Abstract
High-throughput sequencing (HTS) of the T-cell receptor (TCR) has advanced rapidly in recent years, giving the unique opportunity to identify and quantify every distinct T-cell clone present within any biological sample. This sensitive and accurate technique provides the relative frequency of each individual clone within the full T-cell repertoire, allowing to study particular single T-cell clones of interest as well as the concomitant analysis of all different clones of the whole population. HTS is essential to expand our knowledge on the diversity of the TCR repertoire in homeostasis or pathologic conditions, as well as to understand the development and kinetics of antigen-specific T-cell responses that critically determine protective immunity (i.e. vaccination) and immune-related disorders (i.e. auto-immunity and cancer). Interestingly, HTS can be tailored for personalized medicine, having the potential to monitor therapeutic interventions and reveal prognostic and diagnostic biomarkers. In this article, we briefly review the methodology, advances and limitations of HTS, and describe emerging applications of this novel technique in the field of investigative dermatology, including studying the pathogenesis of T cells in allergic dermatitis and its application in diagnosis, early recurrence detection and therapy monitoring of cutaneous T-cell lymphoma.
INTRODUCTION
T lymphocytes form an essential component of the adaptive immune system. Each individual T cell expresses its unique T-cell receptor (TCR) that specifically recognizes a single antigenic determinant. Taking the whole T-cell population together, the adaptive immune system has a spectacular large and highly-diverse TCR repertoire at its disposal, capable of identifying unlimited numbers of antigens, hence providing protection against constant diverse pathogenic treats. TCR are heterodimer molecules consisting of either a combination of α and β chains, being the most common TCR, or a combination of γ and δ chains (Fig. 1). The superb specificity and diversity occurs during lymphocyte development by randomized combinations of DNA from distinct V, D and J (Variable, Diversity, Joining) gene segments, and by deletion and/or insertion of nucleotides at the junctions of these segments, which in particular takes place in the hypervariable complementarity determining region 3 (CDR3). This hypermutation process of the TCR genes can lead to over 1018 different αβ TCR, making it highly improbable to generate two separate TCR with an identical nucleotide CDR3 sequence. Consequently, the TCR nucleotide sequence of each T cell represents an inbuilt bar code that enables us to recognize and track each specific T-cell clone (Fig. 1).
Figure 1. T-cell receptor can function as a unique identifying bar code of T cells.
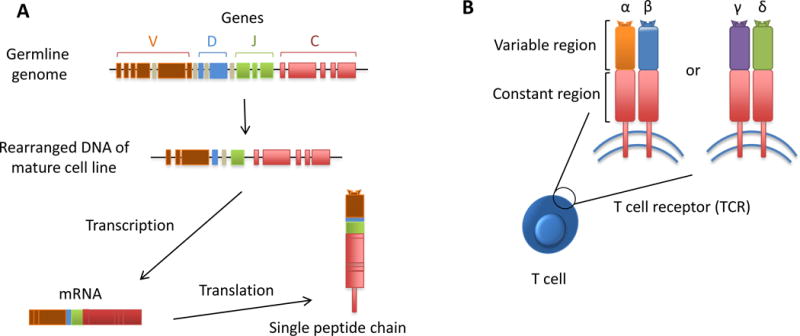
A) In the germline genome, the multiple gene segments are not being expressed and therefore have not been rearranged. The specificity and diversity occurs during lymphocyte development by combining the distinct V, D and J (Variable, Diversity, Joining) gene segments, and by deletion and/or insertion of nucleotides at the junctions of those segments. This randomized process makes it highly improbable to generate two separate T-cell receptors with the same nucleotide CDR3 sequence. Thus, the TCR nucleotide sequence of each T cell is the equivalent of having an inbuilt bar code that enables us to recognize and track each specific T-cell clone. The mRNAs are then translated into the peptide chains of the TCR. B) TCR are heterodimer molecules that can be either a combination of either α and β chains or γ and δ chains.
OVERVIEW OF METHODOLOGY
A recently developed methodology, also known as immunosequencing, combines bias-controlled multiplexed PCR with high-throughput sequencing (HTS) of the CDR3 of the TCR. Subsequent innovative bioinformatic analysis enables the simultaneous identification, quantification and tracking of each individual lymphocyte as well as the entire repertoire within any sample of interest (Fig. 2). To describe the HTS of the TCR in short, the CDR3 region is amplified and sequenced from DNA or complementary DNA (cDNA). Then, in a second PCR step, bias-controlled V and J gene primers are used to amplify the rearranged V(D)J segments which are subjected to HTS. Clustering algorithms are subsequently used to correct the raw data for sequencing errors (Fig. 3). Lastly, the unique CD3 segments and the V, D, and J genes within each rearrangement are identified and quantified (Fig. 2), based on previously described sequences incorporated in data banks (Robins et al. 2009, Lefranc et al. 2009). These results allow identifying somatic allelic mutations, studying the lymphoid clonality and diversity in healthy and malignant tissues, as well as tracking each clone over time (e.g. as a biomarker during disease progression or therapy), among different anatomical sites or within defined populations.
Figure 2. High-throughput TCR sequencing.
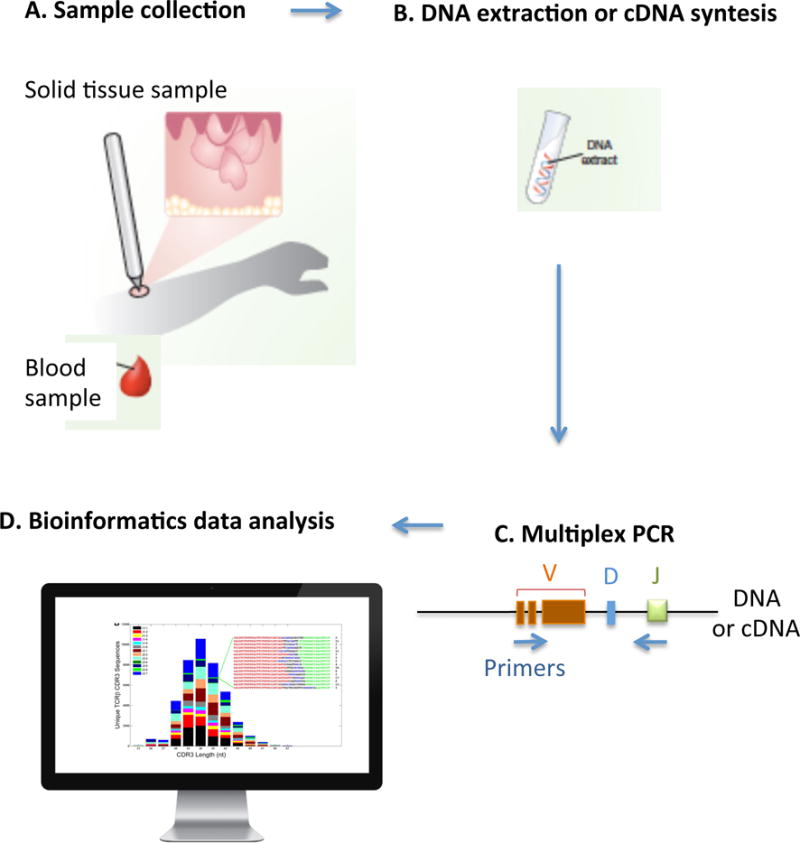
A) The biologic sample of interest is collected. B) DNA is extracted or complementary DNA (cDNA) is synthetized. C) Bias-controlled multiplexed PCR amplifies and sequences the CDR3 from the DNA or cDNA. Then, bias-controlled V and J gene primers are used to amplify the rearranged V(D)J segments. D) Bioinformatics can then be used to identify, quantify and track each individual lymphocyte as well as the entire repertoire within any sample of interest. It is possible to identify and quantify the unique CD3 segments and the V, D, and J genes within each rearrangement, based on previously described sequences incorporated in data banks.
Figure 3. High-throughput TCR CDR3 sequencing captures entire T-cell diversity.
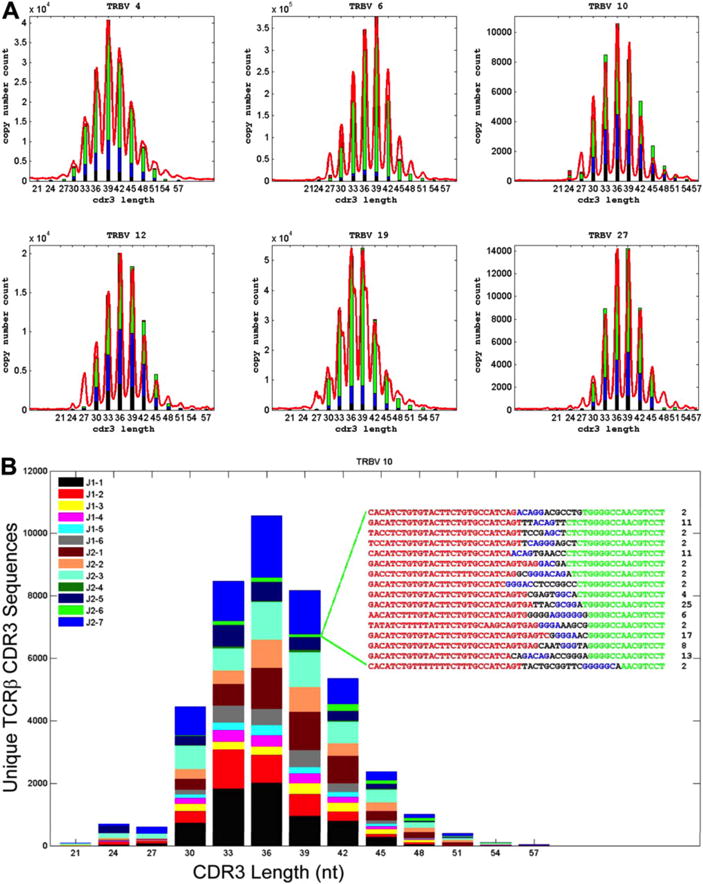
(A) Comparison of standard TCRβ spectratype data and calculated TCRβ CDR3 length distributions for sequences using representative TCR Vβ gene segments. CDR3 length is plotted along the x-axis and the number of unique CDR3 sequences with that length or the relative intensity of the corresponding peak in the spectratype is plotted along the y-axis. The length of the differently colored segments within each bar of the histograms indicates the fraction of unique CDR3 sequences that were observed 1 to 5 times (black), 6 to 10 times (blue), 11 to 100 times (green), or more than 100 times (red). (B) A representative spectratype of TCRβ CDR3 cells that use the Vβ10 gene segment. The CDR3 sequences were sorted by CDR3 length into a frequency histogram, and the sequences within each length were then color-coded on the basis of their Jβ use. The inset represents CDR3 sequences having a length of 39 nucleotides (nt), as well as the number of times that each of these sequences was observed in the data. The origin of the nucleotides in each sequence is color-coded as follows: Vβ gene segment, red; template-independent N nucleotide, black; Dβ gene segment, blue; Jβ gene segment, green. Reprinted from Robins et al. 2009.
There are various terms applied to describe the diversity of T-cell clones in biological samples, including metric terms such as clonality, richness and entropy.
Clonality is a metric of relative abundance that evaluates clonal expansion based on the probability of finding the same sequence in two different replicates, meaning that a higher clonality score reflects greater clonal expansion. Such clonal inflation can be attributed to expansion within the memory compartment, caused by an uneven homeostatic proliferation of naive T-cell repertoire, or may be indicative of a T-cell tumor. For example, cutaneous T-cell lymphoma (CTCL) samples commonly have a high clonality value, since there is a single pathogenic T-cell clone largely predominating in skin lesions (Chitgopeker et al. 2014).
Richness measures how many distinct T-cell clones (unique TCR) are present in a sample. The diversity richness of the repertoire is strongly linked to a healthy immune system, ensuring continuous surveillance and response to unlimited foreign antigens, controlling for acute and chronic infections and preventing mutant cells from unrestricted proliferation.
Entropy provides a theoretic measurement of the probability of a certain clone to be present within the total T-cell repertoire combining information on abundance and richness at the same time. Entropy is the degree of uncertainty associated with identifying clones in the total repertoire based on the total number of clones, their identity and their relative abundances.
APPLICATIONS OF HIGH-THROUGHPUT TCR SEQUENCING IN DERMATOLOGY
High-throughput TCR sequencing is increasingly becoming an important tool in the field of investigative dermatology, including monitoring the immune response to infectious diseases or vaccine, studying pathogenesis of T cell-associated diseases, facilitating diagnosis and early recurrence detection, and assessing T-cell responses during therapies.
Monitoring the immune response to infectious diseases or vaccines
The exposure to pathogens or vaccines induces a specific adaptive immune reaction, which can be monitored by HTS. The study of the TCR of all present T cells allows analysing the efficacy of inducing persistent T-cell memory by tracking unique sequences that were initially present in the naive T-cell population, but appear in the T-cell population with a memory phenotype after an immune response. Moreover, it allows tracking the memory T cells over time unveiling if the vaccine or infection provides long lasting protection. This cumulative knowledge may allow researchers to identify T-cell clones that exclusively react to an antigen of interest, which might eventually be used as a biomarker of exposure or for early detection and prevention of disease spread.
Gaide et al. (2015) used HTS of the TCR to clarify the clonal origin of both central memory T (TCM) cells in lymph nodes and resident memory T (TRM) cells in peripheral tissues after skin immunization. They noted that following skin immunization, a mutual native T-cell precursor gives origin to both antigen-reactive skin TRM and lymph-nodes TCM cells with overlapping TCR repertoires, hence originating memory T cells with different effector properties but with identical antigen specificity in distinct tissue compartments. In addition, TRM cells could rapidly generate a contact hypersensitivity response, while TCM were responsible for a moderate and delayed response. These data indicate that TRM cells mediate allergic contact dermatitis, which explains the site-specificity, recurrence and refractory characteristics of this disease. Similar approaches may help to study human diseases whose clinical features resemble TRM mediated diseases, including psoriasis, vitiligo and fixed drug eruption.
Studying pathogenesis of T-cell diseases
Besides elucidating the pathogenesis of allergic contact dermatitis, HTS also demonstrated that allopurinol-induced severe cutaneous adverse reactions are driven by clonotype-specific T-cells in a dose-dependent response to oxypurinol (Chung et al. 2015). Studies of the TCR γ gene by HTS showed that CTCL is caused by mature memory T cells after undergoing normal thymic maturation and not by lymphoid progenitor cells or immature T cells (Kirsch et al. 2015).
Facilitating diagnosis and early recurrence detection
High-throughput TCR sequencing has been shown to facilitate the diagnosis of CTCL. TCRγ PCR is currently the common diagnostic method of CTCL, however it only detects the malignant T-cell clone in a subgroup of patients. Kirsch et al. (2015) demonstrated that HTS of the TCR β and γ alleles is more sensitive and specific than TCRγ PCR in detecting the pathogenic expanded T-cell clone. HTS can function as an early definitive diagnostic tool for CTCL, and importantly, distinguish early stages of CTCL from benign inflammatory skin diseases (Fig. 4).
Figure 4. High throughput TCRβ CDR3 region sequencing identifies expanded T-cell clones and discriminates CTCL from benign inflammatory skin disorders.
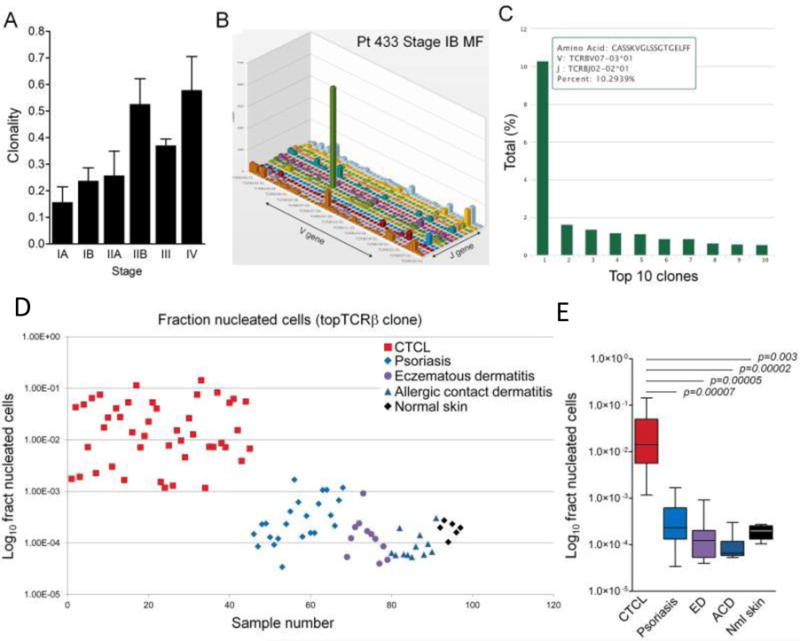
(A) Clonality of lesional skin T cells increased with advanced stage of CTCL. (B, C) TCR sequencing identified expanded populations of clonal malignant T cells in CTCL skin lesions. The V versus J gene usages of T cells from a lesional skin sample are shown (B). The green peak includes the clonal malignant T-cell population. (C) The individual T-cell clone sequence is shown with detailed information on the CDR3 amino acid sequence and V and J gene usage. The nine most frequent TCR sequences of benign infiltrating T cells are also shown. In this patient, the malignant T-cell clone made up 10.3% of the total T-cell population in lesional skin. (D, E) The most frequent T-cell clone expressed as the fraction of total nucleated cells successfully discriminates CTCL from benign inflammatory skin diseases. The most frequent TCR sequence expressed as a fraction of total nucleated cells is shown for individual samples (D) and aggregate data (E). This analysis allowed discrimination of CTCL from benign inflammatory skin diseases and healthy skin. Reprinted from Kirsch et al. 2015.
Assessing immune response to therapy
In a clinical trial testing topical resiquimod gel (an agonist for toll-like receptors 7 and 8) as a treatment for CTCL, it was noted that HTS of the TCR was more specific in assessing malignant T-cell clone clearance than clinical score evaluation (Rook et al. 2015). HTS demonstrated that the malignant T-cell clone reduced in 90% of the treated patients and that the malignant T-cell eradication was correlated with the recruitment and expansion of new benign T cells.
SUMMARY AND FUTURE DIRECTIONS
HTS of the TCR is a highly sensitive and precise technique, providing the relative frequency of each clone within the full T-cell repertoire. It enables to study concomitantly each unique T cell and all clonal populations over time, comparing different biologic tissues of the same individual or among different individuals that may be either healthy of suffering from a certain disease.
HTS technology is based on the specific detection of the CDR3 region on just one of the two chains of the TCR heterodimer. An important next step forward would the establishment of the technology that is able to simultaneously define both chains of the TCR. Currently, the most common way to define both peptide chains of the same TCR is through single cell analysis. However, a recently developed method called pairSEQ, seems to be able to accurately pair TCRα and TCRβ sequences out of hundreds of thousands lymphocytes that were simultaneously analyzed without the need for single-cell technologies (Howie et al. 2015). Once established, this development may significantly contribute to another highly anticipated goal, of being able to link the TCR sequences to their specific target antigenic peptides. These new methods will augment TCR datasets with complementary information on the target epitope recognized by each TCR, and link these TCR sequences to lymphocytic phenotypic markers. In addition, these new methods build upon the current applications of HTS as a monitoring tool for lymphoid malignancies and hematopoietic transplants and potentiate the identification and development of lymphocytes specific for tumor-antigens or self-antigens for anti-cancer or autoimmune therapeutics, respectively.
HTS is meaningfully expanding our understanding of the complex and exquisite role of T cells in the immune system and may lead us in groundbreaking personalized medicine by accurately monitoring and improving therapeutic interventions and possibly discovering diagnostic and prognostic biomarkers.
SUMMARY POINTS.
WHAT HTS OF THE TCR DOES
HTS of the TCR accurately identifies and quantifies each and every T cell present in a certain biologic sample, allowing studying each unique clone and the full T-cell repertoire, including over time, among different tissues and/or individuals.
ADVANTAGES OF HTS OF TCR
-
-
High clone detection sensitivity. Approximately 100-fold greater than other current technologies (e.g. flow cytometry).
-
-
Extremely accurate, with lower rate of false negatives and false positives than analysis of the TCR by PCR.
-
-
Capable of successfully studying any type of tissue and small samples including standard-size punch biopsies or shave biopsies, since it is a cDNA or DNA based technology and the amplification step enables the detection of scarce cells.
-
-
Technique does not require radioactive agents as previously commonly used Southern blotting.
-
-
Can be used for a wide range of applications, from expanding our knowledge of the adaptive immune system, monitor therapeutic interventions and study new diagnostic and prognostic biomarkers.
LIMITATIONS OF HTS OF TCR
-
-
Technology’s sensitivity is only limited by the amount of DNA probed. E.g., if the DNA of a million cells is analyzed, then the clone detection sensitivity is about 1:1,000,000.
-
-
Variations in tissue processing can lead to DNA degradation, section thickness and sample cell size may affect accurate amplification and representation of all gene segments. A certain gene may also be lost as cell undergoes malignant transformation.
-
-
Pairing the α with β or γ with δ chains of a specific TCR is generally only possible when analyzing single cells.
-
-
It is generally still not possible to match the studied TCR sequences to their specific epitope.
QUESTIONS
- What identifies HTS of the TCR?
- Identifies the DNA sequence of the entire TCR
- Identifies the specific epitope of each TCR
- Identifies the variable and constant region of each TCR
-
Identifies and quantifies each and every T cell present in a sampleCorrect answer(D) High-throughput sequencing of the TCR accurately identifies and quantifies each and every T cell present in a certain biologic sample, allowing studying each unique clone and the full T-cell repertoire, including over time, among different tissues and/or individuals.
- All of the following are advantages of HTS of the TCR, except:
- Uses highly accurate radioactive agents
- High clone detection sensitivity
- Can be applied to any biologic tissue and small samples
-
Low rate of false positives and false negativesCorrect answer(A) HTS of the TCR technique does not require radioactive agents as previously commonly used Southern blotting. It has a high clone detection sensitivity; approximately 100-fold greater than other current technologies (e.g. flow cytometry). It is also extremely accurate, with lower rate of false negatives and false positives than analysis of the TCR by PCR. It can be successfully applied to any type of tissue and small samples including standard-size punch biopsies or shave biopsies, since it is a cDNA or DNA based technology and the amplification step enables the detection of scarce cells. Moreover, HTS of the TCR can be used for a wide range of applications, from expanding our knowledge of the adaptive immune system, monitor therapeutic interventions and study new diagnostic and prognostic biomarkers.
- All of the following steps are part of the HTS of the TCR methodology, except:
- Bias-controlled multiplexed PCR
- Western blot
- Advanced bioinformatics
-
HTS of the CDR3 of the TCRCorrect answer(B) HTS of the TCR methodology, also known as immunosequencing, combines bias-controlled multiplexed PCR with HTS of the CDR3 of the TCR and innovative bioinformatics, enables the simultaneous identification, quantification and tracking of each individual lymphocyte as well as the entire repertoire within any sample of interest. In short, the CDR3 region is amplified and sequenced from DNA or complementary DNA (cDNA). Then, bias-controlled V and J gene primers are used to amplify the rearranged V(D)J segments for HTS. Clustering algorithms subsequently correct the raw data for sequencing errors. Lastly, it is possible to identify and quantify the unique CD3 segments and the V, D, and J genes within each rearrangement, based on previously described sequences incorporated in data banks
- Which information provides the clonality score?
- Measures the probability of a clone to be present within the total cell repertoire
- Measures how much a sample is dominated by clonal expansion
- Measures how many distinct clones are present in a sample
-
Measures how many T cells belong to each clonal populationCorrect answer(B) Clonality is a metric of relative abundance that evaluates clonal expansion based on the probability of finding the same sequence in two different replicates, meaning that a higher clonality score reflects greater clonal expansion. Such clonal inflation can be attributed to expansion within the memory compartment or because of an uneven homeostatic proliferation of naive T-cell repertoire. For example, cutaneous T-cell lymphoma (CTCL) samples commonly have a high clonality value, since there is a single pathogenic T cell clone population that largely predominates the skin lesions (Chitgopeker et al. 2014).
- 5. HTS can be used to study:
- The immune response to infectious diseases or vaccine
- Pathogenesis of T cell-associated diseases
- Diagnosis biomarkers and/or immune response to therapies.
-
All of the aboveCorrect answer
- (C) High-throughput TCR sequencing is increasingly becoming an important tool in the field of investigative dermatology, including monitoring the immune response to infectious diseases or vaccine, studying pathogenesis of T cell-associated diseases, facilitating diagnosis and early recurrence detection and assessing responses to therapies.
Acknowledgments
We would like to thank Dr. Jodi L. Johnson for helpful comments, critical reading of the manuscript, and editorial assistance, and Dr. Rachael A. Clark for the valuable scientific advice and guidance. This work was supported by Fondation René Touraine.
Footnotes
Conflict of interests: The authors report no conflicts of interest.
References
- Chitgopeker P, Sahni D. T-Cell Receptor Gene Rearrangement Detection in Suspected Cases of Cutaneous T-Cell Lymphoma. J Invest Dermatol. 2014;134:e19. doi: 10.1038/jid.2014.73. [DOI] [PubMed] [Google Scholar]
- Chung WH, Pan RY, Chu MT, Chin SW, Huang YL, Wang WC, et al. Oxypurinol-Specific T Cells Possess Preferential TCR Clonotypes and Express Granulysin in Allopurinol-Induced Severe Cutaneous Adverse Reactions. J Invest Dermatol. 2015 Sep;135(9):2237–48. doi: 10.1038/jid.2015.165. [DOI] [PubMed] [Google Scholar]
- Gaide O, Emerson RO, Jiang X, Gulati N, Nizza S, Desmarais C, et al. Common clonal origin of central and resident memory T cells following skin immunization. Nat Med. 2015;21:647–653. doi: 10.1038/nm.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Sherwood AM, Berkebile AD, Berka J, Emerson RO, Williamson DW, et al. High-throughput pairing of T cell receptor α and β sequences. Sci Transl Med. 2015 Aug 19;7(301):301ra131. doi: 10.1126/scitranslmed.aac5624. [DOI] [PubMed] [Google Scholar]
- Kirsch IR, Watanabe R, O’Malley JT, Williamson DW, Scott LL, Elco CP, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med. 2015 Oct 7;7(308):308ra158. doi: 10.1126/scitranslmed.aaa9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, et al. IMGT, the international ImMunoGeneTics information system. Nucleic acids research. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. Janeway’s Immunobiology. 8th Garland Science; 2012. [Google Scholar]
- Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook AH, Gelfand JC, Wysocka M, Troxel AB, Benoit B, Surber C, et al. Topical resiquimod can induce disease regression and enhance T-cell effector functions in cutaneous T-cell lymphoma. Blood. 2015 Sep 17;126(12):1452–1461. doi: 10.1182/blood-2015-02-630335. [DOI] [PMC free article] [PubMed] [Google Scholar]


