Abstract
In Arabidopsis, phytochrome A (phyA) is the major photoreceptor both for high irradiance responses to far-red light and broad spectrum very low fluence responses, but little is known of its signaling pathway(s). rsf1 was isolated as a recessive mutant with reduced sensitivity to far-red inhibition of hypocotyl elongation. At the seedling stage rsf1 mutants are affected, to various degrees, in all described phyA-mediated responses. However, in adult rsf1 plants, the photoperiodic flowering response is normal. The rsf1 mutant has wild-type levels of phyA suggesting that RSF1 is required for phyA signaling rather than phyA stability or biosynthesis. RSF1 thus appears to be a major phyA signaling component in seedlings, but not in adult, Arabidopsis plants.
Light is arguably the most important abiotic factor influencing plant growth throughout their life cycle. To detect changes in their light environment plants have evolved several classes of photoreceptors (Kendrick and Kronenberg, 1994). Two families of blue- light photoreceptors, the cryptochromes and the phototropins, have been characterized molecularly (Briggs and Huala, 1999; Cashmore et al., 1999). At the other end of the visible spectrum the phytochromes sense both red (R) and far-red (FR) light (Quail et al., 1995; Whitelam and Devlin, 1997; Neff et al., 2000). In Arabidopsis the apoprotein components of phytochrome are encoded by a small gene family PHYA-PHYE (phytochrome A–E; Quail et al., 1995). Phytochrome apoproteins covalently bind to phytochromobilin, a linear tetrapyrrole chromophore (Lagarias and Rapoport, 1980).
Light responses are classified into very low fluence responses (VLFRs), low fluence responses, and high irradiance responses (HIRs; Kendrick and Kronenberg, 1994). Careful photobiological analysis of phyA and phyB single and double mutants has lead to the conclusion that these two photoreceptors respond to light by different modes of action (Reed et al., 1994; Quail et al., 1995; Shinomura et al., 1996, 1998, 2000; Yanovsky et al., 1997; Casal et al., 1998). PhyB is a major R/FR reversible low fluence response photoreceptor, whereas phyA plays a prominent role both for the broad spectrum VLFR and the FR-HIR. The mechanism of phyA action for those two types of light responses appears to be distinct (Shinomura et al., 1996, 2000; Yanovsky et al., 1997). In addition to numerous seedling phenotypes phyA adult plants are also defective in the perception of daylength extension (Johnson et al., 1994; Reed et al., 1994). Mutants deficient in phytochromobilin biosynthesis are affected in all known phytochrome responses suggesting that all phytochromes bind the same chromophore (Chory et al., 1989; Parks and Quail, 1991; Davis et al., 1999; Muramoto et al., 1999).
Molecular and genetic approaches suggest that signaling downstream of phyA and phyB splits into at least three branches (Deng and Quail, 1999; Neff et al., 2000). A phyB specific branch is altered in mutants such as red1, pef2, pef3, and poc1 (Ahmad and Cashmore, 1996; Wagner et al., 1997; Halliday et al., 1999). Mutants in the psi2 and pef1 genes define a branch implicated in both phyA and phyB signaling (Ahmad and Cashmore, 1996; Genoud et al., 1998). A phyA-specific signaling branch is defined by mutants such as fhy1, fhy3, fin2, spa1, far1, pat1, and eid1 (Whitelam et al., 1993; Hoecker et al., 1998; Soh et al., 1998; Hudson et al., 1999; Bolle et al., 2000; Buche et al., 2000). The existence of both overlapping and specific phyA or phyB signaling pathways has also been deduced from the analysis of phytochrome single and double mutants (Reed et al., 1994; Casal and Mazzella, 1998; Neff and Chory, 1998).
Molecular details of phytochrome signaling are starting to emerge (Neff et al., 2000). The subcellular localization of phytochromes is light regulated. They are cytoplasmic in the dark and appropriate light treatments trigger nuclear translocation of both phyA and phyB (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999). The half-life of phyA is up to 100-fold longer in the dark than in the light. This light lability is believed to result from light-dependent ubiquitination of phyA (Clough et al., 1999). In addition, phyA is a light-regulated Ser/Thr protein kinase (Yeh and Lagarias, 1998). PhyA autophosphorylates and transphosphorylates Cry1, Cry2, and PKS1 (Ahmad et al., 1998; Yeh and Lagarias, 1998; Fankhauser et al., 1999). PKS1, a cytoplasmic protein, is an inhibitor of phyB signaling (Fankhauser et al., 1999). Phytochrome-mediated phosphorylation of the blue-light photoreceptors Cry1 and Cry2 provides a potential molecular link for the co-action of blue- and R-light signaling (Mohr, 1986; Ahmad et al., 1998). In the nucleus phyB interacts with the bHLH transcription factor PIF3; this interaction may directly modulate the expression of light-regulated genes (Martinez-Garcia et al., 2000). It is interesting that two phyA signaling genes SPA1 and FAR1 are also localized in the nucleus (Hoecker et al., 1999; Hudson et al., 1999). However it is unlikely that phytochrome signaling occurs exclusively in the nucleus. The fact that NDPK2, a phytochrome interacting protein, and PAT1, a phyA signaling component, are found in the cytoplasm is in agreement with this view (Choi et al., 1999; Bolle et al., 2000).
To construct a coherent model for phytochrome signaling we need to identify as many components of this network as possible. To this end we have screened for mutants specifically affected in phyA signaling and identified a new mutant rsf1, with reduced sensitivity to FR light. The initial characterization of this mutant shows that not all phyA-mediated responses are affected in these plants. This confirms the view of a branched signaling pathway for phyA-mediated responses (Barnes et al., 1996; Soh et al., 1998).
RESULTS
To identify novel components of the phyA signaling pathway we screened the T-DNA collection described in Weigel et al. (2000) for mutants with a long hypocotyl under FR light. The rsf1 (reduced sensitivity to FR light) mutant was identified after analysis of 26,000 T2 seedlings corresponding to 2,600 independent T-DNA insertion lines. Crosses of rsf1 to Columbia (Col-7) determined that the mutant was recessive and was not caused by a T-DNA insertion since there was no cosegregation of the Basta (ammonium glufosinate) resistance and the rsf1 mutant phenotypes. To map the mutation we crossed the rsf1 mutant (in the Col-7 background) to Landsberg-0 carrying the er mutation. Mutants were scored in FR light in the F2 generation. Using PCR-based markers on DNA prepared from 65 plants we determined that the rsf1 locus maps within 1 centiMorgan of the PVV4 marker on the top of chromosome I (Konieczny and Ausubel, 1993). A number of other FR insensitive mutants have already been described, but none of them is located in the vicinity of PVV4 (Soh et al., 1998; Hudson et al., 1999). Since the map position of the fhy1 mutant is not available wecrossed the rsf1 mutant with fhy1 mutants. The F1 progeny of the cross had a wild-type phenotype in FR light, whereas there was segregation of mutant and wild-type seedlings in the F2 (data not shown). Thus RSF1 is a new locus important for de-etiolation in FR light.
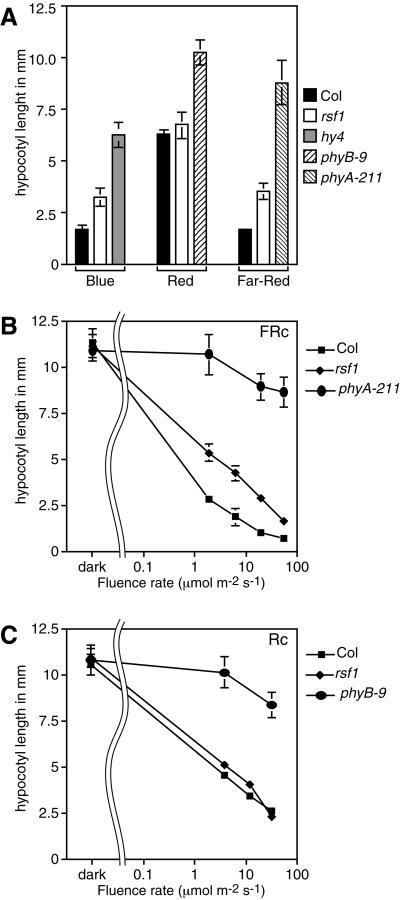
To determine the light specificity of the rsf1 phenotype we measured hypocotyl elongation in the dark and under non-saturating intensities of blue, R, and FR light (Fig. 1). Hypocotyl elongation was unaffected in the dark and rsf1 seedlings responded normally to R light. However, rsf1 hypocotyl elongation was less inhibited in both FR and blue light. Hypocotyl length in rsf1 seedlings was intermediate between the wild type and a photoreceptor null mutant under both light conditions (Fig. 1). PhyA, fhy1, and fhy3 seedlings also show hypocotyl elongation phenotypes in both FR and blue light (Whitelam et al., 1993). Fluence response curves in both R and FR light confirmed that rsf1 seedlings responded normally to R light and are less sensitive to all the tested fluences of FR light (Fig. 1, B and C). However, unlike phyA null mutants (phyA-211), they clearly responded to increasing fluences of FR light.
Figure 1.
rsf1 mutants are impaired in inhibition of hypocotyl elongation in FR and blue light. Data are means ± 2× se of at least 12 seedlings for each light treatment. All seedlings were grown at 22°C in continuous light. A, Col (black bar), rsf1 (white bar), and the appropriate photoreceptor mutants were grown for 6 d in 30 μmol m−2 s−1 blue (hy4, gray bar), 15 μmol m−2 s−1 R (phyB-9, hatched to the left bar), or 10 μmol m−2 s−1 FR (phyA-211, hatched to the right bar). B, Fluence rate response curve in continuous FR light of Col, rsf1, and phyA-211 seedlings. C, Fluence rate response curve in continuous R light of Col, rsf1, and phyB-9 seedlings.
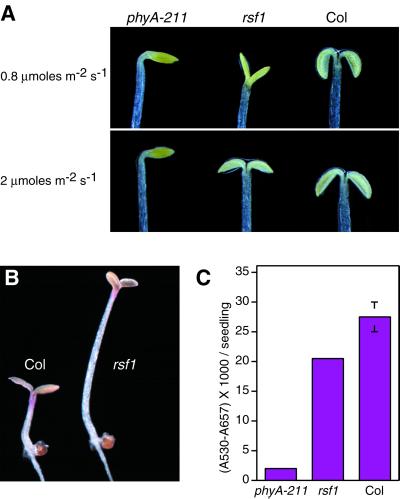
PhyA mutants are also largely impaired in cotyledon unfolding and expansion over a broad range of FR-light fluence rates (Yanovsky et al., 1997; Fig. 2A). In Rsf1 mutants a cotyledon opening phenotype was only apparent under FR fluence rates below 1 μmol m−2 s−1 (Fig. 2A). To test other phyA-dependent FR responses we looked at anthocyanin accumulation in constant FR light on Suc-containing plates (Neff and Chory, 1998). As expected, phyA mutants accumulated minute amounts of anthocyanin; rsf1 mutants accumulated slightly, but significantly lower, levels than the wild type (Fig. 2, B and C). These results show that at the seedling stage rsf1 mutants are deficient in all the FR responses tested, but to a much lower extent than in phyA mutants.
Figure 2.
Seedling phenotypes of rsf1 mutants. A, Wild-type, rsf1, and phyA-211 seedlings were grown for 4 d in continuous FR light 0.8 or 2 μmol m−2 s−1, representative seedlings were photographed. B, Picture of a representative wild-type and rsf1 seedlings grown for 4 d in continuous FR light on Suc-containing plates. Note that the mutant still accumulates anthocyanin. C, Quantification of anthocyanin accumulation in phyA-211, rsf1, and wild-type seedlings. Seedlings were grown on Suc-containing plates under 8 μmol m−2 s−1 FR light for 4 d. The experiment was done in triplicate with 10 seedlings for each measurement (mean ± 2× se).
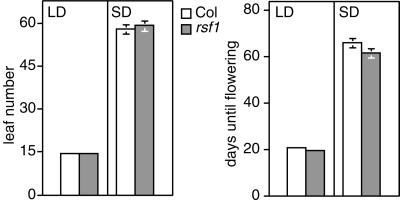
To test if the rsf1 mutation also affects phyA responses later in development we determined flowering time in these plants. PhyA is required to sense daylength extension, which is apparent in long days (LD) where they flower later than the wild type. In contrast, in short days (SD), flowering time is unaffected (Johnson et al., 1994; Reed et al., 1994; Neff and Chory, 1998; Soh et al., 1998). rsf1 plants were indistinguishable from wild type both in LD and SD using two criteria to measure flowering time (days until flowering and leaf number at flowering; Fig. 3). This observation suggests that RSF1 is not required for all phyA responses.
Figure 3.
rsf1 plants have no flowering time phenotype in LD or SD. LD are 16 of h light, 8 h of night; SD are 9 h of light, 15 h of night. Values are the means ± 2× se, with at least 18 plants for each photoperiod condition.
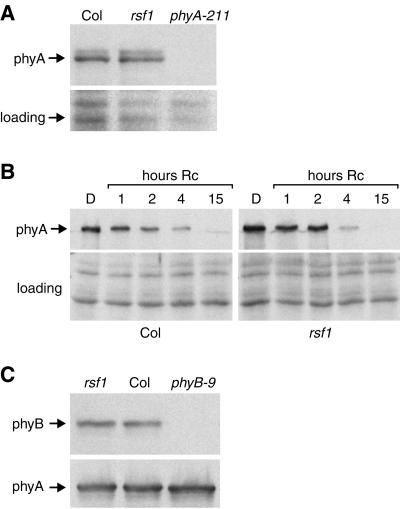
A defect in phyA-mediated responses could arise from altered levels of the active photoreceptor or from a defect in signaling downstream of phyA. Western blotting of total proteins from wild-type and rsf1 seedlings was used to assess the level of phyA in different growth conditions. rsf1 seedlings had wild-type levels of phyA under FR light, whereas rsf1 mutants had the most obvious phenotype (Fig. 4A). Dark-grown rsf1 seedlings exposed to R light for increasing amounts of time had the same phyA degradation kinetics as wild-type seedlings (Fig. 4B). This indicates that the light-dependent stability of phyA is not affected in the rsf1 mutant. Because phyB plays a minor role in FR-light sensing (Neff and Chory, 1998), we also tested the levels of phyB in rsf1 mutants. Western-blot analysis showed that rsf1 seedlings had normal levels of phyB (Fig. 4C). The fact that rsf1 mutants have phenotypes in FR light, but not R light, makes it very unlikely that this mutant is defective in phytochrome chromophore biosynthesis. We therefore conclude that this gene is required for phyA-mediated signaling.
Figure 4.
rsf1 mutants have wild-type levels of phyA and phyB. Proteins were extracted from 6-d-old seedlings, separated on 8% (w/v) SDS-PAGE gels, and blotted onto nitrocellulose. The membrane was probed with mAA1-3 or mBA2, monoclonal antibodies directed against phyA and phyB, respectively (Shinomura et al., 1996). The amido black-stained membrane is shown as a loading control. A, Seedlings were grown for 6 d in continuous 8 μmol m−2 s−1 FR light. B, Seedlings were grown for 6 d in the dark, or for the same total duration of which the last 1, 2, 4, or 15 h was in constant 100 μmol m−2 s−1 R light. C, Seedlings were grown for 6 d in the dark.
DISCUSSION
We have identified a novel locus implicated in a subset of phyA-mediated responses. The most obvious phenotype is the reduced inhibition of hypocotyl elongation in FR light. However, rsf1 seedlings are also affected in de-etiolation in blue light. This observation is not surprising since phyA, fhy1, and fhy3 also have long hypocotyls in both blue and FR light (Whitelam et al., 1993). Thus the blue-light phenotype could be a consequence of reduced phyA signaling, although it is possible that RSF1 is also involved in cryptochrome-mediated blue-light signaling. The analysis of double mutants will allow us to address this point genetically.
rsf1 is most likely involved in phyA signaling rather than the regulation of phyA accumulation. A functional phytochrome photoreceptor comprises the apoprotein and phytochromobilin, a linear tetrapyrrole chromophore. Two results make it unlikely that RSF1 is involved in apoprotein or chromophore biosynthesis. First, the rsf1 mutant has a phenotype in FR light, but not R light. A defect in chromophore biosynthesis would also affect phyB signaling, which would be visible in R-light-grown seedlings and later in development (Chory et al., 1989; Parks and Quail, 1991). The fact that rsf1 mutants have no seedling phenotype in R light and look wild type as adult plants is inconsistent with this idea (Figs. 1 and 3). Second, the rsf1 mutants have wild-type levels of both phyA and phyB (Fig. 4). In contrast, in light-grown chromophore-deficient mutants, the level of phyA is higher than in the wild type, presumably because in the light apoA retains a “Pr-like” conformation, which is more stable than PfrA (Parks and Quail, 1991). Figure 4B shows that this is not the case in rsf1 mutants, since light induced phyA degradation is unaffected in rsf1 seedlings. It is notable that in our hands phyA degradation kinetics in response to R-light irradiation were somewhat slower than those reported by others. This might be due to ecotype differences (Hoecker et al., 1998; Hennig et al., 1999; Hudson et al., 1999).
The observation that rsf1 affects only a subset of phyA-mediated responses (e.g. those at the seedling stage versus flowering time) reveals branching in the signaling pathway downstream of phyA. This possibility has been reported previously (Johnson et al., 1994; Barnes et al., 1996; Soh et al., 1998; Yanovsky et al., 2000). In this respect, rsf1 mutants are similar to fin2 mutants, for example (Soh et al., 1998). In both cases hypocotyl elongation and anthocyanin accumulation are defective, but flowering time is similar to wild type. However, the lack of a flowering time phenotype in rsf1 mutants should be interpreted with caution. In our LD growth conditions phyA mutants only make 20% more leaves than the wild type (Neff and Chory, 1998). Since at the seedling stage the rsf1 mutants have weaker phenotypes than phyA mutants, a flowering phenotype might be difficult to see in our experimental setup. Extending daylength with a period of low-fluence-rate incandescent light could address this issue more rigorously (Johnson et al., 1994). However, it is important to note that the moderate FR-light phenotypes observed in the rsf1 mutant are not the result of a missense allele. We identified a 13-bp deletion in the mutant rsf1 gene, which should result in a truncated protein lacking 60% of the open reading frame (M. Mindrinos, J. Spiegelman, J. Lutes, C. Fankhauser, J. Chory, and P. Oefner, unpublished data).
Far1 null alleles have very obvious defects in both anthocyanin accumulation and hypocotyl elongation, but their flowering time phenotype has not been reported. It is interesting that those mutants are still responsive to FR light. This might be the manifestation of gene redundancy, as FAR1 belongs to a small gene family (Hudson et al., 1999). Probably the best-studied case is fhy3-1, where an elegant set of photobiological experiments have demonstrated that this mutant is affected in FR-HIR, but not phyA-mediated VLFR (Yanovsky et al., 2000). Future characterization of the rsf1 mutant will determine if this locus is also required for phyA-mediated VLFR. A more systematic analysis of available phyA signaling mutants should allow us to determine if the two forms of phyA photoperception require a different set of signaling intermediates. The currently available mutants supports this view (Yanovsky et al., 1997, 2000); however it is possible that some signaling components are required for both.
MATERIAL AND METHODS
Plant Material
The progeny of 2,600 independent Arabidopsis ecotype Col-7 transformants generated by Weigel et al. (2000) were screened in FR light as described (Nagatani et al., 1993). We screened about 10 T2 seeds per original transformant. Seeds were surface sterilized and, except for the anthocyanin measurement experiment, plated on Petri dishes on one-half Murashige and Skoog medium and 0.7% (w/v) phytagar. Plates were stored in the dark at 4°C for 3 d. Germination was induced by a white-light treatment; the seedlings were then grown in the appropriate light conditions for 4 to 6 d depending on the experiment.
Light Sources and Flowering Time Determination
All experiments except the original FR-light screen (see above) were performed in a E-30LED (Percival, Boone, IA) using either the blue (λmax 469 nm), R (λmax 667 nm), or the FR (λmax 739 nm) diodes, at 22°C in continuous light. Light intensities were determined with a spectroradiometer (LI-1800, LiCor, Lincoln, NE) or with an photometer (IL1400A, International Light, Newburyport, MA) equipped with an SEL033 probe with appropriate light filters.
Flowering time was determined as described (Blázquez and Weigel, 1999). In brief, seeds were stratified for 3 d at 4°C and plants were grown at 23°C in LD (16 h of light, 8 h of darkness) or SD (9 h of light, 15 h of darkness) under a mixture 3:1 cool-white:Gro-Lux fluorescent lights.
Hypocotyl Length and Anthocyanin Accumulation
Measurements of hypocotyl length and anthocyanin accumulation were performed as described (Neff and Chory, 1998). For anthocyanin accumulation the seedlings were grown on one-half Murashige and Skoog medium, 1.5% (w/v) Suc, and 0.7% (w/v) phytagar in 8 μmol m−2 s −1 FR light for 4 d.
Western Blotting
About 30 6-d-old seedlings were grown under appropriate light conditions. Proteins were extracted as described in (Haldrup et al., 1999), separated on 8% (w/v) SDS-PAGE gels, and western blotted. The blots were probed with mBA2 or mAA1-3 antibodies as described (Shinomura et al., 1996).
ACKNOWLEDGMENTS
We are grateful to Jason Lutes and Consuelo Salomon for technical support, to Nicolas Roggli for artwork, to Detlef Weigel (The Salk Institute) for providing the T-DNA lines, to Akira Nagatani (University of Kyoto) for the monoclonal antibodies mBA2 and mAA1-3, and to Nicholas Harberd (John Innes Centre, Norwich, UK) for the fhy1 and fhy3 seeds. We thank Michael Neff (Washington University, St. Louis) and Miguel Blázquez (University of Valencia) for critically reading the manuscript.
Footnotes
This work was supported by grants from the Swiss National Science Foundation (no. 63–58–151.99 to C.F.) and the U.S. National Institutes of Health (no. 2RO1 GM52413 to J.C.). C.F. was a postdoctoral fellow of the Swiss National Science Foundation, and J.C. is an Investigator of the Howard Hughes Medical Institute.
LITERATURE CITED
- Ahmad M, Cashmore AR. The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J. 1996;10:1103–1110. doi: 10.1046/j.1365-313x.1996.10061103.x. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The Cry1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- Barnes SA, Quaggio RB, Whitelam GC, Chua N-H. fhy1 defines a branch point in phytochrome A signal transduction pathways for gene expression. Plant J. 1996;10:1155–1161. doi: 10.1046/j.1365-313x.1996.10061155.x. [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Weigel D. Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol. 1999;120:1025–1032. doi: 10.1104/pp.120.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua N-H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Huala E. Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- Buche C, Poppe C, Schäfer E, Kretsch T. eid1: a new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Cerdan PD, Staneloni RJ, Cattaneo L. Different phototransduction kinetics of phytochrome A and phytochrome B in Arabidopsis thaliana. Plant Physiol. 1998;116:1533–1538. doi: 10.1104/pp.116.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Mazzella MA. Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiol. 1998;118:19–25. doi: 10.1104/pp.118.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. Phytochrome signalling is mediated through nucleotide diphosphate kinase 2. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt LH, Ausubel F. Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell. 1989;1:867–880. doi: 10.1105/tpc.1.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough RC, Jordan-Beebe ET, Lohman KN, Marita JM, Walker JM, Gatz C, Vierstra RD. Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 1999;17:155–167. doi: 10.1046/j.1365-313x.1999.00360.x. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA. 1999;96:6541–6546. doi: 10.1073/pnas.96.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Quail PH. Signalling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Genoud T, Millar AJ, Nishizawa N, Kay SA, Schäfer E, Nagatani A, Chua NH. An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldrup A, Naver H, Scheller HV. The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. Plant J. 1999;17:689–698. doi: 10.1046/j.1365-313x.1999.00419.x. [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Hudson M, Ni M, Qin M, Quail PH. poc1: an Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc Natl Acad Sci USA. 1999;96:5832–5837. doi: 10.1073/pnas.96.10.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Buche C, Eichenberg K, Schäfer E. Dynamic properties of endogenous phytochrome A in Arabidopsis seedlings. Plant Physiol. 1999;121:571–577. doi: 10.1104/pp.121.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC. Photoresponses of light-grown phyA mutants of Arabidopsis: phytochrome A is required for the perception of daylength extensions. Plant Physiol. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. Photomorphogenesis in Plants. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schäfer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Lagarias JC, Rapoport H. Chromopeptides from phytochrome: the structure and linkage of the Pr form of the phytochrome chromophore. J Am Chem Soc. 1980;102:4821–4828. [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Mohr H. Coaction between pigment systems. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in Plants. The Netherlands: Martinus Nijhoff; 1986. pp. 547–564. [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A, Reed RW, Chory J. Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Chory J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998;118:27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: an indicator of time and place. Genes Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Hanzawa H, Schäfer E, Furuya M. Mode of phytochrome B action in the photoregulation of seed germination in Arabidopsis thaliana. Plant J. 1998;13:583–590. doi: 10.1046/j.1365-313x.1998.00049.x. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Uchida K, Furuya M. Elementary processes of photoperception by phytochrome A for high irradiance response of hypocotyl elongation in Arabidopsis thaliana. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J. 1998;16:411–419. doi: 10.1046/j.1365-313x.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Wagner D, Hoecker U, Quail PH. RED1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, Nguyen JT, Sato S, Wang ZY, Xia Y, Dixon RA, Harrison MJ, Lamb CJ, Yanofsky MF, Chory J. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1014. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF. Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 1997;20:752–758. [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Casal JJ, Luppi JP. The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J. 1997;12:659–667. doi: 10.1046/j.1365-313x.1997.00659.x. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Whitelam GC, Casal JJ. fhy3-1 retains inductive responses of phytochrome A. Plant Physiol. 2000;123:235–242. doi: 10.1104/pp.123.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]