Abstract
Retrotransposons are genetic elements that are similar in structure and life cycle to retroviruses by replicating via an RNA intermediate and inserting into a host genome. The Saccharomyces cerevisiae (S. cerevisiae) Ty1–5 elements are long terminal repeat (LTR) retrotransposons that are members of the Ty1-copia (Pseudoviridae) or Ty3-gypsy (Metaviridae) families. Four of the five S. cerevisiae Ty elements are inserted into the genome upstream of RNA Polymerase (Pol) III-transcribed genes such as transfer RNA (tRNA) genes. This particular genomic locus provides a safe environment for Ty element insertion without disruption of the host genome and is a targeting strategy used by retrotransposons that insert into compact genomes of hosts such as S. cerevisiae and the social amoeba Dictyostelium. The mechanism by which Ty1 targeting is achieved has been recently solved due to the discovery of an interaction between Ty1 Integrase (IN) and RNA Pol III subunits. We describe the methods used to identify the Ty1-IN interaction with Pol III and the Ty1 targeting consequences if the interaction is perturbed. The details of Ty1 targeting are just beginning to emerge and many unexplored areas remain including consideration of the 3-dimensional shape of genome. We present a variety of other retrotransposon families that insert adjacent to Pol III-transcribed genes and the mechanism by which the host machinery has been hijacked to accomplish this targeting strategy. Finally, we discuss why retrotransposons selected Pol III-transcribed genes as a target during evolution and how retrotransposons have shaped genome architecture.
Keywords: Retrotransposon, S. cerevisiae, Ty element, RNA polymerase III, tRNA, Integrase
Background
Genome evolution and plasticity are impacted by endogenous DNA sequences called transposable elements (TEs), that can mobilize within a genome [1]. TEs, which make up a significant portion of eukaryotic genomes, are divided into two classes: class I retrotransposons that mobilize via an RNA intermediate using a “copy and paste” mechanism and class II DNA transposons that use a “cut and paste” mechanism [2, 3]. Class I retrotransposons can be further divided into five orders: LTR-retrotransposons, DIRS-like elements, Penelope-like elements, long interspersed elements (LINEs) and short interspersed elements (SINEs) [3]. LTR-retrotransposons carry characteristic flanking repetitive sequences and are similar to retroviruses in structure and replication but do not exit the cell. The S. cerevisiae genome contains five types of LTR-retrotransposon elements, known as Ty1–5, that transpose through an RNA intermediate and produce intracellular virus-like particles (VLPs) [4, 5]. The majority of the S. cerevisiae LTR retrotransposons belong to the copia (Pseudoviridiae) family (Ty1, 2, 4, 5) whereas Ty3 belongs to the gypsy (Metaviridae) family [5]. Ty1–4 elements enter the genome in the vicinity of Pol III-transcribed genes, whereas Ty5 elements insert into silent chromatin [5, 6] .
Thirty-two copies of the Ty1 element, which is the most abundant S. cerevisiae TE, as well as 279 solo LTRs, are present in the genome of the commonly used laboratory strain S288C. Ty1 elements are 5.9 kb in length and composed of GAG and POL open reading frames (ORFs) sandwiched in-between 334 bp LTR sequences [7, 8]. GAG encodes the structural protein of the VLP, while POL produces a polyprotein of protease (PR), IN, reverse transcriptase (RT) with ribonuclease H activity (RH) (Fig. 1) [7]. The copia and gypsy families differ in the order of RT/RH and IN such that the Ty3-gypsy element has RT/RH followed by IN (Fig. 1) [9]. Ty1 replication begins with transcription of a genomic Ty1 element using the host RNA Pol II machinery, translation of the Ty1 messenger RNA (mRNA) into the Gag protein or the Gag-Pol fusion protein when a + 1 ribosomal frameshift event places Gag and Pol in frame [7]. The Gag and Pol polypeptide, an initiator methionine tRNA (tRNAimet) and two Ty1 mRNA transcripts, are assembled into VLPs where Gag and Pol undergo processing and maturation by PR [10–12]. Following RT-mediated reverse transcription of the Ty1 mRNA in the VLPs, a pre-integration complex composed minimally of newly synthesized Ty1 cDNA and IN, called the intasome, is generated. The intasome localizes to the nucleus where IN-mediated insertion of the Ty1 cDNA preferentially occurs in a ~ 1 kb window upstream of genes actively transcribed by RNA Pol III including all 275 nuclear tRNA genes and the 5S ribosomal RNA (rRNA) gene [13, 14]. Ty1 cDNA can also enter the genome via homologous recombination with a pre-existing Ty1 element [15, 16]. When Ty1 insertion assays are performed in vitro using purified VLPs and target DNA, targeting is random suggesting that S. cerevisiae host factors are required to target Ty1 elements to Pol III genes [17, 18]. As early as 1979, it was observed that genomic copies of Ty1 are associated with tRNA genes [19]. By 1993, the 5′ region upstream of tRNA genes was defined as the preferred Ty1 element insertion site and the glycine tRNA gene SUF16 was identified as a Ty1 insertion hotspot [20]. Upon completion of the S. cerevisiae genome sequence it was clear that the majority of Ty1–4 elements were located adjacent to tRNA genes or other Pol III-transcribed genes [8, 21]. The Ty2 and Ty4 elements share the same insertion preference as Ty1 elements, whereas the Ty3 element integrates specifically at the RNA Pol III transcription start site (TSS) [5]. To understand the mechanism of Ty insertion at tRNA genes, it is important to briefly describe the RNA Pol III transcription machinery.
Fig. 1.
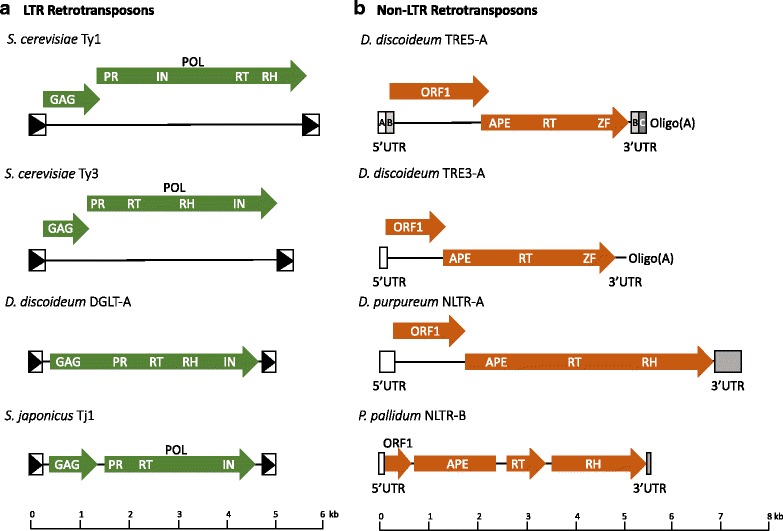
LTR and non-LTR retrotransposons that target to tRNA genes. a. LTR retrotransposons. Ty1, Ty3, DGLT-A and Tj1 elements are depicted in dark green. The boxed black arrows represent the LTRs flanking the two ends of the elements. The first ORF of the Ty1 element encodes Gag and the second ORF encodes a polypeptide (Pol) which is further processed into protease (PR), integrase (IN), and reverse transcriptase (RT)/ ribonuclease H (RH). Ty3 differs in structure from Ty1 by swapping positions of IN and RT/RH. For both Ty1 and Ty3, the Pol polypeptide is generated by a + 1 translational frameshift 38 bp upstream of the 3’end of Gag [169–171]. The D. discoideum DGLT-A element contains one ORF that encodes for both Gag and Pol proteins. DGLT-A belongs to the Ty3-gypsy clade, signified by the arrangement of pol with IN after RT/RH [172]. S. japonicas Tj1 has a similar structural arrangement as Ty3 with GAG and POL as two separate ORFs. The GAG ORF has a stop codon that is thought to be translationally suppressed to allow for translation of the POL ORF which lacks a start codon [121]. The length of each element is depicted by the scale at the bottom in kb. b. non-LTR retrotransposons. D. discoideum TRE5-A and TRE3-A, D. purpureum NLTR-A and P. pallidum NLTR-B are depicted in dark orange and all share a similar structural arrangement. All elements except NLTR-B have two ORFs flanked by untranslated regions (UTR), with TRE5-A and TRE3-A ending with an oligo(A) tail. The 5′ and 3’UTR of TRE5-A are arranged into A- and B-modules, and B- and C-modules respectively. The protein domain arrangement of TRE5-A and TRE3-A ORF2 is the same and encodes a protein containing an apurinic/apyrimidinic endonuclease (APE), RT, and zinc-finger (ZF) domain. Both TRE5-A and TRE3-A require a − 1 frameshift for translation of ORF2 [137, 173]. NLTR-A and NLTR-B have a similar arrangement to the TRE5-A and TRE3-A elements except that an RH domain substitutes for the ZF domain. In addition, NLTR-B has three separate ORFs for APE, RT and RH. It is not yet known if the 5′ and 3’ UTRs of NLTR-A and NLTR-B are arranged into modules. NLTR-A ORF1 overlaps with ORF2 by 13 bp but whether a frameshift occurs or not for translation of ORF2 is not yet known [124]. NLTR-B does not contain overlapping ORFs, however RT does not contain a start codon [124]. The length of each element is depicted by the scale at the bottom in kb
RNA Pol III transcription machinery
RNA Pol III is a 17-subunit complex that, along with TFIIIB and TFIIIC transcription complexes, transcribes all tRNAs, and other essential RNAs including the U6 small nuclear RNA [22, 23]. The 5S rRNA gene, which is also transcribed by RNA Pol III requires the additional TFIIIA transcription factor. For the purposes of this review, we briefly describe tRNA gene promoters because of the frequent use of tRNA genes in Ty1 studies. tRNA genes contain an internal promoter with two highly conserved sequence elements, a proximal box A and a more distal box B, within the transcribed region. tRNA gene activation first requires association of TFIIIC with DNA, followed by TFIIIB, which then recruits RNA Pol III [22, 23]. TFIIIC is a 6-subunit complex with a τA subcomplex that recognizes box A and a τB subcomplex that recognizes box B [24, 25]. TFIIIB is assembled from three proteins in yeast – Brf1, TATA binding protein (TBP)/Spt15 and Bdp1 [26]. Brf1 and TBP assemble first into the transcription complex followed by interaction with Bdp1 [27]. Once TFIIIB is bound, the RNA Pol III transcription complex can assemble onto the promoter [28]. The common features of all types of RNA Pol III promoters is that TFIIIC, TFIIIB and RNA Pol III are recruited to activate transcription. Mutation of the SUF16 tRNA promoter, such as a point mutation in box B, that severely reduces transcription, also dramatically reduces Ty1 element insertion suggesting that active Pol III transcription is required for Ty1 transposition [17].
Mechanism for Ty1 insertion upstream of Pol III-transcribed genes
Two reports have demonstrated that Pol III subunits are essential host factors required for Ty1 intasome targeting upstream of Pol III-transcribed genes [29, 30]. Below we outline the data presented in each study that supports a role for Pol III as the Ty1-IN host factor.
Cheung et al. overexpressed the Ty1 element from an inducible plasmid in yeast cells, purified Ty1-IN using the 8b11 monoclonal anti-IN antibody, then performed mass spectrometry (MS) to identify Ty1-IN co-purifying proteins [18, 30]. Five RNA Pol III subunits were identified by MS (Rpc25, 34, 40, 53, 82) that co-purified with Ty1-IN from two independent purifications [30]. The 17-subunit RNA Pol III complex consists of a ten-subunit core with five subunits shared with all three Pols (Rpb5, Rpb6, Rpb8, Rpb10, Rpb12) and two others shared between Pol I and III (Rpc40 and Rpc19) [31]. The seven remaining subunits are the Rpc53/37 heterodimer, which is the structural counterpart of TFIIF, the Rpc82/34/31 heterotrimer which is related to TFIIE and the Rpc25/17 dimer that is similar to Rpb4/7 [31]. GFP-tagged versions of the two largest subunits of RNA Pol III (Rpc1 and Rpc2) co-purified with Ty1-IN but the homologous Pol II subunits (Rpb1 and Rpb2, respectively) did not, suggesting that Ty1-IN specifically interacts with the Pol III complex [30]. Pol III subunits tagged with either GFP or HA were purified from yeast lysates and Rpc17, 19, 25, 34, 53, and 82 all co-purified with Ty1-IN. However, since the Pol III complex is intact during these pull-downs, it is not possible to pinpoint which Pol III subunit interacts directly with Ty1-IN using this method. Therefore, in vitro binding experiments were also performed and demonstrated that Rpc31, 34 and 53 can interact directly with Ty1-IN using bacterially expressed proteins [30].
There are a few pieces of evidence to support the hypothesis that the Rpc53/37 heterodimer may be directly involved in targeting Ty1-IN. Removal of the N-terminal 280 amino acids from Rpc53 (rpc53Δ2–280) significantly reduced Ty1 element targeting upstream of the SUF16 gene [30]. However, Ty1 mobility in the rpc53Δ2–280 mutant was not significantly impaired (~ 75% of wild type levels) suggesting that the Ty1 element may be targeted elsewhere in the genome. When GFP pull-down experiments were performed with Rpc37-GFP in the rpc53Δ2–280 strain background, Ty1-IN no longer co-purified with Rpc37 [30]. As well, a V5-tagged version of rpc53D2-280 does not interact with Ty-IN in yeast lysates (S.C. and V.M. unpublished data). Since Rpc82-GFP, Rpc19-GFP and Rpc17-GFP interact with Ty1-IN in the rpc53D2-280 mutant, the defect in Ty1 targeting may be due to a loss of interaction between Ty1-IN and the Rpc53/37 heterodimer. However, it is not known which other Ty1-IN and Pol III subunit interactions may be compromised in the rpc53Δ2–280 mutant.
Bridier-Nahmias et al., discovered an interaction between Ty1-IN and the Rpc40 subunit of RNA Pol III using a yeast two-hybrid assay that was confirmed by co-immunoprecipitation (IP) analysis between HA-tagged Rpc40 and Ty1-IN [29]. Using the yeast two-hybrid method, a specific interaction of Rpc40 was detected with only the C-terminal 57 amino acids of Ty1-IN [29]. Cheung et al. found that removal of 75 amino acids from the C-terminus of Ty1-IN abrogated the interaction of Ty1-IN with Rpc82-GFP in pull-down experiments [30]. Therefore, the data from both groups suggests that the C-terminus of Ty1-IN is important for interaction with Pol III. Interestingly, the C-terminus of Ty5-IN interacts with Sir4 to target Ty5 to silent chromatin [32, 33]. To disrupt the interaction of Ty1-IN with RNA Pol III without reducing Pol III transcription, Bridier-Nahmias et al., made clever use of a previous observation that the Schizosaccharomyces pombe (S. pombe) Rpc40 subunit (Rpc40sp) can functionally replace the S. cerevisiae Rpc40 subunit [34]. When Rpc40 was replaced with Rpc40sp, the interaction with Ty1-IN and Ty1 element targeting upstream of Pol III genes was disrupted [29]. Interestingly, overall Ty1 mobility was not impaired in the Rpc40sp strain and genome-wide mapping revealed that Ty1 elements were preferentially targeted to the last 20-30 kb at the ends of each chromosome [29]. This work reveals that Ty1-IN may interact with alternative host factors in the absence of the Rpc40-Ty1-IN interaction. The Ty5 retrotransposon integrates preferentially into heterochromatin at telomeres and silent mating loci [35–37]. It would be interesting to test if Sir4, which targets Ty5-IN to heterochromatin, is able to interact with Ty1-IN in the absence of Rpc40 [32, 33].
Structures of retroviral intasomes, which are INs in complex with their viral cDNA, have revealed that intasomes can be a tetramer, an octamer or even higher order oligomers of IN protomers [38–43]. The structure of Ty1-IN has not been determined yet, nor what type of oligomer structure it may form. Since Ty1-IN is a 636-amino acid protein (predicted molecular weight of 71.5 kDa for a monomer or 286 kDa for a tetramer) it is possible that the Ty1-IN intasome could interact with multiple Pol III subunits as the entire 17-subunit RNA Pol III complex is ~ 690 kDa. In Fig. 2, we provide a structure of RNA Pol III based on recent structural data that highlights the 2 largest Pol III subunits (Rpc1,2) the Pol III specific subunits (Rpc31/34/82 heterotrimer, Rpc53/37 dimer, Rpc17/25 dimer) and Rpc40 [44]. Of the highlighted subunits in Fig. 2, there is evidence that Rpc31, Rpc34, Rpc40 and Rpc53 may interact directly with Ty1-IN [29, 30]. Rpc40 is positioned in the Pol III complex facing the upstream DNA which may be relevant because Ty1 elements are only inserted upstream of Pol III transcribed genes [17, 21]. Future structural studies of Ty1-IN binding to RNA Pol III will help determine precisely how this interaction takes place.
Fig. 2.
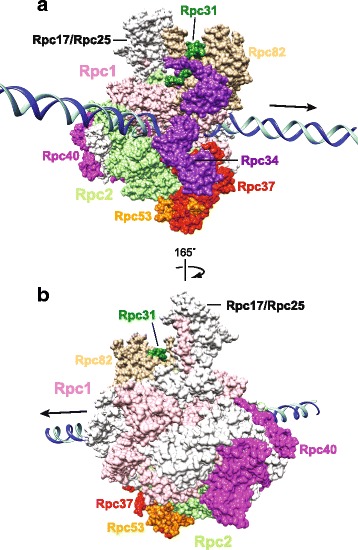
Pol III structure highlighting subunits that may interact with Ty1-IN. The Pol III surface view is based on the cryoelectron microscopy structure of the initially transcribing Pol III complex (Protein Data Bank code 6f41) [44] with TBP, Brf1 and Bdp1 structures excluded. The arrow points to downstream DNA and the DNA template and non-template strands are coloured in light blue and dark blue, respectively. a The highlighted Pol III subunits are Rpc31 (dark green), Rpc34 (purple), Rpc82 (beige), Rpc1 (light pink), Rpc2 (light green), Rpc40 (magenta), Rpc53 (orange) and Rpc37 (red). The N-terminus of Rpc53 (amino acids 1–270) is not depicted due to lack of structural data. b Same as in (a) except rotated 165o
Ty1 targeting into chromatin
Chromatin remodeling
Yeast tRNA genes have an open chromatin structure with strongly ordered upstream nucleosomes and a nucleosome-depleted gene body [45–47]. Ty1 element genome-wide mapping studies demonstrated that Ty1 insertions are targeted to two DNA sites on the same surface of the nucleosome at the H2A/H2B interface [13, 14, 48]. Structural studies of the prototype foamy virus (PFV) intasome, a homotetramer of PFV-IN, attached to a nucleosome have revealed striking similarity to the nucleosome data from the Ty1 genome-wide mapping studies [40, 49]. The PFV intasome also interacts with one H2A/H2B heterodimer and two DNA strands on the same surface of the nucleosome [49]. Therefore, the interaction between homotetramer INs and nucleosomes may be conserved.
Chromatin remodeling complexes, which utilize ATP to mobilize nucleosomal DNA, impact Ty1 transcription and Ty1 genome integration. The SWI/SNF and SAGA chromatin-remodeling complexes are required for Ty1 transcription whereas Isw1 and Isw2 (catalytic subunits of three ISW1 chromatin remodeling enzymes) inhibit Ty1 transcription [50–53]. Deletion of Isw2 disrupts the periodic Ty1 integration pattern upstream of tRNA genes likely because Isw2 is needed to maintain the nucleosome array upstream of all tRNA genes [46, 54, 55]. Isw2 may be recruited by Bdp1, a component of TFIIIB, because removal of the Bdp1 N-terminus (bdp1-Δ240) also results in altered nucleosome positioning and Ty1 insertion upstream of tRNA genes [54]. However, Ty1 elements are still targeted to tRNA genes in the bdp1-Δ240 mutant strain and Bdp1 does not interact with Ty1-IN in yeast lysates [30, 54]. This data suggests that the TFIIIB complex is not a Ty1-IN host targeting factor.
Structural maintenance of chromosomes (Smc) complexes that are essential for chromosome condensation and segregation localize to Pol III-transcribed genes. The Smc2/4 condensin complex, which is required for chromosome compaction, binds to tRNA genes and physically interacts with TFIIIB and TFIIIC [56, 57]. A potential role for condensin in Ty1 targeting has not yet been explored. The Smc1/3 cohesin complex, which holds sister-chromatids together, requires the Scc2/4 complex to load onto chromosomes [58, 59]. Notably, Scc2/4 binds to the same chromosomal locations as condensin and may be recruited by TFIIIC to bind box B sites [56]. Once cohesin loads onto chromosomes at Scc2/4 binding sites, it relocalizes to sites of active transcription [60]. The separation of sister chromatids in mitosis requires cleavage of the cohesin ring by a conserved cysteine protease called separase, or Esp1 in yeast [61]. Interestingly, Esp1 was found to physically interact with Ty1-IN and this interaction is enriched in metaphase cells [62]. An esp1–1 mutant with reduced cleavage activity has reduced Ty1 mobility and Ty1 insertion upstream of the SUF16 tRNA gene [62]. Consistently, mutations in cohesin proteins (including Scc1 which is cleaved by Esp1) cause enhanced Ty1 mobility and increased Ty1 element insertion upstream of the SUF16 tRNA gene [62]. The simplest interpretation of why increased Ty1 mobility is observed upon removal of the cohesin complex is that the Ty1 intasome has increased access to nucleosomes. However, the physical interaction between Ty1-IN and Esp1 could be one mechanism by which Ty1-IN is targeted to chromatin [62].
Histone modification
Chromatin-modifying enzymes, which add or remove post-translational modifications to the core histones, also impact Ty1 targeting. Hos2 and Set3, which are both members of the Set3 histone deacetylase complex, are required for the efficient integration of Ty1 elements upstream of tRNA genes [63]. Although Hos2 is required for Ty1 integration, genome-wide Ty1 mapping studies did not find any difference in the Ty1 insertion pattern of a hos2Δ mutant compared to a wild type strain [13]. Deletion of the Rpd3 histone deacetylase caused reduced Ty1 insertion upstream of the SUF16 tRNAGLY gene [64]. Disruption of other types of complexes that interact with chromatin, such as the Paf1 complex that associates with elongating RNA Pol II, causes an increase in both Ty1 mobility and Ty1 element insertion upstream of SUF16 [64–66]. Paf1 stimulates the monoubiquitylation of histone H2B (H2B K123Ub) by the Bre1-Rad6 ubiquitin ligase complex [67]. Interestingly, genome-wide Ty1 mapping in a rad6Δ mutant demonstrated that Ty1 elements insert more frequently into open reading frames compared to a wild type strain [13]. An attractive hypothesis that emerges from these observations is that modification of nucleosomes by Paf1 associated Bre1-Rad6 restricts insertion of Ty1 elements. A screen for mutants that negatively regulate Ty1 transposition (rtt mutants) identified the Rtt109 histone acetyltransferase and Rtt106 histone chaperone [68]. Rtt109 catalyzes the acetylation of Histone H3 lysine 56 on newly synthesized H3-H4 dimers which interact with Rtt106 to promote replication coupled nucleosome assembly [69]. Stalling of DNA replication in the absence of either Rtt109 or Rtt106 may allow for increased Ty1 mobility. However, genome-wide mapping of Ty1 element insertion in an rtt109Δ mutant strain revealed a similar pattern to wild type strains suggesting that Rtt109 does not directly affect Ty1 targeting [13]. A complete understanding of how chromatin remodelling and histone modifications may impact Ty1 targeting and mobility will be aided by histone mutant libraries. For example, a comprehensive library of H2A and H2B mutants has been generated that could be used for testing Ty1 targeting [70]. A systematic screen of Ty1 targeting in mutants of all chromatin-modifying complexes could also be performed. Ultimately, structural studies of the Ty1 intasome in complex with nucleosomes is a critical step for understanding Ty1 element integration into the genome.
3-dimensional organization of tRNAs in the nucleus
The intranuclear positioning of tRNA genes could potentially affect the dynamics of Ty1 insertion. Multiple S. cerevisiae studies have assessed the localization of tRNA genes in the nucleus and different technical methods reveal different localization patterns. Fluorescence in situ hybridization demonstrated that yeast tRNA genes, although dispersed on linear chromosome maps, cluster in the nucleolus in a condensin-dependant manner [57, 71]. Chromosome conformation capture studies identified a cluster of tRNA genes that co-localized with the nucleolar ribosomal DNA (rDNA) repeats and another cluster that co-localized with centromeres [72–75]. Live-cell imaging of fluorescently labelled tRNA genes in S. cerevisiae demonstrated that tRNA genes can reside at the nucleolus, the nuclear periphery and in the nucleoplasm [76, 77]. In the live-cell imaging studies, the frequency of tRNA association with the nuclear periphery or nucleolus depends on how far the tRNA gene is from a tethering element such as the centromere, telomere, or rDNA. For example, SNR6 is located close to the rDNA and exclusively localizes to the nucleolus whereas SUP53, which is located 23 kb from CENIII, is excluded from the nucleolus [77]. A tRNA gene with no constraints may localize to the nucleolus, nucleolar periphery or nuclear periphery [77]. Fluorescence microscopy and chromatin immunoprecipitation (ChIP) studies demonstrated that tRNA genes are recruited to the nuclear pore complex (NPC) during G2/M phase which also happens to be the peak of tRNA gene expression [78]. These studies highlight the dynamic 3-dimensional positioning of tRNA genes in the nucleus during the yeast cell cycle. Furthermore, evidence is gathering that tRNA genes have broad global effects on genome structure and organization by providing tethers to cellular structures such as the nucleolus, nuclear periphery and mitotic spindle [77–79]. Our group has recently discovered that nuclear basket proteins, which are located on the nuclear side of the NPC, are required for targeting Ty1 elements upstream of tRNA genes [80]. In the absence of the nuclear basket proteins, Ty1 elements are targeted to subtelomeric regions, similar to the Rpc40sp mutant strain described above [80]. HIV-1 viral cDNA is preferentially inserted into transcriptionally active genes that are localized near the nuclear envelope [81]. The HIV-1 intasome also localizes near the nuclear periphery and the chromatin environment on the nuclear basket side of the NPC is favourable for HIV-1 insertion [82, 83]. Chromatin that resides near the nuclear pore may therefore serve as a convenient site for intasomes to insert their cDNA immediately after passage through the NPC.
Comparison of Ty1 and Ty3 targeting
The S. cerevisiae Ty3-gypsy retrotransposon also selectively targets genes transcribed by RNA Pol III, however, unlike Ty1, it has a precise integration site that maps to within 1–4 nucleotides of the Pol III TSS [84–86]. There are two full length Ty3 elements in the S288C S. cerevisiae genome and only one is active [9]. Similar to Ty1, a functional Pol III promoter is required for Ty3 transposition as mutation of the box A or box B promoter sequences prevents insertion of the Ty3 element [85, 87]. However, a tRNA gene with reduced transcriptional activity due to mutations in the transcription initiation region is still an active Ty3 target [85]. The ability of TFIIIC and TFIIIB to load onto the tRNA promoter is essential for Ty3 targeting but a wild type level of tRNA gene transcription is not. In vitro reconstitution with recombinant TFIIIB proteins demonstrated that Ty3-IN, TBP(Spt15) and Brf1 are required for Ty3 insertion while addition of the third component of TFIIIB, Bdp1, enhances integration efficiency [88, 89]. The conserved domain of TBP inserted between the N and C-terminal segments of Brf1, which can function to initiate Pol III transcription, can also mediate Ty3 insertion in vitro [90, 91]. Extra TFIIIC sites in the yeast genome that bind TFIIIC but not TFIIIB or Pol III, are not targeted by Ty3, further strengthening the argument that TFIIIB is the key Ty3 targeting factor [92, 93].
Although TFIIIB is the host factor for Ty3-IN, TFIIIC also influences the Ty3 insertion pattern. The C-terminus of Tfc1 physically interacts with Ty3-IN and enables Ty3 insertion in both orientations [88, 94]. By comparison, no physical interaction was detected between Ty1-IN and Tfc1, Tfc3 or Tfc7 in co-purification experiments from yeast lysates [30]. Another interesting difference between Ty1 and Ty3 targeting is that RNA Pol III, which is required for Ty1 element insertion, is inhibitory to Ty3 insertion in vitro [87, 95]. Genome-wide Ty1 and Ty3 insertion site mapping studies have also discovered interesting targeting differences between the two retrotransposons. For example, Ty3, unlike Ty1, does not target to nucleosomes [13, 14, 93]. Ty3 is capable of inserting at the TSS of the tRNA relic gene ZOD1 which is bound by the Pol III machinery whereas Ty1 is not [13, 14, 93]. The lack of Ty1 targeting to ZOD1 may be due to low ZOD1 transcription levels [13, 14]. Interestingly, the ZOD1 locus is activated upon nucleosome depletion which may also prevent Ty1 targeting [96]. Finally, Ty3 elements only integrate at Pol III-transcribed genes whereas Ty1 elements are capable of integrating at other genomic loci such as within silent mating cassettes, within or near Pol II-transcribed genes and at sub-telomeric regions [29, 97–102]. Ty1-IN may interact with alternative host factors to achieve insertion into such a variety of genomic regions. Although Ty1 and Ty3 are both targeted upstream of Pol III-transcribed genes, they have devised different targeting mechanisms for insertion into the genome.
tRNA targeting TEs in other yeast species
The Saccharomyces sensu stricto genus includes seven natural species: S. arboricolus, S. cerevisiae, S. eubayanus, S. kudriavzevii, S. mikatae, S. paradoxus, S. uvarum, and two hybrid species: S. pastorianus and S. bayanus [103–105]. There is variation in the presence or absence of Ty elements in these species and the abundance of a particular element can vary widely between strains [106–108]. For example, Ty3 and Ty5 elements do not occur in S. uvarum [109]. A novel Ty3-like element, called Ty3p, was discovered in S. paradoxus that shares 82% nucleotide identity with an S. cerevisiae Ty3 element (YGRWTy3–1) and is inserted ~ 6 bp upstream of a tRNA TSS (Table 1) [110]. Degenerate solo LTRs of Ty3p are also present in the S. cerevisiae genome [111]. The targeting of Ty1, Ty2, Ty3, and Ty4 elements upstream of tRNA genes is conserved in the Saccharomyces sensu stricto genus.
Table 1.
Retrotransposons that integrate adjacent to tRNA genes
| Mobile element | Clade | Hosta | Preferred integration site | Host factors mediating this insertion preference |
|---|---|---|---|---|
| LTR Retrotransposons | ||||
| Ty1 | Ty1-copia | Saccharomyces cerevisiae | ~ 1 kb window upstream of RNA Pol III-transcribed genes, including tRNA and 5S rRNA genes [13, 14, 21] | Ty1-IN interaction with Rpc40 [29] and Ty1-IN interaction with Rpc53, Rpc34, Rpc31 [30] |
| Ty2 | Ty1-copia | Saccharomyces cerevisiae | ~ 1 kb window upstream of RNA Pol III-transcribed genes [21] | |
| Ty3 | Ty3-gypsy | Saccharomyces cerevisiae | 1–4 bp upstream of tRNA TSSb [93] | Ty3-IN interaction with TFIIIB [88, 89] |
| Ty3p | Ty3-gypsy | Saccharomyces paradoxus | ~ 6 bp upstream of tRNA TSS [110] | |
| Ty4 | Ty1-copia | Saccharomyces cerevisiae | ~ 1 kb window upstream of RNA Pol III-transcribed genes [21] | |
| Tj1 | Ty3-gypsy | Schizosaccharomyces japonicus | 1–10 bp upstream of tRNA TSS [121] | |
| Beta/Tca8 | Ty3-gypsy | Candida albicans | 6-30 bp upstream of tRNA MCSc [119] | |
| Tyl3 | Ty3-gypsy | Yarrowia lipolytica | ~ 5 bp upstream of tRNA TSS [116] | |
| Ylt1 | Ty3-gypsy | Yarrowia lipolytica | ~ 5 bp upstream of tRNA TSS [117] | |
| Tyl6 | Ty3-gypsy | Yarrowia lipolytica | ~ 5 bp upstream of tRNA TSS [115] | |
| DGLT-A | Ty3-gypsy | Dictyostelium discoideum | 13–33 bp upstream of tRNA MCS [125] | |
| Skipper-2 | Ty3-gypsy | Dictyostelium discoideum | 8–23 bp downstream of tRNA gene [124, 172] | |
| Non-LTR Retrotransposons | ||||
| TRE5 | L1 | Dictyostelium discoideum | 40–54 bp upstream of tRNA MCS; 37–41 bp upstream of extrachromosomal 5S rRNA genes [125, 134, 135, 174] | TRE5 ORF1 interaction with TFIIIB [132] |
| TRE3 | L1 | Dictyostelium discoideum | 40–150 bp downstream tRNA genes [137, 175] | |
| NLTR-A | L1 | Dictyostelium purpureum | 2–6 bp upstream of tRNA MCS [124] | |
| NLTR-B | L1 | Polysphondylium pallidum | 39–64 bp upstream of tRNA MCS [124] | |
aHost that retrotransposon was first identified in
bTSS refers to the tRNA transcription start site which is ~ 10 bp upstream of the mature tRNA
cMCS refers to the tRNA mature coding sequence
The rapid pace of whole genome sequencing in a variety of fungal species has revealed the diversity of retrotransposons [112–114]. Interestingly, a subset of these newly discovered TEs in the fungal Ascomycota phylum are distributed in the genome nearby tRNA genes (Table 1). The genome of the oleaginous yeast, Yarrowia lipolytica contains three Ty3-gypsy-like elements (Tyl3, Ylt1, Tl6) located upstream of Pol-III transcribed genes (Table 1) [115–117]. Candida albicans (C. albicans) is an opportunistic human fungal pathogen that contains 34 LTR-retrotransposon families (alpha, beta, gamma, etc.) in its genome that belong to the Ty1-copia and Ty3-gypsy families [118]. The beta LTR of the Tca8 element, which has partial elements remaining in the genome, is localized within 30 bp upstream of the mature coding sequence (MCS) of tRNA genes (Table 1) [119]. An investigation of the Pol III targets in C. albicans using Rpc82 ChIP-chip revealed that Rpc82 bound tRNA genes at high occupancy and retrotransposon elements at low occupancy [120]. The low occupancy binding of Rpc82 to elements such as Tca8 is likely due to amplification of Rpc82 binding to tRNA genes located adjacent to retrotransposon elements in the C. albicans genome [120].
Whole genome sequencing and comparison of fission yeast genomes revealed that the Schizosaccharomyces japonicus (S. japonicus) genome contains 10 families (Tj1 to Tj10) of Ty3-gypsy related retrotransposons clustered at the centromeres and telomeres [121, 122]. Notably, retrotransposons were dramatically reduced or lost in the other fission yeast genomes likely due to evolutionary change in control of centromere function [122]. Since tRNA genes are clustered at the centromere, the Levin lab hypothesized that the S. japonicus retrotransposons may be specifically targeted to tRNA genes. They tested this hypothesis by cloning the S. japonicus Tj1 retrotransposon and analyzing its integration behaviour in the related fission yeast S. pombe [121]. As predicted, the Tj1 transposon inserted 1–10 bp upstream of the TSS of tRNA genes and also at the Pol III-transcribed 5S rRNA gene (Fig. 1, Table 1) [121]. Therefore, S. japonicus Tj1 targets Pol III-transcribed genes and has similar insertion behaviour to Ty3 retrotransposons.
The diversity of retrotransposons in fungal species now includes Ty1-copia, Ty3-gypsy and LINE elements [112–114, 123]. The target specificity of each of these retrotransposons has not been fully elucidated but it is likely that Pol III-targeting will feature prominently [123].
TEs target RNA pol III transcribed genes in Dictyostelium
Mobile elements in other organisms with compact genomes have also found a safe haven by inserting adjacent to tRNA genes; the social amoeba model organism Dictyostelium discoideum (D. discoideum) is one such organism. D. discoideum has tolerated an expansion of tRNA targeting retrotransposons to 3.8% of its genome whereas 0.9% or less of the genomes of other social amoeba contain tRNA associated retrotransposons [124]. It is not known what selection pressure may have allowed retroelement expansion in D. discoideum [124]. The Dictyostelium gypsy-like transposon (DGLT-A) belongs to the Ty3-gypsy clade of retrotransposons and preferentially inserts 13 to 33 bp upstream of the tRNA MCS in either orientation (Fig. 1, Table 1) [125]. The lack of full length DGLT-A elements in the D. discoideum genome suggests that they are no longer active [124]. Skipper-1 is another LTR retrotransposon in the D. discoideum genome that is related to DGLT-A and the Ty3-gypsy clade. Skipper elements, which accumulate at the centromere, contain a characteristic chromo domain (CHD) in the C-terminus of the Skipper IN protein [126]. The CHD may be important for targeting Skipper-1 into heterochromatin at the centromere [127]. Skipper-2 (previously named DGLT-P) has a diverged CHD and instead of targeting to centromeres is targeted ~ 8-23 bp downstream of tRNA genes (Fig. 3) [124]. Notably, Skipper-2 has also been identified in other amoeba species, including Dictyostelium purpureum (D. purpureum), Dictyostelium fasciculatum (D. fasciculatum), and Polysphondylium pallidum (P. pallidum), where it is located ~ 140 bp downstream of tRNA genes (Table 1) [124]. Skipper-2 is the first LTR retrotransposon that preferentially integrates downstream of a tRNA gene [124]. It will be interesting to determine if the diverged CHD is responsible for targeting Skipper-2 downstream of tRNA genes.
Fig. 3.
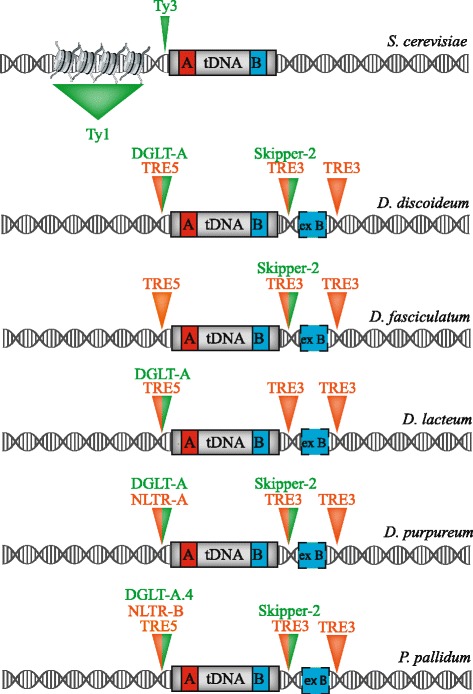
tRNA targeted retrotransposon insertion site profiles. The insertion site preference for S. cerevisiae, Dictyostelium and P. pallidum are shown here upstream and downstream of a tRNA gene. The tRNA gene (gray) contains box A (red) and box B (blue) internal promoters and the external box B (ex B, blue) for social amoeba. LTR-retrotransposons are in green and non-LTR retrotransposons are in orange. Inverted orange or green triangles denote retrotransposon insertion windows ranging from 2 to ~ 1000 bp upstream or 7 to ~ 450 bp downstream of the tRNA gene (not drawn to scale). For the social amoeba, split orange and green inverted triangles denote overlapping insertion footprints for LTR (DGLT-A, Skipper-2) and non-LTR (NLTR-A, NLTR-B, TRE5, TRE3) retrotransposons. For P. pallidum, a specific DLGT-A (DGLT-A.4) is indicated because DGLT-A.1–3 do not target to tRNA genes in this organism [124]. The green triangle with a broader base represents the larger insertion window for S. cerevisiae Ty1 which can insert up to ~ 1 kb upstream of a Pol III-transcribed gene. Nucleosomes are depicted upstream of the S. cerevisiae tRNA gene as Ty1 inserts into nucleosomes
The D. discoideum genome also contains non-LTR retrotransposons called TREs for tRNA gene-targeted retroelements. TRE5 elements preferentially integrate upstream (5′) of tRNA genes, whereas TRE3 elements are targeted downstream (3′) of tRNA genes; the element names are a convenient reminder of their integration preference (Figs. 1,3) [128–130]. There are three TRE5 elements (TRE5-A,B,C) and four TRE3 elements (TRE3-A,B,C,D) in the D. discoideum genome with TRE5-A and TRE3-A in the highest abundance [128]. TRE5 elements insert ~ 44-54 bp upstream of the tRNA MCS in the opposite transcriptional orientation (Table 1, Fig. 3) [130]. The TRE5-A retrotransposon has two ORFs - ORF1 encodes a 51kD protein of unknown function and ORF2 encodes a protein with an apurinic/apyrimidinic endonuclease (APE) domain, an RT domain, and a zinc-finger (ZF) domain (Fig. 1) [129, 131]. Interestingly, protein-protein interactions have been detected between the TRE5-A ORF1 protein and the three D. discoideum TFIIIB proteins TBP, Brf1 and Bdp1 [132]. Despite the similarity to Ty3, which also interacts with TFIIIB, the molecular basis of TRE5-A targeting may differ from Ty3 because of the mechanism by which TRE5-A elements integrate into the genome. Non-LTR retrotransposons such as TRE5-A elements replicate by target-primed reverse transcription whereby the APE domain nicks the target DNA which allows for reverse transcription followed by integration of the element [6]. However, similar to Ty3 elements, mutations of the box B promoter that interfere with binding of TFIIIC abolish the targeting of TRE5-A to the tRNA target gene [133]. TRE5-A insertion profiling demonstrated that TRE5-A can also integrate at the Pol III-transcribed ribosomal 5S gene which is located on a multi-copy extrachromosomal DNA element harboring the rRNA genes [134, 135]. Unlike TRE5, TRE3 has a broader range of insertion that is 40–150 bp downstream of tRNA genes in the same transcription orientation (Fig. 3) [130]. The broader insertion window is because TRE3 can target downstream of either the tRNA internal box B or an external box B (ex B) that is positioned ~ 100 bp downstream of the internal box B and present at ~ 80% of D. discoideum tRNA genes (Fig. 3) [136, 137]. New non-LTR retrotransposons (NLTR) were recently identified in the genomes of D. purpureum (NTLR-A) and P. pallidum (NLTR-B) that are distantly related to TRE elements [124]. P. pallidum NLTR-B inserts upstream of tRNA genes in a similar manner to TRE5 elements however D. purpureum NLTR-A has a unique insertion specificity 2-6 bp upstream of the tRNA MCS (Fig. 3) [124].
Evolutionary selection of pol III transcribed genes as a genomic target for insertion
Survival of mobile elements in the compact Saccharomyces and Dictyostelium genomes necessitated insertion of the element in a locus that minimized host genome damage [138]. During evolution, retrotransposons have independently developed targeting to tRNA genes at least six times in dictyostelids and at least four times (Ty1–4) in S. cerevisiae [124]. Insertion upstream of Pol III-transcribed genes has the advantage that most Pol III-transcribed genes exist in multiple copies, therefore they are an abundant target and insertion into one locus is not likely to be lethal. Furthermore, the promoter elements of tRNA genes are embedded within the coding region and inserting upstream of tRNA genes will not damage promoter activity. The S. cerevisiae genome has 275 copies of tRNA genes for decoding the 20 standard amino acids, and the 5S rRNA exists in a tandem array consisting of 100–200 copies [8]. Therefore, there are plenty of target sites available for Ty1–4 retrotransposon integration. D. discoideum and D. purpureum have an expansion in the number of their tRNA genes (418 and 353, respectively) compared to other dictyostelids [124]. The large number of tRNA genes has allowed amplification of the DGTL-A retrotransposon in D. discoideum but not the other dictyostelids, including D. purpureum [124]. Therefore, an increase in the target site, in this case a tRNA gene, does not always give a retrotransposon freedom to increase in abundance [124]. Insertion of retrotransposons downstream of tRNA genes is only found in dictyostelid genomes (TRE3 and Skipper-2) but not in the S. cerevisiae genome [124]. Integration of retrotransposons downstream of S. cerevisiae tRNA genes may negatively impact tRNA or adjacent gene transcription and overall cell fitness. The insertion of Ty1 or Ty3 elements upstream of tRNA genes does not appear to negatively affect tRNA gene transcription in S. cerevisiae. On the contrary, evidence shows that these elements have a neutral or moderately stimulatory effect on tRNA gene transcription [139, 140]. It has not yet been investigated if tRNA gene expression is affected in D. discoideum when retrotransposons insert nearby [131]. The retrotransposon may benefit however from its targeting preference because the promoter activity of the A module in TRE5-A is enhanced if a tRNA gene is present upstream [141].
Whether or not Ty1 insertion events are advantageous or deleterious to the cell has no simple answer. Single novel Ty1 insertions upstream of Pol III-transcribed genes have no growth advantage or disadvantage compared to a parental strain lacking Ty insertions [142]. This data is consistent with the theory that the insertion site of Ty1 elements has evolved to minimize deleterious effects on the host genome [142]. Ty1 elements also have an internal mechanism of copy number control which likely evolved to prevent retrotransposon bursts that decrease host cell fitness due to genome instability. Expression from an internal promoter of a protein derived from the C-terminal half of Gag inhibits retrotransposition in a dose-dependent manner [143, 144]. Ty1 transposition must be artificially induced to assess the effect of increased Ty1 copy number. As the copy number of novel Ty1 elements doubles, yeast strains develop a wide range of growth phenotypes including insertions that do not affect strain growth, those that confer a negative fitness effect and those that confer a growth advantage [145, 146]. Remarkably, Ty1 copy number can be increased as much as 10-fold and still only modest growth phenotypes are detected [147]. However, with a 10-fold increase in Ty1 elements, the strains become highly sensitive to DNA damaging agents due to increased ectopic recombination [147].
Mechanisms of Ty1-mediated genome evolution
Ty elements can cause genome evolution by a variety of mechanisms [148]. If transcription of the Ty1 element is induced, for example in response to environmental stress (UV light, ionizing radiation) then Ty1-IN mediated insertion events may be a mechanism of genome evolution [149–151]. DNA replication stress, DNA damage and genome damage due to telomere erosion can also activate Ty1 mobility [152–154]. Increased Ty1 mobility is also responsible for chromosome rearrangements in aging yeast populations [155]. Induction of Ty1 transcription and transposition under stress is thought to be a strategy to increase cell survival by inducing adaptive mutations. Ty1 predominantly inserts upstream of Pol III-transcribed genes but can also insert into Pol II-transcribed genes or in subtelomeric regions [13, 14, 29, 80]. Insertion of Ty1 into the URA3 gene can be detected when cells are grown on 5-Fluoroorotic acid which is toxic to cells unless the URA3 locus is mutated and cells are supplemented with uracil [98]. Another classic example of Ty1 insertion into a Pol II-transcribed gene is mutation of the CAN1 locus which results in resistance to the arginine analogue canavanine [102].
Repetitive elements such as Ty retrotransposons and tRNA genes are fragile genomic sites because they are prone to genome rearrangement. Experimental evolution of S. cerevisiae in a glucose-limited environment caused chromosomal rearrangements due to ectopic recombination between tRNA genes, entire Ty elements or solo LTRs on different chromosomes [156]. Double-strand breaks (DSBs) induced by ionizing radiation or perturbations of essential DNA replication proteins cause chromosome breakage at repetitive Ty elements and chromosome translocations due to ectopic recombination with Ty elements on other chromosomes [157–161]. DSBs can also be repaired by ectopic recombination using Ty elements that are located up to ~ 50 kb away from the break site [162]. Interestingly, DSB repair has also been shown to occur at NPCs, where active transcription tRNA genes occurs [163, 164]. Pol III-transcribed genes are also prone to RNA:DNA hybrid formation (R-loops) that are susceptible to DNA damage due to exposure of single stranded DNA [165, 166]. In the absence of RNAse H, which removes RNA:DNA hybrids, Ty1 cDNA also forms R-loops likely during reverse transcription, and is elevated ~ 3-fold resulting in increased Ty1 mobility [166]. Taken together, tRNA and Ty repetitive elements are dynamic regions of genetic movement contributing to the evolutionary flux of the eukaryotic genome.
Conclusions
Retrotransposons and retroviruses have successfully utilized the Pol III transcription machinery and Pol III-transcribed genes to replicate in eukaryotic cells. Both retrotransposons and retroviruses use a tRNA priming system for reverse transcription. SINE elements, which constitute ~ 11% of the human genome, evolved from tRNA priming of retroviral genomes and contain box A and box B elements in their 5′ regions [167, 168]. Both yeast and social amoeba retrotransposons with different structures and ORFs have found a safe haven near tRNA genes (Fig. 3). The ongoing search for new TEs that are targeted adjacent to Pol III-transcribed genes and the host factors required for their insertion will allow better understanding of the mechanisms used by retrotransposons and retroviruses to gain access to host genomes. Future studies on how mobile elements contribute to the maintenance of the global architecture of the genome will provide novel evolutionary insights into the importance of these abundant elements.
Acknowledgements
We would like to thank Dr. Hung-Ta Chen for generating Fig. 2.
Funding
SC holds a University of British Columbia 4-Year Fellowship. VM is supported by a grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2016-04261) and the Canadian Institutes of Health Research (HOP-131559).
Author’s contributions
SC, SM and VM shared equal roles in writing the manuscript. SC generated Fig. 1 and Table 1, SM generated Fig. 3 and Dr. Hung-Ta Chen generated Fig. 2.
Abbreviations
- APE
Apurinic/apyrimidinic endonuclease
- C. albicans
Candida albicans
- ChIP
Chromatin immunoprecipitation
- D. discoideum
Dictyostelium discoideum
- DGLT-A
Dictyostelium gypsy-like transposon
- DSB
Double-strand break
- ex B
External box B
- IN
Integrase
- IP
Immunoprecipitation
- LINE
Long interspersed element
- LTR
Long terminal repeat
- MCS
Mature coding sequence
- mRNA
Messenger RNA
- NLTR
Non-LTR retrotransposon
- NPC
Nuclear pore complex
- ORF
Open reading frame
- P. pallidum
Polysphondylium pallidum
- PFV
Prototype foamy virus
- Pol
Polymerase
- PR
Protease
- rDNA
Ribosomal DNA
- RH
Ribonuclease H
- rRNA
Ribosomal RNA
- RT
Reverse transcriptase
- S.cerevisiae
Saccharomyces cerevisiae
- S.japonicus
Schizosaccharomyces japonicus
- S.pombe
Schizosaccharomyces pombe
- SINEs
Short interspersed elements
- TBP
TATA binding protein
- TE
Transposable elements
- TOR
Target of rapamycin
- TRE
tRNA gene targeted retroelement
- tRNA
Transfer RNA
- TSS
Transcription start site
- UTR
Untranslated region
- VLP
Virus-like particle
- ZF
Zinc-finger
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stephanie Cheung, Email: stephanieth.cheung@ubc.ca.
Savrina Manhas, Email: savrinam@outlook.com.
Vivien Measday, Phone: (604) 827-5744, Email: vmeasday@mail.ubc.ca.
References
- 1.Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5(4):103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 3.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 4.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 5.Lesage P, Todeschini AL. Happy together: the life and times of ty retrotransposons and their hosts. Cytogenet Genome Res. 2005;110(1–4):70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- 6.Sultana T, Zamborlini A, Cristofari G, Lesage P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat Rev Genet. 2017;18(5):292–308. doi: 10.1038/nrg.2017.7. [DOI] [PubMed] [Google Scholar]
- 7.Curcio MJ, Lutz S, Lesage P. The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol Spectr. 2015;3(2):1–35. doi: 10.1128/microbiolspec.MDNA3-0053-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274(5287):546. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 9.Sandmeyer S, Patterson K, Bilanchone V. Ty3, a position-specific retrotransposon in budding yeast. Microbiol Spectr. 2015;3(2):1–29. doi: 10.1128/microbiolspec.MDNA3-0057-2014. [DOI] [PubMed] [Google Scholar]
- 10.Garfinkel DJ, Hedge AM, Youngren SD, Copeland TD. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65(9):4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merkulov GV, Lawler JF, Jr, Eby Y, Boeke JD. Ty1 proteolytic cleavage sites are required for transposition: all sites are not created equal. J Virol. 2001;75(2):638–644. doi: 10.1128/JVI.75.2.638-644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merkulov GV, Swiderek KM, Brachmann CB, Boeke JD. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 gag protein. J Virol. 1996;70(8):5548–5556. doi: 10.1128/jvi.70.8.5548-5556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baller JA, Gao J, Stamenova R, Curcio MJ, Voytas DF. A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res. 2012;22(4):704–713. doi: 10.1101/gr.129585.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mularoni L, Zhou Y, Bowen T, Gangadharan S, Wheelan SJ, Boeke JD. Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 2012;22(4):693–703. doi: 10.1101/gr.129460.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melamed C, Nevo Y, Kupiec M. Involvement of Cdna in homologous recombination between ty elements in Saccharomyces-cerevisiae. Mol Cell Biol. 1992;12(4):1613–1620. doi: 10.1128/mcb.12.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharon G, Burkett TJ, Garfinkel DJ. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14(10):6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10(5):620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 18.Eichinger DJ, Boeke JD. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54(7):955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 19.Cameron JR, Loh EY, Davis RW. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- 20.Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, Natsoulis G, Boeke JD. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73(5):1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 21.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8(5):464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 22.Acker J, Conesa C, Lefebvre O. Yeast RNA polymerase III transcription factors and effectors. Biochim Biophys Acta. 2013;1829(3–4):283–295. doi: 10.1016/j.bbagrm.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Turowski TW, Tollervey D. Transcription by RNA polymerase III: insights into mechanism and regulation. Biochem Soc Trans. 2016;44(5):1367–1375. doi: 10.1042/BST20160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ducrot C, Lefebvre O, Landrieux E, Guirouilh-Barbat J, Sentenac A, Acker J. Reconstitution of the yeast RNA polymerase III transcription system with all recombinant factors. J Biol Chem. 2006;281(17):11685–11692. doi: 10.1074/jbc.M600101200. [DOI] [PubMed] [Google Scholar]
- 25.Male G, von Appen A, Glatt S, Taylor NM, Cristovao M, Groetsch H, Beck M, Muller CW. Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat Commun. 2015;6:7387. doi: 10.1038/ncomms8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310(1):1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 27.Kassavetis GA, Joazeiro CA, Pisano M, Geiduschek EP, Colbert T, Hahn S, Blanco JA. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor. TFIIIB Cell. 1992;71(6):1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 28.Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP. S. Cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 29.Bridier-Nahmias A, Tchalikian-Cosson A, Baller JA, Menouni R, Fayol H, Flores A, Saib A, Werner M, Voytas DF, Lesage P. Retrotransposons. An RNA polymerase III subunit determines sites of retrotransposon integration. Science. 2015;348(6234):585–588. doi: 10.1126/science.1259114. [DOI] [PubMed] [Google Scholar]
- 30.Cheung S, Ma L, Chan PH, Hu HL, Mayor T, Chen HT, Measday V. Ty1 integrase interacts with RNA polymerase III-specific subcomplexes to promote insertion of Ty1 elements upstream of polymerase (pol) III-transcribed genes. J Biol Chem. 2016;291(12):6396–6411. doi: 10.1074/jbc.M115.686840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 32.Gai X, Voytas DF. A single amino acid change in the yeast retrotransposon Ty5 abolishes targeting to silent chromatin. Mol Cell. 1998;1(7):1051–1055. doi: 10.1016/s1097-2765(00)80105-7. [DOI] [PubMed] [Google Scholar]
- 33.Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, Voytas DF. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol. 2001;21(19):6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shpakovski GV, Shematorova EK. Rpc19 and Rpc40, two alpha-like subunits shared by nuclear RNA polymerases I and III, are interchangeable between the fission and budding yeasts. Curr Genet. 1999;36(4):208–214. doi: 10.1007/s002940050492. [DOI] [PubMed] [Google Scholar]
- 35.Zou S, Ke N, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10(5):634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 36.Zou S, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 influences the organization of chromosome ends. Nucleic Acids Res. 1996;24(23):4825–4831. doi: 10.1093/nar/24.23.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou S, Voytas DF. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc Natl Acad Sci U S A. 1997;94(14):7412–7416. doi: 10.1073/pnas.94.14.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballandras-Colas A, Brown M, Cook NJ, Dewdney TG, Demeler B, Cherepanov P, Lyumkis D, Engelman AN. Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature. 2016;530(7590):358–361. doi: 10.1038/nature16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballandras-Colas A, Maskell DP, Serrao E, Locke J, Swuec P, Jonsson SR, Kotecha A, Cook NJ, Pye VE, Taylor IA, et al. A supramolecular assembly mediates lentiviral DNA integration. Science. 2017;355(6320):93–95. doi: 10.1126/science.aah7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464(7286):232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468(7321):326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passos DO, Li M, Yang R, Rebensburg SV, Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R, Lyumkis D. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science. 2017;355(6320):89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Z, Shi K, Banerjee S, Pandey KK, Bera S, Grandgenett DP, Aihara H. Crystal structure of the Rous sarcoma virus intasome. Nature. 2016;530(7590):362–366. doi: 10.1038/nature16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vorlander MK, Khatter H, Wetzel R, Hagen WJH, Muller CW. Molecular mechanism of promoter opening by RNA polymerase III. Nature. 2018;553(7688):295–300. doi: 10.1038/nature25440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486(7404):496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar Y, Bhargava P. A unique nucleosome arrangement, maintained actively by chromatin remodelers facilitates transcription of yeast tRNA genes. BMC Genomics. 2013;14:402. doi: 10.1186/1471-2164-14-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18(7):1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bridier-Nahmias A, Lesage P. Two large-scale analyses of Ty1 LTR-retrotransposon de novo insertion events indicate that Ty1 targets nucleosomal DNA near the H2A/H2B interface. Mob DNA. 2012;3(1):22. doi: 10.1186/1759-8753-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523(7560):366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15(5):619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petty E, Pillus L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 2013;29(11):621–629. doi: 10.1016/j.tig.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pollard KJ, Peterson CL. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17(11):6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winston F, Durbin KJ, Fink GR. The SPT3 gene is required for normal transcription of ty elements in S. Cerevisiae. Cell. 1984;39(3 Pt 2):675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 54.Bachman N, Gelbart ME, Tsukiyama T, Boeke JD. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. Cerevisiae tDNAs. Genes Dev. 2005;19(8):955–964. doi: 10.1101/gad.1299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19(8):942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22(16):2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22(16):2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5(2):243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 59.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91(1):35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 60.Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430(6999):573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400(6739):37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 62.Ho KL, Ma L, Cheung S, Manhas S, Fang N, Wang K, Young B, Loewen C, Mayor T, Measday V. A role for the budding yeast separase, Esp1, in Ty1 element retrotransposition. PLoS Genet. 2015;11(3):e1005109. doi: 10.1371/journal.pgen.1005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mou Z, Kenny AE, Curcio MJ. Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics. 2006;172(4):2157–2167. doi: 10.1534/genetics.105.054072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics. 2008;178(1):197–214. doi: 10.1534/genetics.107.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799(5–6):379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta. 2013;1829(1):116–126. doi: 10.1016/j.bbagrm.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Oss SB, Cucinotta CE, Arndt KM. Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem Sci. 2017;42(10):788–798. doi: 10.1016/j.tibs.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159(4):1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dahlin JL, Chen X, Walters MA, Zhang Z. Histone-modifying enzymes, histone modifications and histone chaperones in nucleosome assembly: lessons learned from Rtt109 histone acetyltransferases. Crit Rev Biochem Mol Biol. 2015;50(1):31–53. doi: 10.3109/10409238.2014.978975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang S, Liu Y, Xu C, Wang Y, Gong J, Shen Y, Wu Q, Boeke JD, Dai J. Dissecting nucleosome function with a comprehensive histone H2A and H2B mutant library. G3 (Bethesda) 2017;7(12):3857–3866. doi: 10.1534/g3.117.300252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302(5649):1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465(7296):363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodley CD, Bertels F, Jones B, O'Sullivan JM. Global identification of yeast chromosome interactions using genome conformation capture. Fungal Genet Biol. 2009;46(11):879–886. doi: 10.1016/j.fgb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 74.Rodley CD, Pai DA, Mills TA, Engelke DR, O'Sullivan JM. tRNA gene identity affects nuclear positioning. PLoS One. 2011;6(12):e29267. doi: 10.1371/journal.pone.0029267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rutledge MT, Russo M, Belton JM, Dekker J, Broach JR. The yeast genome undergoes significant topological reorganization in quiescence. Nucleic Acids Res. 2015;43(17):8299–8313. doi: 10.1093/nar/gkv723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Albert B, Mathon J, Shukla A, Saad H, Normand C, Leger-Silvestre I, Villa D, Kamgoue A, Mozziconacci J, Wong H, et al. Systematic characterization of the conformation and dynamics of budding yeast chromosome XII. J Cell Biol. 2013;202(2):201–210. doi: 10.1083/jcb.201208186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belagal P, Normand C, Shukla A, Wang R, Leger-Silvestre I, Dez C, Bhargava P, Gadal O. Decoding the principles underlying the frequency of association with nucleoli for RNA polymerase III-transcribed genes in budding yeast. Mol Biol Cell. 2016;27(20):3164–3177. doi: 10.1091/mbc.E16-03-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen M, Gartenberg MR. Coordination of tRNA transcription with export at nuclear pore complexes in budding yeast. Genes Dev. 2014;28(9):959–970. doi: 10.1101/gad.236729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Snider CE, Stephens AD, Kirkland JG, Hamdani O, Kamakaka RT, Bloom K. Dyskerin, tRNA genes, and condensin tether pericentric chromatin to the spindle axis in mitosis. J Cell Biol. 2014;207(2):189–199. doi: 10.1083/jcb.201405028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manhas S, Ma L, Measday V. The yeast Ty1 retrotransposon requires components of the nuclear pore complex for transcription and genomic integration. Nucleic Acids Res. 2018;46(7):3552–78. doi: 10.1093/nar/gky109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marini B, Kertesz-Farkas A, Ali H, Lucic B, Lisek K, Manganaro L, Pongor S, Luzzati R, Recchia A, Mavilio F, et al. Nuclear architecture dictates HIV-1 integration site selection. Nature. 2015;521(7551):227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 82.Albanese A, Arosio D, Terreni M, Cereseto A. HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS One. 2008;3(6):e2413. doi: 10.1371/journal.pone.0002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lelek M, Casartelli N, Pellin D, Rizzi E, Souque P, Severgnini M, Di Serio C, Fricke T, Diaz-Griffero F, Zimmer C, et al. Chromatin organization at the nuclear pore favours HIV replication. Nat Commun. 2015;6:6483. doi: 10.1038/ncomms7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chalker DL, Sandmeyer SB. Transfer RNA genes are genomic targets for de novo transposition of the yeast retrotransposon Ty3. Genetics. 1990;126(4):837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chalker DL, Sandmeyer SB. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6(1):117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 86.Sandmeyer SB, Bilanchone VW, Clark DJ, Morcos P, Carle GF, Brodeur GM. Sigma elements are position-specific for many different yeast tRNA genes. Nucleic Acids Res. 1988;16(4):1499–1515. doi: 10.1093/nar/16.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kirchner J, Connolly CM, Sandmeyer SB. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267(5203):1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 88.Yieh L, Hatzis H, Kassavetis G, Sandmeyer SB. Mutational analysis of the transcription factor IIIB-DNA target of Ty3 retroelement integration. J Biol Chem. 2002;277(29):25920–25928. doi: 10.1074/jbc.M202729200. [DOI] [PubMed] [Google Scholar]
- 89.Yieh L, Kassavetis G, Geiduschek EP, Sandmeyer SB. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J Biol Chem. 2000;275(38):29800–29807. doi: 10.1074/jbc.M003149200. [DOI] [PubMed] [Google Scholar]
- 90.Kassavetis GA, Soragni E, Driscoll R, Geiduschek EP. Reconfiguring the connectivity of a multiprotein complex: fusions of yeast TATA-binding protein with Brf1, and the function of transcription factor IIIB. Proc Natl Acad Sci U S A. 2005;102(43):15406–15411. doi: 10.1073/pnas.0507653102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qi X, Sandmeyer S. In vitro targeting of strand transfer by the Ty3 retroelement integrase. J Biol Chem. 2012;287(22):18589–18595. doi: 10.1074/jbc.M111.326025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol. 2004;24(10):4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qi X, Daily K, Nguyen K, Wang H, Mayhew D, Rigor P, Forouzan S, Johnston M, Mitra RD, Baldi P, et al. Retrotransposon profiling of RNA polymerase III initiation sites. Genome Res. 2012;22(4):681–692. doi: 10.1101/gr.131219.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aye M, Dildine SL, Claypool JA, Jourdain S, Sandmeyer SB. A truncation mutant of the 95-kilodalton subunit of transcription factor IIIC reveals asymmetry in Ty3 integration. Mol Cell Biol. 2001;21(22):7839–7851. doi: 10.1128/MCB.21.22.7839-7851.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Connolly CM, Sandmeyer SB. RNA polymerase III interferes with Ty3 integration. FEBS Lett. 1997;405(3):305–311. doi: 10.1016/s0014-5793(97)00200-7. [DOI] [PubMed] [Google Scholar]
- 96.Guffanti E, Percudani R, Harismendy O, Soutourina J, Werner M, Iacovella MG, Negri R, Dieci G. Nucleosome depletion activates poised RNA polymerase III at unconventional transcription sites in Saccharomyces cerevisiae. J Biol Chem. 2006;281(39):29155–29164. doi: 10.1074/jbc.M600387200. [DOI] [PubMed] [Google Scholar]
- 97.Eibel H, Philippsen P. Preferential integration of yeast transposable element ty into a promoter region. Nature. 1984;307(5949):386–388. doi: 10.1038/307386a0. [DOI] [PubMed] [Google Scholar]
- 98.Natsoulis G, Thomas W, Roghmann MC, Winston F, Boeke JD. Ty1 transposition in Saccharomyces cerevisiae is nonrandom. Genetics. 1989;123(2):269–279. doi: 10.1093/genetics/123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rose M, Winston F. Identification of a ty insertion within the coding sequence of the S. Cerevisiae URA3 gene. Mol Gen Genet. 1984;193(3):557–560. doi: 10.1007/BF00382100. [DOI] [PubMed] [Google Scholar]
- 100.Simchen G, Winston F, Styles CA, Fink GR. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc Natl Acad Sci U S A. 1984;81(8):2431–2434. doi: 10.1073/pnas.81.8.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weinstock KG, Mastrangelo MF, Burkett TJ, Garfinkel DJ, Strathern JN. Multimeric arrays of the yeast retrotransposon ty. Mol Cell Biol. 1990;10(6):2882–2892. doi: 10.1128/mcb.10.6.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wilke CM, Heidler SH, Brown N, Liebman SW. Analysis of yeast retrotransposon ty insertions at the CAN1 locus. Genetics. 1989;123(4):655–665. doi: 10.1093/genetics/123.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Borneman AR, Pretorius IS. Genomic insights into the Saccharomyces sensu stricto complex. Genetics. 2015;199(2):281–291. doi: 10.1534/genetics.114.173633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Libkind D, Hittinger CT, Valerio E, Goncalves C, Dover J, Johnston M, Goncalves P, Sampaio JP. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci U S A. 2011;108(35):14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 (Bethesda) 2011;1(1):11–25. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edwards-Ingram LC, Gent ME, Hoyle DC, Hayes A, Stateva LI, Oliver SG. Comparative genomic hybridization provides new insights into the molecular taxonomy of the Saccharomyces sensu stricto complex. Genome Res. 2004;14(6):1043–1051. doi: 10.1101/gr.2114704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liti G, Peruffo A, James SA, Roberts IN, Louis EJ. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast. 2005;22(3):177–192. doi: 10.1002/yea.1200. [DOI] [PubMed] [Google Scholar]
- 109.Bon E, Neuveglise C, Casaregola S, Artiguenave F, Wincker P, Aigle M, Durrens P. Genomic exploration of the hemiascomycetous yeasts: 5. Saccharomyces bayanus var. uvarum. FEBS Lett. 2000;487(1):37–41. doi: 10.1016/s0014-5793(00)02276-6. [DOI] [PubMed] [Google Scholar]
- 110.Fingerman EG, Dombrowski PG, Francis CA, Sniegowski PD. Distribution and sequence analysis of a novel Ty3-like element in natural Saccharomyces paradoxus isolates. Yeast. 2003;20(9):761–770. doi: 10.1002/yea.1005. [DOI] [PubMed] [Google Scholar]
- 111.Carr M, Bensasson D, Bergman CM. Evolutionary genomics of transposable elements in Saccharomyces cerevisiae. PLoS One. 2012;7(11):e50978. doi: 10.1371/journal.pone.0050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bleykasten-Grosshans C, Neuveglise C. Transposable elements in yeasts. C R Biol. 2011;334(8–9):679–686. doi: 10.1016/j.crvi.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 113.Castanera R, Borgognone A, Pisabarro AG, Ramirez L. Biology, dynamics, and applications of transposable elements in basidiomycete fungi. Appl Microbiol Biotechnol. 2017;101(4):1337–1350. doi: 10.1007/s00253-017-8097-8. [DOI] [PubMed] [Google Scholar]
- 114.Donnart T, Piednoel M, Higuet D, Bonnivard E. Filamentous ascomycete genomes provide insights into Copia retrotransposon diversity in fungi. BMC Genomics. 2017;18(1):410. doi: 10.1186/s12864-017-3795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kovalchuk A, Senam S, Mauersberger S, Barth G. Tyl6, a novel Ty3/gypsy-like retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. Yeast. 2005;22(12):979–991. doi: 10.1002/yea.1287. [DOI] [PubMed] [Google Scholar]
- 116.Magnan C, Yu J, Chang I, Jahn E, Kanomata Y, Wu J, Zeller M, Oakes M, Baldi P, Sandmeyer S. Sequence assembly of Yarrowia lipolytica strain W29/CLIB89 shows transposable element diversity. PLoS One. 2016;11(9):e0162363. doi: 10.1371/journal.pone.0162363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schmid-Berger N, Schmid B, Barth G. Ylt1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. J Bacteriol. 1994;176(9):2477–2482. doi: 10.1128/jb.176.9.2477-2482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang L, Yan L, Jiang J, Wang Y, Jiang Y, Yan T, Cao Y. The structure and retrotransposition mechanism of LTR-retrotransposons in the asexual yeast Candida albicans. Virulence. 2014;5(6):655–664. doi: 10.4161/viru.32180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Perreau VM, Santos MA, Tuite MF. Beta, a novel repetitive DNA element associated with tRNA genes in the pathogenic yeast Candida albicans. Mol Microbiol. 1997;25(2):229–236. doi: 10.1046/j.1365-2958.1997.4521816.x. [DOI] [PubMed] [Google Scholar]
- 120.Sellam A, Hogues H, Askew C, Tebbji F, van Het Hoog M, Lavoie H, Kumamoto CA, Whiteway M, Nantel A. Experimental annotation of the human pathogen Candida albicans coding and noncoding transcribed regions using high-resolution tiling arrays. Genome Biol. 2010;11(7):R71. doi: 10.1186/gb-2010-11-7-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo Y, Singh PK, Levin HL. A long terminal repeat retrotransposon of Schizosaccharomyces japonicus integrates upstream of RNA pol III transcribed genes. Mob DNA. 2015;6:19. doi: 10.1186/s13100-015-0048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332(6032):930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Neuveglise C, Feldmann H, Bon E, Gaillardin C, Casaregola S. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res. 2002;12(6):930–943. doi: 10.1101/gr.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Spaller T, Kling E, Glockner G, Hillmann F, Winckler T. Convergent evolution of tRNA gene targeting preferences in compact genomes. Mob DNA. 2016;7(1):17. doi: 10.1186/s13100-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hofmann J, Schumann G, Borschet G, Gosseringer R, Bach M, Bertling WM, Marschalek R, Dingermann T. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5′ and 3′-flanking regions. J Mol Biol. 1991;222(3):537–552. doi: 10.1016/0022-2836(91)90495-r. [DOI] [PubMed] [Google Scholar]
- 126.Glockner G, Heidel AJ. Centromere sequence and dynamics in Dictyostelium discoideum. Nucleic Acids Res. 2009;37(6):1809–1816. doi: 10.1093/nar/gkp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18(3):359–369. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Winckler T, Dingermann T, Glockner G. Dictyostelium mobile elements: strategies to amplify in a compact genome. Cell Mol Life Sci. 2002;59(12):2097–2111. doi: 10.1007/s000180200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Winckler T, Schiefner J, Spaller T, Siol O. Dictyostelium transfer RNA gene-targeting retrotransposons: studying mobile element-host interactions in a compact genome. Mob Genet Elements. 2011;1(2):145–150. doi: 10.4161/mge.1.2.17369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Winckler T, Szafranski K, Glockner G. Transfer RNA gene-targeted integration: an adaptation of retrotransposable elements to survive in the compact Dictyostelium discoideum genome. Cytogenet Genome Res. 2005;110(1–4):288–298. doi: 10.1159/000084961. [DOI] [PubMed] [Google Scholar]
- 131.Malicki M, Iliopoulou M, Hammann C. Retrotransposon domestication and control in Dictyostelium discoideum. Front Microbiol. 2017;8:1869. doi: 10.3389/fmicb.2017.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chung T, Siol O, Dingermann T, Winckler T. Protein interactions involved in tRNA gene-specific integration of Dictyostelium discoideum non-long terminal repeat retrotransposon TRE5-a. Mol Cell Biol. 2007;27(24):8492–8501. doi: 10.1128/MCB.01173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Siol O, Boutliliss M, Chung T, Glockner G, Dingermann T, Winckler T. Role of RNA polymerase III transcription factors in the selection of integration sites by the dictyostelium non-long terminal repeat retrotransposon TRE5-a. Mol Cell Biol. 2006;26(22):8242–8251. doi: 10.1128/MCB.01348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Siol O, Spaller T, Schiefner J, Winckler T. Genetically tagged TRE5-a retrotransposons reveal high amplification rates and authentic target site preference in the Dictyostelium discoideum genome. Nucleic Acids Res. 2011;39(15):6608–6619. doi: 10.1093/nar/gkr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Spaller T, Groth M, Glockner G, Winckler T. TRE5-a retrotransposition profiling reveals putative RNA polymerase III transcription complex binding sites on the Dictyostelium extrachromosomal rDNA element. PLoS One. 2017;12(4):e0175729. doi: 10.1371/journal.pone.0175729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Szafranski K, Glockner G, Dingermann T, Dannat K, Noegel AA, Eichinger L, Rosenthal A, Winckler T. Non-LTR retrotransposons with unique integration preferences downstream of Dictyostelium discoideum tRNA genes. Mol Gen Genet. 1999;262(4–5):772–780. doi: 10.1007/s004380051140. [DOI] [PubMed] [Google Scholar]
- 137.Winckler T, Tschepke C, de Hostos EL, Jendretzke A, Dingermann T. Tdd-3, a tRNA gene-associated poly(a) retrotransposon from Dictyostelium discoideum. Mol Gen Genet. 1998;257(6):655–661. doi: 10.1007/s004380050694. [DOI] [PubMed] [Google Scholar]
- 138.Boeke JD, Devine SE. Yeast retrotransposons: finding a nice quiet neighborhood. Cell. 1998;93(7):1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 139.Bolton EC, Boeke JD. Transcriptional interactions between yeast tRNA genes, flanking genes and ty elements: a genomic point of view. Genome Res. 2003;13(2):254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kinsey PT, Sandmeyer SB. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991;19(6):1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schumann G, Zundorf I, Hofmann J, Marschalek R, Dingermann T. Internally located and oppositely oriented polymerase II promoters direct convergent transcription of a LINE-like retroelement, the Dictyostelium repetitive element, from Dictyostelium discoideum. Mol Cell Biol. 1994;14(5):3074–3084. doi: 10.1128/mcb.14.5.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Blanc VM, Adams J. Ty1 insertions in intergenic regions of the genome of Saccharomyces cerevisiae transcribed by RNA polymerase III have no detectable selective effect. FEMS Yeast Res. 2004;4(4–5):487–491. doi: 10.1016/S1567-1356(03)00199-5. [DOI] [PubMed] [Google Scholar]
- 143.Garfinkel DJ, Tucker JM, Saha A, Nishida Y, Pachulska-Wieczorek K, Blaszczyk L, Purzycka KJ. A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces. Curr Genet. 2016;62(2):321–329. doi: 10.1007/s00294-015-0550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pachulska-Wieczorek K, Blaszczyk L, Gumna J, Nishida Y, Saha A, Biesiada M, Garfinkel DJ, Purzycka KJ. Characterizing the functions of Ty1 gag and the gag-derived restriction factor p22/p18. Mob Genet Elements. 2016;6(2):e1154637. doi: 10.1080/2159256X.2016.1154637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Boeke JD, Eichinger DJ, Natsoulis G. Doubling Ty1 element copy number in Saccharomyces cerevisiae: host genome stability and phenotypic effects. Genetics. 1991;129(4):1043–1052. doi: 10.1093/genetics/129.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wilke CM, Adams J. Fitness effects of ty transposition in Saccharomyces cerevisiae. Genetics. 1992;131(1):31–42. doi: 10.1093/genetics/131.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Scheifele LZ, Cost GJ, Zupancic ML, Caputo EM, Boeke JD. Retrotransposon overdose and genome integrity. Proc Natl Acad Sci U S A. 2009;106(33):13927–13932. doi: 10.1073/pnas.0906552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Garfinkel DJ. Genome evolution mediated by ty elements in Saccharomyces. Cytogenet Genome Res. 2005;110(1–4):63–69. doi: 10.1159/000084939. [DOI] [PubMed] [Google Scholar]
- 149.McClanahan T, McEntee K. Specific transcripts are elevated in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1984;4(11):2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rolfe M, Spanos A, Banks G. Induction of yeast ty element transcription by ultraviolet-light. Nature. 1986;319(6051):339–340. [Google Scholar]
- 151.Sacerdot C, Mercier G, Todeschini AL, Dutreix M, Springer M, Lesage P. Impact of ionizing radiation on the life cycle of Saccharomyces cerevisiae Ty1 retrotransposon. Yeaste. 2005;22(6):441–455. doi: 10.1002/yea.1222. [DOI] [PubMed] [Google Scholar]
- 152.Curcio MJ, Kenny AE, Moore S, Garfinkel DJ, Weintraub M, Gamache ER, Scholes DT. S-phase checkpoint pathways stimulate the mobility of the retrovirus-like transposon Ty1. Mol Cell Biol. 2007;27(24):8874–8885. doi: 10.1128/MCB.01095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maxwell PH, Coombes C, Kenny AE, Lawler JF, Boeke JD, Curcio MJ. Ty1 mobilizes subtelomeric Y' elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol Cell Biol. 2004;24(22):9887–9898. doi: 10.1128/MCB.24.22.9887-9898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Scholes DT, Kenny AE, Gamache ER, Mou Z, Curcio MJ. Activation of a LTR-retrotransposon by telomere erosion. Proc Natl Acad Sci U S A. 2003;100(26):15736–15741. doi: 10.1073/pnas.2136609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Maxwell PH, Burhans WC, Curcio MJ. Retrotransposition is associated with genome instability during chronological aging. Proc Natl Acad Sci U S A. 2011;108(51):20376–20381. doi: 10.1073/pnas.1100271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99(25):16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Argueso JL, Westmoreland J, Mieczkowski PA, Gawel M, Petes TD, Resnick MA. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci U S A. 2008;105(33):11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Casper AM, Greenwell PW, Tang W, Petes TD. Chromosome aberrations resulting from double-strand DNA breaks at a naturally occurring yeast fragile site composed of inverted ty elements are independent of Mre11p and Sae2p. Genetics. 2009;183(2):423–439. doi: 10.1534/genetics.109.106385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cheng E, Vaisica JA, Ou J, Baryshnikova A, Lu Y, Roth FP, Brown GW. Genome rearrangements caused by depletion of essential DNA replication proteins in Saccharomyces cerevisiae. Genetics. 2012;192(1):147–160. doi: 10.1534/genetics.112.141051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120(5):587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 161.Song W, Dominska M, Greenwell PW, Petes TD. Genome-wide high-resolution mapping of chromosome fragile sites in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2014;111(21):E2210–E2218. doi: 10.1073/pnas.1406847111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hoang ML, Tan FJ, Lai DC, Celniker SE, Hoskins RA, Dunham MJ, Zheng Y, Koshland D. Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet. 2010;6(12):e1001228. doi: 10.1371/journal.pgen.1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chung DK, Chan JN, Strecker J, Zhang W, Ebrahimi-Ardebili S, Lu T, Abraham KJ, Durocher D, Mekhail K. Perinuclear tethers license telomeric DSBs for a broad kinesin- and NPC-dependent DNA repair process. Nat Commun. 2015;6:7742. doi: 10.1038/ncomms8742. [DOI] [PubMed] [Google Scholar]
- 164.Freudenreich CH, Su XA. Relocalization of DNA lesions to the nuclear pore complex. FEMS Yeast Res. 2016;16(8) [DOI] [PMC free article] [PubMed]
- 165.Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10(4):e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10(10):e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3(5):370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 168.Okada N, Ohshima K. A model for the mechanism of initial generation of short interspersed elements (SINEs) J Mol Evol. 1993;37(2):167–170. doi: 10.1007/BF02407352. [DOI] [PubMed] [Google Scholar]
- 169.Clare J, Farabaugh P. Nucleotide sequence of a yeast ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Farabaugh PJ, Zhao H, Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993;74(1):93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mellor J, Fulton SM, Dobson MJ, Wilson W, Kingsman SM, Kingsman AJ. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature. 1985;313(5999):243–246. doi: 10.1038/313243a0. [DOI] [PubMed] [Google Scholar]
- 172.Glockner G, Szafranski K, Winckler T, Dingermann T, Quail MA, Cox E, Eichinger L, Noegel AA, Rosenthal A. The complex repeats of Dictyostelium discoideum. Genome Res. 2001;11(4):585–594. doi: 10.1101/gr.162201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Marschalek R, Hofmann J, Schumann G, Gosseringer R, Dingermann T. Structure of DRE, a retrotransposable element which integrates with position specificity upstream of Dictyostelium discoideum tRNA genes. Mol Cell Biol. 1992;12(1):229–239. doi: 10.1128/mcb.12.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Beck P, Dingermann T, Winckler T. Transfer RNA gene-targeted retrotransposition of Dictyostelium TRE5-a into a chromosomal UMP synthase gene trap. J Mol Biol. 2002;318(2):273–285. doi: 10.1016/S0022-2836(02)00097-9. [DOI] [PubMed] [Google Scholar]
- 175.Marschalek R, Borschet G, Dingermann T. Genomic organization of the transposable element Tdd-3 from Dictyostelium discoideum. Nucleic Acids Res. 1990;18(19):5751–5757. doi: 10.1093/nar/18.19.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]


