Abstract
Granulocyte differentiation and immune response function is a dynamic process governed by a highly coordinated transcriptional program that regulates cellular fate and function, often in a context-dependent manner. Advances in high-throughput technologies and bioinformatics have allowed us to better understand complex biological processes at the genomic and proteomic levels. Components of the environmental milieu, along with the molecular mechanisms that drive the development, activation, and regulation of granulocytes, have since been elucidated. In this chapter, we present the intricate network in which these elements come together and influence one another. In particular, we describe the critical roles of transcription factors like PU.1, CCAAT/enhancer-binding protein (C/EBPα; alpha), C/EBPε (epsilon), and growth factor independent-1 (Gfi-1). We also review granulocyte colony-stimulating factor (G-CSF) receptor-induced signal transduction pathways, their influence on proliferation and differentiation, and the cooperativity of cytokines and chemokines in this process.
Keywords: Systems biology, Phagocyte, Granulocyte, Neutrophil, Macrophage, Transcription factor, Granulopoiesis, Chemotaxis, Phagocytosis, Apoptosis
Phagocytes constitute the primary line of host defense through the highly coordinated process of chemotaxis; ingestion of microbes, particles, and cells; and production and secretion of peptides, lipids, and reactive oxygen species (ROS). This biological function has been critical for the success of multicellular organisms and is found in elementary forms such as dichtyostelium. Phagocytes provide the cornerstone of the innate immune system. Unlike the adaptive immune response that requires prior microbe exposure and time to develop specific antigen recognition, phagocytes are critical for the rapid, nonspecific targeting and elimination of infectious pathogens. These events involve complex interactions between the host, pathogen recognition, and phagocytic effector cells that must be tightly regulated. Efficient innate immune responses must be balanced against prevention of unabated inflammation linked to autoimmune and inflammatory disease states. For example, the recognition and subsequent phagocytosis of apoptotic neutrophils by macrophages is a key homeostatic event in the resolution of inflammation. The components and molecular mechanisms governing the development, activation, and regulation of these cells have been established, and they present the basis for this chapter and future work with a systems analysis.
From a common myeloid progenitor (CMP) cell, phagocytic cells develop into highly specialized cells. Granulocytes, also known as polymorphonuclear leukocytes (PMNs), consist of neutrophils, basophils, and eosinophils. Of these, the neutrophil is the most predominant circulating leukocyte in humans, whereas lymphocytes predominate in mice. Peripheral blood monocytes undergo a process of activation and differentiation to become resident tissue macrophages. Both cell types are descended from a common hematopoietic progenitor cell, the colony-forming unit granulocyte/macrophage (CFU-GM).
Granulocyte production must be sufficient and dynamic to protect the host against infection, but not excessive as to cause chronic inflammation and tissue damage. Production begins at the earliest stage when the hematopoietic stem cell (HSC) is recruited from a pool of dormant HSCs (Figs. 6.1 and 6.2). A granulocyte requires 12 days in the bone marrow before it leaves to eventually reside in tissues. In peripheral blood, neutrophil levels are finely controlled (2–8 × 103/mm3) and their circulating half-life is brief (~ 6h). Most of the bone marrow activity (~ 67% of the cells belong to the myeloid, nonerythroid lineage) is directed toward a continuous, prodigious degree of neutrophil production (~ 5–10 × 1010/day) [1]. Life-threatening sepsis occurs when absolute neutrophil counts are less than 0.5 × 103/mm3. Granulocyte production can respond quickly to severe infection with a log-fold increase in circulating neutrophil counts (~ 3–5 × 104/mm3) within 48–72h. When healthy adult volunteers received a single dose of granulocyte colony-stimulating factor (GCSF), neutrophil counts increased rapidly, peaking at 12 h and returning to baseline by 48–72 h [2]. Bone marrow reserve is critical. The neonatal neutrophil bone marrow storage pool is decreased compared to the adult counterpart [3, 4]. Moreover, there is delayed neutrophil response to infection (3–4 h in the neonate compared to 30–90 min in the adult) [5]. Thus, neonates are especially susceptible to neutrophil exhaustion when stressed by severe infection (sepsis). Understanding neutrophil production and kinetics has led to improved survival in stressed neonates [6, 7].
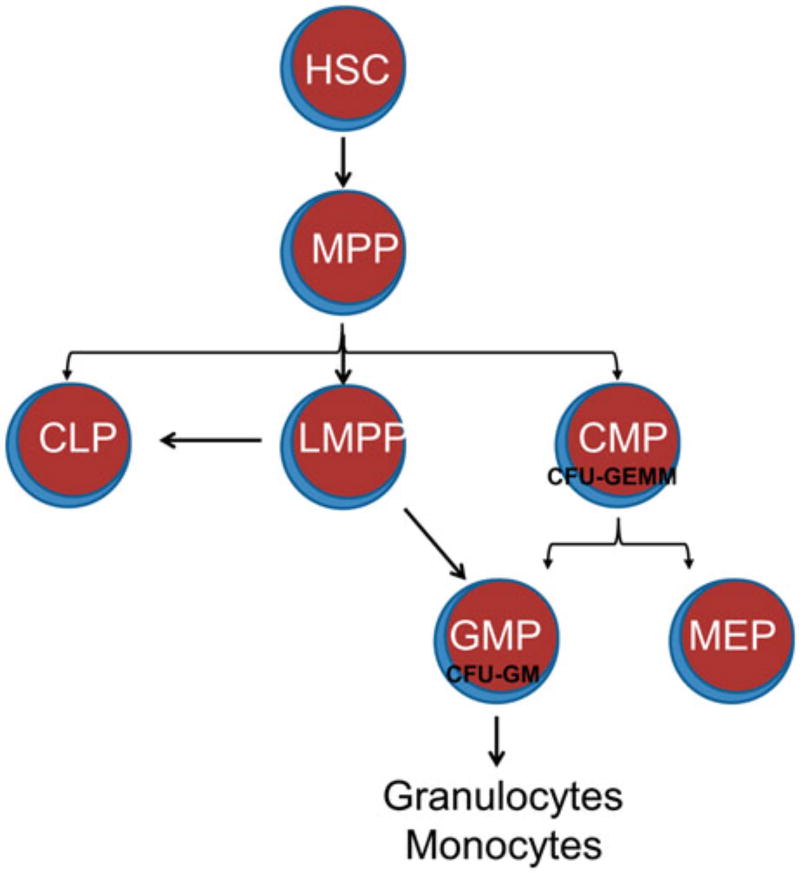
Figure 6.1.
Overview of derivation of granulocytes and monocytes from hematopoietic stem cells. Hematopoietic stem cells (HSC) give rise to multipotent progenitors (MPP) which can produce three lineage-committed progenitors, which are the classical common lymphoid progenitors (CLP) and common myeloid progenitor (CMP) and a nonclassical lymphoid/myeloid multipotent progenitor (LMPP). CMPs produce colony-forming units which consist of granulocytes erythrocytes, monocytes, and megakaryocytes (CFU-GEMM). Granulocyte and monocyte progenitors (GMP) and megakaryocyte and erythroid progenitors (MEP) descend from CMP. However, GMP can also be derived from LMPP. CFU-GM colony forming unit-granulocyte and monocyte
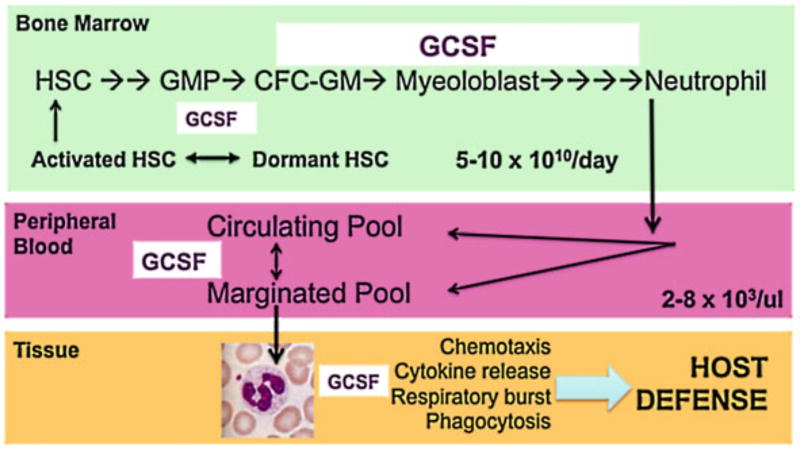
Figure 6.2.
Multiscale analysis of granulopoiesis. Granulocytes are produced in the bone marrow through self-renewal and differentiation of hematopoietic stem cells (HSC). These pluripotential HSC initiate granulopoiesis by becoming granulocyte/macrophage progenitor cells (GMP) and colony-forming cells of granulocytes/macrophages (CFC-GM). Soon, they become myeloblasts, which are easily identifiable precursors in the bone marrow. This prodigious amount of stem cell renewal and differentiation produces approximately 10 billion cells per day in each adult person. The number of circulating granulocytes is kept within a narrow range of 2000–8000 per μl. The bone marrow is able to respond quickly to infectious stimuli and amplify granulocyte numbers by several fold. The granulocytes leave the blood vessels and migrate (chemotaxis) to the tissues where they release cytokines, engulf microbes, and produce reactive oxygen species (ROS) through a respiratory burst involving the NADPH oxidase. Granulocyte colony-stimulating factor (G-CSF) is the primary cytokine responsible for stem cell expansion toward the granulocyte, inducer of differentiation of the myeloblasts, and enhancer of granulocyte function. NADPH nicotinamide adenine dinucleotide phosphate
Granulopoiesis
Granulopoiesis is a complex process by which a CMP, under the stimulation of cytokines interleukin-3 (IL3), GCSF, and/or granulocyte macrophage colony-stimulating facto GMCSF, induces CFU-granulocytes erythrocytes, monocytes, and megakaryocytes (GEMM) to differentiate into CFU-GM, the common precursor for both neutrophils and monocytes. Myelopoiesis involves stem and progenitor cells that generate also megakaryocytes (and platelets) and erythrocytes (Fig 6.1). This hierarchy has been challenged and a newer paradigm has emerged to reflect evidence that the cells of both the innate (neutrophils and macrophages) and adaptive (T and B cells) immune system are derived from a lymphoid/myeloid multipotent progenitor (LMPP; Fig 6.1), which does not give rise to megakaryocytes or erythrocytes [8, 9]. Other cytokines involved in multipotential lineage progenitors include thrombopoietin and Flt3. The precise combination(s) of growth factors and stromal factors that lead to the production of a specific granulocyte remains poorly understood. The CFU-GM stem cell differentiates into either a CFU-G or CFU-M stem cell. The development of mature granulocytes from hematopoietic precursor cells is controlled by a small number of transcription factors and complex gene regulatory networks, including those encoding growth factors and their receptors, enzymes, adhesion molecules, and transcription factors. In particular, PU.1, CCAAT/enhancer-binding protein (C/EBPα; alpha), C/EBPε (epsilon), and growth factor independent (Gfi)-1 have emerged as critical players, master regulators of myeloid development [10], and constitute a gene regulatory networking for granulopoiesis. A systematic study of the regulatory components and their complex interactions will enable higher-order understanding of how granulocytes are produced, how they modulate other immune responses and themselves, and how aberrations in these pathways lead to disease states.
GCSF is the most important hematopoietic growth factor that drives the production, proliferation, and differentiation of myeloid progenitor and precursor cells, beginning with the bone marrow HSC and terminating as a mature neutrophil released into the periphery. It also acts to enhance the survival and function of mature neutrophils by delaying apoptosis [1, 2] and acting cooperatively with other cytokines (e.g., IL-8 and tumor necrosis factor (TNF)) to activate or “prime” the neutrophil, in a dose-dependent manner [3, 4]. The clinical utility of GCSF is well evidenced by its use in the treatment and survival of patients with congenital and chemotherapy-induced neutropenias.
GCSF acts through its cognate receptor (GCSFR) via multiple signal transduction pathways, including the Janus kinase (JAK)/signal transducer and activator of transcription (STAT), Ras/mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathways, described in detail below. Seven isoforms of GCSF receptor, termed class I through VII, have been identified by screening placental and myeloid leukemia complementary DNA (cDNA) libraries [11–14]. These isoforms result from the alternative spicing of the GCSFR mRNA. Although multiple isoforms of GCSFR were identified, only the class I and class IV isoforms appear to be important for granulopoiesis, as these were the only two isoforms validated to be expressed in normal and leukemic hematopoietic cells using quantitative PCR [15]. The class I isoform represents the wild type since it is the predominantly expressed GCSFR isoform. The class IV GCSFR is an alternatively spliced, truncated isoform lacking the distal 87 amino acids, which are replaced by a novel 34 amino acid sequence [13]. Quantitative analysis of class I and class IV isoform expression revealed very low relative levels of class IV in mature circulating neutrophils compared to class I [15]. However, in CD34+ cells from adult bone marrow, a higher ratio of class IV to I expression was observed, suggesting that class IV expression drops during differentiation and is therefore developmentally regulated [16]. Interestingly increased class IV to class I ratio was also observed in leukemic cell lines and patient samples from acute myeloid leukemia (AML) patients [15]. Functional studies of the class IV isoform demonstrated that the class IV GCSFR as differentiation defective, but able to promote proliferation [17]. The class IV isoform is similar in structure to a series of truncated GCSFR mutants resulting from somatic nonsense mutations identified in patients with severe congenital neutropenia that developed acute myelogenous leukemia [18–20]. Similar to the class IV GCSFR, these truncated forms of GCSFR also conferred a maturation arrest with enhanced proliferation [19, 20].
Ligand-induced dimerization of the GCSFR rapidly triggers downstream signal transduction pathways including JAK/STAT, Ras/MAPK, and Lyn-PI3K/Akt signal transduction pathways [21–29]. The proximal cytoplasmic domain of the GCSFR contains Box 1 and Box 2, which are conserved in the hematopoietic cytokine receptor superfamily. The distal domain also contains a di-leucine receptor internalization signal and four tyrosine residues (Y704, Y729, Y744, and Y764 in the human receptor sequence). The tyrosine residues can be phosphorylated and serve as docking sites for SH2-containing proteins. The truncated forms of GCSFR lack three of four tyrosine residues (Y729, Y744, Y769) in the distal domain, which strongly implicate them in promoting differentiation signaling. The distal domain also contains a dileucine receptor internalization signal and is also a target of suppressor of cytokine signaling (SOCS) protein binding which serves to target the receptor for ubiquitination. Together, they contribute to signaling termination via receptor degradation [30]. Thus, the loss of the distal domain results in increased receptor signaling promoted by both reduced internalization and also increased recycling of internalized receptor to the surface. In the full-length class I GCSFR, cell signaling and receptor internalization synergize to promote granulopoiesis by coupling differentiation signaling with attenuation of long-term proliferation.
The GCSFR dimerization-induced JAK transautophosphorylation promotes activation of STAT5 and STAT3 protein phosphorylation which then translocate to the nucleus and promote gene expression. STAT5 is primarily implicated in promoting proliferation, whereas STAT3 is implicated in promoting both proliferation and differentiation. STAT3 can bind to multiple sites on the GCSFR, which is dependent on the GCSF dose, resulting in STAT3 activation. Activation of STAT3 promotes differentiation indirectly by inducing growth arrest, but does not induce the differentiation program, demonstrating the role of GCSFR signaling in maintenance of granulocyte precursors and cell fate determination. STAT3 induces cell cycle termination by inducing expression of feedback inhibitors such as SOCS3 that inhibits the JAK/STAT pathway and promotes signal termination by ubiquitin-mediated receptor degradation. STAT3 also promotes expression of p27kip1, an inhibitor of cyclin-dependent kinases, and promoting cell cycle arrest and, perhaps, differentiation. Another important regulator of granulocyte maturation is SHP2.A protein tyrosine phosphatase, SHP2 favors granulopoiesis over monopoiesis by promoting the expression of the transcription factor C/EBPα [31–33]. One possible target of SHP2 is runt-related transcription factor 1 (RUNX1), a transcription factor that induces the expression of C/EBPα [34]. SHP2 also represents a nonconventional downstream target of JAK2 for the class IV isoform of GCSFR. Class IV-mediated proliferation was identified to be mediated by a nonclassical JAK2-SHP2 pathway as opposed to the classical JAK/STAT pathway [16].
The transcription factors PU.1 and globin transcription factor (GATA)-1 act to inhibit each other and are implicated in early determination of the MPP to either LMPP or megakaryocyte erythrocyte progenitor (MEP) [35, 36]. Higher PU.1 levels inhibit the activity of GATA-1, which inhibits the erythrocyte development and by default promotes LMPP generation [37]. Thus, PU.1 is deterministic during the early stage of granulocyte/monocyte formation, which depends on inactivation of GATA-1. Development along the LMPP lineage progresses to formation of CLP and granulocyte macrophage progenitor (GMP). At this stage of development, PU.1 and C/EBPα appear to act in concert to promote GMP formation over CLP. C/EBPα expression is observed in CMP and GMP but not in CLP and MEP [38] and thus its expression would direct the decision to generate GMP from LMPP. Along with C/EBPα expression, graded expression of PU.1 also determines lymphoid versus myeloid decision making in LMPP. High PU.1 expression is observed in macrophages and lower expression in B cells, which leads to PU.1 dose-dependent activation of signals that guide macrophage development whereas low PU.1 levels promote B cell development [39].
Determination of cell fate in GMP to form either macrophages or granulocytes is dependent on the interplay of C/EBPα and PU.1. High PU.1 expression promotes increased expression of Egr1,2 and Nab2, which collectively promote monopoiesis by both promoting expressing monocyte specific genes and also inhibit neutrophilspecific genes [10, 39]. An important determinant of neutrophil formation, Gfi-1, is inhibited by Egr2/Nab complex. However, Gfi-1 itself can repress Egrs in addition to repressing PU.1 expression [40]. Expression of Gfi-1 is promoted by C/EBPα. Thus, Gfi-1 and Egrs represent secondary regulators of late-stage granulocyte and monocyte lineage commitment by counteractive regulation of gene expression. The secondary regulators along with the master regulators help make cell fate decisions between granulocytes and monocyte lineages [40]. Gfi-1 and C/EBPε (epsilon) have been identified as regulators of terminal neutrophil differentiation.
Gene Expression Analysis of Neutrophil Development
System-level studies such as microarray and proteomic analysis using mass spectrometry have revealed distinct patterns of gene expression, defining the phagocytic precursor cell at different stages of differentiation. Microarray analysis indicated that neutrophil development can be divided into two segments: early and terminal differentiation. Gene expression analysis of early differentiation revealed expected upregulation of cell cycle proteins in the order HSC < MPP < CLP/CMP along with selective expression of myeloid genes in CMPs but not CLPs supporting existence of lineage specific genes and patterning [41]. Additionally, genes associated with HSCs were downregulated in MPP, CMP, and CLP. Multiple studies have been carried out in both microarray and proteomic scales to identify cellular differences during terminal differentiation. The milieu of genes and proteins identified has been characterized under functional groups to simplify their role during terminal differentiation.
Granule proteins are functional components of the terminally differentiated neutrophils that are released either into phagosomes or to the extracellular space. Granule proteins are stored in azurophilic, specific, and gelatinase granules, also classified as primary, secondary, and tertiary granules based on their sequential production [42]. Microarray-based analysis of morphologically defined stages in terminal neutrophil differentiation identified 16 new proteins which involved proteases, protease inhibitors, and signaling molecules [43]. Proteomic analysis of granule proteins was performed by subcellular fractionation of the three different types of granules and identified 286 proteins [44]. Additional proteomic studies have further identified proteins specific to granules or plasma membrane as well as their localization in lipid rafts [45–48].
Cell surface protein expression follows developmental patterns, which allows them to be used as cell surface markers for differentiation. Identification of cell surface proteins also provides the mechanisms by which phagocytic cells or their precursors can interact with the environment and define cell fate or help gauge changes in the extracellular environment. Microarray analysis showed low-level expression of receptors involved in inflammatory responses such as some IL, interferon, transforming growth factor, and chemokine receptors during early terminal differentiation stages. Expression of these receptors increase in terminally differentiated neutrophils [43]. An increase in GCSFR and GMCSFR was also observed along with a reduction in MCSFR expression. Thus, increased receptor expression profile shows priming of the cell to detect inflammatory responses.
Microarray gene expression patterns obtained from highly purified subsets of cells representing terminal differentiation stages of neutrophil differentiation demonstrated expected expression of cell cycle and apoptosis proteins [49]. A hallmark of differentiation is cell cycle arrest and downregulation of cell cycle proteins during an early terminal differentiation stage. The cell cycle promoting proteins cyclin-dependent kinases (cdk) 2, 4, and 6 and E2F target genes were upregulated. However, E2F expression was not downregulated, suggesting that other transcription factors were downregulating E2F targets. Inhibitor of cell cycle p27kip1 was increased, a target of GCSF-mediated STAT3 and also under the control of C/EBPα (alpha) and C/EBPε (epsilon). Differential expression of apoptosis-related genes during terminal differentiation stages shows a difference in the mechanism of apoptosis at early and late stages, indicative of the cellular function. Early stages involve upregulation of p53-mediated apoptosis pathway, which surveys DNA damage, thus preventing proliferation of mutations. However, gene expression profile at the nonproliferative stage of differentiation showed an upregulation in ligand–receptor-mediated apoptosis pathway genes, with a concurrent reduction in p53-induced apoptotic genes. Thus, the profile suggests apoptosis in neutrophils is mediated upon activation of the neutrophil and in response to inflammatory cytokines.
Monocytopoiesis
Monocytes and macrophages are important effectors of the innate immune response and inflammation. Monopoiesis proceeds from the monoblast in the bone marrow to the circulating monocyte in the periphery, and eventually matures without proliferation to the tissue macrophage. However, monocyte development may be less linear than classically understood, and not necessarily a mere developmental intermediate between bone marrow precursors and tissue macrophages. Evidence suggests the possibility of a macrophage dendritic cell progenitor (MDP) and additionally shows that some subsets of both dendritic cells and tissue macrophages do not originate from monocytes in a steady state. Furthermore, monocytes may carry out specific effector functions during inflammation without further differentiation to macrophage or dendritic cell [50–53]. However, this dichotomy of thought between the classical hierarchy of monopoiesis and the novel existence of an MDP bone marrow progenitor has yet to be resolved.
GMCSF (or CSF2) is different from GCSF in that it acts on all granulocytes, monocytes, and macrophages. Because of this broad activity and the vast expression of the GMCSF receptor on hematopoietic cells, it was originally thought that the action of GMCSF was critical to the regulation and maintenance of the granulocyte and monocyte populations. However, deletion of neither the gene for GMCSF nor the GMCSF receptor had a significant impact on myelopoiesis but revealed an unexpected role for GMCSF in pulmonary homeostasis [54–56] 56. Additional evidence suggests GMCSF plays a vital role in stress or emergency myelopoiesis, with resultant increased production of granulocytes and monocytes in the bone marrow and stimulation of their survival and function in the tissues where they are recruited [57, 58].
Function of Neutrophils and Monocytes
Monocytes circulate in the bone marrow, blood, and spleen and are thought not to proliferate at steady state. In the setting of infection, monocytes are released from the bone marrow into the peripheral blood and migrate to sites of inflammation or injury where they mature to express distinct effector phenotypes [59]. A large portion of undifferentiated monocytes are also contained in the spleen, and serve as a storage reservoir for additional rapid deployment to sites of injury or infection [50]. Monocytes express chemokine receptors and pathogen associated pattern recognition receptors (e.g., toll-like receptors; TLRs) that mediate this process. Migration to tissues and further differentiation to inflammatory macrophages or dendritic cells is likely determined by the inflammatory milieu and the nature of the invading pathogen and TLR [59].
Macrophages are resident tissue phagocytes important for maintenance of tissue health via the clearance of apoptotic cells and other debris. Like their monocyte predecessors, macrophages also express a wide range of pattern recognition receptors that make them efficient effectors of the innate immune response in addition to their role in tissue homeostasis [60]. However, macrophages also play a vital role in initiating the adaptive immune response as antigen-presenting cells via MHC II. The developmental origin and more detailed function of tissue macrophage subsets remain poorly understood.
Neutrophils and monocytes released from the bone marrow can circulate for 24 h and 1–3 days respectively. The process of recruitment of neutrophils and monocytes involves recruitment by chemoattractants, IL-8, and bacterial proteins (N-formylmethionyl-leucyl-phenylalanine (fMLF), peptidoglycans). The delivery of neutrophils to the site starts off with “rolling” along the blood vessel walls and is mediated by low-affinity interactions between the selectin family of proteins [61, 62]. L-selectins are expressed on neutrophils which interact with transient and sequentially expressed P- and E-selectins on the inflammatory endothelial cells. Interaction between the selectins is followed by interaction of β2 (beta2)-integrins on neutrophils and intercellular adhesion molecule (ICAM) 1 and ICAM2 on endothelial cell wall (tethering). Integrin-binding affinity is increased in neutrophils upon activation by chemokines, which results in opening up of the integrin receptor conformation and of the ligand-binding pocket [63]. Increased affinity of the integrins brings the rolling of neutrophils to a stop, followed by transmigration across the vascular wall to the tissues. Microarray analysis of neutrophils exposed to fMLF showed an increase in pro-inflammatory molecules such as IL-8, TNF, IL1B, and both CXC and CC type chemokine [64]. Increased expression of pro-inflammatory cytokines contributes to delaying of apoptosis, which is essential for neutrophil function. In support, downregulation of apoptotic proteins was also observed. Additionally, in agreement with other studies, an upregulation of cytoskeletal reorganization proteins and adhesion-mediating molecules was observed [64, 65].
Priming of neutrophils is a process that activates the neutrophil and increases expression of proteins that are required for increased activity and also delay apoptosis. Priming agents include GCSF, GMCSF, IL-8, bacterial lipopolysaccharides (LPS) and TNF-α (alpha) [66–69]. Priming with GCSF enhances chemotaxis and mobilization of neutrophils to the site of injury, whereas GMCSF is involved in promoting a more robust response that is involved in both delaying apoptosis and increasing the bactericidal activity of neutrophils by promoting expression of antiapoptotic proteins and cell surface receptors involved in recognition of antigens [70]. A role in antigen presentation was also evident from the increased expression of major histocompatibility complex II (MHC II). Priming with LPS enhances the bactericidal activity, with recruitment of components necessary for the assembly of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex. In addition, priming with LPS also induced expression of proteins required for the NF-κB pathway [64, 71].
Phagocytosis is the penultimate step in neutrophil and macrophage function, wherein they ingest to get rid of the invading bacteria or apoptotic cells. The process of phagocytosis involves recognition of the bacteria or antigen via opsonization with antibodies or complement that is recognized by receptors on neutrophils. Opsonized bacteria are recognized by receptors against the Fc region of the antibody. Bacteria opsonized by complement bind are then able to bind to receptors like CD11b/CD18 on the neutrophil surface. Binding of opsonized bacteria to the activated neutrophil surface initiates changes in cytoskeleton and membrane to promote ingestion of the organism as a phagosome. Finally, the phagosome after sequential integration with the neutrophil granules turns into a phagolysosome and the invading organism is killed by exposure to products derived from ROS and antimicrobial granule proteins such as proteases, gelatinase, peroxidase, and other degradative enzymes.
Transcriptome analysis of phagocytosis identified expression of several hundred messenger RNAs (mRNAs) within 2 h of exposure [72, 73]. The changes in expression are divided into an early response and late response. Early response included cytokines and chemokines that act as pro-inflammatory molecules and aid in further recruitment of monocytes and neutrophils. Late-stage transcriptional changes involve upregulation of proapoptotic proteins of the receptor-mediated apoptotic pathway, such as TNFα, TNFR1, and tumor necrosis factor related apoptosis inducing ligand receptor (TRAILR). Other changes include proteins that are involved in the signal transduction pathway that involve TLRs. The downregulation of proteins that are involved in antibody- and complement-opsonized microbe recognition parallels the apoptotic expression.
ROS play a very important role in phagocytosis-mediated killing of bacteria by neutrophils. The production of ROS in neutrophils is mediated by an enzyme complex NADPH oxidase, which is composed of several components: oxidase specific (p22phox, p47phox, p67phox, and gp91phox) and guanosine triphosphate (GTPase; Rac1/2) [74]. Components of NADPH oxidase are present either in the cytoplasm or in either the plasma membrane or secretory vesicle membrane. Priming with GMCSF, TNFα, and LPS triggers phosphorylation of oxidase components and recruitment of the cytosolic components to the phagocyte membrane. Assembled NADPH oxidase mediates transfer of electrons from extracellular NADPH to oxygen within the phagosome, resulting in formation of a superoxide anion [75]. The superoxide anion dismutates to form hydrogen peroxide, which then oxidizes chloride anion to form hypochlorous acid. The reaction is catalyzed by myeloperoxidase, which resides in azurophilic granules and is released into the phagosome upon degranulation. Other products formed by hydrogen peroxide-mediated oxidation include hydroxyl radical. Together they have strong microbicidal activity.
Neutrophils contain highly toxic components used to kill microbes, but these molecules do not differentiate between host and pathogen. Neutrophils undergo apoptosis 24 h after they leave the bone marrow. The transcriptome analysis of neutrophils in the bone marrow and in peripheral blood show an upregulation of proapoptotic genes which indicates that neutrophils are destined to die as soon as they differentiate [43]. Priming of neutrophils delays the apoptotic response by upregulating antiapoptotic genes; however, upon phagocytosis, the transcriptional program now directs apoptosis of the neutrophils, termed as delayed apoptosis. The delayed apoptosis program is mediated by death receptors and is accompanied by a decreased inflammatory response. The decreased inflammatory response and mediation of apoptosis promotes resolution of the immune response and removal of the infection by macrophages. A downregulation of NF-κB expression is observed which halts the antiapoptotic response [76].
Contributor Information
Hrishikesh M. Mehta, Feinberg School of Medicine, Northwestern University, 303 E. Superior Street, Lurie 5-220, Chicago, IL 60611, USA
Taly Glaubach, Department of Pediatrics, Lurie Children’s Hospital of Chicago, Chicago, IL 60611, USA, Tel: 312-227-4000.
Seth Joel Corey, Departments of Pediatrics and Cell & Molecular Biology, Northwestern University Feinberg, School of Medicine and Lurie Children’s Hospital of Chicago, 303 E. Superior Street, Lurie 5-107, Chicago, IL 60611, USA.
References
- 1.Summers C, Rankin SM, Condliffe AM, Singh N, Peters AM, Chilvers ER. Neutrophil kinetics in health and disease. Trends Immunol. 2010;31:318–24. doi: 10.1016/j.it.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerst JM, de Haas M, van der Schoot CE, Slaper-Cortenbach IC, Kleijer M, von dem Borne AE, van Oers RH. Recombinant granulocyte colony-stimulating factor administration to healthy volunteers: induction of immunophenotypically and functionally altered neutrophils via an effect on myeloid progenitor cells. Blood. 1993;82:3265–72. [PubMed] [Google Scholar]
- 3.Ohls RK, Li Y, Abdel-Mageed A, Buchanan G, Jr, Mandell L, Christensen RD. Neutrophil pool sizes and granulocyte colony-stimulating factor production in human mid-trimester fetuses. Pediatric Res. 1995;37:806–11. doi: 10.1203/00006450-199506000-00022. [DOI] [PubMed] [Google Scholar]
- 4.Erdman SH, Christensen RD, Bradley PP, Rothstein G. Supply and release of storage neutrophils. A developmental study. Biol Neonate. 1982;41:132–7. doi: 10.1159/000241541. [DOI] [PubMed] [Google Scholar]
- 5.al-Mulla ZS, Christensen RD. Neutropenia in the neonate. Clin Perinatol. 1995;22:711–39. [PubMed] [Google Scholar]
- 6.Cairo MS, Christensen R, Sender LS, Ellis R, Rosenthal J, van de Ven C, Worcester C, Agosti JM. Results of a phase I/II trial of recombinant human granulocyte-macrophage colony-stimulating factor in very low birthweight neonates: significant induction of circulatory neutrophils, monocytes, platelets, and bone marrow neutrophils. Blood. 1995;86:2509–15. [PubMed] [Google Scholar]
- 7.Uguz A, Coskun M, Yuzbey S, Kizilors A, Karadogan I, Gura A, Yoldas B, Oygur N, Yegin O. Apoptosis of cord blood neutrophils and their response to colony-stimulating factors. Am J Perinatol. 2002;19:427–34. doi: 10.1055/s-2002-36838. [DOI] [PubMed] [Google Scholar]
- 8.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–8. [PMC free article] [PubMed] [Google Scholar]
- 9.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–7. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 10.Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBP[alpha] ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–36. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 11.Fukunaga R, Seto Y, Mizushima S, Nagata S. Three different mRNAs encoding human granulocyte colony-stimulating factor receptor. Proc Natl Acad Sci U S A. 1990;87:8702–6. doi: 10.1073/pnas.87.22.8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki H, Shimoda K, Okamura S, Otsuka T, Nagafuji K, Harada N, Ohno Y, Miyamoto T, Akashi K, Harada M, et al. Production of Soluble Granulocyte Colony-Stimulating Factor Receptors from Myelomonocytic Cells. J Immunol. 1999;163:6907–11. [PubMed] [Google Scholar]
- 13.Larsen A, Davis T, Curtis BM, Gimpel S, Sims JE, Cosman D, Park L, Sorensen E, March CJ, Smith CA. Expression cloning of a human granulocyte colony-stimulating factor receptor: a structural mosaic of hematopoietin receptor, immunoglobulin, and fibronectin domains. J Exp Med. 1990;172:1559–70. doi: 10.1084/jem.172.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard T, Gale RE, Linch DC. Analysis of granulocyte colony stimulating factor receptor isoforms, polymorphisms and mutations in normal haemopoietic cells and acute myeloid leukaemia blasts. Br J Haematol. 1996;93:527–33. doi: 10.1046/j.1365-2141.1996.d01-1696.x. [DOI] [PubMed] [Google Scholar]
- 15.White SM, Ball ED, Ehmann WC, Rao AS, Tweardy DJ. Increased expression of the differentiation-defective granulocyte colony-stimulating factor receptor mRNA isoform in acute myelogenous leukemia. Leukemia. 1998;12:899–906. doi: 10.1038/sj.leu.2401062. [DOI] [PubMed] [Google Scholar]
- 16.Mehta HM, Futami M, Glaubach T, Lee DW, Andolina JR, Yang Q, Whichard Z, Quinn M, Lu HF, Kao WM, et al. Alternatively spliced, truncated gcsf receptor promotes leukemogenic properties and sensitivity to jak inhibition. Leukemia. 2014;28:1041–51. doi: 10.1038/leu.2013.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White SM, Alarcon MH, Tweardy DJ. Inhibition of granulocyte colony-stimulating factor-mediated myeloid maturation by low level expression of the differentiation-defective class IV granulocyte colony-stimulating factor receptor isoform. Blood. 2000;95:3335–40. [PubMed] [Google Scholar]
- 18.Dong F, Brynes RK, Tidow N, Welte K, Lowenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333:487–93. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 19.Dong F, Hoefsloot LH, Schelen AM, Broeders CA, Meijer Y, Veerman AJ, Touw IP, Lowenberg B. Identification of a nonsense mutation in the granulocyte-colony-stimulating factor receptor in severe congenital neutropenia. Proc Natl Acad Sci U S A. 1994;91:4480–4. doi: 10.1073/pnas.91.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong F, van Paassen M, van Buitenen C, Hoefsloot LH, Lowenberg B, Touw IP. A point mutation in the granulocyte colony-stimulating factor receptor (G-CSF-R) gene in a case of acute myeloid leukemia results in the overexpression of a novel G-CSF-R isoform. Blood. 1995;85:902–11. [PubMed] [Google Scholar]
- 21.Nicholson SE, Oates AC, Harpur AG, Ziemiecki A, Wilks AF, Layton JE. Tyrosine kinase JAK1 is associated with the granulocyte-colony-stimulating factor receptor and both become tyrosine-phosphorylated after receptor activation. Proc Natl Acad Sci U S A. 1994;91:2985–8. doi: 10.1073/pnas.91.8.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimoda K, Iwasaki H, Okamura S, Ohno Y, Kubota A, Arima F, Otsuka T, Niho Y. G-CSF induces tyrosine phosphorylation of the JAK2 protein in the human myeloid G-CSF responsive and proliferative cells, but not in mature neutrophils. Biochem Biophysical Res Commun. 1994;203:922–8. doi: 10.1006/bbrc.1994.2270. [DOI] [PubMed] [Google Scholar]
- 23.Tian SS, Tapley P, Sincich C, Stein RB, Rosen J, Lamb P. Multiple signaling pathways induced by granulocyte colony-stimulating factor involving activation of JAKs, STAT5, and/or STAT3 are required for regulation of three distinct classes of immediate early genes. Blood. 1996;88:4435–44. [PubMed] [Google Scholar]
- 24.Corey SJ, Burkhardt AL, Bolen JB, Geahlen RL, Tkatch LS, Tweardy DJ. Granulocyte colony-stimulating factor receptor signaling involves the formation of a three-component complex with Lyn and Syk protein-tyrosine kinases. Proc Natl Acad Sci U S A. 1994;91:4683–7. doi: 10.1073/pnas.91.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corey SJ, Dombrosky-Ferlan PM, Zuo S, Krohn E, Donnenberg AD, Zorich P, Romero G, Takata M, Kurosaki T. Requirement of Src kinase Lyn for induction of DNA synthesis by granulocyte colony-stimulating factor. J Biol Chem. 1998;273:3230–5. doi: 10.1074/jbc.273.6.3230. [DOI] [PubMed] [Google Scholar]
- 26.Futami M, Zhu QS, Whichard ZL, Xia L, Ke Y, Neel BG, Feng GS, Corey SJ. G-CSF receptor activation of the Src kinase Lyn is mediated by Gab2 recruitment of the Shp2 phosphatase. Blood. 2011;118:1077–86. doi: 10.1182/blood-2009-12-261636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu QS, Robinson LJ, Roginskaya V, Corey SJ. G-CSF-induced tyrosine phosphorylation of Gab2 is Lyn kinase dependent and associated with enhanced Akt and differentiative, not proliferative, responses. Blood. 2004;103:3305–12. doi: 10.1182/blood-2003-06-1861. [DOI] [PubMed] [Google Scholar]
- 28.de Koning JP, Soede-Bobok AA, Schelen AM, Smith L, van Leeuwen D, Santini V, Burgering BM, Bos JL, Lowenberg B, Touw IP. Proliferation signaling and activation of Shc, p21Ras, and Myc via tyrosine 764 of human granulocyte colony-stimulating factor receptor. Blood. 1998;91:1924–33. [PubMed] [Google Scholar]
- 29.Wang L, Xue J, Zadorozny EV, Robinson LJ. G-CSF stimulates Jak2-dependent Gab2 phosphorylation leading to Erk1/2 activation and cell proliferation. Cell Signal. 2008;20:1890–9. doi: 10.1016/j.cellsig.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfler A, Irandoust M, Meenhuis A, Gits J, Roovers O, Touw IP. Site-specific ubiquitination determines lysosomal sorting and signal attenuation of the granulocyte colony-stimulating factor receptor. Traffic. 2009;10:1168–79. doi: 10.1111/j.1600-0854.2009.00928.x. [DOI] [PubMed] [Google Scholar]
- 31.de Bruin AM, Libregts SF, Valkhof M, Boon L, Touw IP, Nolte MA. IFNgamma induces monopoiesis and inhibits neutrophil development during inflammation. Blood. 2012;119:1543–54. doi: 10.1182/blood-2011-07-367706. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Friedman AD. SHP2 tyrosine phosphatase stimulates CEBPA gene expression to mediate cytokine-dependent granulopoiesis. Blood. 2011;118:2266–74. doi: 10.1182/blood-2011-01-331157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 34.Guo H, Ma O, Speck NA, Friedman AD. Runx1 deletion or dominant inhibition reduces Cebpa transcription via conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis. Blood. 2012;119:4408–18. doi: 10.1182/blood-2011-12-397091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orkin SH, Shivdasani RA, Fujiwara Y, McDevitt MA. Transcription factor GATA–1 in megakaryocyte development. Stem Cells. 1998;2(16 Suppl):79–83. doi: 10.1002/stem.5530160710. [DOI] [PubMed] [Google Scholar]
- 36.Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, et al. Reciprocal activation of GATA–1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–27. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU.1 inhibits GATA–1 function and erythroid differentiation by blocking GATA–1 DNA binding. Blood. 2000;96:2641–8. [PubMed] [Google Scholar]
- 38.Traver D, Miyamoto T, Christensen J, Iwasaki-Arai J, Akashi K, Weissman IL. Fetal liver myelopoiesis occurs through distinct, prospectively isolatable progenitor subsets. Blood. 2001;98:627–35. doi: 10.1182/blood.v98.3.627. [DOI] [PubMed] [Google Scholar]
- 39.Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–66. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 40.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh HA. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–86. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akashi K, He X, Chen J, Iwasaki H, Niu C, Steenhard B, Zhang J, Haug J, Li L. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–89. doi: 10.1182/blood-2002-06-1780. [DOI] [PubMed] [Google Scholar]
- 42.Borregaard N, Cowland JB. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 1997;89:3503–21. [PubMed] [Google Scholar]
- 43.Theilgaard-Monch K, Jacobsen LC, Borup R, Rasmussen T, Bjerregaard MD, Nielsen FC, Cowland JB, Borregaard N. The transcriptional program of terminal granulocytic differentiation. Blood. 2005;105:1785–96. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- 44.Lominadze G, Powell DW, Luerman GC, Link AJ, Ward RA, McLeish KR. Proteomic analysis of human neutrophil granules. Mol Cell Proteomics. 2005;4:1503–21. doi: 10.1074/mcp.M500143-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Jethwaney D, Islam MR, Leidal KG, de Bernabe DB, Campbell KP, Nauseef WM, Gibson BW. Proteomic analysis of plasma membrane and secretory vesicles from human neutrophils. Proteome Sci. 2007;5:12. doi: 10.1186/1477-5956-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uriarte SM, Powell DW, Luerman GC, Merchant ML, Cummins TD, Jog NR, Ward RA, McLeish KR. Comparison of proteins expressed on secretory vesicle membranes and plasma membranes of human neutrophils. J Immunol. 2008;180:5575–81. doi: 10.4049/jimmunol.180.8.5575. [DOI] [PubMed] [Google Scholar]
- 47.Feuk-Lagerstedt E, Movitz C, Pellme S, Dahlgren C, Karlsson A. Lipid raft proteome of the human neutrophil azurophil granule. Proteomics. 2007;7:194–205. doi: 10.1002/pmic.200600482. [DOI] [PubMed] [Google Scholar]
- 48.Nebl T, Pestonjamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ. Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J Biol Chem. 2002;277:43399–409. doi: 10.1074/jbc.M205386200. [DOI] [PubMed] [Google Scholar]
- 49.Rosmarin AG, Yang Z, Resendes KK. Transcriptional regulation in myelopoiesis: hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp Hematol. 2005;33:131–43. doi: 10.1016/j.exphem.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 50.Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, Figueiredo JL, Kohler RH, Chudnovskiy A, Waterman P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–16. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–92. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 52.Yona S, Jung S. Monocytes: subsets, origins, fates and functions. Curr Opin Hematol. 2010;17:53–9. doi: 10.1097/MOH.0b013e3283324f80. [DOI] [PubMed] [Google Scholar]
- 53.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nature reviews. Immunology. 2005;5:953–64. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 54.Dranoff G, Mulligan RC. Activities of granulocyte-macrophage colony-stimulating factor revealed by gene transfer and gene knockout studies. Stem Cells. 1994;1(12 Suppl):173–82. (discussion 182-174) [PubMed] [Google Scholar]
- 55.Lieschke GJ, Stanley E, Grail D, Hodgson G, Sinickas V, Gall JA, Sinclair RA, Dunn AR. Mice lacking both macrophage- and granulocyte-macrophage colony-stimulating factor have macrophages and coexistent osteopetrosis and severe lung disease. Blood. 1994;84:27–35. [PubMed] [Google Scholar]
- 56.Stanley E, Lieschke GJ, Grail D, Metcalf D, Hodgson G, Gall JA, Maher DW, Cebon J, Sinickas V, Dunn AR. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc Natl Acad Sci U S A. 1994;91:5592–96. doi: 10.1073/pnas.91.12.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhan Y, Lieschke GJ, Grail D, Dunn AR, Cheers C. Essential roles for granulocyte-macrophage colony-stimulating factor (GM-CSF) and G-CSF in the sustained hematopoietic response of Listeria monocytogenes-infected mice. Blood. 1998;91:863–9. [PubMed] [Google Scholar]
- 58.Nakata K, Akagawa KS, Fukayama M, Hayashi Y, Kadokura M, Tokunaga T. Granulocyte-macrophage colony-stimulating factor promotes the proliferation of human alveolar macrophages in vitro. J Immunol. 1991;147:1266–72. [PubMed] [Google Scholar]
- 59.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. 2008;26:421–52. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. 2002;111:927–30. doi: 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 61.Lawrence MB, Springer TA. Leukocytes roll on a selectin at physiologic flow rates: distinction from and prerequisite for adhesion through integrins. Cell. 1991;65:859–73. doi: 10.1016/0092-8674(91)90393-d. [DOI] [PubMed] [Google Scholar]
- 62.Lawrence MB, Springer TA. Neutrophils roll on E-selectin. J Immunol. 1993;151:6338–46. [PubMed] [Google Scholar]
- 63.Chigaev A, Zwartz G, Graves SW, Dwyer DC, Tsuji H, Foutz TD, Edwards BS, Prossnitz ER, Larson RS, Sklar LA. Alpha4beta1 integrin affinity changes govern cell adhesion. J Biol Chem. 2003;278:38174–82. doi: 10.1074/jbc.M210472200. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Kluger Y, Nakayama Y, Poddar R, Whitney C, DeTora A, Weissman SM, Newburger PE. Gene expression in mature neutrophils: early responses to inflammatory stimuli. J Leukoc Biol. 2004;75:358–72. doi: 10.1189/jlb.0903412. [DOI] [PubMed] [Google Scholar]
- 65.Boldt K, Rist W, Weiss SM, Weith A, Lenter MC. FPRL–1 induces modifications of migration-associated proteins in human neutrophils. Proteomics. 2006;6:4790–99. doi: 10.1002/pmic.200600121. [DOI] [PubMed] [Google Scholar]
- 66.DeLeo FR, Renee J, McCormick S, Nakamura M, Apicella M, Weiss JP, Nauseef WM. Neutrophils exposed to bacterial lipopolysaccharide upregulate NADPH oxidase assembly. J Clin Invest. 1998;101:455–63. doi: 10.1172/JCI949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guichard C, Pedruzzi E, Dewas C, Fay M, Pouzet C, Bens M, Vandewalle A, Ogier-Denis E, Gougerot-Pocidalo MA, Elbim C. Interleukin–8-induced priming of neutrophil oxidative burst requires sequential recruitment of NADPH oxidase components into lipid rafts. J Biol Chem. 2005;280:37021–32. doi: 10.1074/jbc.M506594200. [DOI] [PubMed] [Google Scholar]
- 68.Dang PM, Dewas C, Gaudry M, Fay M, Pedruzzi E, Gougerot-Pocidalo MA, El Benna J. Priming of human neutrophil respiratory burst by granulocyte/macrophage colony-stimulating factor (GM-CSF) involves partial phosphorylation of p47(phox) J Biol Chem. 1999;274:20704–8. doi: 10.1074/jbc.274.29.20704. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki S, Kobayashi M, Chiba K, Horiuchi I, Wang J, Kondoh T, Hashino S, Tanaka J, Hosokawa M, Asaka M. Autocrine production of epithelial cell-derived neutrophil attractant–78 induced by granulocyte colony-stimulating factor in neutrophils. Blood. 2002;99:1863–5. [PubMed] [Google Scholar]
- 70.Kobayashi SD, Voyich JM, Whitney AR, DeLeo FR. Spontaneous neutrophil apoptosis and regulation of cell survival by granulocyte macrophage-colony stimulating factor. J Leukoc Biol. 2005;78:1408–18. doi: 10.1189/jlb.0605289. [DOI] [PubMed] [Google Scholar]
- 71.Tsukahara Y, Lian Z, Zhang X, Whitney C, Kluger Y, Tuck D, Yamaga S, Nakayama Y, Weissman SM, Newburger PE. Gene expression in human neutrophils during activation and priming by bacterial lipopolysaccharide. J Cell Biochem. 2003;89:848–61. doi: 10.1002/jcb.10526. [DOI] [PubMed] [Google Scholar]
- 72.Subrahmanyam YV, Yamaga S, Prashar Y, Lee HH, Hoe NP, Kluger Y, Gerstein M, Goguen JD, Newburger PE, Weissman SM. RNA expression patterns change dramatically in human neutrophils exposed to bacteria. Blood. 2001;97:2457–68. doi: 10.1182/blood.v97.8.2457. [DOI] [PubMed] [Google Scholar]
- 73.Kobayashi SD, Voyich JM, Buhl CL, Stahl RM, DeLeo FR. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proc Natl Acad Sci U S A. 2002;99:6901–6. doi: 10.1073/pnas.092148299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quinn MT, Gauss KA. Structure and regulation of the neutrophil respiratory burst oxidase: comparison with nonphagocyte oxidases. J Leukoc Biol. 2004;76:760–81. doi: 10.1189/jlb.0404216. [DOI] [PubMed] [Google Scholar]
- 75.Babior BM. NADPH oxidase. Curr Opin Immunol. 2004;16:42–7. doi: 10.1016/j.coi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 76.Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM, DeLeo FR. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci U S A. 2003;100:10948–53. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]