Abstract
Biosensors are becoming increasingly important and implemented in various fields such as pathogen detection, molecular diagnosis, environmental monitoring, and food safety control. In this context, we used β-lactamases as efficient reporter enzymes in several protein-protein interaction studies. Furthermore, their ability to accept insertions of peptides or structured proteins/domains strongly encourages the use of these enzymes to generate chimeric proteins. In a recent study, we inserted a single-domain antibody fragment into the Bacillus licheniformis BlaP β-lactamase. These small domains, also called nanobodies, are defined as the antigen-binding domains of single chain antibodies from camelids. Like common double chain antibodies, they show high affinities and specificities for their targets. The resulting chimeric protein exhibited a high affinity against its target while retaining the β-lactamase activity. This suggests that the nanobody and β-lactamase moieties remain functional. In the present work, we report a detailed protocol that combines our hybrid β-lactamase system to the biosensor technology. The specific binding of the nanobody to its target can be detected thanks to a conductimetric measurement of the protons released by the catalytic activity of the enzyme.
Keywords: Bioengineering, Issue 132, β-lactamase, hybrid protein technology (BHP), conductimetric biosensor, nanobodies, molecular interactions, lysozyme
Introduction
Biosensors are analytical devices that combine a bio-molecular interaction with physical or chemical signaling devices referred to as transducers1. The recorded signals can then be interpreted and converted to monitor the interactions between the immobilized and free partners. Most of the biosensors involve the use of an antibody to detect analytes such as hormones or different pathogen markers2. Different sensor formats can be used and include mass-based, magnetic, optical or electrochemical biosensors. The latter are among the most commonly used sensors, and function by converting a binding event into an electrical signal. The performances and sensitivities of all antibody-based biosensors are strongly dependent on essentially two parameters: i) the quality of the antibody and ii) the properties of the system used to generate the signal2.
Antibodies are high-molecular mass dimeric proteins (150–160 kDa) that are composed of two heavy chains and two light chains. The interaction between the light and heavy chains is mostly stabilized by hydrophobic interactions as well as a conserved disulfide bond. Each chain includes a variable domain that interacts with the antigen essentially via three hypervariable regions named Complementary Determining Regions (CDR1-2-3). Despite numerous advances in the field, the large-scale expression of full-length antibodies with low-cost expression systems (e.g., E. coli) often leads to the production of unstable and aggregated proteins. This is why various antibody fragments have been engineered such as single-chain variable fragments3 (ScFvs ≈ 25 kDa). They consist of the variable domains of respectively one heavy and one light chains that are covalently linked by a synthetic amino acid sequence. However, these fragments often display a poor stability and have the tendency to aggregate, since they expose a large portion of their hydrophobic regions to the solvent4. In this context, single chain camelid antibody fragments, referred to as nanobodies or VHHs, seem to be excellent alternatives to ScFvs. These domains correspond to the variable domains of camelid single-chain antibodies. In contrast to conventional antibodies, camelid antibodies are devoid of light chains and only contain two heavy chains5. Therefore, nanobodies are the smallest monomeric antibody fragments (12 kDa) able to bind to an antigen with an affinity similar to that of conventional antibodies6. In addition, they present improved stability and solubility compared to other full-length antibodies or antibody fragments. Finally, their small sizes and their extended CDR3 loops allow them to recognize cryptic epitopes and bind to enzyme active sites7,8. Nowadays, these domains are receiving considerable attention and have been combined to the biosensor technology. For example, Huang et al. have developed a nanobody-based biosensor for the detection and quantification of human prostate-specific antigen (PSA)9.
As mentioned-above, an important parameter in biosensor assays is the efficiency of the system used to generate the electric signal. For this reason, enzyme-based electrochemical biosensors have attracted ever-increasing attention and have been used widely for various applications such as health care, food safety, and environmental monitoring. These biosensors rely on the catalytic hydrolysis of a substrate by an enzyme to generate the electrical signal. In this context, β-lactamases were shown to be more specific, more sensitive and easier to implement experimentally than many other enzymes such as alkaline phosphatase or horseradish peroxidase10. β-lactamases are enzymes that are responsible for bacterial resistance to β-lactam antibiotics by hydrolyzing them. They are monomeric, very stable, efficient, and of small size. Moreover, domain/peptide insertions into β-lactamases generate bi-functional chimeric proteins that were shown to be efficient tools to study protein-ligand interactions. Indeed, recent studies have shown that insertion of antibody variable fragments into the TEM1 β-lactamase results in a chimeric protein that remains able to bind with high affinity to its target antigen. Interestingly, the antigen binding was shown to induce allosteric regulation of TEM1 catalytic activity11,12. Furthermore, we showed in several studies that protein domain insertion into a permissive loop of the Bacillus licheniformis BlaP β-lactamase generates functional chimeric proteins that are well suited to monitor protein-ligand interactions13,14. We recently inserted a nanobody, named cAb-Lys3, into this permissive insertion site of BlaP15. This nanobody was shown to bind to hen-egg-white lysozyme (HEWL) and to inhibit its enzymatic activity16. We showed that the generated hybrid protein, named BlaP-cAb-Lys3, retained a high specificity / affinity against HEWL while the β-lactamase activity remained unchanged. Then we successfully combined the hybrid β-lactamase technology to an electrochemical biosensor and showed that the amount of generated electric signal was dependent of the interaction between BlaP-cAb-Lys3 and HEWL immobilized on an electrode. Indeed, hydrolysis of β-lactam antibiotics by BlaP induces a proton release that can be converted into a quantitative electric signal. This combination of the hybrid β-lactamase technology with an electrochemical biosensor is fast, sensitive, quantitative, and allows real-time measurement of the generated signal. This methodology is described herein.
Protocol
1. Protein Sample Preparation
Produce and purify the hybrid protein BlaP-cAb-Lys3 as reported in our previous study15. Store the protein in 50 mM phosphate buffer pH 7.4 with the following composition: 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4 and 0.24 g of KH2PO4 dissolved in 800 mL of distilled water Fix the pH of the solution to 7.4 before adjusting the final volume of the solution to 1 L. Filter sterilize the protein solution.
Prepare a hen egg white lysozyme (HEWL) stock solution. Dissolve 100 mg (40,000 units/mg) of commercially purchased HEWL in 10 mL of phosphate buffer saline (PBS see step 2.1.1). Sterilize the protein solution by filtration using filters with a 0.22 μm cutoff.
2. Biosensor Assays
- Solution and buffer preparation
- Prepare 50 mM PBS by dissolving 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 in 800 mL of distilled water. Adjust to pH 7.4 with 1 N HCl or 1 N NaOH before adjusting the volume of the solution to 1 L. Filter sterilize and store at 4 °C.
- Prepare a saturation/blocking solution by dissolving 3 g of casein hydrolysate in 100 mL of PBS prepared as described above (see step 2.1.1). Filter sterilize and store at 4 °C.
- Prepare a binding solution by dissolving 1 g of casein hydrolysate in 100 mL of PBS prepared as described above (see step 2.1.1). Filter sterilize and store at 4 °C.
- Prepare a washing solution (0.1% Tween - PBS) by adding 100 μL of Tween 20 (100%) in 100 mL of PBS prepared as described above (see step 2.1.1). Store at 4 °C.
- Prepare an electrode preparation solution (1% Triton X-100 - PBS) by adding 1 mL of Triton X-100 (100%) in 100 mL of PBS prepared as described above (see step 2.1.1). Store at 4 °C.
- Prepare an electrode regeneration solution (3.5 M KCl) by dissolving 26 g of KCl in distilled water to a final volume 100 mL. Filter sterilized and store at 4 °C.
- Prepare a 5 mM NaCl solution by dissolving 0.29 g of NaCl in distilled water to a final volume of 1 L. Filter sterilize and store at 4 °C. Then, prepare a detection solution (4 mM benzylpenicillin) by dissolving 26.7 mg of benzylpenicillin in 20 mL of 5 mM NaCl solution. Filter sterilize and store at -20 °C.
- Sensor preparation and regeneration Note: The polyaniline coated sensor chips were developed and kindly provided by Dr. P. Bogaerts, Dr. S. Yunus and Prof. Y. Glupczynski’s (Catholic University of Louvain-la-Neuve - CHU Mont-Godinne). The description of the sensor as well as the polyaniline electro-polymerization protocols used to synthesize these sensors are detailed in their previous work17. Briefly, this system uses re-usable sensors of eight individual chips that were manufactured by classical printed circuit board (PCB) techniques. Individual chips are composed of three electrode round spots. The top one is the working electrode on which polyaniline was electro-synthesized. The middle one is the reference electrode and the bottom electrode constitutes the counter electrode. Both, the reference and the counter electrodes are functionalized using solid Ag/AgCl amalgam on top of the carbon layer.
- Perform 3 washes of the electrodes by dipping the tips into wells of a 96-well plate containing 300 µL/well of electrode preparation solution (1% Triton X-100 - PBS, see step 2.1.5.). Perform each wash for 2 min with gentle mixing at room temperature.
- Rinse the electrodes by dipping the tips into wells of a 96-well plate containing 300 µL/well of distilled water for 2 min with gentle mixing at room temperature.
- Regenerate the electrodes by dipping the tips into wells of a 96-well plate containing 300 µL/well of regeneration solution (3.5 M KCl, see step 2.1.6) overnight at 4 °C or 1 h at room temperature.
- Perform 3 washes of the electrodes by dipping the tips into wells of a 96-well plate containing 300 µL/well of phosphate buffer saline (see step 2.1.1.). Perform each wash for 2 minutes with gentle mixing at room temperature.
- Binding assay performed on the sensor
- Coat HEWL onto the PANI (polyaniline) surface of the electrode by depositing a 15 µL drop of 40 µg/mL HEWL prepared in PBS onto the electrode surface. Incubate overnight at 4 °C or 1 hour at room temperature.
- Perform three washes of the electrodes with phosphate buffer saline (see step 2.1.1) by dipping the electrode parts of the sensor chips into wells of a 96-well plate containing 300 µL/well of phosphate buffer saline. Perform each wash for 2 min with gentle mixing at room temperature.
- Saturate the electrodes by adding a 50 µL drop of the blocking solution (see step 2.1.2) onto the electrode surface. Incubate for 1 h at room temperature. Then wash three times as described in the previous step (see step 2.3.2).
- Dilute the BlaP-cAb-Lys3 solution to 20 µg/mL in binding solution (see step 2.1.3) and apply a 15 µL drop of this diluted solution onto the electrodes. Incubate for 10 min at room temperature. After antigen-nanobody reaction, wash three times as described in the previous step using the wash solution (see step 2.1.4). Then rinse the electrode once with PBS (see step 2.1.1).
- For detection, plug the sensor chip via the copper-circuitry part to a digital multimeter. Then, initiate the sensor response by applying a 50 µL drop of detection solution (see step 2.1.7) onto the positive electrodes and applying a 50 µL drop of NaCl 5 mM solution onto the negative electrodes (see step 2.1.7). Incubate for 30 min at room temperature. Monitor the conductance with a digital multimeter. Note: The multimeter was provided by Dr. P. Bogaerts, Dr. S. Yunus and Prof. Y. Glupczynski’s (Catholic University of Louvain-la-Neuve - CHU Mont-Godinne. This potentiostat is computer-controlled via a USB port and analyzes the eight different chips of the sensor simultaneously. The software created by Yunus and colleagues17 creates a real-time plot that represents the measurements of the conductance difference between the reference and sample electrodes against time.
Representative Results
Design and engineering of the chimeric protein BlaP-cAb-Lys3
Figure 1 represents the insertion of cAb-Lys3 into a permissive loop of the BalP class A β-lactamase from Bacillus licheniformis. The insertion was performed between residues Asp198 and Lys199. A thrombin cleavage site was introduced on each side of cAb-Lys3. Cells transformed with a constitutive expression plasmid encoding the BlaP-cAb-Lys3 chimeric protein were able to grow in the presence of a small concentration of ampicillin. This result indicates that the hybrid β-lactamase is soluble, correctly folded and can be successfully excreted into the periplasmic space of the bacteria where it can efficiently provide resistance against ampicillin. We further analyzed the bifunctionality of our chimeric protein. As reported in our previous studies, our data showed that the β-lactamase as well as the cAb-Lys3 moieties of the chimeric protein retained their biological activities15.
Conductimetric biosensor assay
The sensor chips used for this assay is shown in Figure 2. The sensors used for these experiments contain 8 chips. Each chip includes three electrodes: one working electrode, one counter-electrode, and one reference electrode. These 8 chips are organized as 4 pairs in the sensor. For each pair, one of the chips labeled “-“ corresponds to a negative control, whereas the chip labeled “+” corresponds to the tested sample.
Figure 3 represents the schematic description of the experimental setup used in this work that combines the hybrid β-lactamase technology to a potentiometric biosensor. In this setup, two electrodes are used: i) a reference electrode and ii) a polyaniline (PANI) coated electrode. HEWL is adsorbed on the PANI coated electrode as reported in other studies18,19. After adequate washes, BlaP-cAb-Lys3 (for “+” labeled chips) or BlaP without inserted nanobody (for “-“ labeled chips) are applied on the electrodes. Following the addition of β-lactam (benzylpenicillin) on the electrodes, changes in electrode conductance are measured. Indeed, β-lactam hydrolysis by β-lactamases generates a release of protons; which was shown to create significant changes in electrode conductance with a very short response time. The biosensor assays presented in Figure 4 indicate that the binding of BlaP-cAb-lys3 to HEWL immobilized on the PANI electrode can be detected and monitored by measuring the proton release resulting from the immobilized β-lactamase activity. In contrast, no conductance difference was detected when the experiment was performed with BlaP without any inserted nanobody.
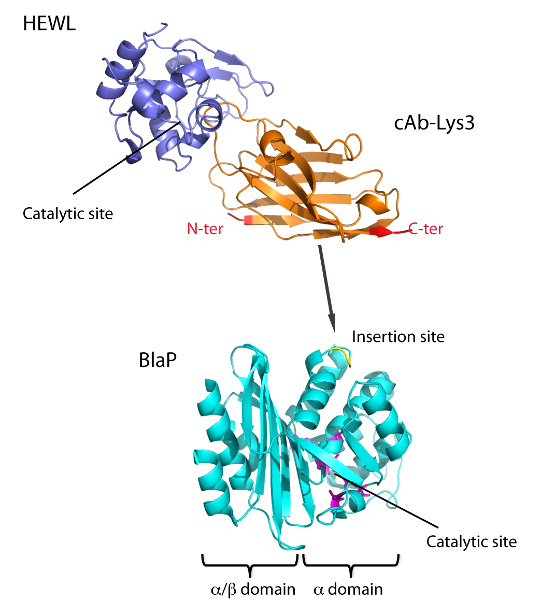 Figure 1: Scheme representing the chimeric protein BlaP-cAb-Lys3 interacting with HEWL. BlaP is shown in cyan, cAb-Lys3 in orange and HEWL in blue. This figure was obtained by combining the tridimensional structures of BlaP (PDB ID: 4BLM) and the cAb-Lys3/HEWL complex (PDB ID: 1MEL). The β-lactamase contains 2 domains: the α/β domain and the α domain, the catalytic site is located at the interface between both domains. The loop used for the insertion is highlighted in yellow and located in a solvent-exposed loop in the α domain. The N- and C-terminal parts of cAb-Lys3 are shown in red. The CDR3 loop of cAb-Lys3 makes most of the contacts with the HEWL catalytic site. The binding of cAB-Lys3 to HEWL inhibits its enzymatic activity. This figure and the results presented herein have been published in our previous work15. Please click here to view a larger version of this figure.
Figure 1: Scheme representing the chimeric protein BlaP-cAb-Lys3 interacting with HEWL. BlaP is shown in cyan, cAb-Lys3 in orange and HEWL in blue. This figure was obtained by combining the tridimensional structures of BlaP (PDB ID: 4BLM) and the cAb-Lys3/HEWL complex (PDB ID: 1MEL). The β-lactamase contains 2 domains: the α/β domain and the α domain, the catalytic site is located at the interface between both domains. The loop used for the insertion is highlighted in yellow and located in a solvent-exposed loop in the α domain. The N- and C-terminal parts of cAb-Lys3 are shown in red. The CDR3 loop of cAb-Lys3 makes most of the contacts with the HEWL catalytic site. The binding of cAB-Lys3 to HEWL inhibits its enzymatic activity. This figure and the results presented herein have been published in our previous work15. Please click here to view a larger version of this figure.
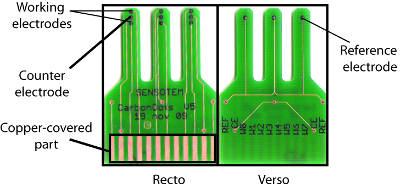 Figure 2: Picture of the sensor used in the experiment. Each sensor includes a group of 8 individual chips. On each chip, there are three electrodes: one working electrode, a reference electrode, and a counter ion electrode. The copper-covered part is plugged to a digital multimeter connected to a computer. The individual chips are organized as 4 pairs where the “-“ labeled chips are negative control electrodes and the “+” labeled chips are the sample electrodes. This setup allows different experimental replicates or HEWL / β-lactamase concentrations to be tested simultaneously. Please click here to view a larger version of this figure.
Figure 2: Picture of the sensor used in the experiment. Each sensor includes a group of 8 individual chips. On each chip, there are three electrodes: one working electrode, a reference electrode, and a counter ion electrode. The copper-covered part is plugged to a digital multimeter connected to a computer. The individual chips are organized as 4 pairs where the “-“ labeled chips are negative control electrodes and the “+” labeled chips are the sample electrodes. This setup allows different experimental replicates or HEWL / β-lactamase concentrations to be tested simultaneously. Please click here to view a larger version of this figure.
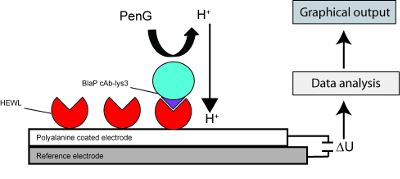 Figure 3: Schematic representation of the experimental setup used in our biosensor assay. HEWL was immobilized onto a PANI (polyaniline) coated electrode. The chimeric protein BlaP-cAb-Lys3 was then applied onto the electrode. The immobilized β-lactamase activity, which is determined by measuring the release of protons induced by the hydrolysis of benzylpenicillin, is directly proportional to the interaction between HEWL and the chimeric protein. The proton release induces electrical conductance changes that are converted to a signal that can be interpreted by the user. Evolution of the conductance difference observed between the PANI coated and the reference electrode as a function of time. This figure and the results presented herein have been published in our previous work15. Please click here to view a larger version of this figure.
Figure 3: Schematic representation of the experimental setup used in our biosensor assay. HEWL was immobilized onto a PANI (polyaniline) coated electrode. The chimeric protein BlaP-cAb-Lys3 was then applied onto the electrode. The immobilized β-lactamase activity, which is determined by measuring the release of protons induced by the hydrolysis of benzylpenicillin, is directly proportional to the interaction between HEWL and the chimeric protein. The proton release induces electrical conductance changes that are converted to a signal that can be interpreted by the user. Evolution of the conductance difference observed between the PANI coated and the reference electrode as a function of time. This figure and the results presented herein have been published in our previous work15. Please click here to view a larger version of this figure.
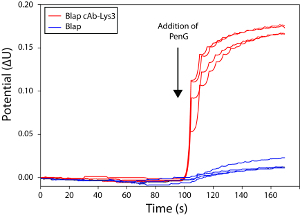 Figure 4: Graphical representation of the conductimetricmeasurements showing the specific interaction of BlaP-cAb-Lys3 to HEWL. The addition of benzylpenicillin is indicated by an arrow. This potential difference results from the proton release occurring upon antibiotic hydrolysis. The measurements were performed with the hybrid protein BlaP-cAb-Lys3 (red) and the native β-lactamase BlaP without any inserted nanobody (blue), as a negative control. The different curves represent independent measurements performed on different chips. This figure and the results presented herein have been published in our previous work15. Please click here to view a larger version of this figure.
Figure 4: Graphical representation of the conductimetricmeasurements showing the specific interaction of BlaP-cAb-Lys3 to HEWL. The addition of benzylpenicillin is indicated by an arrow. This potential difference results from the proton release occurring upon antibiotic hydrolysis. The measurements were performed with the hybrid protein BlaP-cAb-Lys3 (red) and the native β-lactamase BlaP without any inserted nanobody (blue), as a negative control. The different curves represent independent measurements performed on different chips. This figure and the results presented herein have been published in our previous work15. Please click here to view a larger version of this figure.
Discussion
In this work we present a method to functionalize a nanobody using the BlaP β-lactamase as a carrier protein and we show that we can successfully implement the resulting hybrid protein in a potentiometric sensor assay. The main innovation aspect of our work compared to other biosensor assays is the covalent coupling of the antibody part to the enzymatic activity that generates the electrical signal. This so-called protein insertion technology presents advantages and limitations that will be the main focus of this section.
Advantages of the hybrid β-lactamase technology.
β-lactamases are efficient enzymes One of the most important parameters that influence the sensitivity and signal-to-noise ratio of a biosensor is the enzymatic activity used to generate the signal. In this context, β-lactamases present several advantages: they are small (≈29 kDa), monomeric, very stable, and more importantly, exhibit high specific activity and high turnover compared to other enzymes used in potentiometric sensor assays such as glucose oxidase, urease, lipase, peroxydase or alkaline phosphatase20. For all these reasons, β-lactamases are enzymes of choice for various biosensor assays.
Direct Insertion into the β-lactamase can improve the production yield and stability of the inserted protein/domain. One of the limiting aspects of many immunosensor assays is the quality (e.g. stability, purity) of the antibody used to detect the analyte2. Currently, low cost production systems of antibodies or antibody fragments (e.g. E. coli) remain challenging and often lead to aggregated proteins with poor solubility and stability21. Our hybrid β-lactamase technology seems to be a good approach to overcome these difficulties since we previously showed that this strategy improves the expression yield and the stability of the inserted protein domains14. In particular, in the present study, using our hybrid β-lactamase system, the chimeric protein named BlaP-cAb-Lys3 was successfully expressed in E. coli with a very good yield (≈10 mg of pure protein per liter of culture) and purified to homogeneity.
Covalent coupling between the antibody and enzymatic moieties makes cheaper and faster sensor assays In conventional immunosensors, the detection of the analyte requires the utilization of primary antibodies that are immobilized on the sensor chip. Subsequently, a secondary antibody that is coupled to an enzyme or a labeled probe is also required to generate a measurable signal. This approach involves several incubations and washing steps and therefore can be time consuming. In addition, this protocol is expensive since several antibodies are required and covalent coupling between the secondary antibody and an enzyme/probe is also necessary. In contrast, our system only uses a hybrid protein to detect and quantify the analyte, and therefore allows real-time monitoring without the use of secondary antibodies.
Furthermore, it is important to note that domain insertion into class A β-lactamases was shown to generate hybrid proteins exhibiting allosteric switch-like behavior22,23. Such switches could find numerous applications in biosensor-based assays.
Limitations of the hybrid β-lactamase technology.
Limitations induced by protein engineering. In this system, the main difficulty is to design and obtain a hybrid protein by inserting the antibody moiety into the β-lactamase. This resulting chimeric protein must be bifunctional: the enzymatic moiety must remain able to efficiently hydrolyse β-lactam antibiotics to generate the electrical signal, whereas the antibody moiety must bind to the targeted analyte with high affinity and specificity. To obtain bifunctional chimeric proteins, various parameters need to be considered in order to avoid steric constraints. The first critical point is the position of the insertion site. Although it has been shown that multiple insertion points are possible24,25,26, insertion positions are often located in solvent exposed loops far away from the active site of the carrier protein. This minimizes potential conformational changes or steric hindrance induced by the inserted protein. For the same reasons, it is also recommended that the active site of the inserted protein be located far away from the insertion site in order to prevent alterations of its activity. Finally, insertion of proteins that present flexible or neighbouring N- and C-terminal extremities will be better tolerated. Indeed, distant and rigid extremities can impose steric constraints on both partners of the chimeric protein, and thus alter their respective biological activities. Therefore, it is important to mention that the size of the inserted protein has only little impact on the resulting chimeric protein. Indeed, it has been shown that large structured domains can be successfully inserted into β-lactamases, as long as their N- and C-terminal extremities are flexible or close to each other13,14,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29. In the present study, the insertion site in BlaP is located far away from its active site and the inserted nanobody has relatively long and flexible extremities (not visible in the X-ray structure) that are located far away from the paratope. These specific features ensure that minimal steric constraints are imposed on the biologically active surfaces of both partners of the hybrid protein. However, when the inserted protein does not present the recommended criteria for optimum insertion into a carrier protein, it is possible to engineer linker regions in order to lower potential steric constraints. Indeed, the presence of a flexible linker (e.g. Gly-Ser repetitions) to connect the carrier and the inserted protein was shown to dramatically increase the tolerance towards the insertion of large and structured proteins into a carrier one30,12.
Limitations inherent to electrochemicalbiosensors. Although the development of electrochemical/potentiometric biosensors has become an ever-growing field, these assays have important limitations that need to be considered when designing the biosensor experiment. First, all biosensors that involve H+ release or uptake require the use of very weakly buffered solutions (i.e. <5 mM)31 in order to measure significant potential differences. The pH variation induced upon H+ release can impact the protein properties and the enzymatic activity used to generate the signal. Secondly, pH and ionic strength in biofluids can vary significantly and consequently result in important variations in the response and increase the background noise of the biosensors32. This is why, various research groups have tried to develop nanotechnologies to decrease the sizes of electrochemical sensor elements in order to increase the signal-to-noise ratio for the processes that occur at the interface of the device33,34,35. As a consequence, it is also possible to develop antibody molecules labeled with several molecules of the same enzyme to increase the signal resulting from the binding of one molecule to its target32. However, despite these limitations, conductometric/biosensors remain highly efficient and sensitive devices with estimated limits of detection (LODs) in the range of 10-8 to 10-11 M36.
In this study, we have shown that we can successfully insert a nanobody that targets HEWL into a class A β-lactamase named BlaP, and that the generated hybrid protein retains both biological activities: i) tight binding of HEWL and ii) the capacity to hydrolyse β-lactam antibiotics. This study constitutes a proof of concept for insertion of various nanobodies or antibody fragments into BlaP and the implementation of this hybrid protein technology in potentiometric sensor assays. This technology could potentially be used to detect various protein epitopes and implemented in numerous diagnostic tools. Indeed, the development and effective use of such assays have become crucial in our society and are essentially driven by social and economic needs for low-cost and easy to operate technologies in various fields such as food and health systems, especially in developing countries. Furthermore, nanotechnologies play an increasing role in the development of such sensors. Nowadays, signal processing technologies are available on portable devices such as smartphones and tablets and different smartphone applications already exist for signal processing37. For example, recently, a potentiometric sensor device has been integrated into a smartphone to allow glucose concentration monitoring38. The health experts are confident that these kind of innovative devices will become more important health solutions in the coming years39,40.
In conclusion, this work represents an example that shows the potential and advantages of our protein insertion technology. We hope that this work will contribute to develop innovative and useful technologies for research and medical purposes.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors acknowledge the Walloon Region of Belgium within the framework of the SENSOTEM and NANOTIC research projects as well as the National Funds For the Scientific Research (F.R.S.-F.N.R.S) for their financial support.
References
- Higgins IJ, Lowe CR. Introduction to the principles and applications of biosensors. Philos Trans R Soc Lond B Biol Sci. 1987;316:3–11. doi: 10.1098/rstb.1987.0013. [DOI] [PubMed] [Google Scholar]
- Byrne B, Stack E, Gilmartin N, O'Kennedy R. Antibody-based sensors: principles, problems and potential for detection of pathogens and associated toxins. Sensors (Basel) 2009;9:4407–4445. doi: 10.3390/s90604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston JS, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechaly A, Zahavy E, Fisher M. Development and implementation of a single-chain Fv antibody for specific detection of Bacillus anthracis spores. Appl Environ Microbiol. 2008;74:818–822. doi: 10.1128/AEM.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Sheriff S, Constantine KL. Redefining the minimal antigen-binding fragment. Nat Struct Biol. 1996;3:733–736. doi: 10.1038/nsb0996-733. [DOI] [PubMed] [Google Scholar]
- Stijlemans B, et al. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J Biol Chem. 2004;279:1256–1261. doi: 10.1074/jbc.M307341200. [DOI] [PubMed] [Google Scholar]
- Thanongsaksrikul J, et al. A V H H that neutralizes the zinc metalloproteinase activity of botulinum neurotoxin type A. J Biol Chem. 2010;285:9657–9666. doi: 10.1074/jbc.M109.073163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, et al. Prostate-specific antigen immunosensing based on mixed self-assembled monolayers, camel antibodies and colloidal gold enhanced sandwich assays. Biosens. Bioelectron. 2005;21:483–490. doi: 10.1016/j.bios.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Yolken RH, Wee SB, Van Regenmortel M. The use of beta-lactamase in enzyme immunoassays for detection of microbial antigens. J Immunol Methods. 1984;73:109–123. doi: 10.1016/0022-1759(84)90036-x. [DOI] [PubMed] [Google Scholar]
- Kojima M, et al. Activation of circularly permutated beta-lactamase tethered to antibody domains by specific small molecules. Bioconjug Chem. 2011;22:633–641. doi: 10.1021/bc1004125. [DOI] [PubMed] [Google Scholar]
- Iwai H, Kojima-Misaizu M, Dong J, Ueda H. Creation of a Ligand-Dependent Enzyme by Fusing Circularly Permuted Antibody Variable Region Domains. Bioconjug Chem. 2016;27:868–873. doi: 10.1021/acs.bioconjchem.6b00040. [DOI] [PubMed] [Google Scholar]
- Vandevenne M, et al. The Bacillus licheniformis BlaP beta-lactamase as a model protein scaffold to study the insertion of protein fragments. Protein Sci. 2007;16:2260–2271. doi: 10.1110/ps.072912407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandevenne M, et al. Rapid and easy development of versatile tools to study protein/ligand interactions. Protein Eng Des Sel. 2008;21:443–451. doi: 10.1093/protein/gzn021. [DOI] [PubMed] [Google Scholar]
- Crasson O, et al. Enzymatic functionalization of a nanobody using protein insertion technology. Protein Eng Des Sel. 2015;28:451–460. doi: 10.1093/protein/gzv020. [DOI] [PubMed] [Google Scholar]
- Yunus S, Attout A, Vanlancker G, Bertrand P, Ruth N, Galleni G. A method to probe electrochemically active material state in portable sensor applications. Sensors and Actuators B: Chemical. 2011;156:35–42. [Google Scholar]
- Bogaerts P, Yunus S, Massart M, Huang TD, Glupczynski Y. Evaluation of the BYG Carba Test, a New Electrochemical Assay for Rapid Laboratory Detection of Carbapenemase-Producing Enterobacteriaceae. J Clin Microbiol. 2016;54:349–358. doi: 10.1128/JCM.02404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LP, Wang W, Di L, Lu YN, Wang JY. Protein adsorption under electrical stimulation of neural probe coated with polyaniline. Colloids Surf B Biointerfaces. 2010;80:72–78. doi: 10.1016/j.colsurfb.2010.05.034. [DOI] [PubMed] [Google Scholar]
- Piletsky S, Piletska E, Bossi A, Turner N, Turner A. Surface functionalization of porous polypropylene membranes with polyaniline for protein immobilization. Biotechnol. Bioeng. 2003;82:86–92. doi: 10.1002/bit.10544. [DOI] [PubMed] [Google Scholar]
- Khatkhatay MI, Desai M. A comparison of performances of four enzymes used in ELISA with special reference to beta-lactamase. J Immunoassay. 1999;20:151–183. doi: 10.1080/01971529909349349. [DOI] [PubMed] [Google Scholar]
- Worn A, et al. Correlation between in vitro stability and in vivo performance of anti-GCN4 intrabodies as cytoplasmic inhibitors. J Biol Chem. 2000;275:2795–2803. doi: 10.1074/jbc.275.4.2795. [DOI] [PubMed] [Google Scholar]
- Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein Eng Des Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- Choi JH, Laurent AH, Hilser VJ, Ostermeier M. Design of protein switches based on an ensemble model of allostery. Nat Commun. 2015;6:6968. doi: 10.1038/ncomms7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinet B, et al. Functionally accepted insertions of proteins within protein domains. J Biol Chem. 2000;275:17428–17433. doi: 10.1074/jbc.M000666200. [DOI] [PubMed] [Google Scholar]
- Betton JM, Jacob JP, Hofnung M, Broome-Smith JK. Creating a bifunctional protein by insertion of beta-lactamase into the maltodextrin-binding protein. Nat Biotechnol. 1997;15:1276–1279. doi: 10.1038/nbt1197-1276. [DOI] [PubMed] [Google Scholar]
- Ay J, Gotz F, Borriss R, Heinemann U. Structure and function of the Bacillus hybrid enzyme GluXyn-1: native-like jellyroll fold preserved after insertion of autonomous globular domain. Proc Natl Acad Sci U S A. 1998;95:6613–6618. doi: 10.1073/pnas.95.12.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruth N, et al. DNA vaccination for the priming of neutralizing antibodies against non-immunogenic STa enterotoxin from enterotoxigenic Escherichia coli. Vaccine. 2005;23:3618–3627. doi: 10.1016/j.vaccine.2004.11.080. [DOI] [PubMed] [Google Scholar]
- Zervosen A, et al. Characterization of the cattle serum antibody responses against TEM beta-lactamase and the nonimmunogenic Escherichia coli heat-stable enterotoxin (STaI) FEMS Immunol Med Microbiol. 2008;54:319–329. doi: 10.1111/j.1574-695X.2008.00482.x. [DOI] [PubMed] [Google Scholar]
- Chevigne A, et al. Use of bifunctional hybrid beta-lactamases for epitope mapping and immunoassay development. J Immunol Methods. 2007;320:81–93. doi: 10.1016/j.jim.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Ke W, et al. Structure of an engineered beta-lactamase maltose binding protein fusion protein: insights into heterotropic allosteric regulation. PloS One. 2012;7:39168. doi: 10.1371/journal.pone.0039168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeedfar K, Heng LY, Ling TL, Rezayi M. Potentiometric urea biosensor based on an immobilised fullerene-urease bio-conjugate. Sensors (Basel) 2013;13:16851–16866. doi: 10.3390/s131216851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazio P. Biosensors in clinical chemistry. Clin Chim Acta. 2003;334:41–69. doi: 10.1016/s0009-8981(03)00241-9. [DOI] [PubMed] [Google Scholar]
- Szucs J, Pretsch E, Gyurcsanyi RE. Potentiometric enzyme immunoassay using miniaturized anion-selective electrodes for detection. Analyst. 2009;134:1601–1607. doi: 10.1039/b904321g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Wang X, Qin W. Pulsed galvanostatic control of a polymeric membrane ion-selective electrode for potentiometric immunoassays. ACS Appl Mater Interfaces. 2013;5:9488–9493. doi: 10.1021/am402245f. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. A polymeric liquid membrane electrode responsive to 3,3',5,5'-tetramethylbenzidine oxidation for sensitive peroxidase/peroxidase mimetic-based potentiometric biosensing. Anal Chem. 2014;86:4416–4422. doi: 10.1021/ac500281r. [DOI] [PubMed] [Google Scholar]
- Grieshaber D, MacKenzie R, Voros J, Reimhult E. Electrochemical Biosensors - Sensor Principles and Architectures. Sensors (Basel) 2008;8:1400–1458. doi: 10.3390/s80314000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E, Pretsch E. Nanoscale potentiometry. Trends Analyt Chem. 2008;27:612–618. doi: 10.1016/j.trac.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liu Q. Biosensors and bioelectronics on smartphone for portable biochemical detection. Biosens Bioelectron. 2016;75:273–284. doi: 10.1016/j.bios.2015.08.037. [DOI] [PubMed] [Google Scholar]
- Nemiroski A, et al. Universal mobile electrochemical detector designed for use in resource-limited applications. Proc Natl Acad Sci U S A. 2014;111:11984–11989. doi: 10.1073/pnas.1405679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socio-economic impact of mHealth- An assessment report for the European Union. Price Waterhouse Coopers; 2013. Commission, T.E. [Google Scholar]


