (ProteoGenix, France). Rabbits were subcutaneously immunized with 10 × 106 colony-forming units (CFUs) of ultraviolet-killed S. mitis type strain (CCUG31611) twice at an interval of 2 weeks, and the antisera were collected at day 28 after the first immunization. We also purchased rabbit antisera raised against whole cell of S. pneumoniae serotypes; (1) Type serum 2 against 1, 2, 4, 5, 11, 18, 20, 22, and 33 serotypes (SSI Diagnostica, Denmark) and (2) pooled antisera against S. pneumoniae 3, 4, 6, 9, 14, 18, 19, and 23 surface antigens (Meridian Life Sciences, TN, USA), for specific experiments. Pre- and post-inoculation plasma samples from human volunteers intranasally inoculated with S. pneumoniae were received from the EHPC collaboration at the Liverpool School of Tropical Medicine, Liverpool, UK (24). In brief, 13 healthy individuals of 18–50 years of age were inoculated with 8 × 104 CFU per 100 µl of S. pneumoniae 6B (BHN418) per naris, which eventually developed pneumococcal carriage, as previously described (24). Plasma samples were collected at baseline and at day 21 post-inoculation and stored at −80°C (24). The individuals naturally colonized with pneumococcus at baseline or in regular contact with at-risk individuals were excluded. Of note, both informed and written consent was obtained from the participants. Ethical approvals were obtained from the National Health Service Research Ethics Committee (12/NW/0873).
Western Blot and Immunoinhibition Assay
SDS-PAGE-separated proteins were transferred from gradient gels (4–20%) to nitrocellulose membranes using a constant voltage of 100 V for 2 h using the BioRad Midi-PROTEAN Western blotting module according to the manufacturer’s instructions (BioRad, CA, USA). The membranes were blocked with blocking solution [Tris-buffered saline (TBS) (pH 7.4) containing 0.1% Tween 20 (Sigma-Aldrich) and 5% BSA] for 1 h at room temperature, washed three times with TBS-Tween, and incubated overnight with antisera at 4°C. Following the incubation, the membranes were washed and finally incubated with AP-conjugated goat anti-rabbit or antihuman IgG secondary antibodies and later developed with the substrate (4-chloro-1-naphthol). Of note, the exposure time was similar for the membranes treated with pre- and post-inoculation serum/plasma samples. For immunoinhibition assays, lysates from S. mitis CCUG31611 and S. pneumoniae 6B were immobilized on CNBr-activated Sepharose 4B beads according to the instructions of the manufacturer (GE Healthcare, Germany). After blocking with 0.2 M glycine (pH 8), the beads were washed and used for absorption of rabbit anti-S. mitis antibodies. Following absorption, the unabsorbed serum antibodies were used to perform immunoblotting.
ELISA
Bacterial whole-cell ELISAs were performed to measure the levels of antibodies. Briefly, each well of a 96-well plate (Maxisorb, Nunc, Thermo Scientific) was coated overnight with 100 µl of bacterial suspension (OD600 0.5), which was washed and then fixed with 10% formalin. The plate was washed four times (200 µl PBS + 0.05% Tween) before addition of 100 µl blocking buffer (PBS + 0.05% Tween + 1% BSA) and incubation for 1 h at 37°C. Serially diluted plasma samples were added to wells in duplicate, incubated for 2 h at room temperature before addition of the anti-IgG/HRP secondary antibody diluted 1:10,000, followed by incubation for 2 h at room temperature, washing and addition of 100 µl of TMB substrate (ThermoFisher Scientific, Rockford, IL, USA) to each well. The plates were then incubated in the dark at room temperature for 15 min, after which stop solution (ThermoFisher Scientific, Rockford, IL, USA) was added to each well to terminate the reaction, and absorbance was measured by reading the plates at 450 nm using a cell imaging Multi-Mode reader (BioTek™ Cytation™ 3; ThermoFischer Scientific). For evaluation, titration curves were established for each measurement and cutoffs applied to convert serum activities into titers. For estimation of titers, cutoffs were placed at OD450 = 0.4 for the rabbit hyperimmune sera and OD450 = 0.2 for the human sera because those values were within the linear parts of the titration curves. As an example, an ELISA titration curve has been provided as Figure S2 in Supplementary Material.
Production and Overexpression of Recombinant Proteins
E. coli clones for expression of the S. pneumoniae TIGR4 recom-binant proteins were generated at John Craig Venter Institute, USA, whereas expression clones for the S. mitis type strain recombinant proteins were constructed at the Department of Oral Biology, University of Oslo, Norway. Gateway cloning strategy was used to generate expression clones for the production of recombinant proteins. Briefly, S. mitis target genes were amplified using the oligonucleotide primers listed in Table S1 in Supplementary Material. S. pneumoniae and S. mitis genes cloned in entry clone pDONR221 were transferred into the destination vector T02, which encodes a hexa-histidine tag at the amino terminal as described previously (25). The cloned inserts were sequence validated before transformation of T02 destination vector into E. coli expression strain BLR (DE3). For overexpression of recombinant proteins, frozen stock of E. coli BLR (DE3)-containing expression clones were inoculated in 1 ml 2× YT medium in deep 96-well blocks with ampicillin (100 µg/ml) and tetracycline (12.5 µg/ml). The bacterial cultures were incubated at 37°C with shaking at 800 rpm. Upon reaching an OD600 of 0.7–0.9, protein expression was induced by addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 1 mM, and incubation proceeded overnight at 25°C at 900 rpm. The overnight cultures were then collected by centrifugation at 3,500 rpm at 4°C for 20 min. The pellets were resuspended in 90 µl low salt lysis buffer (50 mM Tris, 100 mM NaCl, 10 mM imidazole, pH 7.3), 1 mM DTT, 1 µl lysonase (Novagen), and 200 µM aminoethyl benzenesulfonyl fluoride hydrochloride (AEBSF) protease inhibitor and incubated for 20 min at room temperature. To complete the lysis process, 10 µl of pop culture (Novagen) was added and incubated for 30 min at room temperature. The bacterial lysate was run on the Nu-PAGE gel (Invitrogen Nu-PAGE 12% Bis–Tris gel) to validate expression and size of proteins.
On-Chip Purification of Protein Microarray and Testing With Antiserum
High-density Cu++ chelated slides (MicroSurfaces, Inc., Englewood, NJ, USA) were used for on-chip purification of hexa-His tagged recombinant proteins as previously described (26). Slides were incubated at room temperature in a humidity chamber for 30 min before printing. Cell lysates were diluted 1:20 in printing buffer PBST (10 mM phosphate buffer, 2.7 mM KCl, 140 mM NaCl, 0.05% Tween 20, pH 7.4) and printed manually on chips with Microcaster array tool (Whatman). Slides were dried in 70% humidity chamber at room temperature. The printed slides were assembled in the fast frame incubation chamber (Whatman) and blocked with blocking buffer (RayBiotech) for 30 min at room temperature. The antisera were cleaned up with E. coli lysate and protease inhibitor cocktail by incubation and spinning down. Blocking buffer was removed, the cleaned and diluted 1:100–1:1,000 antisera in blocking buffer were added on the slides and incubated for 1 h. After incubation, the arrayed slides were washed with 1 ml PBST four times by incubating for 2 min between each wash. Alexa-647 conjugated goat anti-rabbit antibody was also cleaned with E. coli cell lysate and applied on the slide and the secondary antibody on the array was incubated for 1 h. The arrays were washed with PBST four times for 2 min each wash. The array assembly was departed, and the array slide was put into a 30 ml washing tube and washed with PBST for 15 min. Then, the array slides were rinsed with water and dried by centrifugation. To detect immobilized recombinant proteins, Alexa-555 conjugated anti-hexa-His tag antibody (ThermoFisher Scientific) was cleaned with E. coli lysate, diluted to 1:1,000 in blocking buffer, applied on the array slide and incubated for 1 h. After incubating, the array slides were washed and dried as described earlier. Slides were scanned using Genepix 4000B and quantified using Genepix Pro 4.0.
Purification of PsaA Protein of S. mitis and S. pneumoniae
The PsaA protein of S. mitis and S. pneumoniae was purified using Dynabeads TALON which utilizes cobalt based immobilized metal affinity chromatography. E. coli BLR (DE3) expression clones containing PsaA gene from S. mitis and S. pneumoniae were grown in 50 ml 2× YT medium supplemented with ampicillin (100 µg/ml) and tetracycline (12.5 µg/ml). Protein expression was induced by IPTG at OD600 0.6 followed by incubation at 25°C for 4 h at 200 rpm. Cells were harvested by centrifugation, and cell pellets were stored overnight at −80°C. For cell lysis, pellets were resuspended in B-PER complete bacterial protein extraction reagent (ThermoFisher Scientific). Supernatant was collected by centrifugation of cell lysates at 16,000 g for 20 min and subjected to His-tag protein purification using Dynabeads TALON (Dynal biotech) according to the manufacturer’s instructions. The purified proteins were analyzed by SDS-PAGE gel electrophoresis, and reactivity to S. mitis and S. pneumoniae rabbit antisera was assessed using Western blot.
Statistical Analysis
Wilcoxon Signed Rank tests were applied using the SigmaPlot v13, Systat Software Inc., London, UK. p Values < 0.05 were considered to be significant.
Results
IgG Antibodies Specific to S. mitis Cross-React With Different S. pneumoniae Serotypes
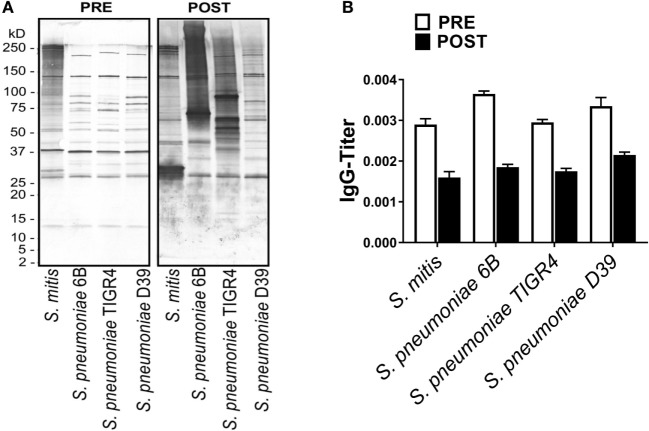
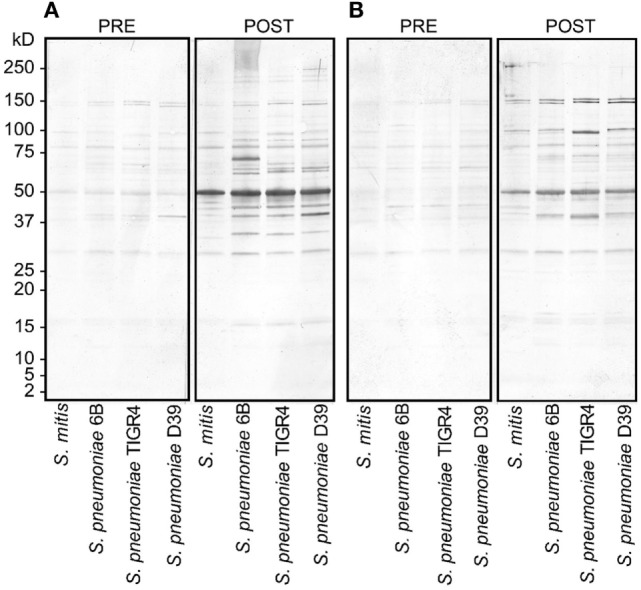
To assess the reactivity of antibodies raised against S. mitis with pneumococcal antigens, we used antisera from rabbits that received S. mitis type strain (rabbit 1 and rabbit-2), and performed SDS-PAGE and immunoblotting. We found an enhanced binding of the IgG antibodies from S. mitis antisera (rabbit-1 and rabbit-2), compared with the pre-immunized sera, to the proteins from S. pneumoniae serotypes (6B, TIGR4, and D39), as shown by multiple stronger bands (Figures 1A,B). Most of the strongest bands were present at the level of molecular weight 50 kDa for all the S. pneumoniae serotypes (Figures 1A,B). Interestingly, a clear and thick band of molecular weight between 50–75 kDa was found in the lane receiving 6B pneumococcal protein, but absent from all other lanes including the one that had S. mitis proteins (Figure 1A). Of note, we did not find any appreciable bands while examining the binding of IgA antibodies to pneumococcal proteins (data not shown). After having immunoblotting data on the cross-reactivity of S. mitis antisera, we sought to examine the titer of anti-S. mitis rabbit IgG antibodies reactive to pneumococcal proteins. We found an increased titer of the IgG antibodies reactive to S. pneumoniae and S. mitis compared with the antibodies from unimmunized rabbits (Figure 2; nota bene as titers were used, lower numbers indicate higher antibody concentration, please see legend). To further confirm the cross-reactivity between S. mitis and S. pneumoniae, we performed an immunoinhibition assay by incubating rabbit anti-S. mitis sera with immobilized S. pneumoniae 6B or S. mitis lysates. Upon immunoblotting, the S. mitis antisera incubated with S. pneumoniae 6B or S. mitis exhibited significant inhibition of bands for S. mitis and S. pneumoniae compared with the antisera without antibody absorption (Figure 3). Similar findings were observed when we looked for the bacteria-specific IgG titer in the antisera with different incubations (Figure 3). Overall, these findings show that IgG antibodies specific to S. mitis cross-react with S. pneumoniae.
Figure 1.
Reactivity of rabbit antisera raised against Streptococcus mitis with Streptococcus pneumoniae strains (6B, TIGR4, and D39) by SDS-PAGE in gradient gels and immunoblotting. Each lane was loaded with 50 µg of protein from the indicated bacterial species. The pre- and post-inoculation sera were diluted 1:1,000. (A,B) Antisera from rabbit-1 and rabbit-2, which were immunized with S. mitis CCUG31611. The experiment was performed twice. Abbreviations: PRE, pre-inoculation sera; POST, post-inoculation sera.
Figure 2.
Levels of Streptococcus pneumoniae-reactive IgG antibodies from pre- or post Streptococcus mitis-immunized rabbits. The titer of IgG antibodies was determined using whole-cell ELISA. Antisera were used from rabbit-1 and rabbit-2, immunized with S. mitis CCUG31611. The serum samples were serially diluted, and the OD values converted to titers, with a cutoff of OD value of 0.40 (see Materials and Methods; low numbers indicate high antibody concentration, e.g., 0.002 = 1/500 and 0.004 = 1/250). The bars show averages of two replicates. Abbreviations: PRE, pre-inoculation sera; POST, post-inoculation sera.
Figure 3.
Immunoinhibition for IgG cross-reactivity between Streptococcus mitis and Streptococcus pneumoniae. For SDS-PAGE, wells were loaded with 50 µg of protein from S. mitis CCUG31611 and S. pneumoniae strains. After blotting, untreated rabbit S. mitis antiserum or the same antiserum pre-incubated with immobilized S. mitis or S. pneumoniae 6B lysates were used to detect immunoreactivity. Levels of S. mitis or S. pneumoniae-reactive IgG antibodies in the same antiserum preparations. The levels of IgG antibodies were determined using whole-cell ELISA. The OD values were converted to titers, with a cutoff of OD value of 0.40 (see legend to Figure 2). The bars show averages of two replicates.
Enhanced Reactivity of Antisera Raised Against S. pneumoniae to S. mitis
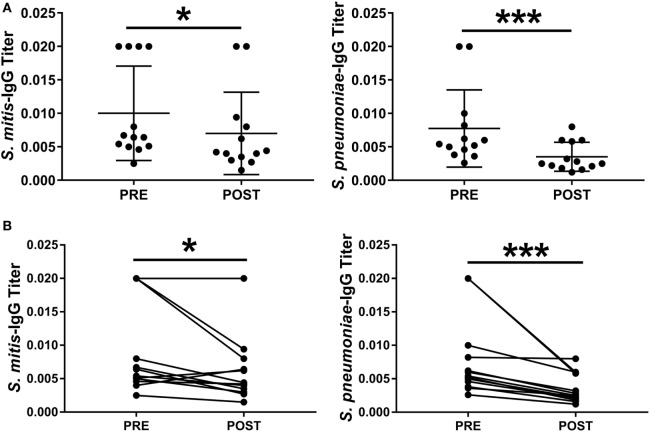
To understand whether antibodies specific to S. pneumoniae can in turn cross-react with S. mitis, we used rabbit antisera raised against S. pneumoniae to examine the reactivity of the antisera IgG antibodies with S. mitis proteins. Our findings demonstrated an enhanced binding pattern of anti-S. pneumoniae IgG antibodies to S. mitis and S. pneumoniae strains, particularly 6B and TIGR4, when compared with the control antibodies from unimmunized rabbits (Figure 4A). However, the post-inoculation binding pattern for S. mitis had some differences compared with S. pneumoniae 6B and TIGR4 (Figure 4A). Similarly, the levels of anti-S. pneumoniae IgG titer reactive to S. mitis increased compared with the antibody levels of pre-immunized serum (Figure 4B). To validate these rabbit findings to humans, we used plasma samples from 13 healthy adults intranasally inoculated with S. pneumoniae 6B, which eventually acquired pneumococcal carriage (24). Our whole-cell ELISA experiments revealed that plasma IgG antibodies from humans inoculated with S. pneumoniae show increased reactivity to S. mitis and S. pneumoniae 6B compared with the pre-inoculation samples (Figure 5). Taken together, these data indicate that cross-reactivity is conferred by pneumococcal antibodies against S. mitis antigens.
Figure 4.
Reactivity of rabbit antiserum raised against Streptococcus pneumoniae with Streptococcus mitis. (A) Reactivity of a rabbit antiserum raised against a pool of S. pneumoniae serotypes (1, 2, 4, 5, 11, 18, 20, 22, and 33), with S. mitis and S. pneumoniae strains (6B, TIGR4, and D39) using SDS-PAGE and immunoblotting. Each lane was loaded with 50 µg of protein from each bacterial species. The pre- and post-inoculation sera were diluted 1:1,000. (B) Pre- and post-immunization levels of S. mitis-reactive IgG antibodies from a S. pneumoniae-immunized rabbit. The titer of IgG antibodies was determined by converting the OD values to titers, with a cutoff of OD value of 0.40 (see legend to Figure 2). The bars show averages of two replicates. Abbreviations: PRE, pre-inoculation sera; POST, post-inoculation sera.
Figure 5.
Levels of Streptococcus mitis- and Streptococcus pneumoniae-reactive IgG antibodies isolated from S. pneumoniae 6B-inoculated adults. Pre- and post-inoculation plasma samples were collected from 13 individuals inoculated with S. pneumoniae 6B. Serially diluted plasma samples were added to wells in duplicate. The OD values were converted to titers, with a cutoff value of 0.20 (see legend to Figure 2). The levels of IgG antibody titers against S. mitis and S. pneumoniae strains are shown. OD values are shown for each subject at pre- and post-challenge time points. (A) Pre- and post-challenge IgG levels to S. mitis and S. pneumoniae 6B. (B) Pre- and post-challenge IgG levels to S. mitis and S. pneumoniae 6B linked for each subject. Wilcoxon Signed Rank Test, *p < 0.05; ***p < 0.001. Abbreviations: PRE, pre-inoculation sera; POST, post-inoculation sera.
Identification of Pneumococcal Proteins That Cross-React With S. mitis Antisera
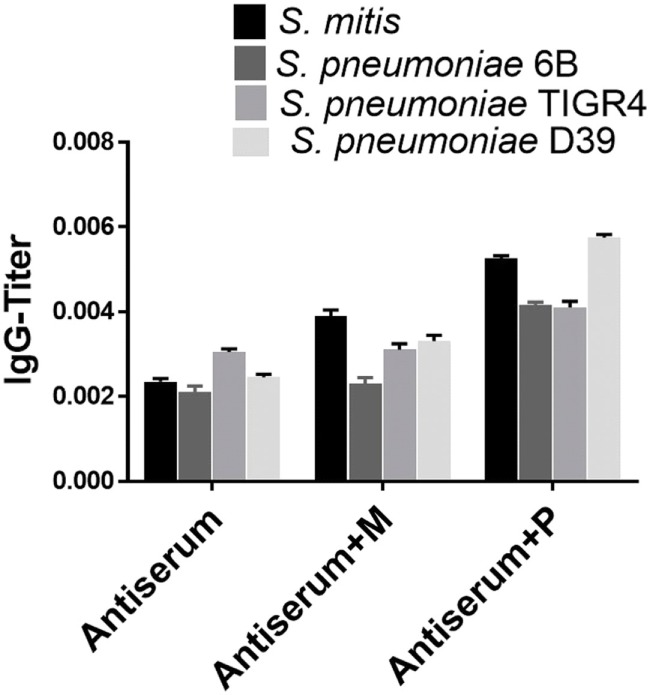
Pneumococcal surface proteins have been shown to induce host responses to S. pneumoniae, underscoring the potential of these proteins as potent vaccine candidates (6, 7). Our Western blot data show that some of the most prominent bands, which developed due to reaction between S. pneumoniae/S. mitis proteins and anti-S. mitis IgG antibodies had a molecular weight around 50 kDa (Figure 1). To identify some of the bacterial proteins that cross-react with anti-S. mitis IgG antibodies, we selected a panel of 30 predicted extracellular and membrane proteins that have around 50 kDa molecular weight (Table 1) and analyzed them using on-chip high quality protein microarray. An array map has been included that shows the position of S. pneumoniae TIGR4 proteins printed and immobilized on high-density Cu++ chelated slides (Table S2 in Supplementary Material). The immobilized proteins on microarray chips were visualized using Alexa-555 conjugated anti-His-tag antibody. Data analysis identified three different pneumococcal proteins with signal ratio (Fsample/FE. coli) > 1.5—choline-binding protein D (CbpD; SP2201), cell division protein (FtsH; SP0013), and manganese ABC transporter or manganese-binding adhesion lipoprotein (PsaA; SP1650)—upon treatment with antisera (IgG) against S. mitis (Figure 6; Table 2). Maltose/maltodextrin-binding protein (SP2108) was identified using antisera against S. pneumoniae, but not with antisera against S. mitis (Figure 6A). S. pneumoniae PsaA, in particular, is a protein with immunogenic activity that has been investigated in several studies as a potential vaccine against pneumococcal infections (27–29). Thus, we cloned and expressed the S. mitis PsaA in E. coli to check it for cross-reactivity with antibodies raised against S. mitis and S. pneumoniae. Our Western blot analysis revealed a stronger binding of rabbit S. mitis and S. pneumoniae antisera to PsaA compared with the pre-inoculation serum (Figure 6B). The original immunoblot has been provided (Figure S1 in Supplementary Material). Thus, we identified three common proteins, CbpD, FtsH, and PsaA, derived from S. pneumoniae reactive to antisera raised against S. mitis or S. pneumoniae. We also found that purified PsaA from S. mitis reacted with the antisera against S. mitis, suggesting similar cross-reactive responses for protein antigens from S. mitis and S. pneumoniae.
Table 1.
List of Streptococcus pneumoniae proteins selected for on-chip protein microarray experiments.
| Locus ID S. pneumoniae TIGR4 | Protein | Subcellular location | Molecular weight (Da) | Ortholog in Streptococcus mitis NCTC12261 | % Identity |
|---|---|---|---|---|---|
| SP0013 | Cell division protein FtsH | Membrane | 71,326 | NAa | 93 |
| SP0091 | ABC transporter; permease protein | Membrane | 34,545 | SM12261_0720 | 93 |
| SP0092 | ABC transporter substrate-binding protein | Extracellular | 54,485 | SM12261_0719 | 92 |
| SP0102 | Glycosyl transferase | Membrane | 40,703 | No significant similarity | – |
| SP0107 | LysM domain protein | Extracellular | 20,921 | SM12261_0703 | 96 |
| SP0117 | Surface protein A | Membrane | 82,764 | No significant similarity | – |
| SP0385 | Hypothetical protein | Membrane | 27,192 | SM12261_1022 | 95 |
| SP0523 | ABC transporter, permease protein, putative | Membrane | 40,505 | SM12261_0538 | 94 |
| SP0704 | Hypothetical protein | Membrane | 28,261 | No significant similarity | – |
| SP0749 | Branched-chain amino acid ABC transporter; amino acid-binding protein (livJ) | Extracellular | 40,414 | SM12261_0255 | 94 |
| SP0751 | Branched-chain amino acid ABC transporter; permease protein (livM) | Membrane | 33,564 | SM12261_0253 | 96 |
| SP0954 | Competence protein CelA | Membrane | 23,181 | SM12261_1388 | 92 |
| SP1002 | Adhesion lipoprotein (lmb) | Extracellular | 33,951 | SM12261_0792 | 93 |
| SP1067 | Cell division protein FtsW; putative | Membrane | 45,449 | No significant similarity | – |
| SP1069 | Conserved hypothetical protein | Extracellular | 36,428 | SM12261_1418 | 90 |
| SP1264 | Conserved domain protein | Membrane | 38,825 | No significant similarity | – |
| SP1400 | Phosphate ABC transporter, phosphate-binding protein, putative | Extracellular | 31,201 | SM12261_0153 | 78 |
| SP1601 | Conserved hypothetical protein | Membrane | 24,619 | SM12261_0284 | 92 |
| SP1604 | Hypothetical protein | Membrane | 16,812 | SM12261_0287 | 79 |
| SP1628 | Hypothetical protein | Membrane | 8,493 | NA | 97 |
| SP1650 | Manganese ABC transporter; manganese-binding adhesion lipoprotein (PsaA) | Extracellular | 34,593 | SM12261_0346 | 95 |
| SP1684 | PTS system; IIBC components | Membrane | 54,471 | SM12261_0375 | 98 |
| SP1839 | ABC transporter ATP-binding protein/permease | Membrane | 65,551 | NA | 92 |
| SP1952 | Hypothetical protein | Membrane | 36,169 | No significant similarity | – |
| SP1964 | DNA-entry nuclease (endA) | Membrane | 29,890 | SM12261_0490 | 93 |
| SP2013 | Conserved hypothetical protein | Membrane | 35,985 | No significant similarity | – |
| SP2051 | Competence protein CglC (cglC) | Membrane | 12,177 | SM12261_0632 | 91 |
| SP2108 | Maltose/maltodextrin ABC transporter maltose/maltodextrin-binding protein | Extracellular | 45,337 | SM12261_0678 | 96 |
| SP2201 | Choline-binding protein D | Extracellular | 50,349 | SM12261_0760 | 89 |
| SP2239 | Serine protease | Membrane | 41,842 | SM12261_0790 | 94 |
aNA, not annotated in the NCBI database, but present in the genome sequences.
Figure 6.
Identification of pneumococcal proteins that cross-react with rabbit Streptococcus mitis antisera. (A) Immunogenic screening of on-chip purified Streptococcus pneumoniae TIGR4 protein microarray. Lysates of 30 recombinant proteins overexpressed in E. coli were printed on the chelated Cu++/PEG surface slides in triplicates. The slides were probed with S. mitis and S. pneumoniae antiserum to identify immunoreactive pneumococcal antigens. Pre- and post-immune serum from rabbits challenged with S. mitis type strain, and post-immune serum from rabbits challenged with S. pneumoniae were used on the slides. Values were obtained by ratio of fluorescence intensity of spot to fluorescence intensity of E. coli cell lysate. E. coli, His-tag MBP; ~50% similarity to SP2108. Abbreviations: PRE-SM, pre-inoculation S. mitis serum; POST-SM, post-inoculation S. mitis serum; POST-SP, post-inoculation S. pneumoniae serum; CbpD, choline-binding protein D; FtsH, cell division protein; PsaA, manganese ABC transporter adhesion lipoprotein; MBP, maltose-binding protein. (B) Reactivity of rabbit anti-S. mitis IgG antisera with S. pneumoniae TIGR4 and S. mitis PsaA by SDS-PAGE in gradient gels and immunoblotting. Each lane was loaded with 50 µg of protein from the indicated bacterial species. The pre- and post-inoculation sera were diluted 1:1,000. Abbreviations: M, S. mitis CCUG31611; P, S. pneumoniae TIGR4.
Table 2.
Quantification of fluorescence signal ratio of on-chip purified Streptococcus pneumoniae proteins obtained by ratio of fluorescence intensity of spot to fluorescence intensity of E. coli lysate.
|
S. pneumoniae |
Streptococcus mitis |
|||
|---|---|---|---|---|
| Average | SD | Average | SD | |
| Buffer | 0.9 | 0.1 | 1.0 | 0.3 |
| E. coli_lysate | 1.0 | 0.2 | 1.0 | 0.4 |
| SP0013a | 2.1 | 0.4 | 1.9 | 0.5 |
| SP0091 | 1.0 | 0.1 | 1.0 | 0.3 |
| SP0092 | 1.4 | 0.2 | 1.1 | 0.3 |
| SP0102 | 1.2 | 0.2 | 1.1 | 0.3 |
| SP0107 | 0.9 | 0.1 | 1.1 | 0.3 |
| SP0117 | 2.0 | 0.4 | 1.4 | 0.4 |
| SP0385 | 1.2 | 0.2 | 1.4 | 0.4 |
| SP0523 | 1.2 | 0.2 | 1.0 | 0.3 |
| SP0704 | 1.1 | 0.3 | 0.9 | 0.3 |
| SP0749 | 1.1 | 0.1 | 1.0 | 0.3 |
| SP0751 | 1.1 | 0.1 | 1.1 | 0.4 |
| SP0954 | 1.2 | 0.2 | 1.0 | 0.3 |
| SP1002 | 1.3 | 0.2 | 1.2 | 0.3 |
| SP1067 | 1.0 | 0.2 | 1.1 | 0.4 |
| SP1069 | 1.0 | 0.1 | 1.2 | 0.4 |
| SP1264 | 2.1 | 0.4 | 1.2 | 0.4 |
| SP1400 | 1.1 | 0.1 | 1.0 | 0.4 |
| SP1601 | 1.2 | 0.2 | 1.1 | 0.6 |
| SP1604 | 1.5 | 0.2 | 1.0 | 0.4 |
| SP1628 | 1.3 | 0.2 | 1.2 | 0.3 |
| SP1650a | 41.3 | 6.4 | 2.4 | 0.7 |
| SP1684 | 1.3 | 0.2 | 1.1 | 0.5 |
| SP1839 | 1.1 | 0.1 | 1.1 | 0.3 |
| SP1952 | 1.4 | 0.3 | 1.3 | 0.5 |
| SP1964 | 1.3 | 0.4 | 1.2 | 0.6 |
| SP2013 | 1.3 | 0.1 | 1.3 | 0.4 |
| SP2108 | 2.7 | 0.9 | 1.2 | 0.3 |
| SP2201a | 2.7 | 0.6 | 1.6 | 0.5 |
| SP2239 | 1.1 | 0.2 | 1.0 | 0.4 |
aSignal intensity ratio > 1.5 for both S. pneumoniae and S. mitis.
Average values were calculated from three replicates.
Discussion
The aim of this study was to examine the antibody-mediated cross-reactivity between S. pneumoniae and the commensal S. mitis and to identify common cross-reactive protein antigens using multiple experimental approaches. The major findings of the study include the following: (1) rabbit IgG antibodies raised with S. mitis cross-react with S. pneumoniae; (2) rabbit anti-S. pneumoniae IgG antibodies show cross-reactivity with S. mitis; (3) IgG antibodies from humans inoculated with S. pneumoniae demonstrate increased reactivity to S. mitis; and (4) antisera raised against S. mitis recognize specific S. pneumoniae and S. mitis surface proteins.
Our findings demonstrated that antisera raised with S. mitis in rabbits cross-react with S. pneumoniae serotypes (6B, TIGR4 of different serotypes, and D39). Cross-reactions have been documented among microbial proteins with similar evolutionary development and struc-ture, e.g., antibody cross-reactivity between proteins from the commensal Neisseria lactamica and the pathogenic Neisseria meningitidis (20, 30). Previous studies suggest that S. mitis and S. pneumoniae share a close genetic relatedness with around 80% genome similarity, and that several pneumococcal proteins are ubiquitously present in S. mitis (31, 32). Therefore, it is understandable why many S. mitis and S. pneumoniae proteins are reactive to antisera against S. mitis as shown in this study. These findings are in line with human studies where salivary antibodies (IgA) were found to react with S. mitis (15–17). Since we did not find significant differences between the post-inoculation antibody levels reacting to S. mitis and S. pneumoniae strains, it may be inferred that the pneumococcal cross-reactive antigens and their ability to bind with S. mitis antisera were similar. Another important finding in our study is that IgG antibodies from humans inoculated with S. pneumoniae show increased reactivity to S. mitis as well as S. pneumoniae 6B, as demonstrated by whole-cell ELISA data. This is in accordance with previous studies showing that S. pneumoniae carriage promotes an increase in IgG responses specific to protein and capsular antigens of S. pneumoniae 6B (8). Overall, these findings support the concept that natural immunity against pneumococcal diseases could be partially due to nasopharyngeal and oral colonization by S. mitis, and this may have consequences in the design of better prophylactic strategies for containing infections for all pneumococcal serotypes.
In the on-chip protein microarray analysis, we evaluated 30 different proteins with a molecular weight of around 50 kDa, as major pneumococcal bands in the immunoblotting experiments were found within this range. We identified three pneumococcal proteins that showed reactivity with antibodies (IgG) against both S. mitis and S. pneumoniae. These proteins included CbpD, PsaA, and FtsH, which are present in the two species and show more than 90% similarity. CbpD and PsaA are extracellular proteins that belong to the family of choline-binding proteins (CBP) and are shown to contribute to nasopharyngeal colonization by S. pneumoniae and competence-induced cell lysis (27, 33). So far, several studies on pneumococcal infections conducted in animal models have demonstrated an antibody-mediated protection induced by CBP members, such as PspC and PsaA (27–29). However, the immunogenic potential of CbpD is still unclear. Given the fact that CbpD facilitates nasopharyngeal colonization by S. pneumoniae and that it possesses ~90% similarity with its ortholog (SM12261_0760) in S. mitis, CbpD holds promise as an immunogenic candidate against S. pneumoniae serotypes, warranting further exploration to assess its efficacy to mount protective immunity that is independent of pneumococcal serotypes. A combined formulation of a vaccine containing PsaA and PspA has been shown to prevent bacterial colonization and otitis media in animal models (34, 35). It would be interesting to investigate the immunogenic potential of a combination of PsaA, FtsH, and CbpD, which may confer an additive protective effect as well as limit the occurrence of allelic variation within these individual proteins.
In conclusion, our findings demonstrate that antibodies raised against S. mitis and S. pneumoniae cross-react with S. pneumoniae and S. mitis proteins, respectively. These findings are in accordance with our hypothesis that antibodies elicited against S. mitis and S. pneumoniae show cross-reactivity to S. pneumoniae and S. mitis proteins, respectively. It is important to note that reactivity of S. mitis-specific antibodies with pneumococcal proteins provides a platform upon which the development a serotype-independent protein-based pneumococcal vaccine with extended coverage can be carried out. Future studies need to assess whether antibody responses due to nasopharyngeal colonization by S. mitis generate cross-protection against S. pneumoniae infections. In addition, it is worth testing the effect of cross-reactive antigens on rendering protection against diverse S. pneumoniae serotypes.
Ethics Statement
Pre- and post-inoculation plasma samples from human volunteers intranasally inoculated with Streptococcus pneumoniae were received from the EHPC collaboration at the Liverpool School of Tropical Medicine, Liverpool, UK. Ethical approvals were obtained from the National Health Service Research Ethics Committee, UK (12/NW/0873).
Author Contributions
SS designed research studies, conducted experiments, acquired and analyzed data, and wrote the paper. RK designed and conducted experiments, acquired and analyzed data, and wrote the paper. DF, EM, and EG provided serum samples from S. pneumoniae (6B) inoculated individuals. GR conducted experiments and analyzed data. DF, DB, and KS contributed to the design of experiments and revision of the manuscript. KK designed and conducted experiments, acquired and analyzed data, and revised the manuscript. FP designed research studies, analyzed data, and wrote the paper. All the authors revised and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to extend their sincere thanks to Ali-Oddin Naemi and Olav Schreurs for their technical assistance.
Footnotes
Funding. This work was supported by a grant from the Norwegian Research Council (Grant number—241011). EHPC studies and sample collections were supported by the Bill and Melinda Gates Foundation (OPP1117728) and Medical Research Council (MR/M011569/1).
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.00747/full#supplementary-material.
References
- 1.Mitchell TJ. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat Rev Microbiol (2003) 1:219–30. 10.1038/nrmicro771 [DOI] [PubMed] [Google Scholar]
- 2.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol (2003) 30:644–54. 10.1034/j.1600-051X.2003.00376.x [DOI] [PubMed] [Google Scholar]
- 3.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis (2005) 5:685–94. 10.1016/S1473-3099(05)70267-X [DOI] [PubMed] [Google Scholar]
- 4.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet (2009) 374:893–902. 10.1016/S0140-6736(09)61204-6 [DOI] [PubMed] [Google Scholar]
- 5.Shigayeva A, Rudnick W, Green K, Chen DK, Demczuk W, Gold WL, et al. Invasive pneumococcal disease among immunocompromised persons: implications for vaccination programs. Clin Infect Dis (2016) 62:139–47. 10.1093/cid/civ803 [DOI] [PubMed] [Google Scholar]
- 6.Daniels CC, Rogers PD, Shelton CM. A review of pneumococcal vaccines: current polysaccharide vaccine recommendations and future protein antigens. J Pediatr Pharmacol Ther (2016) 21:27–35. 10.5863/1551-6776-21.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Hill A, Beitelshees M, Shao S, Lovell JF, Davidson BA, et al. Directed vaccination against pneumococcal disease. Proc Natl Acad Sci U S A (2016) 113:6898–903. 10.1073/pnas.1603007113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira DM, Neill DR, Bangert M, Gritzfeld JF, Green N, Wright AK, et al. Controlled human infection and rechallenge with Streptococcus pneumoniae reveals the protective efficacy of carriage in healthy adults. Am J Respir Crit Care Med (2013) 187:855–64. 10.1164/rccm.201212-2277OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright AK, Bangert M, Gritzfeld JF, Ferreira DM, Jambo KC, Wright AD, et al. Experimental human pneumococcal carriage augments IL-17A-dependent T-cell defence of the lung. PLoS Pathog (2013) 9:e1003274. 10.1371/journal.ppat.1003274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCool TL, Cate TR, Moy G, Weiser JN. The immune response to pneumococcal proteins during experimental human carriage. J Exp Med (2002) 195:359–65. 10.1084/jem.20011576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen JM, Khandavilli S, Camberlein E, Hyams C, Baxendale HE, Brown JS. Protective contributions against invasive Streptococcus pneumoniae pneumonia of antibody and Th17-cell responses to nasopharyngeal colonisation. PLoS One (2011) 6:e25558. 10.1371/journal.pone.0025558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen JM, Chimalapati S, de Vogel C, van Belkum A, Baxendale HE, Brown JS. Contributions of capsule, lipoproteins and duration of colonisation towards the protective immunity of prior Streptococcus pneumoniae nasopharyngeal colonisation. Vaccine (2012) 30:4453–9. 10.1016/j.vaccine.2012.04.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson R, Cohen JM, Jose RJ, de Vogel C, Baxendale H, Brown JS. Protection against Streptococcus pneumoniae lung infection after nasopharyngeal colonization requires both humoral and cellular immune responses. Mucosal Immunol (2015) 8:627–39. 10.1038/mi.2014.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Clarke TB, Weiser JN. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest (2009) 119:1899–909. 10.1172/JCI36731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nogueira RD, Sesso MLT, Borges MCL, Mattos-Graner RO, Smith DJ, Ferriani VPL. Salivary IgA antibody responses to Streptococcus mitis and Streptococcus mutans in preterm and fullterm newborn children. Arch Oral Biol (2012) 57:647–53. 10.1016/j.archoralbio.2011.11.011 [DOI] [PubMed] [Google Scholar]
- 16.Borges MCL, Sesso MLT, Roberti LR, Oliveira MAHD, Nogueira RD, Geraldo-Martins VR, et al. Salivary antibody response to streptococci in preterm and fullterm children: a prospective study. Arch Oral Biol (2015) 60:116–25. 10.1016/j.archoralbio.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 17.Wirth KA, Bowden GH, Kirchherr JL, Richmond DA, Sheridan MJ, Cole MF. Humoral immunity to commensal oral bacteria in human infants: evidence that Streptococcus mitis biovar 1 colonization induces strain-specific salivary immunoglobulin A antibodies. ISME J (2008) 2:728–38. 10.1038/ismej.2008.26 [DOI] [PubMed] [Google Scholar]
- 18.Engen SA, Rukke HV, Becattini S, Jarrossay D, Blix IJ, Petersen FC, et al. The oral commensal Streptococcus mitis shows a mixed memory Th cell signature that is similar to and cross-reactive with Streptococcus pneumoniae. PLoS One (2014) 9(8):e104306. 10.1371/journal.pone.0104306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trama AM, Moody MA, Alam SM, Jaeger FH, Lockwood B, Parks R, et al. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe (2014) 16:215–26. 10.1016/j.chom.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troncoso G, Sanchez S, Moreda M, Criado MT, Ferreiros CM. Antigenic cross-reactivity between outer membrane proteins of Neisseria meningitidis and commensal Neisseria species. FEMS Immunol Med Microbiol (2000) 27:103–9. 10.1111/j.1574-695X.2000.tb01419.x [DOI] [PubMed] [Google Scholar]
- 21.Lund E. Antigenic relationship between pneumococci and non-hemolytic streptococci. Acta Pathol Microbiol Scand (1950) 27:110–8. 10.1111/j.1699-0463.1950.tb05200.x [DOI] [PubMed] [Google Scholar]
- 22.Lee CJ, Koizumi K, Henrichsen J, Perch B, Lin CS, Egan W. Capsular polysaccharides of nongroupable streptococci that cross-react with pneumococcal group-19. J Immunol (1984) 133:2706–11. [PubMed] [Google Scholar]
- 23.Kilian M, Poulsen K, Blomqvist T, Håvarstein LS, Bek-Thomsen M, Tettelin H, et al. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One (2008) 3(7):e2683. 10.1371/journal.pone.0002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins AM, Wright AD, Mitsi E, Gritzfeld JF, Hancock CA, Pennington SH, et al. First human challenge testing of a pneumococcal vaccine double-blind randomized controlled trial. Am J Respir Crit Care Med (2015) 192:853–8. 10.1164/rccm.201503-0542OC [DOI] [PubMed] [Google Scholar]
- 25.Kwon K, Pieper R, Shallom S, Grose C, Kwon E, Do Y, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins. Protein Expr Purif (2007) 55:368–78. 10.1016/j.pep.2007.06.006 [DOI] [PubMed] [Google Scholar]
- 26.Kwon K, Grose C, Pieper R, Pandya GA, Fleischmann RD, Peterson SN. High quality protein microarray using in situ protein purification. BMC Biotechnol (2009) 9:72. 10.1186/1472-6750-9-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajam G, Anderton JM, Carlone GM, Sampson JS, Ades EW. Pneumococcal surface adhesin A (PsaA): a review. Crit Rev Microbiol (2008) 34(3-4):131–42. 10.1080/10408410802275352 [DOI] [PubMed] [Google Scholar]
- 28.Paton JC, Andrew PW, Boulnois GJ, Mitchell TJ. Molecular analysis of the pathogenicity of Streptococcus pneumoniae – the role of pneumococcal proteins. Annu Rev Microbiol (1993) 47:89–115. 10.1146/annurev.mi.47.100193.000513 [DOI] [PubMed] [Google Scholar]
- 29.Ogunniyi AD, Woodrow MC, Poolman JT, Paton JC. Protection against Streptococcus pneumoniae elicited by immunization with pneumolysin and CbpA. Infect Immun (2001) 69:5997–6003. 10.1128/IAI.69.10.5997-6003.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Troncoso G, Sanchez S, Criado MT, Ferreiros CM. Analysis of Neisseria lactamica antigens putatively implicated in acquisition of natural immunity to Neisseria meningitidis. FEMS Immunol Med Microbiol (2002) 34:9–15. 10.1111/j.1574-695X.2002.tb00597.x [DOI] [PubMed] [Google Scholar]
- 31.Madhour A, Maurer P, Hakenbeck R. Cell surface proteins in S. pneumoniae, S. mitis and S. oralis. Iran J Microbiol (2011) 3:58–67. [PMC free article] [PubMed] [Google Scholar]
- 32.Denapaite D, Bruckner R, Nuhn M, Reichmann P, Henrich B, Maurer P, et al. The genome of Streptococcus mitis B6-what is a commensal? PLoS One (2010) 5:e9426. 10.1371/journal.pone.0009426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kausmally L, Johnsborg O, Lunde M, Knutsen E, Havarstein LS. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J Bacteriol (2005) 187:4338–45. 10.1128/JB.187.13.4338-4345.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogunniyi AD, Folland RL, Briles DE, Hollingshead SK, Paton JC. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect Immun (2000) 68:3028–33. 10.1128/IAI.68.5.3028-3033.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briles DE, Ades E, Paton JC, Sampson JS, Carlone GM, Huebner RC, et al. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun (2000) 68:796–800. 10.1128/IAI.68.2.796-800.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.