Abstract
Innate immune receptors have a key role in the sensing of malaria and initiating immune responses. As a consequence of infection, systemic inflammation emerges and is directly related to signs and symptoms during acute disease. We have previously reported that Plasmodial DNA is the primary driver of systemic inflammation in malaria, both within the phagolysosome and in the cytosol of effector cells. Here we demonstrate that Plasmodium falciparum (Pf) genomic DNA (gDNA) delivered to the cytosol of human monocytes binds and activates the cyclic GMP-AMP synthase (cGAS). Activated cGAS synthesizes 2′3′-cGAMP, which we subsequently can detect using liquid chromatography-tandem mass spectrometry. 2′3′-cGAMP acts as a second messenger for STING activation and triggers STING/TBK1/IRF3 activation, resulting in type I interferon (IFN) production in human cells. This induction of type I IFN was independent of IFI16. Access of DNA to the cytosolic compartment is mediated by hemozoin (Hz), as incubation of purified malaria pigment with DNase abrogated IFNβ induction. Collectively, these observations implicate cGAS as an important cytosolic sensor of Pf gDNA and reveal the role of the cGAS/STING pathway in the induction of type I IFN in response to malaria parasites.
Introduction
Malaria remains a major cause of morbidity and mortality worldwide. The World Health Organization has estimated that there were ~212 million cases of malaria globally in 2016 and about 429,000 deaths, primarily (~70%) occurring in children under age 5 (1). Despite many gains against the disease, the problems associated with malaria eradication remain significant. These include the increasing resistance of insect vectors to insecticides and the emerging resistance of Plasmodium to the most efficacious antimalarial drugs (2). Current evidence indicates that drug resistance to artemisinin derivatives, the last generation treatment for asexual blood-stage infection, has developed in Southeast Asia and Africa (3–5). Despite these setbacks, efforts continue with the objective of achieving the global elimination of malaria.
Our understanding of the pathogenesis of malaria is still limited (6). Therefore, a top priority in basic research is to dissect the mechanisms involved in malaria disease development and provide new approaches for therapeutic and prophylactic interventions. An important component of the pathogenesis of malaria is the host innate immune response to the parasite. The activation of innate immune cells and the associated systemic inflammation leads to the initial signs and symptoms of disease and can influence the development of severe disease (7). Inflammatory mediators during malaria infection are produced as a result of direct recognition of Plasmodial PAMPs (pathogen-associated molecular patterns) by innate immune receptors, including Toll-Like Receptors (TLRs) (7), Nod-Like Receptors (NLRs) and nucleic acid sensors (8). Concomitant with TLR activation, expression of sensor proteins including NLRs is augmented and NLR-inflammasomes are assembled. Pro-inflammatory cytokines and mediators like TNFα, IL-12, Caspase-1 and IL-1β are then released (9). Elevated expression of interferon-stimulated genes (ISGs) in innate immune cells is also characteristic during Plasmodium infection (8, 10).
The recognition of microbial DNA by the immune system provides a general mechanism for the detection of pathogens (11, 12). Delivery of foreign or self DNA into the cytoplasm (which is largely free of self DNA) through microbial infection activates innate cytosolic nucleic acid sensors (13). Plasmodial DNA represents a major trigger of innate immunity during infection (7, 8, 14). The Plasmodial genome contains highly stimulatory CpG motifs, which are thought to activate TLR9 when carried into the phagolysosomal compartment by the malaria pigment Hz (15–17). However, CpG-rich motifs are relatively rare in P. falciparum (Pf). In contrast, AT-rich DNA motifs are present in abundance and induce type I IFN via a pathway that is independent of TLRs, DNA-dependent activator of IFN-regulatory factors (DAI), RNA polymerase III and interferon gamma-inducible protein 16 (IFI16/p204) but dependent on Stimulator of Interferon Genes (STING) (18, 19), Tank-binding kinase 1 (TBK1) (20, 21) and the Interferon Regulatory Factor 3 and 7 (IRF3 and IRF7) (8, 22).
The enzyme cyclic GMP-AMP synthase (cGAS) has been identified as a cytosolic DNA sensor whose activation results in the subsequent activation of STING (23, 24). Specifically, in the presence of cytosolic double-stranded DNA, cGAS catalyzes the synthesis of cyclic GMP-AMP (2′3′-cGAMP) from ATP and GTP. 2′3′-cGAMP then functions as a second messenger that binds to and activates STING. Activated STING leads to IRF3 phosphorylation, type I IFN production and expression of ISGs (12, 25). Recently, the rodent pathogen Plasmodium yoelii (P. yoelii) was reported to active the negative regulator SOCS1 in a cGAS dependent manner; under these conditions, TLR7 was found to drive IFNα/β production (26). Here, we report that cGAS has an important role as a sensor of Pf gDNA. Our data suggest that 2′3′-cGAMP acts as a second messenger after the sensing of Pf gDNA and other malaria PAMPs to induce IFN via the cGAS/STING pathway. We suggest that cGAS detection of cytosolic malaria DNA is an important molecular feature of malaria pathogenesis.
Materials and methods
Ethics statement
The protocol and consent forms for experiments with human samples were approved by the Institutional Research Boards from the University of Massachusetts Medical School (IRB H-14839) and CIDEIM (CIEIH-1249). All experiments involving animals were performed in accordance with guidelines set forth by the American Association for Laboratory Animal Science and were approved by the Institutional Animal Care and Use Committee (A-1332) at the University of Massachusetts Medical School.
Subjects
Participants were healthy males and females between 18 and 60 years of age with no prior exposure to malaria or residence in malaria-endemic regions. Individuals with any comorbidity at the moment of enrollment, recent or concurrent treatment with anti-inflammatory or immunosuppressive drugs or pregnancy were excluded. Sixty to 100 ml of total blood were collected from healthy donors by phlebotomy.
Culture of parasites and natural Hz preparation
Pf parasites (3D7 strain) were cultured as described previously (14). Briefly, plate cultures were prepared with human erythrocytes at a 5% hematocrit and about 1% parasitemia in Malaria Culture Medium (27). Plates were put in a candle jar to produce low oxygen and placed at 37°C. Pf culture stage and parasitemia was assessed daily by Giemsa staining. Where indicated, infected red blood cells (iRBCs) were purified from Pf cultures at ~8% parasitemia and trophozoite or schizont stages were recovered as described (14, 28). The iRBCs suspension was loaded onto LD columns (Miltenyi Biotec), placed in a magnetic cell separator and eluted with endotoxin-free Dulbecco’s PBS. Natural Hz was extracted from the parasite cultures as has been described (16). Briefly, supernatants of Pf cultures were pelleted and loaded onto LD columns as described above. Hz was eluted with Dulbecco’s PBS, quantified and frozen at −20°C.
Isolation of Pf genomic DNA
Pf culture (~30% parasitemia) at the trophozoite stage was harvested and parasites from iRBCs were released by treatment with 0.2% saponin. The released parasites were pelleted at 3000 × g for 20 min, washed with ice cold Dulbecco’s PBS, suspended in Dulbecco’s PBS supplemented with proteinase K (25 μg/ml) and incubated at 56 °C for 10 min. Then, the resultant Pf gDNA containing solution was extracted with phenol/chloroform/isoamyl alcohol, and centrifuged at 10.000 × g for 10 min. The parasite DNA was precipitated overnight with NaOAc and absolute ethanol at −80°C, then washed with 70% ethanol and suspended in nuclease and endotoxin free water. The DNA concentration was estimated by measuring absorption at 260 nm, and Pf gDNA stored at −20°C. The purity of Pf gDNA was confirmed by PCR using primers for Plasmodium 18S RNA as described previously (29) and human TLR7 genes.
Cell culture and stimulation
PBMCs from healthy donors were obtained as described previously using Ficoll gradient separation (10). Human primary CD14+ monocytes were purified from PBMCs using the Pan Monocyte isolation kit (Milteny Biotec) and MACS according to the manufacturer’s instructions. Monocyte derived macrophages (MDMs) were obtained by adherence and differentiation of CD14+ monocytes in RPMI 1640-10% FBS medium for 7 days. Human pro-monocyte THP-1 cells (wild-type-WT, cGAS+/+, cGAS−/−, IFI16+/+ and IFI16−/−) cells were generated as previously described (30). WT and knockout (KO) THP-1 cells were grown in RPMI 1640/glutamine supplemented with 10% FBS. Primary Bone marrow derived macrophages (BMDMs) from WT, STING−/− and cGAS−/− C57BL/6 mice were generated as described previously (31) and cultured in Dulbecco’s modified Eagle’s medium (GIBCO-BRL) supplemented with 4 mM glutamine and 10% FBS. For stimulations, poly(deoxyadenylic-deoxythymidylic) [p(dA:dT)], immuno-stimulatory DNA (ISD), Pf gDNA, natural Hz, and AT-rich oligodeoxynucleotides (ODNs) were transfected at the indicated concentrations using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After transfection, cell death was monitored by Trypan blue staining (Corning). Sendai virus (SeV, Cantrell strain, 20 U/ml) was used as a control where indicated. DNase digestion of Pf gDNA and Hz was performed using DNase I (Qiagen) according to the recommended protocol. Primary cells and cell lines were stimulated as stated and collected after the described time points for RNA extraction or cell lysate preparation.
Quantitative real-time PCR and ELISA
Total RNA was extracted using Trizol (Invitrogen) or the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Five hundred nanograms of total RNA were used for cDNA synthesis using the iScript Select cDNA synthesis kit (BioRad). Quantitative PCR was performed using iQ SYBR Green Supermix (BioRad). Levels of human IFNβ mRNA were normalized relative to levels of β-actin mRNA and expressed as a fold induction compared with unstimulated controls. ELISA for mouse IFNβ protein was performed as described in detail previously (32).
Detection of IRF3 phosphorylation
Cell lysates were prepared in RIPA lysis buffer as described previously (33), subjected to SDS-PAGE and visualized by Western Blotting using antibodies against phospho-IRF3 (S386) (Abcam), total IRF3 (D614C) (Cell Signaling Technology), and monoclonal anti-β-actin (Sigma). To accurate determine levels of IRF3 phosphorylation, the intensity signal of the bands was measured by densitometry using the Image Studio Lite software.
Preparation of cytosolic extracts for analysis of endogenous cGAMP
THP-1 cells were transfected with Pf gDNA and cytosolic extracts from ~1.8 × 107 cells were prepared by hypotonic lysis. Briefly, cells were incubated in hypotonic buffer (10mM Tris-HCl (pH 7.4), 10 mM KCl, 1.5 mM MgCl2) and for 30 minutes and then dounce-homogenized × 100 strokes. Cells lysates were heated at 95°C for 5 min and then centrifuged at 17,000 × g for 10 min to remove denatured proteins. The heat resistant supernatant was recovered and stored at −80°C until tested for 2′3′-cGAMP.
Quantification of 2′3′-cGAMP by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
Quantification of 2′3′-cGAMP was performed by LC-MS/MS as described previously (30). Extraction of cGAMP from cell lysates was performed with acetonitrile/methanol/water (2/2/1, vol/vol/vol) buffer. 3′3′ cGAMP (500 pg, BioLog) was added to each sample as an internal standard. Extracts of untreated and Pf gDNA stimulated THP-1 cells were spiked with 3′3′-cGAMP and the levels of 2′3′-cGAMP (endogenous) and 3′3′-cGAMP internal standard were measured in parallel. As an additional control, extracts of untreated (medium control) THP-1 cells with an added spike of synthetic 2′3′-cGAMP (BioLog) were also prepared and used as positive controls.
Statistical analysis
Differences in groups were analyzed with Student’s t-test or the Wilcoxon rank test. Analyzes were performed with GraphPad Prism 7 software (GraphPad Inc., San Diego, CA). P values of <0.05 were considered statistically significant. Otherwise, data were analyzed using an unpaired, two-tailed Student’s t test with a 95% confidence interval, with non-parametric Mann-Whitney U test or a non-parametric ANOVA (Kruskal-Wallis).
Results
Type I IFN are induced in human primary cells in response to Pf gDNA
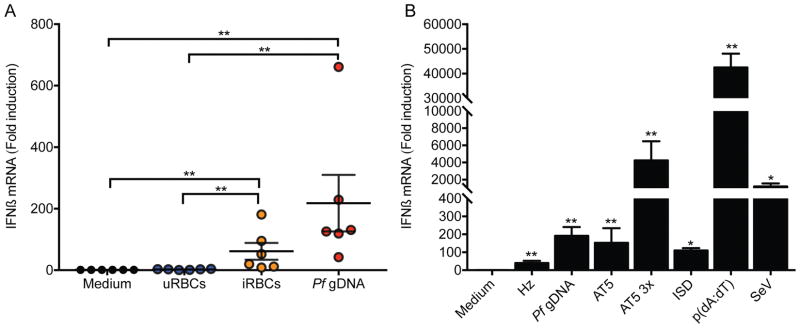
Previously, our group demonstrated that Pf infected RBCs (iRBCs), but not uninfected RBCs (uRBCs), stimulated IFNβ expression in human PBMCs. Pf gDNA also stimulated IFNβ when transfected into PBMC (8). We extended these studies to determine the direct effect of Pf on type I IFN induction in purified human CD14+ monocytes. CD14+ cells were stimulated with iRBCs, uRBCs or transfected with Pf gDNA as a control. A significant induction of IFNβ mRNA was observed with iRBCs. As expected, uRBCs did not induce IFNβ mRNA (Fig. 1A). The induction of IFNβ by iRBCs was similar to that elicited by transfected Pf gDNA (Fig. 1A). IFNα was also induced at the mRNA level (data not shown).
Figure 1. Human CD14+ monocytes produce IFN in response to Pf gDNA and infected RBCs.
(A) CD14+ monocytes were isolated from PBMCs from human healthy donors by MACS and transfected with Pf gDNA (10 μg/ml) using Lipofectamine 2000 or incubated with uninfected RBCs (uRBCs) or Pf infected RBCs (iRBCs) at a 20 to 1 RBC per monocyte ratio. RNA was prepared after 6 h incubation or transfection and IFNβ and β-actin levels were measured by qRT-PCR. (B) Human MDMs from healthy donors were transfected with natural Hz (50 μM), AT5 and AT5 3X ODNs (3 μM) or Pf gDNA (0.5 ml total at 1 μg/ml). IFNβ and β-actin levels measured as in (A). ISD, p(dA:dT) and Sendai virus (SeV) were used as positive controls. All samples were compared to cells incubated with medium alone or between specific groups when indicated and analyzed by Mann-Whitney U test. Data are presented as mean ± SD and are representative of 3 independent experiments. * indicates a p value of < 0.05, ** indicates a p value of < 0.001.
Previous results demonstrated that Hz presents Plasmodium DNA to TLR9 (16) and induces substantial amounts of IFNβ (8). Like CD14+ monocytes, human monocyte-derived macrophages (MDMs) expressed IFNβ mRNA in response to transfected Pf gDNA (Fig. 1B). We also determined if other malarial products would induce IFNβ in MDMs. Hz and Pf gDNA strongly stimulated IFNβ when delivered into the cytosol of MDMs (Fig. 1B). Likewise, other known type I IFN inducers (p(dA:dT), ISD and SeV) as well as the AT-rich ODNs AT5 and AT5 3x containing stem-loops (whose design was based on sequences from the Pf genome (8)), significantly promoted IFNβ expression in MDMs (Fig. 1B). Collectively, these data indicate that indeed malaria parasites and Plasmodial PAMPs (Pf gDNA, Hz, AT-rich oligos) activate type I IFN in human monocytes and MDMs.
Type I IFN are induced after cytosolic delivery of malaria DNA
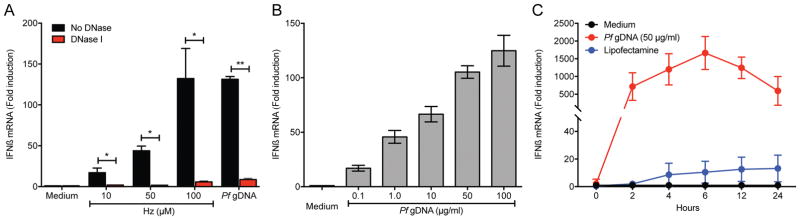
To understand the effect of cytosolic location of Pf gDNA, we determined the role of cytosolic delivery of DNA in the ability of Hz to induce IFNβ expression in WT THP-1 cells. Hz significantly induced IFNβ expression in a dose-dependent manner when transfected into cells. This activity was abolished when Hz was pretreated with DNase I (Fig. 2A). Similarly, purified Pf gDNA induced IFNβ when delivered to the cytosol of THP-1 cells in a dose and time-dependent manner (Figs. 2B and 2C).
Figure 2. Type I IFN are induced by Pf gDNA.
(A) The effect of Pf Hz in the induction of IFNβ mRNA was assessed by the treatment with DNase I and qRT-PCR. THP-1 cells were transfected with Lipofectamine 2000 and the indicated amounts of Hz for 6 h followed by measurement of IFNβ mRNA by qRT-PCR. Data were analyzed for the difference in IFNβ mRNA induction in cells treated with DNase vs untreated by paired Student’s t-test (B) Cells were transfected as indicated in (A) and scaled concentrations (μg/ml) of Pf gDNA for 6 h followed by measurement of IFNβ mRNA by qRT-PCR. (C) Time course of induction of IFNβ mRNA in THP-1 cells transfected with 50 μg/ml of Pf gDNA. THP-1 cell incubated with Lipofectamine 2000 or medium alone were used as controls. Data are presented as mean ± SD and are representative of 3 independent experiments. * indicates a p value of < 0.05, ** indicates a p value of < 0.001.
Type I IFN induction by Pf gDNA is dependent on cGAS
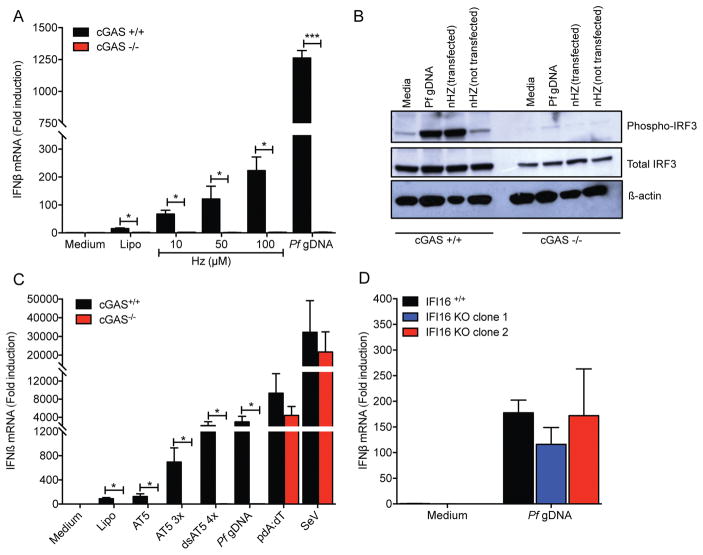
Cyclic GMP-AMP synthase (cGAS) is a DNA sensor that catalyzes the synthesis of cGAMP to drive activation of STING, resulting in the induction of type I IFN and other inflammatory genes (23–25). In order to determine the involvement of cGAS as a sensor for Plasmodium DNA, IFNβ expression was assessed in cGAS+/+ and cGAS−/− THP-1 cells. Transfection of cultured Hz or Pf gDNA induced expression of IFNβ mRNA in cGAS+/+ but not in cGAS KO THP-1 cells (Fig. 3A). Detection of phosphorylated IRF3, an indicator of STING activation and an upstream readout for type I IFN induction, revealed phosphorylation of IRF3 in cGAS+/+ but not cGAS−/− cells when transfected with Hz or Pf gDNA (Fig. 3B). Cytosolic delivery of AT-rich ODNs also induced IFNβ in cGAS+/+ THP-1 cells but did not induce IFNβ expression in cGAS−/− cells (Fig. 3C). Finally, transfection of IFI16-deficient cells with Pf gDNA induced equivalent levels of IFNβ mRNA to IFI16+/+ cells (Fig. 3D), suggesting that IFI16 is not required for detection of Pf gDNA. Altogether, these results indicate that cGAS is the cytosolic sensor of Pf gDNA and is required for the induction of IFNβ by malaria Hz as carrier of Pf gDNA.
Figure 3. Pf gDNA induces IFNβ expression through the cGAS-STING pathway.
(A) cGAS+/+ and cGAS−/− THP-1 cells were transfected with 10, 50 or 100 μM of Hz for 6 h followed by detection of IFNβ mRNA by qRT-PCR. Pf gDNA was used as control. Data were analyzed for the difference in IFNβ mRNA induction in WT cells vs KO cells by paired Student’s t-test. (B) Phosphorylation of IRF3 was detected by SDS-PAGE followed by western-blot. Detection of total IRF3 and β-actin was used as loading control. (C) Transfection of AT-rich ODNs (AT5, AT5 3x and dsAT5 4x, 3 μM each) results in a cGAS dependent induction of type I IFN. Sendai virus (SeV) was used as control for cGAS-independent induction of IFNβ. (D) IFI16+/+ (EV-empty vector) and IFI16 KO (clones 1 and 2) THP-1 cells were transfected with 10 μg/ml of Pf gDNA for 6 h, then IFNβ mRNA was measured by qRT-PCR. Data were analyzed for the difference in IFNβ mRNA induction in WT cells vs KO cells by Wilcoxon rank test. Data are presented as mean ± SD and are representative of 3 independent experiments. * indicates a p value of < 0.05, ** indicates a p value of < 0.001.
The cGAS-STING pathway is involved in the induction of type I IFN in response to malaria DNA
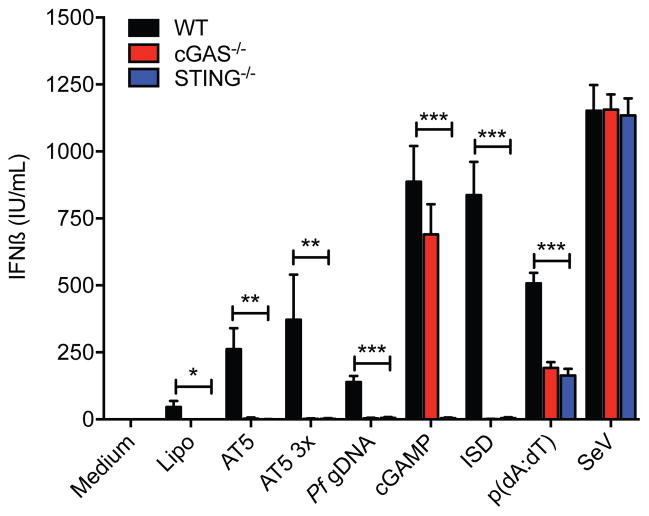
To corroborate the involvement of the cGAS-STING pathway in the production of IFNβ, we measured IFNβ protein secreted by mouse BMDMs stimulated with Pf gDNA. Cytosolic delivery of Pf gDNA or AT-rich ODNs induced the secretion of IFNβ from WT BMDMs (Fig. 4). This induction of IFNβ protein secretion was not observed in either STING or cGAS KO macrophages (Fig. 4). These data indicate the involvement of the cGAS-STING pathway in the induction of type I IFN in response to the sensing of Pf DNA.
Figure 4. Type I IFN are produced through the activation of the cGAS-STING pathway in response to Pf gDNA.
BMDM from WT, STING−/− and cGAS −/− mice were transfected with Pf gDNA (1 μg/ml), AT5 and AT5 3x ODNs (3 μM), 2′3′ cGAMP (10 nM). Supernatants were recovered after 18 h stimulation and levels of mouse IFNβ were measured by ELISA. Synthetic 2′3′-cGAMP, ISD, p(dA:dT) and Sendai virus (SeV) were used as controls. Data were analyzed for the difference in IFNβ mRNA induction in WT cells vs KO cells by unpaired t-test corrected for multiple comparisons using the Holm-Sidak method. Data are presented as mean ± SD and are representative of 3 independent experiments. * indicates a p value of < 0.05, ** indicates a p value of < 0.001, *** indicates a p value of < 0.0001.
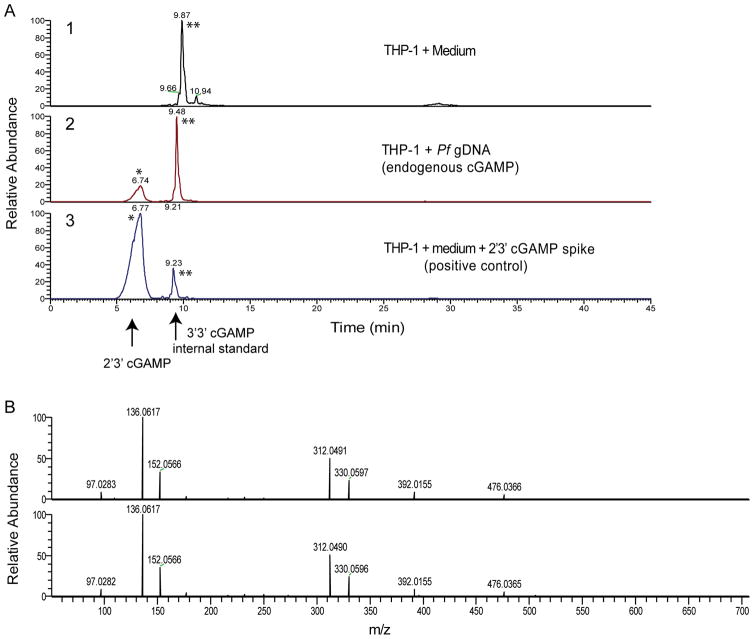
2′3′-cGAMP is induced after sensing of Pf gDNA
cGAS catalyzes the synthesis of cyclic GMP-AMP (2′3′-cGAMP) in the presence of cytosolic DNA (24). To further characterize this activity in response to Plasmodium DNA, we transfected THP-1 cells with Pf gDNA and measured 2′3′-cGAMP production. We began by heat-treating total extracts of transfected cells, as cGAMP is heat-stable. We confirmed the induction of 2′3′-cGAMP in cells transfected with Pf gDNA by LS-MS/MS, consistent with the cGAS-dependent induction of IFNβ shown before in response to Pf gDNA (Fig. 3). 2′3′-cGAMP was produced in Pf gDNA-transfected THP-1 cells (Fig. 5A, panel 2) but not in control THP-1 cells (Fig. 5A, panel 1; 2′3′-cGAMP peak is represented by one asterisk). As a positive control, the lysate from untreated THP-1 cells was spiked with synthetic 2′3′-cGAMP (Fig. 5A, panel 3). All lysates also included an internal standard of 3′3′-cGAMP (500 pg per sample; represented by two asterisks). MS/MS of the 2′3′-cGAMP peak revealed several fragmented ions with the expected m/z values for product ions of 2′3′-cGAMP and were observed in similar ratios in DNA transfected samples and spiked controls (Fig. 5B), confirming the induction of 2′3′-cGAMP by transfected Pf gDNA. Together our data indicate that the cGAS-Pf DNA complex activates 2′3′-cGAMP synthesis and consequently the induction of IFNβ. The data also suggest that cGAS is an important sensor for Pf gDNA.
Figure 5. 2′3′-cGAMP is induced by Pf gDNA.
Lysates from THP-1 cells left untreated (medium) or transfected and incubated for 6 h with 50 μg/ml of Pf gDNA were heated at 95°C for 5 min to denature proteins. The heat-resistant supernatants were recovered after 17,000 × g centrifugation and analyzed by LC-MS/MS. As positive control, isolate from THP-1 cells untreated was spiked with 100 nM 2′3′-cGAMP. (A) 1. THP-1 cells untreated (medium), 2. THP-1 cells transfected with Pf gDNA and 3. THP-1 cells untreated and spiked with synthetic 2′3′-cGAMP. Reconstructed ion chromatograms of cGAMP fragment ion at m/z 312.049 following LC-MS/MS fragmentation of cGAMP protonated ion. Peaks marked with a single asterisk are from endogenous or positive control 2′3′-cGAMP; double asterisks indicate 3′3′ cGAMP spiked at 500 pg to each isolate as internal standard. (B) Tandem mass spectra (MS/MS) of the peak observed at 6.7 min (from corresponding panels A2 and A3, respectively). Peaks are 2′3′-cGAMP fragment ions and are observed in similar ratios.
Discussion
Much progress has been made in our understanding of how phagocytes sense Plasmodium and their associated host receptors that elicit inflammation (7). Three major Pf PAMPs have been described: glycosylphosphatidylinositol anchors (GPI anchors) (34), Hz crystals (9, 16) and immune stimulatory DNA (8, 16). Still, the receptor or family of receptors for Plasmodium DNA-driven IFN responses has been elusive. In previous reports, we described “unknown” cytosolic DNA sensor(s) that coupled to STING, TBK1 and IRF3-IRF7 signaling pathway. This receptor or family of receptors acted as a sensor for Plasmodium DNA, iRBCs and oligonucleotides containing the AT-rich motif, leading to type I IFN production (7, 8). We also revealed that the induction of type I IFN is driven by a pathway that did not involve TLR9, DAI, RNA polymerase-III, IFI16/p204 (8) or DDX41 and IFI203 as reported previously (26). Recently, cGAS has been described as a DNA sensor for detecting malaria DNA in the context of P. yoelii infection. The role of cGAS in P. yoelii was somewhat complex, as cGAS activation appeared to down regulate IFNα/β production rather than enhance it through induction of the negative regulator SOCS1 (26). In this report, we demonstrate that Pf gDNA and AT-rich ODNs induce type I IFN production through cGAS and the synthesis of 2′3′-cGAMP, which drives the activation of the STING pathway, raising the role of IFN-α/β in malaria pathogenesis.
The sensing of pathogen-derived DNA is a central strategy used by the innate immune system to initiate immune responses following microbial invasion (11). cGAS acts as cytosolic DNA sensor that activates innate immune responses through production of the second messenger 2′3′-cGAMP, which in turn activates the adaptor STING and leads to type I IFN production (12). The importance of this cytosolic DNA sensor and type I IFN release is now being appreciated in the context of various infectious diseases beyond their traditional roles in antiviral immunity. A growing list of pathogens as diverse as Neisseria gonorrhoeae (30), Cytomegalovirus (35), Mycobacterium tuberculosis (36), Human Immunodeficiency Virus (37), Streptococcus agalactiae (38), Listeria monocytogenes (39), Chlamydia trachomatis (40) all induce type I IFN through the cGAS/STING pathway. The data in this paper show this is also true for Pf.
Exogenous DNA that gains access to the cytosol is a particularly potent and clear danger signal for the innate immune system (12). We have previously demonstrated that natural Hz activates TLR9 due the delivery of Plasmodial DNA to the endosomal compartment (16). However, a TLR9-independent response also occurs when purified Hz acts as vehicle to deliver Plasmodium DNA into the cytosol leading to IFNβ production (8). Experimentally, we addressed this hypothesis transfecting Pf gDNA and Hz with lipofectamine. In contrast to our experimental in vitro conditions, Hz crystals appear to destabilize the phagolysosome during infection, which allows the delivery of phagosomal contents including DNA to the cytosol (9). This highlights a mechanism by which Hz-associated cargo such as Plasmodial DNA might access the cytosol and suggest that Pf gDNA can drive IFNβ production upon access to the cytosolic compartment.
The large content of AT motifs in the Pf genome (present over 6,000 times) raised the question of their impact in the cGAS/STING-dependent type I IFN response. Our previous studies showed the immune stimulatory effect of this unique motif containing stem-loop secondary structure in both human and mouse cells (8). cGAS has been described as a sensor of double-stranded DNA, in a nucleotide length-dependent, sequence-independent manner (41). This DNA sensor has also been linked to the recognition of stem-loop DNA structures formed from the HIV-1 genome reverse-transcribed into ssDNA triggering type I IFN production (42). Collectively, our results suggest that cGAS sensing of AT-rich ODNs is involved in the type I IFN response elicited during Pf infection.
Data presented in this study identify the basis of type I IFN production mediated by the sensing of cytosolic Plasmodial DNA by cGAS and subsequent activation of the cGAS/STING pathway. This conclusion is supported by a recent study showing that Tmem173gt mice (coding for a null allele of STING) were completely resistant and Mb21d1−/− mice (coding for cGAS) showed partial protection in a lethal model of P. yoelii infection (26). This study highlights cGAS as sensor for detection of P. yoelii gDNA by reporting a decrease in IFNβ mRNA induction after cGAS knockdown (cGAS siRNA in RAW264.7 cells) and an increase in IFNβ mRNA induction in HEK293T cells after transfection with cGAS. The current study adds to our knowledge of Plasmodial infections as we have focused on the major human pathogenic species of Plasmodium, ie, P. falciparum. Importantly, by using LC-MS/MS, we provide direct evidence that 2′3′-cGAMP is produced in response to Pf gDNA.
The consequences of type I IFN induction during malarial infection are currently under investigation in numerous laboratories. Several reports have revealed the ability of Plasmodium spp., to induce type I IFN and a type I IFN gene signature (8, 10, 15, 43–45). Previous studies have shown that exogenous recombinant IFNα can even inhibit experimental cerebral malaria and reduce parasite burden in mice infected with P. berghei ANKA (PbA) (43). However, other studies demonstrate that signaling via the type I IFN receptor impairs DC function and T cell responses that control parasitemia in mouse models of malaria (46). Our studies in the PbA model have demonstrated that the TBK1, IRF3-IRF7 dependent production of type I IFN is central to the progression of cerebral malaria, given that, mice deficient in these factors survived far longer than WT mice (8). In contrast, resistance of mice in the lethal model of P. yoelii has been linked to early robust IFNα/β production by pDCs, showing that STING and cGAS-deficient mice were more resistant compared to WT mice (26). A recent report identifies an immunological regulatory effect of type I IFN in patients during blood-stage Pf infection. Type I IFN were found to suppress IL-6 but not TNFα production by blood monocytes, promote IL-10-producing CD4+ T cells and generate regulatory Tr1 cells (45). Altogether, these data indicate the relevant and complex involvement of type I IFN in the pathogenesis of malaria.
The activation of innate immune cells and consequent systemic inflammation lead to the initial signs and symptoms of malaria, and can also influence the development of the more severe forms of the disease. The data presented in this paper corroborate the role of Plasmodium DNA as a potent PAMP during malaria infection and identify cGAS as an important cytosolic sensor that activates the second messenger 2′3′-cGAMP. We also reveal the role of the cGAS/STING pathway in the consequent type I IFN production. Our results suggest that any attempts at immunomodulatory therapy for patients with severe malaria will need to take into account the role of DNA-induced type I IFN and its impact on malaria pathogenesis.
Acknowledgments
Grant support:
This work received support by the National Institute of Allergy and Infectious Diseases grant R21AI124171 (to E.A.K-J., D.T.G. and C.G.) and R01AI079293 (K.A.F. and D.T.G.). A.M.L and L.F.G were supported by the Departamento de Ciencia y Tecnologia – Colciencias-CIDEIM grant 0234-2014 and 0552-2015 and the NIH/Fogarty International Center training grant D43TW006589.
The authors are grateful to Elizabeth J. Thatcher for assistance with sample preparation and data analysis, and to Maria Adelaida Gomez for critically reviewing this manuscript.
Abbreviations used in this article
- gDNA
genomic DNA
- Pf
Plasmodium falciparum
- cGAS
cyclic GMP-AMP synthase
- 2′3′-cGAMP
cyclic GMP-AMP
- Hz
hemozoin
- IFI16
interferon gamma-inducible protein 16
- STING
Stimulator of Interferon Genes
- IRF3
Interferon Regulatory Factor 3
- AT-rich ODNs
Adenine Thymine-rich oligodeoxynucleotides
Bibliography
- 1.World Malaria Report 2016. Geneva: World Health Organization; 2016. [Google Scholar]
- 2.Cotter C, Sturrock HJ, Hsiang MS, Liu J, Phillips AA, Hwang J, Gueye CS, Fullman N, Gosling RD, Feachem RG. The changing epidemiology of malaria elimination: new strategies for new challenges. Lancet. 2013;382:900–911. doi: 10.1016/S0140-6736(13)60310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu F, Culleton R, Zhang M, Ramaprasad A, von Seidlein L, Zhou H, Zhu G, Tang J, Liu Y, Wang W, Cao Y, Xu S, Gu Y, Li J, Zhang C, Gao Q, Menard D, Pain A, Yang H, Zhang Q, Cao J. Emergence of Indigenous Artemisinin-Resistant Plasmodium falciparum in Africa. The New England journal of medicine. 2017;376:991–993. doi: 10.1056/NEJMc1612765. [DOI] [PubMed] [Google Scholar]
- 4.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. Spread of artemisinin resistance in Plasmodium falciparum malaria. The New England journal of medicine. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattarai A, Ali AS, Kachur SP, Martensson A, Abbas AK, Khatib R, Al-Mafazy AW, Ramsan M, Rotllant G, Gerstenmaier JF, Molteni F, Abdulla S, Montgomery SM, Kaneko A, Bjorkman A. Impact of artemisinin-based combination therapy and insecticide-treated nets on malaria burden in Zanzibar. PLoS medicine. 2007;4:e309. doi: 10.1371/journal.pmed.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA, 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazzinelli RT, Kalantari P, Fitzgerald KA, Golenbock DT. Innate sensing of malaria parasites. Nature reviews Immunology. 2014;14:744–757. doi: 10.1038/nri3742. [DOI] [PubMed] [Google Scholar]
- 8.Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, Chan J, Bartholomeu DC, Lauw F, Hall JP, Barber GN, Gazzinelli RT, Fitzgerald KA, Golenbock DT. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalantari P, DeOliveira RB, Chan J, Corbett Y, Rathinam V, Stutz A, Latz E, Gazzinelli RT, Golenbock DT, Fitzgerald KA. Dual engagement of the NLRP3 and AIM2 inflammasomes by plasmodium-derived hemozoin and DNA during malaria. Cell reports. 2014;6:196–210. doi: 10.1016/j.celrep.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirako IC, Gallego-Marin C, Ataide MA, Andrade WA, Gravina H, Rocha BC, de Oliveira RB, Pereira DB, Vinetz J, Diamond B, Ram S, Golenbock DT, Gazzinelli RT. DNA-Containing Immunocomplexes Promote Inflammasome Assembly and Release of Pyrogenic Cytokines by CD14+ CD16+ CD64high CD32low Inflammatory Monocytes from Malaria Patients. mBio. 2015;6:e01605–01615. doi: 10.1128/mBio.01605-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annual review of immunology. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nature immunology. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 13.Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends in immunology. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Gowda NM, Kumar S, Gowda DC. Protein-DNA complex is the exclusive malaria parasite component that activates dendritic cells and triggers innate immune responses. J Immunol. 2010;184:4338–4348. doi: 10.4049/jimmunol.0903824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichyangkul S, Yongvanitchit K, Kum-arb U, Hemmi H, Akira S, Krieg AM, Heppner DG, Stewart VA, Hasegawa H, Looareesuwan S, Shanks GD, Miller RS. Malaria blood stage parasites activate human plasmacytoid dendritic cells and murine dendritic cells through a Toll-like receptor 9-dependent pathway. J Immunol. 2004;172:4926–4933. doi: 10.4049/jimmunol.172.8.4926. [DOI] [PubMed] [Google Scholar]
- 16.Parroche P, Lauw FN, Goutagny N, Latz E, Monks BG, Visintin A, Halmen KA, Lamphier M, Olivier M, Bartholomeu DC, Gazzinelli RT, Golenbock DT. Malaria hemozoin is immunologically inert but radically enhances innate responses by presenting malaria DNA to Toll-like receptor 9. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1919–1924. doi: 10.1073/pnas.0608745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gowda NM, Wu X, Gowda DC. The nucleosome (histone-DNA complex) is the TLR9-specific immunostimulatory component of Plasmodium falciparum that activates DCs. PloS one. 2011;6:e20398. doi: 10.1371/journal.pone.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. The Journal of experimental medicine. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 22.Gun SY, Claser C, Tan KS, Renia L. Interferons and interferon regulatory factors in malaria. Mediators of inflammation. 2014;2014:243713. doi: 10.1155/2014/243713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X, Cai B, Wang M, Tan P, Ding X, Wu J, Li J, Li Q, Liu P, Xing C, Wang HY, Su XZ, Wang RF. Cross-Regulation of Two Type I Interferon Signaling Pathways in Plasmacytoid Dendritic Cells Controls Anti-malaria Immunity and Host Mortality. Immunity. 2016;45:1093–1107. doi: 10.1016/j.immuni.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 28.Baratin M, Roetynck S, Lepolard C, Falk C, Sawadogo S, Uematsu S, Akira S, Ryffel B, Tiraby JG, Alexopoulou L, Kirschning CJ, Gysin J, Vivier E, Ugolini S. Natural killer cell and macrophage cooperation in MyD88-dependent innate responses to Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14747–14752. doi: 10.1073/pnas.0507355102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. The American journal of tropical medicine and hygiene. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 30.Andrade WA, Agarwal S, Mo S, Shaffer SA, Dillard JP, Schmidt T, Hornung V, Fitzgerald KA, Kurt-Jones EA, Golenbock DT. Type I Interferon Induction by Neisseria gonorrhoeae: Dual Requirement of Cyclic GMP-AMP Synthase and Toll-like Receptor 4. Cell reports. 2016;15:2438–2448. doi: 10.1016/j.celrep.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charrel-Dennis M, Latz E, Halmen KA, Trieu-Cuot P, Fitzgerald KA, Kasper DL, Golenbock DT. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell host & microbe. 2008;4:543–554. doi: 10.1016/j.chom.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. The Journal of experimental medicine. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-kappaB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Zhu J, Krishnegowda G, Gowda DC. Induction of proinflammatory responses in macrophages by the glycosylphosphatidylinositols of Plasmodium falciparum: the requirement of extracellular signal-regulated kinase, p38, c-Jun N-terminal kinase and NF-kappaB pathways for the expression of proinflammatory cytokines and nitric oxide. J Biol Chem. 2005;280:8617–8627. doi: 10.1074/jbc.M413539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lio CW, McDonald B, Takahashi M, Dhanwani R, Sharma N, Huang J, Pham E, Benedict CA, Sharma S. cGAS-STING Signaling Regulates Initial Innate Control of Cytomegalovirus Infection. Journal of virology. 2016;90:7789–7797. doi: 10.1128/JVI.01040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiens KE, Ernst JD. The Mechanism for Type I Interferon Induction by Mycobacterium tuberculosis is Bacterial Strain-Dependent. PLoS pathogens. 2016;12:e1005809. doi: 10.1371/journal.ppat.1005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrade WA, Firon A, Schmidt T, Hornung V, Fitzgerald KA, Kurt-Jones EA, Trieu-Cuot P, Golenbock DT, Kaminski PA. Group B Streptococcus Degrades Cyclic-di-AMP to Modulate STING-Dependent Type I Interferon Production. Cell host & microbe. 2016;20:49–59. doi: 10.1016/j.chom.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA, Jakobsen MR, Paludan SR. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. The EMBO journal. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. J Immunol. 2014;193:2394–2404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kubler K, Wittmann S, Gramberg T, Andreeva L, Hopfner KP, Mertens C, Zillinger T, Jin T, Xiao TS, Bartok E, Coch C, Ackermann D, Hornung V, Ludwig J, Barchet W, Hartmann G, Schlee M. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nature immunology. 2015;16:1025–1033. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vigario AM, Belnoue E, Gruner AC, Mauduit M, Kayibanda M, Deschemin JC, Marussig M, Snounou G, Mazier D, Gresser I, Renia L. Recombinant human IFN-alpha inhibits cerebral malaria and reduces parasite burden in mice. J Immunol. 2007;178:6416–6425. doi: 10.4049/jimmunol.178.10.6416. [DOI] [PubMed] [Google Scholar]
- 44.Rocha BC, Marques PE, Leoratti FM, Junqueira C, Pereira DB, Antonelli LR, Menezes GB, Golenbock DT, Gazzinelli RT. Type I Interferon Transcriptional Signature in Neutrophils and Low-Density Granulocytes Are Associated with Tissue Damage in Malaria. Cell reports. 2015;13:2829–2841. doi: 10.1016/j.celrep.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montes de Oca M, Kumar R, Rivera FL, Amante FH, Sheel M, Faleiro RJ, Bunn PT, Best SE, Beattie L, Ng SS, Edwards CL, Boyle GM, Price RN, Anstey NM, Loughland JR, Burel J, Doolan DL, Haque A, McCarthy JS, Engwerda CR. Type I Interferons Regulate Immune Responses in Humans with Blood-Stage Plasmodium falciparum Infection. Cell reports. 2016;17:399–412. doi: 10.1016/j.celrep.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haque A, Best SE, Ammerdorffer A, Desbarrieres L, de Oca MM, Amante FH, de Labastida Rivera F, Hertzog P, Boyle GM, Hill GR, Engwerda CR. Type I interferons suppress CD4(+) T-cell-dependent parasite control during blood-stage Plasmodium infection. European journal of immunology. 2011;41:2688–2698. doi: 10.1002/eji.201141539. [DOI] [PubMed] [Google Scholar]