Abstract
Objectives
Investigate clinical and epidemiological factors of pediatric GII.4 norovirus infections in children with acute gastroenteritis (AGE) in Nicaragua between 1999 and 2015.
Methods
We retrospectively analyzed laboratory and epidemiologic data from 1,790 children ≤ 7 years with AGE from 6 hospitals in Nicaragua (n = 538), and 3 community clinics (n = 919) and households (n = 333) in León, between 1999 and 2015. Moreover, asymptomatic children from community clinics (n = 162) and households (n = 105) were enrolled. Norovirus was detected by real-time PCR and genotyped by sequencing the N-terminal and shell region of the capsid gene.
Results
Norovirus was found in 19% (n = 338) and 12% (n = 32) of children with and without AGE, respectively. In total, 20 genotypes including a tentatively new genotype were detected. Among children with AGE, the most common genotypes were GII.4 (53%), GII.14 (7%), GII.3 (6%) and GI.3 (6%). In contrast, only one (1.4%) GII.4 was found in asymptomatic children. The prevalence of GII.4 infections was significantly higher in children between 7 and 12 months of age. The prevalence of GII.4 was lowest in households (38%), followed by community clinics (50%) and hospitals (75%). Several different GII.4 variants were detected and their emergence followed the global temporal trend.
Conclusions
Overall our study found the predominance of pediatric GII.4 norovirus infections in Nicaragua mostly occurring in children between 7 and 12 months of age, implicating GII.4 as the main norovirus vaccine target.
Keywords: Norovirus, Nicaragua, GII.4, Pediatric, Gastroenteritis, Asymptomatic, Community
1. Introduction
Globally, norovirus is a key pathogen associated with nearly a fifth of all cases of acute gastroenteritis [Ahmed et al., 2014]. In countries where infant vaccination has reduced the incidence of rotavirus disease, norovirus has become the most common cause of pediatric gastroenteritis [Bucardo et al., 2014; Payne et al., 2013]. Despite efforts to understand norovirus disease epidemiology in low- and middle-income countries (LMIC), there is still limited comprehensive epidemiological studies from these areas [Ayukekbong et al., 2015; da Silva et al., 2016]. A recent review and meta-analysis from Latin American studies showed a norovirus prevalence in acute gastroenteritis (AGE) cases of 15% in the community, 14% in outpatient settings, 16% in hospital locations and 8% among asymptomatic subjects [O'Ryan et al., 2017].
Viruses belonging to the Norovirus genus in the Caliciviridae family can be divided into at least six genogroups (GI to GVI), of which GII viruses cause the majority (> 80%) of disease in humans and GI viruses being less common detected(< 11%) [Pringle et al., 2015; Vinje, 2015]. By phylogenetic analysis of the major capsid protein VP1, 22 genotypes have been recognized in GII and 9 in GI [Kroneman et al., 2013; Vinje, 2015]. Among the GII genotypes, GII.4 is the single most common genotype infecting humans worldwide, associated with approximately 60% of all reported norovirus outbreaks [Siebenga et al., 2009] and 70% of sporadic norovirus gastroenteritis in children [Hoa Tran et al., 2013].
Since 1995, six different GII.4 pandemic variants have emerged and each of them replaced a previous predominant GII.4 variant. Studies suggest that the antigenic changes in the new GII.4 variants result both in loss of neutralizing epitopes as well as changing ligand in the host over time, both which may contribute to escape of herd immunity [Lindesmith et al., 2012]. In late 1995, GII.4 US95_96 emerged causing the first reported norovirus pandemic [Fankhauser et al., 2002]. In 2002, GII.4 Farmington Hills viruses emerged followed by GII.4 Hunter in 2004–2005 in Australia, Europe, Asia and Central America [Bucardo et al., 2008; Bull et al., 2006; Kroneman et al., 2006; Lopman et al., 2004; Phan et al., 2006]. In 2006, GII.4 Den Haag emerged and became the predominant variant [CDC, 2007; Kroneman et al., 2006; Siebenga et al., 2008] followed by GII.4 New Orleans in 2009 which gradually replaced GII.4 Den Haag viruses [Vinje, 2015]. In 2012, GII.4 Sydney viruses emerged and became pandemic [Hoa Tran et al., 2013].
The reason why GII.4 is the dominant genotype worldwide is unclear, but a combination of host factors and increased virulence of viruses belonging to this genotype have been suggested. GII.4 noroviruses infect secretor positive individuals, which express α1,2-linked fucose residue on surface epithelial cells of the gut and in body fluids [Le Pendu et al., 2006]. Secretors represent ≥80% of several populations worldwide, whereas other genotypes could have a more narrow host specificity [Nordgren et al., 2016]. GII.4 strains are undergoing antigenic variation at a faster rate as compared to other genotypes, likely in response to herd immunity [Bull et al., 2010; Debbink et al., 2013; Lindesmith et al., 2012; Lindesmith et al., 2011]. Moreover, observations of increased viral shedding in children infected with GII.4 compared to other genotypes, suggest that GII.4 genotypes could have higher replication rates and thus higher transmission between individuals [Bucardo et al., 2008].
This study aims to demonstrate the long term predominance and clinical severity of pediatric gastroenteritis associated with one particular norovirus genotype (GII.4), by obtaining new genotypes data of archived specimens and complementing with genotypes data of studies published elsewhere [Bucardo et al., 2011; Bucardo et al., 2010; Bucardo et al., 2008].
2. Material and methods
2.1. Subjects and specimens
We retrospectively analyzed laboratory and epidemiologic data from 1790 children ≤ 7 years (median = 14; IQR = 8–25) with AGE from 6 hospitals in Nicaragua (n = 538), and 3 community clinics (n = 919) and households (n = 333) in León, between 1999 and 2015. Moreover, 267 asymptomatic children (median age= 18; IQR = 5–30) from community clinics (n = 162) and households (n = 105) were also enrolled. Setting data and samples were not consistently available from every year, thus, material from the hospital represent 2002–2013 and from the community 1999–2010 and 2015, with exception of 2007 and 2008 for both settings. Household represents 2010–2011 and asymptomatic 2005–2006, 2010–2011. All children with AGE were clinically evaluated by pediatricians or general practitioners following the World Health Organization (WHO) strategy for diarrhea management. Accordingly, only children with signs of dehydration and not tolerating oral rehydration were treated at the hospital, otherwise they were treated at the community clinics in the oral rehydration unit. Sampling at the hospital was performed within 24 h after admission. All samples were collected following a standard procedure that involved collection in sterile plastic containers and transportation at 4 °C to the laboratory of Microbiology and the Faculty of Medical Science of UNAN-León. A 10% (wt/vol) suspension of the stools was prepared with phosphate-buffered saline (pH = 7.2), and two aliquots were frozen at −20 °C for virus analysis.
2.2. RNA extraction and reverse transcription
Viral RNA was extracted from 200 µl of 1:10 stool suspensions using High Pure Viral RNA Kit (Roche Diagnostics) following the manufacturer's instructions. A total of 50 µl of RNA was collected and stored at −20 °C until reverse transcription. Reverse transcription (RT) was carried out as described previously and cDNA was stored at −20 °C until used [Bucardo et al., 2008]. RNA from household and asymptomatic samples from 2010 and 2011 were purified by using automated RNA purification (either EZ1 robot (QIAGEN) or KingFisher magnetic particle processor (Thermo Fisher Scientific).
2.3. Real-time PCR assays for detection of norovirus
SYBR real-time RT-PCR was used for norovirus screening of samples collected at hospital and community between 2009 and 2013 and 2015 from symptomatic children. In brief, 2.5 µl of cDNA was added to a reaction mixture consisting of 12.5 µl of FastStart Universal SYBR Green Master (ROX) (Roche Applied Science, IN, USA), 0.4 pmol of each GII primers (NVG2f2 and COG2R) or each GI primers (NVGIF1b and NVG1r) [Nordgren et al., 2008], and 8 µl of RNAse free water, to final volume of 25 µl. The real-time PCR reactions were performed in a 96-well reaction plate using the ABI 7500 Real Time PCR System (Applied Biosystems, Foster, CA). PCR was performed under the following conditions: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 30 s and 72 °C for 1 min. Melting curve analysis, to confirm amplicon specificity, was performed immediately after PCR completion. A sample was considered norovirus-positive for GI and/or GII if the Ct value was ≤40 and Tm of 76.1 ± 0.6 °C for GI and 77.1 ± 0.6 °C for GII. A Taqman assay previously described by Nordgren and coworkers [Nordgren et al., 2013] was used for norovirus screening of samples collected at hospital and community between 1999 and 2006 from symptomatic and asymptomatic children. Household and asymptomatic samples from 2010 and 2011 were analyzed by using the TaqMan assay previously described Kageyama and coworkers [Kageyama et al., 2003].
2.4. Norovirus genotyping
Nucleotide sequencing of the N-terminal and shell (NS) region of the capsid gene was performed by Macrogen Inc. (Seoul, South Korea). The sequencing reaction was based on BigDye chemistry; NVG1f1b or NVG2f2 forward primers and G1SKR or G2SKR reverse primers were used as sequencing primers for norovirus GI (381 bp) or GII (378 bp), respectively [Kojima et al., 2002; Nordgren et al., 2008]. Genotypes were determined in norovirus positive samples by submitting the sequences to either the Norovirus Genotyping Tool Version 1.0 or to CaliciNet, both systems can determine genotype or GII.4 variant upon analyzing the NS sequences [Kroneman et al., 2011; Vega et al., 2011].
2.5. Statistical analysis
We first generated descriptive statistics to characterize norovirus infections by genotypes and GII.4 variants, stratified by setting and year of collection. We then estimated odds ratios (OR) and 95% confidence intervals (CI) to investigate the associations between GII.4 infections and setting, age group, and quarter using logistic regression. Adjusted Odds ratios and 95% CIs (variables setting, age group, and quarter) was calculated using multivariable logistic regression in SPSS. Statistical significance of temporal peaks of norovirus incidence was determined by logistic multivariate analysis of norovirus incidence by bimester. An alpha of 0.05 was used to determine statistical significance. All analyses were performed using SPSS (Statistical Program for Social Science version 14.0.0 for Windows; Chicago, IL).
3. Results
3.1. One fifth of pediatric AGE in Nicaragua was associated with norovirus
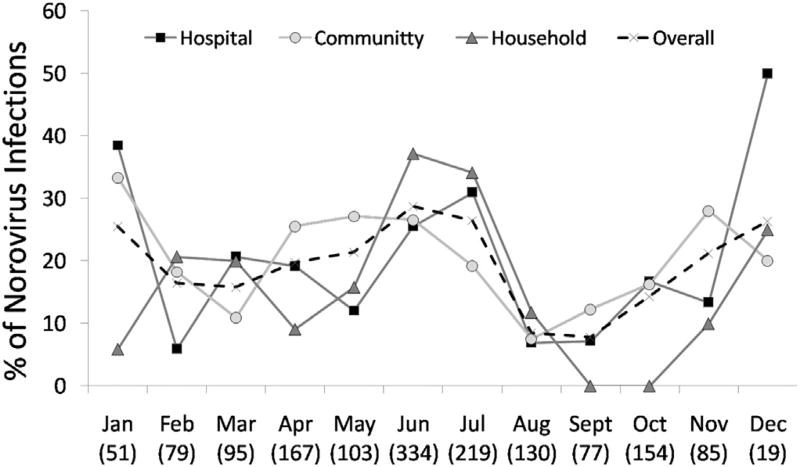
A total of 338 (19%) of the 1790 symptomatic and 32 (12%) of the 267 asymptomatic children tested norovirus-positive (Table 1). The median age for asymptomatic norovirus infection was 15 months (IQR = 4–28), as compared to symptomatic infections that was 13 months (IQR = 8–20). There were no association between norovirus infections and gender. A similar prevalence of symptomatic norovirus detection was observed among the different settings, but with differences between years. The norovirus yearly prevalence rate ranged from 3% to 27% (median= 20%) in hospitalized children to 12% to 43% (median =19%) in children attending the community clinics (Table 1) (setting/years with < 30 samples were excluded from these analysis). Although norovirus was detected year round, statistically well supported (p < 0.01) peak of symptomatic norovirus infections occurred in the rainy season (June – July), and a second peak during December– January (early dry season), but frequencies of infection were low in this period warranting careful interpretation (Fig. 1).
Table 1.
Frequency of asymptomatic and symptomatic norovirus infections among children < 7 years of age at different settings in Nicaragua, 1999–2015.
| Clinical status/Year/setting | No. of children | Norovirus screening by PCR n (%)e |
Genogroups n (%) | Genotypes n (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| GI | GII | GI:GII | No. of genotyped samplesf |
Any GI type |
GII.4 | Other GII types |
|||
| Asymptomatica | |||||||||
| 2005 | 58 | 4 (7) | 1 | 3 | 0 | 4 | 1 | 1 | 2 |
| 2006 | 104 | 14 (13) | 4 | 9 | 1 | 11 | 4 | 0 | 7 |
| 2010 | 4 | 3 (75) | 0 | 3 | 0 | 3 | 0 | 0 | 3 |
| 2011 | 101 | 11 (11) | 0 | 11 | 0 | 9 | 0 | 0 | 9 |
| All years | 267 | 32 (12) | 5 (16) | 26 (81) | 1 (3) | 27 | 5 (19) | 1 (4) | 21 (78) |
| Symptomatic | |||||||||
| Household | |||||||||
| 2010 | 322 | 67 (21) | 17 | 46 | 4 | 62 | 15 | 23 | 24 |
| 2011 | 11 | 1 (9) | 0 | 1 | 0 | 1 | 0 | 0 | 1 |
| Both years | 333 | 68 (20) | 17 (25) | 47 (69) | 4 (6) | 63 | 15 (24) | 23 (36) | 25 (40) |
| Communityb | |||||||||
| 1999 | 38 | 5 (13) | 2 | 3 | 0 | 2 | 1 | 0 | 1 |
| 2002 | 55 | 8 (15) | 2 | 5 | 1 | 4 | 2 | 1 | 1 |
| 2003 | 112 | 48 (43) | 10 | 31 | 7 | 34 | 10 | 7 | 17 |
| 2005c | 392 | 48 (12) | 5 | 43 | 0 | 21 | 2 | 12 | 7 |
| 2006 | 17 | 5 (29) | 0 | 5 | 0 | 4 | 0 | 4 | 0 |
| 2009 | 59 | 16 (27) | 0 | 16 | 0 | 6 | 0 | 5 | 1 |
| 2010 | 142 | 27 (19) | 5 | 22 | 0 | 19 | 2 | 12 | 5 |
| 2015 | 104 | 19 (18) | 1 | 17 | 1 | 15 | 0 | 12 | 3 |
| All years | 919 | 176 (19) | 25 (14) | 142 (81) | 9 (5) | 105 | 17 (16) | 53 (50) | 35 (33) |
| Hospitald | |||||||||
| 2002 | 63 | 2 (3) | 1 | 0 | 1 | 2 | 2 | 0 | 0 |
| 2003 | 40 | 8 (20) | 1 | 7 | 0 | 7 | 0 | 2 | 5 |
| 2004 | 48 | 4 (8) | 3 | 0 | 1 | 3 | 3 | 0 | 0 |
| 2005 | 128 | 26 (20) | 6 | 20 | 0 | 13 | 2 | 10 | 1 |
| 2006 | 7 | 0 (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2009 | 2 | 1 (50) | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 2010 | 160 | 36 (23) | 2 | 33 | 1 | 26 | 1 | 24 | 1 |
| 2011 | 30 | 5 (17) | 0 | 5 | 0 | 2 | 0 | 1 | 1 |
| 2012 | 30 | 8 (27) | 1 | 7 | 0 | 7 | 0 | 7 | 0 |
| 2013 | 30 | 4 (13) | 0 | 4 | 0 | 4 | 0 | 4 | 0 |
| All years | 538 | 94 (17) | 14 (15) | 77 (82) | 3 (3) | 64 | 8 (13) | 48 (75) | 8 (12) |
| All symptomatic | 1790 | 338 (19) | 56 (17) | 266 (79) | 16 (5) | 232 | 40 (17) | 124 (53) | 68 (29) |
Asymptomatic norovirus cases were not analyzed by setting due to low sample size.
Community refers to enrolment at primary care health facilities at the community.
Hospital and community norovirus frequencies between 2005 and 2010 have been published elsewhere.
Hospital refers to enrolment at either the emergency or the pediatric ward of the hospital.
Percentages of norovirus, genogroups and genotypes were calculated per line in the table; thus for the total number of children, for the total norovirus, for the total genogroups and for the total genotyped samples, respectively.
Due to low PCR product or bad quality sequences, the genotype was not established a subset of 111 norovirus-positive symptomatic (n =106) and asymptomatic (n = 5).
Fig. 1.
Seasonal distribution of pediatric norovirus AGE at different settings in Nicaragua between 1999 and 2015. In the X axis, the number in parenthesis indicates the total of AGE cases enrolled by each month. In the Y axis, each marker present the proportion of AGE that was norovirus positive in each setting.
3.2. Genogroup and genotype distribution
GII noroviruses were more predominant than GI, in both symptomatic (79%, 266/338) and asymptomatic (81%, 26/32) norovirus-positive children (Table. 1). GII viruses were detected less in the household setting (69%, 47/68) compared to hospital (82%, 77/94) and community (81%, 142/176) settings.
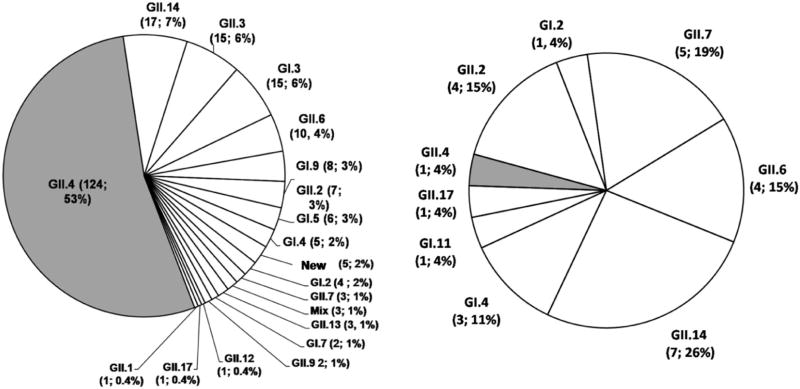
In total, 20 different genotypes were determined in 259 of the 370 norovirus-positive samples, of which 232 were symptomatic and 27 asymptomatic, all of them collected between 1999 and 2015. These included 13 different GII genotypes including a tentatively new genotype (KU306738), and 7 GI genotypes. In symptomatic children the distribution of genotypes was GII.4 (53%) followed by GII.14 (7%), GII.3 (6%), GI.3 (6%) and other genotypes in frequencies ranging from 4% to 0.4% (Fig. 2A). In contrast, in asymptomatic children, the most frequently detected genotypes were GII.14 (26%), GII.7 (19%), GII.6 (15%), GII.2 (15%), with only one sample (1.4%) being GII.4 (Fig. 2B). Of note, only 3 of the 24 (12%) GII.14 strains, the second most common genotype in this study, were found in hospitalized children. Co-infection with different genotypes (GI.3/GII.14 (n = 2) and GI.3/GII.8 (n = 1)) were also observed in children with AGE.
Fig. 2.
Distribution of norovirus genotypes in symptomatic (2A) and asymptomatic (2B) children from Nicaragua. 1999–2015. The numbers in brackets represent frequencies and percentages for each genotype respectively.
3.3. Infections with GII.4 norovirus were associated with age, setting and season of child enrolment
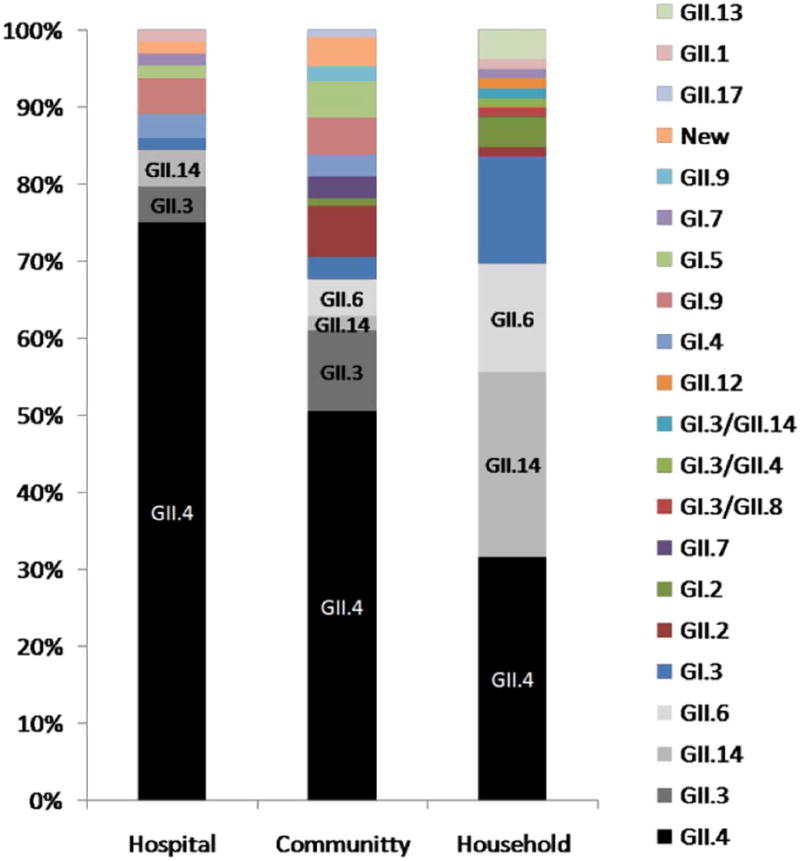
Overall, similar prevalence of GII.4 (124/232; 53%) and non-GII.4 (108/232; 47%) were observed in children with norovirus AGE, but non-GII.4 genotypes were significantly more common in asymptomatic norovirus-positive children (26/27 vs 1/27; OR =30 (95% CI: 5–623) (Fig. 2B). GII.4 was the most predominant genotype in children enrolled at the hospital (75%, OR 4.9 compared to household level p < 0.001), while in community clinics similar frequency of GII.4 and non-GII.4 were found which was not significantly different from frequencies observed at the household level (OR 1.7 p = 0.12) (Table 2) (Fig. 3). In children with AGE enrolled at the household, GII.4 viruses constituted only 38% compared to 62% of non-GII.4 genotypes. Symptomatic GII.4 infections occurred with the highest detection rate in children 7 to 12 months of age in all settings (OR 8.2 compared to children > 24 month of age (Table 2)). The detection frequency decreased significantly in older children, which were more frequently infected with non-GII.4 genotypes. Children aged ≤ 18 months were significantly more likely to be infected with GII.4 than children ≥ 24 months (Table 2). The median age for symptomatic GII.4 compared to non-GII.4 infections was 11 months (IQR= 8–16) and 17 months (IQR = 9–27), respectively (p < 0.001). The prevalence of GII.4 detection varied by season of the year. The proportion of genotype GII.4 was significantly higher in the quarters Aug – Oct and Nov – Jan, but not in May – Jul, compared to Feb – Apr (OR 3.0, 2.8 and 1.5, respectively, p < 0.05) (Table 2). A multivariate logistic regression analysis was performed of the variables associated with symptomatic infections in Table 2 (setting, age-group and quarter). The adjusted odds ratios did not differ substantially from the univariate analysis in most cases, and did not change interpretations of the results. Thus, as observed in Table 2, enrollment in the hospital setting between August and January, at an age up to 18 months were all associated with a higher likelihood of being infected with the GII.4 genotype. For statistical analysis, year of sample collection was not considered because genotype data was not consistently available from every year. Of note, GII.4 was detected at higher frequencies than non-GII.4 viruses for most years in community and hospital, except for 2003 when the GII.3 genotype was more commonly detected than GII.4 at community (11/34 vs 7/34) and hospital (3/7 vs 2/7) setting.
Table 2.
Predictors of GII.4 infection in symptomatic children by setting, age group, and time of year in Leon, Nicaragua, 1999–2015 (N = 232).
| Parameter | Genotyped strains | OR (95% C·I) | P-value | Adjusted ORa (95% C·I) | ||
|---|---|---|---|---|---|---|
|
|
||||||
| All genotypes | GII.4 (%) | Non-GII.4 (%) | ||||
| Setting | ||||||
| Hospital | 64 | 48 (75) | 16 (25) | 4.9 (2.3–10) | < 0.001 | 6.4 (2.4–17) |
| Community | 105 | 53 (50) | 52 (49) | 1.7 (0.9–3.1) | 0.12 | 1.0 (0.5–2.1) |
| Household | 63 | 24 (38) | 39 (62) | 1 | ||
| Total | 232 | 125 (54) | 107 (46) | |||
| Age groups (months) | ||||||
| ≤ 6 | 33 | 19 (58) | 14 (42) | 3.8 (1.4–10) | < 0.01 | 2.8 (0.9–8.5) |
| 7 to 12 | 71 | 53 (75) | 18 (25) | 8.2 (3.4–20) | < 0.001 | 5.6 (2.0–15) |
| 13 to 18 | 51 | 30 (59) | 21 (41) | 4.0 (1.6–10) | < 0.01 | 3.1 (1.2–8.3) |
| 19 to 24 | 29 | 13 (45) | 16 (55) | 2.3 (0.8–6.4) | 0.12 | 1.8 (0.6–5.5) |
| > 24 | 38 | 10 (26) | 28 (74) | 1 | ||
| Missing age | 10 | 0 (0) | 10 (100) | |||
| Total | 232 | 125 (54) | 107 (46) | |||
| Quarter | ||||||
| May – Jul | 130 | 69 (53) | 61 (47) | 1.5 (0.8–2.9) | 0.22 | 1.4 (0.6–3.0) |
| Aug – Oct | 26 | 18 (69) | 8 (31) | 3.0 (1.1–8.2) | 0.03 | 2.9 (0.9–9.1) |
| Nov – Jan | 25 | 17 (68) | 8 (32) | 2.8 (1.0–7.8) | 0.04 | 2.7 (0.85–8.5) |
| Feb – Apr | 49 | 21 (43) | 28 (57) | 1 | ||
| Missing collection date | 2 | 0 (0) | 2 (100) | |||
| Total | 232 | 125 (54) | 107 (46) | |||
Multivariable logistic regression adjusted for all other variables in the table.
Fig. 3.
Distribution of norovirus genotypes found in children with gastroenteritis at the hospital, community clinics and household, respectively from Nicaragua, between 1999 and 2015.
3.4. Emergence, selection and replacement of norovirus GII.4 variants
During the study period of 1999 to 2015, 9 different GII.4 variants were observed to circulate in children with AGE in Nicaragua. Each variant predominated for 1 to 4 years until it was replaced by a new variant; the replaced variant was subsequently not detected in the following years (Table 3). Thus, Farmington Hills circulated in 2002 and co-circulated with Lanzou in 2003, Hunter was the only variant detected in 2005, Den Haag appeared in 2009 and co-circulated with New Orleans in 2010, which was detected until 2013. The Sydney variant was detected in samples collected in 2012, had a higher prevalence in 2013, and was the only GII.4 variant detected in 2015. Other less common GII.4 variants such as Osaka and Yerseke were also detected at low frequencies (≤4%) in 2006 and 2010, respectively.
Table 3.
Frequencies of norovirus GII.4 variants in children with gastroenteritis from the hospital, community clinics and household from Nicaragua between 2002 and 2015 (N = 126).
| GII.4 Variant | Years of norovirus surveillance, n (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| 2002 | 2003 | 2005 | 2006 | 2009 | 2010 | 2011 | 2012 | 2013 | 2015 | All years (%) | |
| Farmington_Hills_2002 | 1 (100) | 4 (44) | 5 (4) | ||||||||
| Lanzou_2002 | 4 (44) | 1 (5) | 5 (4) | ||||||||
| Oxfordshire-Like | 1 (11) | 1 (1) | |||||||||
| Hunter_2004 | 21 (95)a | 21 (17) | |||||||||
| Osaka_2007 | 4 (100) | 4 (3) | |||||||||
| Den_Haag_2006b | 5 (100) | 38 (62) | 1 (100) | 3 (43) | 47 (37) | ||||||
| New_Orleans_2009 | 22 (36) | 3 (43) | 1 (25) | 26 (21) | |||||||
| Yerseke_2006a | 1 (2) | 1 (1) | |||||||||
| Sydney_2012 | 1 (14) | 3 (75) | 12 (100) | 16 (13) | |||||||
| All variants | 1 | 9 | 22 | 4 | 5 | 61 | 1 | 7 | 4 | 12 | 126 |
In total, only1 GII.4 was observed in asymptomatic children and it as Hunter_2004.
3.5. Detection and characterization of a novel genotype
Between April and June 2005, 5 symptomatic children (4 from the community and 1 from the hospital) with ages ranging from 5 to 25 months were infected with a GII norovirus of unknown genotype, according to preliminary sequence analysis of the regions C (EU780736), D and P2 (KR075984) of the capsid gene. Near full genome sequence (7233 nt) analysis of one of these strains was obtained by next generation sequencing and submitted to GenBank (KU306738). The VP1 protein sequences were compared with the existing norovirus reference sequences and based on the criteria to assign new genotypes could be tentatively classified as a new genotype after revision by the international norovirus working group [Kroneman et al., 2013].
3.6. Norovirus re-infections
During the cohort study carried out from 2010 to 2011 to examine the incidence of infectious causes of childhood diarrhea at the household level in Nicaragua a total of 8 of 68 norovirus-positive children were found to experience secondary norovirus re-infections, mainly symptomatic, and one child experienced an asymptomatic tertiary norovirus infection. The shortest and longest periods for re-infection were 28 and 256 days (median = 98), respectively. Of note, half of these children were re-infected with norovirus of the same genogroup but of different genotype, with exception of one child who experienced two consecutive symptomatic infections with GI.3.
4. DISCUSION
The current study highlights the significant contribution of genotype GII.4 norovirus infections to the burden of pediatric AGE in the period of 1999 to 2015 in Nicaragua. We found a significant association between setting of the children with norovirus gastroenteritis and proportion of GII.4 viruses, with a lower prevalence found at the household level (38%), which represent mainly mild AGE, followed by 50% in children at the community clinics, which represent mild to moderate AGE, and 75% in hospitalized children (OR = 4.9 compared to children enrolled at household), which represent mainly AGE cases who required intravenous rehydration. Moreover, while genotype GII.4 viruses predominated in symptomatic infections, they were virtually non-existent in asymptomatic cases. These results strengthen previous data that GII.4 noroviruses are associated with higher clinical severity as compared to other norovirus genotypes [Huhti et al., 2011]. Similar suggestions have been proposed for patients involved in GII.4 outbreaks [Friesema et al., 2009; Mai et al., 2013; Trivedi et al., 2012]. Possible explanations are host genetic factors or, as of yet unknown virulence factors or higher replication rates of the GII.4 genotypes [Bucardo et al., 2008; Nordgren et al., 2016]. We have previously observed that children infected with GII.4 viruses who required intravenous rehydration, shed the highest viral load as compared with children infected with other genotypes [Bucardo et al., 2008], suggesting a link between viral load and clinical severity.
Several others studies have shown that GII.4 viruses are more common in hospital settings. Data from Finland, Thailand, Israel, Italy, Malawi, Iraq, Brazil, Bolivia and Guatemala also describe GII.4 as the most common genotype found in hospitalized children [Al-Mashhadani et al., 2008; Estevez et al., 2013; Huhti et al., 2014; Kittigul et al., 2010; Leshem et al., 2015; McAtee et al., 2016; Medici et al., 2006; Trainor et al., 2013; Victoria et al., 2007]. The high prevalence of GII.4 genotypes in hospitalized children is however seemingly a relatively recent phenomenon. For example, GII.3 and not GII.4 viruses were the most prevalent genotype for 34 years (1974–1991) in children seeking care at the Children's Hospital in Washington, USA [Bok et al., 2009]. Also, the global spread of norovirus GII.17 between 2014 and 2016 have raised concern about the potential of this emerging genotype to replace GII.4[Chan et al., 2017].
However, there are limited data on the norovirus genotype distribution at the household or community level. Interestingly, our data showed that the prevalence of non-GII.4 noroviruses increased from 25% in the hospitalized children to 49% in community clinics and 62% in households, a trend inversely correlated to clinical severity. Huhti and coworkers also found that non-GII.4 genotypes were predominant (54%) at the community level in Finland between 1998 and 2007, but important variations were observed over time [Huhti et al., 2014]. Similarly, Bruggink and coworkers found a predominance of non-GII.4 viruses (65/87) during a community-based study of sporadic gastroenteritis incidents in Australia [Bruggink et al., 2015]. Non-GII.4 viruses were also found predominantly in birth cohort studies (children up to 3 years of age) from Bolivia (75%), Peru (59%) and India (47%) [Lopman et al., 2015; Menon et al., 2016; Saito et al., 2014]. Moreover, in our study non-GII.4 noroviruses were predominantly detected in asymptomatic children and more common in symptomatic children ≥ 18 months of age. Our results add to other data that suggest that non-GII.4 infections are associated with milder clinical symptoms and also suggest that might be more often found in symptomatic older children [Huhti et al., 2011].
In our study, all age groups up to 18 month of age were significantly more likely to have GII.4 NoV compared to children > 2 years of age; with the highest prevalence in children between 7 and 12 months of age (OR = 8.2 compared to > 24 month of age), which coincide with weaning. In Nicaragua, breastfeeding is highly prevalent (approximately 97%) during the first 6 months of life, therefore breastfed children could be either protected by maternal norovirus IgA or virus decoying human milk oligosaccharides [Colombara et al., 2015; Khodayar-Pardo et al., 2014; Weichert et al., 2016]. After weaning, children might both be more exposed to norovirus infections, and have less protection, factors that might explain the high GII.4 burden in that age group.
A multivariate analysis of setting, age group and season was performed which yielded similar results as univariate analysis (Table 2). The model thus shows that the hospital setting, an age of ≤18 month and infection in the time of August to January in Nicaragua are factors associated with GII.4 NoV infections, with highest odds ratios observed for the hospital setting and in the age-group 7–12 month of age.
Early human challenge studies with Norwalk virus (genotype GI.1) have demonstrated that many subjects were susceptible to re-infections, pre-challenge norovirus antibodies were not protective and short-term resistance lasted for about 6 months after challenge [Johnson et al., 1990; Parrino et al., 1977]. Data from recent birth cohort studies have shown that re-infection can occur in up to 40% of children during the first 3 years of life [Lopman et al., 2015; Menon et al., 2016]. In the current study, 8 (12%) of the 68 norovirus-positive children from the household cohort were found to experience re-infections, some of them with the same genogroup but only 1 out 8 with the same genotype (GI.3). Despite differences in study design, repeated norovirus infections with the same genotype were also rare in a birth cohort of Peruvian children, but repeated infections by viruses from the same genogroup were common [Saito et al., 2014]. Our observations strengthen the hypothesis that immunity, at least against symptomatic infection, induced by natural norovirus infections is type-specific and of short duration, at least in young children [Debbink et al., 2012].
Rapid antigenic changes in predominant strains may be another factor for the increased persistence of GII.4 in our pediatric population since new variants emerge under the influence of population immunity and each variant is replaced within 3–4 years by a new GII.4 variant [Siebenga et al., 2009]. Studies of the antigenic properties of contemporary GII.4 Sydney viruses compared to previously circulating GII.4 strains suggest that the emergence of new pandemic GII.4 variants correlates with escape from herd immunity [Debbink et al., 2013].
This study has several limitations. First, the data analyzed originated from different settings (hospital, health clinic, households) over a long time-period without a uniform and systematic sampling strategy for cases and matching controls. For example, there were only 55 cases of AGE in children < 7 years old enrolled from community clinics in 2002 and none in 2004 asymptomatic sampling was performed in only two time frames (2005–2006 and 2010–2011).Therefore, our data should be interpreted with caution and no definitive conclusions can be drawn. Furthermore part of the samples were tested with SYBR real-time PCR while others were tested by TaqMan PCR, might also be considered a limitation. However, both used the same primers for the same region, so likely differing little in sensitivity and no tendencies of differences in frequencies of norovirus were observed at time frames (1999–2006) when TaqMan was used compared to 2009–2015 when SYBR green was used (Table 1). The major strengths of this study include a long observation period which allowed the investigators to monitor genotypes distribution from 1999 to 2015. Further, this study contributes to the evidence for the high prevalence and severity of GII.4 infections, implicating this genotype as a primary target for pediatric norovirus vaccine development.
In summary, we retrospectively analyzed norovirus laboratory data and available epidemiologic data from children with AGE and for some years from healthy controls from children < 7 years of AGE in the hospital, clinics and the community. Our data demonstrate the predominance and clinical severity of pediatric GII.4 norovirus infections in Nicaragua with the majority of infections occurring in children between 7 months and 2 years of age. Our results also show that re-infection with the same genotype is uncommon, which together with a low prevalence of GII.4 genotype in older children indicate type specific immunity.
Our data from Nicaragua suggest that a future vaccine formulation should at least include a GII.4 strain and the first immunization should be offered before 6 months of age to prevent the majority of pediatric norovirus AGE cases and reduce epidemic cycles. In the same line Bruggink and coworkers suggest that vaccination before the age of one would appear to be the most efficacious and Shioda and coworkers suggest that an immunization schedule completed by 6 months could have the potential to prevent about 85% of pediatric norovirus cases [Bruggink et al., 2017; Shioda et al., 2015].
Acknowledgments
Financial support
This work was supported by the Swedish Research Council (grant number dnr-348-2011-7420 to L. S. and F. B.). The University of North Carolina, Institute for Global Health & Infectious Diseases (to N.B.), and the Fogarty International Center at the National Institutes of Health (grant 5K01TW008401-04 to S.B.D.).
To Nadja Vielot, for help with the statistical analysis and design of tables and figures.
Footnotes
Disclaimer
The findings and conclusion in this report are those of the authors and not necessarily represent the official position of the CDC.
References
- Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14(8):725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mashhadani MN, Nakagomi O, Dove W, Ahmed H, Nakagomi T, Hart CA, Cunliffe NA. Norovirus gastroenteritis among children in Iraqi Kurdistan. J. Med. Virol. 2008;80(3):506–509. doi: 10.1002/jmv.21099. [DOI] [PubMed] [Google Scholar]
- Ayukekbong JA, Mesumbe HN, Oyero OG, Lindh M, Bergstrom T. Role of noroviruses as aetiological agents of diarrhoea in developing countries. J. Gen. Virol. 2015;96(8):1983–1999. doi: 10.1099/vir.0.000194. [DOI] [PubMed] [Google Scholar]
- Bok K, Abente EJ, Realpe-Quintero M, Mitra T, Sosnovtsev SV, Kapikian AZ, Green KY. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 2009;83(22):11890–11901. doi: 10.1128/JVI.00864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggink LD, Dunbar NL, Marshall JA. Norovirus genotype diversity in community-based sporadic gastroenteritis incidents: a five-year study. J. Med. Virol. 2015;87(6):961–969. doi: 10.1002/jmv.24154. [DOI] [PubMed] [Google Scholar]
- Bruggink LD, Moselen JM, Marshall JA. Genotype analysis of noroviruses associated with gastroenteritis outbreaks in childcare centres, Victoria, Australia, 2012–2015. Epidemiol. Infect. 2017;145(9):1933–1941. doi: 10.1017/S0950268817000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucardo F, Nordgren J, Carlsson B, Paniagua M, Lindgren PE, Espinoza F, Svensson L. Pediatric norovirus diarrhea in Nicaragua. J. Clin. Microbiol. 2008;46(8):2573–2580. doi: 10.1128/JCM.00505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucardo F, Nordgren J, Carlsson B, Kindberg E, Paniagua M, Mollby R, Svensson L. Asymptomatic norovirus infections in Nicaraguan children and its association with viral properties and histo-blood group antigens. Pediatr. Infect. Dis. J. 2010;29(10):934–939. doi: 10.1097/INF.0b013e3181ed9f2f. [DOI] [PubMed] [Google Scholar]
- Bucardo F, Lindgren PE, Svensson L, Nordgren J. Low prevalence of rotavirus and high prevalence of norovirus in hospital and community wastewater after introduction of rotavirus vaccine in Nicaragua. PLoS One. 2011;6(10):e25962. doi: 10.1371/journal.pone.0025962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucardo F, Reyes Y, Svensson L, Nordgren J. Predominance of norovirus and sapovirus in Nicaragua after implementation of universal rotavirus vaccination. PLoS One. 2014;9(5):e98201. doi: 10.1371/journal.pone.0098201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, ET Tu, McIver CJ, Rawlinson WD, White PA. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 2006;44(2):327–333. doi: 10.1128/JCM.44.2.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, Eden JS, Rawlinson WD, White PA. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 2010;6(3):e1000831. doi: 10.1371/journal.ppat.1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Norovirus activity—United States, 2006–2007. MMWR Morb. Mortal. Wkly Rep. 2007;56(33):842–846. [PubMed] [Google Scholar]
- Chan MCW, Hu Y, Chen H, Podkolzin AT, Zaytseva EV, Komano J, Sakon N, Poovorawan Y, Vongpunsawad S, Thanusuwannasak T, Hewitt J, Croucher D, Collins N, Vinje J, Pang XL, Lee BE, de Graaf M, van Beek J, Vennema H, Koopmans MPG, Niendorf S, Poljsak-Prijatelj M, Steyer A, White PA, Lun JH, Mans J, Hung TN, Kwok K, Cheung K, Lee N, Chan PKS. Global spread of Norovirus GII.17 Kawasaki 308, 2014–2016. Emerg. Infect. Dis. 2017;23(8):1359–1354. doi: 10.3201/eid2308.161138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombara DV, Hernandez B, Gagnier MC, Johanns C, Desai SS, Haakenstad A, McNellan CR, Palmisano EB, Rios-Zertuche D, Schaefer A, Zuniga-Brenes P, Zyznieuski N, Iriarte E, Mokdad AH. Breastfeeding practices among poor women in Mesoamerica. J. Nutr. 2015;145(8):1958–1965. doi: 10.3945/jn.115.213736. [DOI] [PubMed] [Google Scholar]
- da Silva, Polo T, Peiro JR, Mendes LC, Ludwig LF, de Oliveira-Filho EF, Bucardo F, Huynen P, Melin P, Thiry E, Mauroy A. Human norovirus infection in Latin America. J. Clin. Virol. 2016;78:111–119. doi: 10.1016/j.jcv.2016.03.016. [DOI] [PubMed] [Google Scholar]
- Debbink K, Lindesmith LC, Donaldson EF, Baric RS. Norovirus immunity and the great escape. PLoS Pathog. 2012;8(10):e1002921. doi: 10.1371/journal.ppat.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbink K, Lindesmith LC, Donaldson EF, Costantini V, Beltramello M, Corti D, Swanstrom J, Lanzavecchia A, Vinje J, Baric RS. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J Infect Dis. 2013;208(11):1877–1887. doi: 10.1093/infdis/jit370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez A, Arvelo W, Hall AJ, Lopez MR, Lopez B, Reyes L, Moir JC, Gregoricus N, Vinje J, Parashar UD, Lindblade KA. Prevalence and genetic diversity of norovirus among patients with acute diarrhea in Guatemala. J. Med. Virol. 2013;85(7):1293–1298. doi: 10.1002/jmv.23578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Ando T, Glass RI. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J Infect Dis. 2002;186(1):1–7. doi: 10.1086/341085. [DOI] [PubMed] [Google Scholar]
- Friesema IH, Vennema H, Heijne JC, de Jager CM, Teunis PF, van der Linde R, Duizer E, van Duynhoven YT. Differences in clinical presentation between norovirus genotypes in nursing homes. J. Clin. Virol. 2009;46(4):341–344. doi: 10.1016/j.jcv.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Hoa Tran TN, Trainor E, Nakagomi T, Cunliffe NA, Nakagomi O. Molecular epidemiology of noroviruses associated with acute sporadic gastroenteritis in children: global distribution of genogroups, genotypes and GII.4 variants. J. Clin. Virol. 2013;56(3):185–193. doi: 10.1016/j.jcv.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Huhti L, Szakal ED, Puustinen L, Salminen M, Huhtala H, Valve O, Blazevic V, Vesikari T. Norovirus GII-4 causes a more severe gastroenteritis than other noroviruses in young children. J Infect Dis. 2011;203(10):1442–1444. doi: 10.1093/infdis/jir039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhti L, Blazevic V, Puustinen L, Hemming M, Salminen M, Vesikari T. Genetic analyses of norovirus GII.4 variants in Finnish children from 1998 to 2013. Infect. Genet. Evol. 2014;26:65–71. doi: 10.1016/j.meegid.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Johnson PC, Mathewson JJ, DuPont HL, Greenberg HB. Multiple-challenge study of host susceptibility to Norwalk gastroenteritis in US adults. J Infect Dis. 1990;161(1):18–21. doi: 10.1093/infdis/161.1.18. [DOI] [PubMed] [Google Scholar]
- Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41(4):1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodayar-Pardo P, Martinez-Costa C, Carmona-Vicente N, Buesa J. Norovirus GII.4 antibodies in breast milk and serum samples: their role preventing virus-like particles binding to their receptors. Pediatr. Infect. Dis. J. 2014;33(6):554–559. doi: 10.1097/INF.0000000000000207. [DOI] [PubMed] [Google Scholar]
- Kittigul L, Pombubpa K, Taweekate Y, Diraphat P, Sujirarat D, Khamrin P, Ushijima H. Norovirus GII-4 2006b variant circulating in patients with acute gastroenteritis in Thailand during a 2006–2007 study. J. Med. Virol. 2010;82(5):854–860. doi: 10.1002/jmv.21746. [DOI] [PubMed] [Google Scholar]
- Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods. 2002;100(1–2):107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- Kroneman A, Vennema H, Harris J, Reuter G, von Bonsdorff CH, Hedlund KO, Vainio K, Jackson V, Pothier P, Koch J, Schreier E, Bottiger BE, Koopmans M. Increase in norovirus activity reported in Europe. Euro Surveill. 2006;11(12):E061214–061211. doi: 10.2807/esw.11.50.03093-en. [DOI] [PubMed] [Google Scholar]
- Kroneman A, Vennema H, Deforche K, v d Avoort H, Penaranda S, Oberste MS, Vinje J, Koopmans M. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 2011;51(2):121–125. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Kroneman A, Vega E, Vennema H, Vinje J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013;158(10):2059–2068. doi: 10.1007/s00705-013-1708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pendu J, Ruvoen-Clouet N, Kindberg E, Svensson L. Mendelian resistance to human norovirus infections. Semin. Immunol. 2006;18(6):375–386. doi: 10.1016/j.smim.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem E, Givon-Lavi N, Vinje J, Gregoricus N, Parashar U, Dagan R. Differences in Norovirus-associated hospital visits between Jewish and Bedouin children in Southern Israel. Pediatr. Infect. Dis. J. 2015;34(9):1036–1038. doi: 10.1097/INF.0000000000000786. [DOI] [PubMed] [Google Scholar]
- Lindesmith LC, Donaldson EF, Baric RS. Norovirus GII.4 strain antigenic variation. J. Virol. 2011;85(1):231–242. doi: 10.1128/JVI.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith LC, Beltramello M, Donaldson EF, Corti D, Swanstrom J, Debbink K, Lanzavecchia A, Baric RS. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 2012;8(5):e1002705. doi: 10.1371/journal.ppat.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopman B, Vennema H, Kohli E, Pothier P, Sanchez A, Negredo A, Buesa J, Schreier E, Reacher M, Brown D, Gray J, Iturriza M, Gallimore C, Bottiger B, Hedlund KO, Torven M, von Bonsdorff CH, Maunula L, Poljsak-Prijatelj M, Zimsek J, Reuter G, Szucs G, Melegh B, Svennson L, van Duijnhoven Y, Koopmans M. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet. 2004;363(9410):682–688. doi: 10.1016/S0140-6736(04)15641-9. [DOI] [PubMed] [Google Scholar]
- Lopman BA, Trivedi T, Vicuna Y, Costantini V, Collins N, Gregoricus N, Parashar U, Sandoval C, Broncano N, Vaca M, Chico ME, Vinje J, Cooper PJ. Norovirus infection and disease in an Ecuadorian birth cohort: association of certain norovirus genotypes with host FUT2 secretor status. J Infect Dis. 2015;211(11):1813–1821. doi: 10.1093/infdis/jiu672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai H, Jin M, Guo X, Liu J, Liu N, Cong X, Gao Y, Wei L. Clinical and epidemiologic characteristics of norovirus GII.4 Sydney during winter 2012–13 in Beijing, China following its global emergence. PLoS One. 2013;8(8):e71483. doi: 10.1371/journal.pone.0071483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAtee CL, Webman R, Gilman RH, Mejia C, Bern C, Apaza S, Espetia S, Pajuelo M, Saito M, Challappa R, Soria R, Ribera JP, Lozano D, Torrico F. Burden of norovirus and rotavirus in children after rotavirus vaccine introduction, Cochabamba, Bolivia. Am. J. Trop. Med. Hyg. 2016;94(1):212–217. doi: 10.4269/ajtmh.15-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici MC, Martinelli M, Abelli LA, Ruggeri FM, Di Bartolo I, Arcangeletti MC, Pinardi F, De Conto F, Izzi G, Bernasconi S, Chezzi C, Dettori G. Molecular epidemiology of norovirus infections in sporadic cases of viral gastroenteritis among children in Northern Italy. J. Med. Virol. 2006;78(11):1486–1492. doi: 10.1002/jmv.20723. [DOI] [PubMed] [Google Scholar]
- Menon VK, George S, Sarkar R, Giri S, Samuel P, Vivek R, Saravanabavan A, Liakath FB, Ramani S, Iturriza-Gomara M, Gray JJ, Brown DW, Estes MK, Kang G. Norovirus Gastroenteritis in a Birth Cohort in Southern India. PLoS One. 2016;11(6):e0157007. doi: 10.1371/journal.pone.0157007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren J, Bucardo F, Dienus O, Svensson L, Lindgren PE. Novel light-upon-extension real-time PCR assays for detection and quantification of genogroup I and II noroviruses in clinical specimens. J. Clin. Microbiol. 2008;46(1):164–170. doi: 10.1128/JCM.01316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren J, Nitiema LW, Ouermi D, Simpore J, Svensson L. Host genetic factors affect susceptibility to norovirus infections in Burkina Faso. PLoS One. 2013;8(7):e69557. doi: 10.1371/journal.pone.0069557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren J, Sharma S, Kambhampati A, Lopman B, Svensson L. Innate Resistance and Susceptibility to Norovirus Infection. PLoS Pathog. 2016;12(4):e1005385. doi: 10.1371/journal.ppat.1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Ryan M, Riera-Montes M, Lopman B. Norovirus in Latin America: systematic review and meta-analysis. Pediatr. Infect. Dis. J. 2017;36(2):127–134. doi: 10.1097/INF.0000000000001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrino TA, Schreiber DS, Trier JS, Kapikian AZ, Blacklow NR. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N. Engl. J. Med. 1977;297(2):86–89. doi: 10.1056/NEJM197707142970204. [DOI] [PubMed] [Google Scholar]
- Payne DC, Vinje J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT, Wikswo M, Shirley SH, Hall AJ, Lopman B, Parashar UD. Norovirus and medically attended gastroenteritis in U.S. children. N. Engl. J. Med. 2013;368(12):1121–1130. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Kuroiwa T, Kaneshi K, Ueda Y, Nakaya S, Nishimura S, Yamamoto A, Sugita K, Nishimura T, Yagyu F, Okitsu S, Muller WE, Maneekarn N, Ushijima H. Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J. Med. Virol. 2006;78(7):971–978. doi: 10.1002/jmv.20649. [DOI] [PubMed] [Google Scholar]
- Pringle K, Lopman B, Vega E, Vinje J, Parashar UD, Hall AJ. Noroviruses: epidemiology, immunity and prospects for prevention. Future Microbiol. 2015;10(1):53–67. doi: 10.2217/fmb.14.102. [DOI] [PubMed] [Google Scholar]
- Saito M, Goel-Apaza S, Espetia S, Velasquez D, Cabrera L, Loli S, Crabtree JE, Black RE, Kosek M, Checkley W, Zimic M, Bern C, Cama V, Gilman RH. Multiple norovirus infections in a birth cohort in a Peruvian periurban community. Clin. Infect. Dis. 2014;58(4):483–491. doi: 10.1093/cid/cit763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda K, Kambhampati A, Hall AJ, Lopman BA. Global age distribution of pediatric norovirus cases. Vaccine. 2015;33(33):4065–4068. doi: 10.1016/j.vaccine.2015.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenga J, Kroneman A, Vennema H, Duizer E, Koopmans M. Food-borne viruses in Europe network report: the norovirus GII.4 2006b (for US named Minervalike, for Japan Kobe034-like, for UK V6) variant now dominant in early seasonal surveillance. Euro Surveill. 2008;13(2) [PubMed] [Google Scholar]
- Siebenga JJ, Vennema H, Zheng DP, Vinje J, Lee BE, Pang XL, Ho EC, Lim W, Choudekar A, Broor S, Halperin T, Rasool NB, Hewitt J, Greening GE, Jin M, Duan ZJ, Lucero Y, O'Ryan M, Hoehne M, Schreier E, Ratcliff RM, White PA, Iritani N, Reuter G, Koopmans M. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200(5):802–812. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- Trainor E, Lopman B, Iturriza-Gomara M, Dove W, Ngwira B, Nakagomi O, Nakagomi T, Parashar U, Cunliffe N. Detection and molecular characterisation of noroviruses in hospitalised children in Malawi, 1997–2007. J. Med. Virol. 2013;85(7):1299–1306. doi: 10.1002/jmv.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi TK, DeSalvo T, Lee L, Palumbo A, Moll M, Curns A, Hall AJ, Patel M, Parashar UD, Lopman BA. Hospitalizations and mortality associated with norovirus outbreaks in nursing homes, 2009–2010. JAMA. 2012;308(16):1668–1675. doi: 10.1001/jama.2012.14023. [DOI] [PubMed] [Google Scholar]
- Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinje J. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg. Infect. Dis. 2011;17(8):1389–1395. doi: 10.3201/eid1708.101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria M, Carvalho-Costa FA, Heinemann MB, Leite JP, Miagostovich M. Prevalence and molecular epidemiology of noroviruses in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil, 2004. Pediatr. Infect. Dis. J. 2007;26(7):602–606. doi: 10.1097/INF.0b013e3180618bea. [DOI] [PubMed] [Google Scholar]
- Vinje J. Advances in laboratory methods for detection and typing of norovirus. J. Clin. Microbiol. 2015;53(2):373–381. doi: 10.1128/JCM.01535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert S, Koromyslova A, Singh BK, Hansman S, Jennewein S, Schroten H, Hansman GS. Structural basis for norovirus inhibition by human milk oligosaccharides. J. Virol. 2016;90(9):4843–4848. doi: 10.1128/JVI.03223-15. [DOI] [PMC free article] [PubMed] [Google Scholar]