Abstract
Excessive alcohol consumption remains a major public health problem that affects millions of people worldwide. Accumulative experimental evidence has suggested an important involvement of gut microbiota in the modulation of host’s immunological and neurological functions. However, it is previously unknown whether enteric microbiota is implicated in the formation of alcohol withdrawal-induced anxiety. Using a murine model of chronic alcoholism and withdrawal, we examined the impact of alcohol consumption on the possible alterations of gut microbiota as well as alcohol withdrawal-induced anxiety and behavior changes. The 16S rRNA sequencing revealed that alcohol consumption did not alter the abundance of bacteria, but markedly changed the composition of gut microbiota. Moreover, the transplantation of enteric microbes from alcohol-fed mice to normal healthy controls remarkably shaped the composition of gut bacteria, and elicited behavioral signs of alcohol withdrawal-induced anxiety. Using quantitative real-time polymerase chain reaction, we further confirmed that the expression of genes implicated in alcohol addiction, BDNF, CRHR1 and OPRM1, was also altered by transplantation of gut microbes from alcohol-exposed donors. Collectively, our findings suggested a possibility that the alterations of gut microbiota composition might contribute to the development of alcohol withdrawal-induced anxiety, and reveal potentially new etiologies for treating alcohol addiction.
Keywords: Alcohol withdrawal-induced anxiety, Gut microbiota, Intestinal bacterial composition, Faecal microbiota transplantation
1. Introduction
According to the World Health Organization, excessive alcohol abuse brings about approximately 3.3 million of deaths in the world each year, and contributes to more than two hundred types of diseases. Individuals who consume alcohol engage in repeated and excessive episodic drinking are considered to be “problem drinkers” (Gorini et al., 2014). In the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM), alcohol abuse and dependence are now gathered in alcohol use disorders (AUDs) that comprise different severity stages (mild, moderate and severe) depending on the number of encountered criterions. AUDs could be identified by alcohol craving, seeking, a loss of control in alcohol consumption as well as alcohol tolerance and withdrawal symptoms, covering anxiety, depressive episodes, social withdrawal, insomnia, nausea and seizures, which can be lethal (Ron and Barak, 2016). A series of genetic (Levey et al., 2014), neurobiological (Levey et al., 2014), environmental (Salvatore et al., 2014) and psychosocial (Whelan et al., 2014) risk factors may contribute to the development of alcoholism, but the molecular mechanisms underlying alcohol addiction remain poorly understood. It has been suggested that long-term alcohol exposure induces changes in microRNA (miRNA), gene and protein expression levels in specific brain regions (Gorini et al., 2014). The expression of several genes, such as BDNF, CRHR1 and OPRM1 have been suggested as possible biomarkers of alcoholism with some diagnostic and prognostic value (Bierut, 2011; Treutlein et al., 2006; Pandey, 2003; Uhl et al., 2001), however, the molecular mechanisms underlying the pathogenesis of alcoholism remain largely unknown. It is thus a major challenge to develop effective therapeutic strategies to clinically manage alcoholism and to alleviate alcohol withdrawal syndrome, partly because of the heterogeneity clinical patient population (Addolorato et al., 2012). Another limitation is that a high level of relapse is observed in patients suffering from AUDs, even after protracted abstinence (Seo et al., 2013). Thus, detailed investigations are urgently needed for better understanding the underlying pathogenic mechanisms of alcoholism and alcohol withdrawal syndrome.
It is now increasingly recognized that animals share an intimated and life-long partnership with a myriad of resident microbial species, collectively referred to as the microbiota. Gut microbiota governs multiple pathophysiological processes and even regulates the function of distant organs. For instance, intestinal microbes have ascended to prominence as key modulators of host immunity (Belkaid and Hand, 2014), metabolic disease (Moreno-Indias et al., 2014), cardiovascular disease (Aron-Wisnewsky and Clément, 2015), host radiosensitivity (Cui et al., 2017b; Cui et al., 2016) and other diseases (Cui et al., 2017a). There is emerging evidence that the enteric microbiome extends its influence to the brain through various pathways connecting the gut to the central nervous system which formed the gut – microbiota–brain axis (Sampson and Mazmanian, 2015). Moreover, gut microbes impact neurological outcomes altering behavior and potentially affecting the onset and severity of nervous system disorders (Arentsen et al., 2015). Meanwhile, divergent factors, such as genetic predisposition, diet and inflammation states, can differently affect enteric bacterial flora. In this regard, exploring factors that alter the composition and function of gut microbiota are important to understand the processes of alcohol withdrawal-induced anxiety and behavior changes, and to identify new targets for treatment. Heretofore, it is important to assess whether a possible alteration of host gut microbiota contributes to the development of alcohol withdrawal-induced anxiety.
In the present study, we investigate the effect of gut microbiota on the development of alcohol withdrawal-induced anxiety in a rodent model, and obtain that gut microbiota is implicated in alcohol withdrawal-induced anxiety and behavior changes. Our findings suggested a possibility that the alteration of gut microbiota composition may contribute to the development of alcoholism, and revealed potentially therapeutic approaches to mitigate alcohol withdrawal syndrome.
2. Materials and methods
2.1. Animals
Six- to 8-week-old male C57BL/6 mice were kept at the temperature of 21–23 °C under a twelve-hour regular light/dark cycle. Mice were given access to food and water excluding the temporary time of being removed from their cages for conducting the tests. All animals were treated according to the NIH guidelines for use and care of live animals and approved by our Institutional Animal Care and Use Committee (IACUC). All the mice in this study were of a pure C57BL/6 genetic background and separated into groups randomly. All procedures and animals handlings were performed following the ethical guidelines for animal studies.
2.2. Donor stool preparation and administration
The donor’s faecal droppings from alcohol-exposed mice (Alcohol group) were collected under SPF conditions. Donor stool was freshly prepared on the day of transplant and that in all cases was prepared and transplanted within 4 h. Donor stool was weighed and diluted with 1 ml of saline per 0.1 g of stool. Briefly, the stool was steeped in saline for about 15 min, shaken and then centrifuged at 800 rpm for 3 min. The supernatant was obtained for transplantation.
2.3. Experimental protocol
The animals were separated into 4 groups: (1) Water group (n = 12): mice were treated with normal water during all experiments; (2) Alcohol group (n = 12): 5% alcoholic solution 0.2 ml was force-fed into the mice’s stomach using gavage for one time per day during the first week. Moreover, 5% alcoholic solution was added in their drinking water. For the second week, mice were treated 10% alcoholic solution, 20% alcoholic solution during the third week and 35% alcoholic solution during the fourth week. After withdrawal from alcohol solution for one day (7 day, 14 day, 21 day, 28 day), forced swim test and tail suspension test were performed. (3) Saline group: saline 0.2 ml was force-fed into the mice’s stomach using gavage for one time per day which continued for 14 days. (4) FMT group: mice were administered with 0.2 ml donor stool supernatant by gavage for one time per day which continued for 14 days.
2.4. Forced swim test
Forced swim test experiments were performed on day 0, day 7, day 14, day 21, and day 28 of treatment. Mice were placed one at a time in an open cylinder-shaped flask (45 cm height and 20 cm diameter), with water depth of 20 cm and temperature of 22 ± 2 °C. Time spent immobile in a 6 min interval was scored live by the same observer blinded to the genotype and treatment group of the animals. Being allowed to swim for 6 min for habituation with the condition, each mouse was regarded as immobile when stopped struggling and floated still in the water. The duration of remaining immobile within the last 4 min of the test was recorded. Increased immobility has been proposed as an index of behavioral despair in animals (Kordjazy et al., 2015; Le-Niculescu et al., 2011).
2.5. Tail suspension test
This test relies on the fact that animals when facing the inescapable stress of being suspended by their tail will develop an immobile posture. In detail, each mouse was suspended by its tail, about 50 cm above the ground for 6 min, placed approximately 1 cm from the tip of the tail. The duration of immobility was manually scored for a 6 min observation period. Mice were considered immobile only when they hung down passively and were completely motionless (Cryan et al., 2005).
2.6. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was separated from mouse hippocampus using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. cDNA was produced by using poly(A)-tailed total RNA and reverse transcription primer with ImPro-II Reverse Transcriptase (Promega, Madison, WI, USA), according to the manufacturer’s instructions. The qRT-PCR was performed according to the instructions of Fast Start Universal SYBR Green Master (Rox) (Roche Diagnostics GmbH Mannheim, Germany). The primers were listed in Supplementary Table. GAPDH was used as control.
2.7. Bacterial diversity analysis
Stool samples were freshly collected and stored at −80 °C until use. DNA was extracted from the stool using the Power Fecal® DNA Isolation Kit (MoBio Carlsbad, CA USA). The DNA was recovered with 30 ml of buffer in the kit. PCR products were mixed in equidensity ratios. Then, mixture PCR products were purified with Qiagen Gel Extraction Kit (Qiagen, Germany). The 16S ribosomal RNA (rRNA) V4 gene was analyzed to evaluate the bacterial diversity using Illumina HiSeq (Novogene Bioinformatics Technology Co., Ltd.). Sequences analysis was performed by Uparse software (Uparse v7.0.1001, http://drive5.com/uparse/). Sequences with ≥97% similarity were assigned to the same OTUs. Representative sequence for each OTU was screened for further annotation. For each representative sequence, the Silva123 Database was used based on RDP classifier (Version 2.2, http://sourceforge.net/projects/rdp-classifier/) algorithmto annotate taxonomic information. Briefly, each cohort contains 12 mice, and 6 mice share one cage. For gut microbiota analysis, we collected 2 faecal pellets from one cage and 3 from the other cage to avoid cage effects. Five male C57BL/6 mice from Alcohol group before treating alcohol solution were grouped as Alcohol 0D. When those mice have treated with alcohol solution for two weeks, their stool samples were collected and grouped as Alcohol 14D. For FMT performance, five healthy male C57BL/6 mice before FMT treatment were grouped as FMT 0D. Then those mice have been treated with donor stool supernatant from Alcohol group mice for two weeks, their stool samples were collected and grouped as FMT 14D. The primers are listed in Supplementary Table.
2.8. Statistical analysis
The data are presented as the means ± SEM with respect to the number of samples (n) in each group. Statistical significance between multiple treatment groups was determined by analysis of variance (ANOVA) and Tukey’s t-test. Results with p < 0.05 were considered statistically significant.
3. Results
3.1. Excessive alcohol consumption did not affect the abundance of enteric bacteria
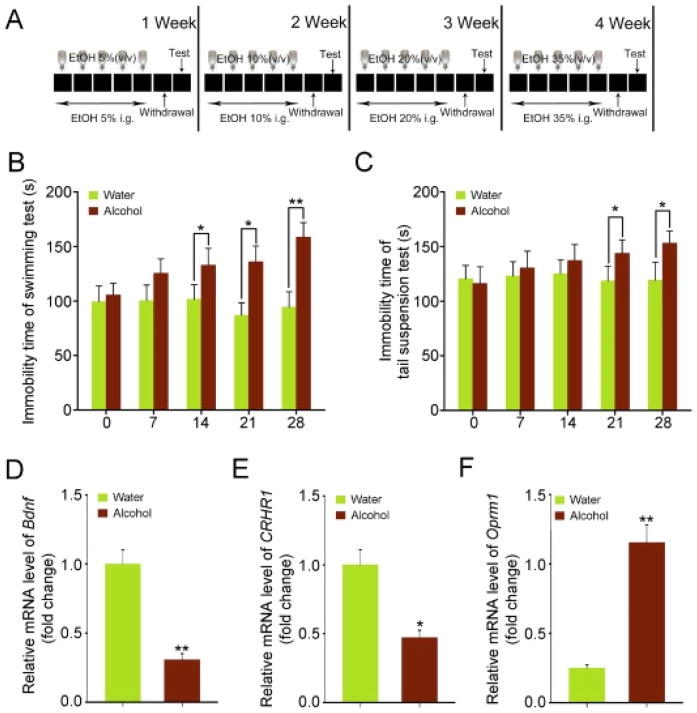
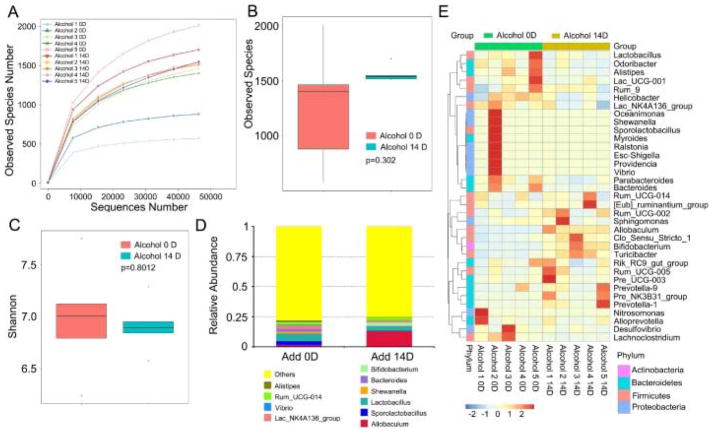
To confirm the signs of alcohol withdrawal-induced anxiety, an array of behavioral tests was performed as Fig. 1A showed. As expected, the mice of alcohol-challenged group showed obvious behavioral signs of alcohol withdrawal-induced anxiety, covering continuous and significant increase of the immobility time in the forced swimming and tail suspension tests from 14th day and 21th day, with increasing drinking days and withdrawal times (Fig. 1B and C). Quantitative real-time polymerase chain reactionassay revealed that alcohol consumption caused a significant decrease of Bdnf and CRHR1 mRNA levels, but a contrarily increase of Oprm1 mRNA levels in the hippocampus (Fig. 1D–F). Intriguingly, 16 s rRNA sequencing revealed that alcohol consumption did not change the species numbers of enteric bacteria (Fig. 2A and B), which was confirmed by the Shannon analysis (Fig. 2C). However, alcohol consumption did reduce the relative abundance of Lactobacillus (or Sporolactobacillus) but elevated the relative abundance of Allobaculum at the genus level (Fig. 2D and E), implying that alcohol consumption might educate the composition of gut microbiota. Together, our observations demonstrate that excessive alcohol consumption has no effect on the abundance of enteric bacteria in this alcohol addiction mouse model.
Fig. 1. The successful establishment of murine model of chronic alcoholism.
(A) Scheme for experimental protocol. (B, C) The effects of chronic alcohol consumption on cognitive functions. The forced swimming test (B) and tail suspension test (C) were performed at day 7, 14, 21 and 28. (D– F) The expression levels of Bdnf (D), CRHR1 (E) and Oprm1 (F) in the hippocampus were assessed at day 28 by qRT-PCR, n = 6 per group. Statistically significant differences are indicated: *P < 0.05; **P < 0.01; Student’s t-test.
Fig. 2. Excessive alcohol consumption did not affect the abundance of enteric bacteria.
(A–C) The observed species number (A, B) and Shannon diversity index (C) of intestinal bacteria in mice before (Alcohol 0D) and after (Alcohol 14D) alcohol treatment was assessed by 16S RNAsequencing, n = 5 per group. The top and bottom boundaries of each box indicate the 75th and 25th quartile values, respectively, and lines within each box represent the 50th quartile (median) values. Ends of whiskers mark the lowest and highest diversity values in each instance. (D, E) The alteration of intestinal bacterial patterns at the genus level in mice before (Alcohol 0D) and after (Alcohol 14D) alcohol treatment were assessed using 16S high throughput sequencing, n = 5 per group. Statistically significant differences are indicated: Student’s t-test. The heatmap is colour-based on row Z-scores.
3.2. Excessive alcohol consumption shaped the intestinal bacterial composition pattern
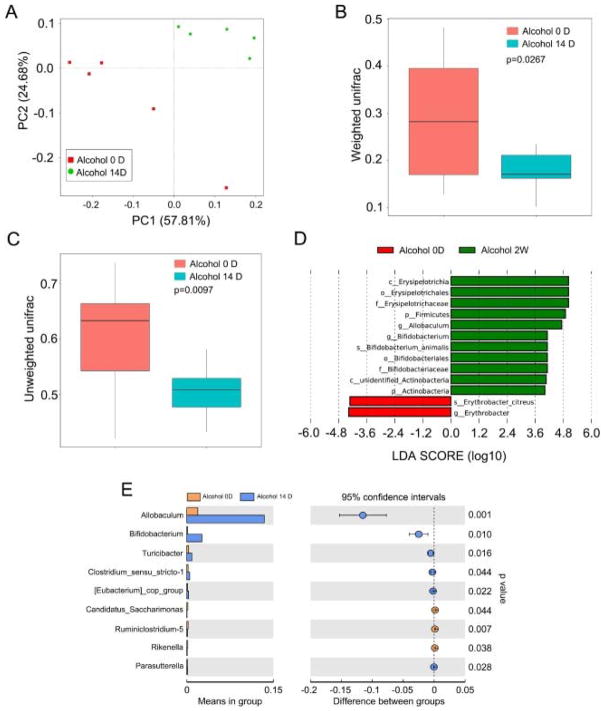
Next, we employed Principal Component Analysis (PCA) to further determine the impact of alcohol consumption on the intestinal bacterial flora profile. As shown in Fig. 3A, the intestinal bacterial composition profile was substantially changed after 14 days alcohol consumption, which was similarly demonstrated by the weighted and unweighted Unifrac analyses (Fig. 3B and C). Comparing the gut bacteria at the genus level using LDA effect size (LEfSe) calculation, many taxa were found to be different in relative abundance after 14 days alcohol consumption. For instance, the Erysipelotrichia became more abundant after alcohol exposure compared to control vehicle group, which was more abundant in Erythrobacter (Fig. 3D). Moreover, alcohol consumption reduced the relative abundance of Allobaculum at the genus level (Fig. 3E). Together, our data indicate that excessive alcohol consumption has altered the intestinal bacterial composition.
Fig. 3. Excessive alcohol consumption shaped the intestinal bacterial composition.
(A) Principal component analysis was performed in mice before (Alcohol 0D) and after (Alcohol 14D) alcohol treatment by 16S high throughput sequencing, n = 5 per group. (B, C) The β diversity of intestinal bacteria was compared by the weighted and unweighted Unifrac analysis between Alcohol 0D and Alcohol 14D group. The top and bottom boundaries of each box indicate the 75th and 25th quartile values, respectively, and lines within each box represent the 50th quartile (median) values. Ends of whiskers mark the lowest and highest diversity values in each instance. (D, E) Linear discriminant analysis (LDA) effect size (LEfSe) results showed bacteria were significantly different in the abundance between Alcohol 0D and Alcohol 14D group. Statistically significant differences are indicated: t-test.
3.3. Transplantation of faecal microbiota from alcohol-exposed mice did not affect the abundance of gut bacteria in the recipients
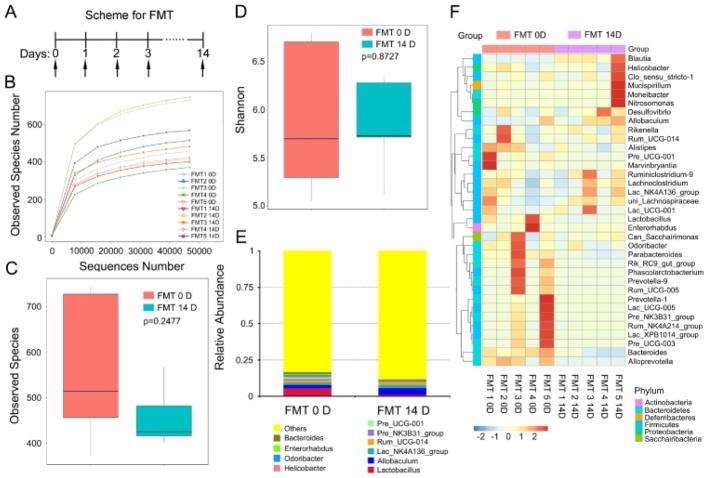
To understand the significance of the alcohol-induced change of gut microbiota composition, we performed faecal microbiota transplantation (FMT) from alcohol-exposed (for 14 days) mice to healthy controls (Fig. 4A). Subsequently, we examined the gut bacterial profile using high throughput sequencing, but did not find a significant difference in the species numbers of enteric bacteria before and after FMT (Fig. 4B and C), which was confirmed by the Shannon analysis (Fig. 4D). However, at the genus level, the relative abundance of Allobaculum and Blautia was elevated, but the relative abundance of Lactobacillus and Bacteroides was reduced in the FMT 14D group after faecal transplantation (Fig. 4E and F). Thus, transplantation of enteric microbes from alcohol-exposed mice fails to affect the abundance of gut bacteria in healthy control recipients.
Fig. 4. Faecal microbiota transplantation from the alcohol-fed mice did not affect the abundance of gut bacterial in the recipients.
(A) Scheme for faecal microbiota transplantation. (B–D) The observed species number (B, C) and Shannon diversity index (D) of intestinal bacteria in mice before (FMT 0D) and after (FMT 14D) alcohol-fed mice’s stool supernatant treatment was assessed by 16S RNA sequencing, n = 5 per group. The top and bottom boundaries of each box indicate the 75th and 25th quartile values, respectively, and lines within each box represent the 50th quartile (median) values. Ends of whiskers mark the lowest and highest diversity values in each instance. (E, F) The alteration of intestinal bacterial patterns at the genus level in mice before (FMT 0D) and after (FMT 14D) alcohol-addicted mice’s stool supernatant treatment was assessed by 16S RNA sequencing, n = 5 per group. Statistically significant differences are indicated: Student’s t-test. The heatmap is colour-based on row Z-scores.
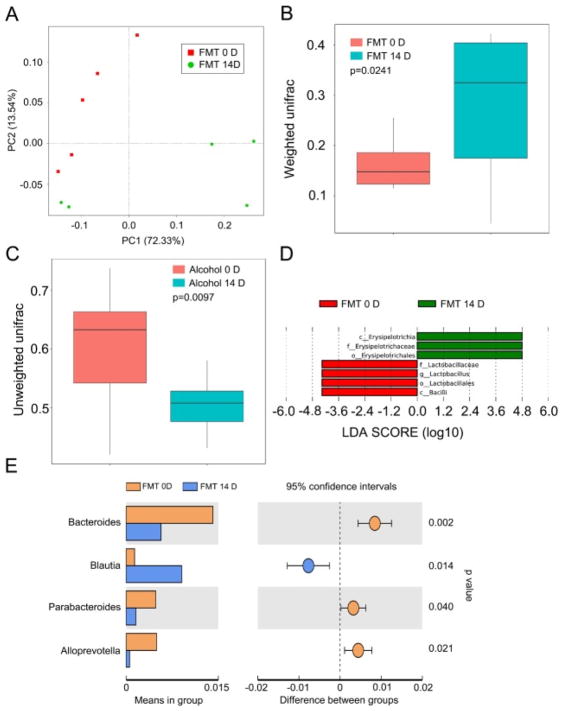
3.4. FMT altered gut microbiota structure of healthy control recipients
To evaluate the effect of FMT on the gut microbiota structure, we analyzed the gut bacterial composition using Principal Component Analysis (PCA). As displayed in Fig. 5A, FMT for 14 days overtly shifted the gut bacterial composition structure. Statistical analysis revealed a significant change in microbial composition following prolonged and repetitive alcohol exposure, as judged by both the weighted (p = 0.0241) and unweighted Unifrac analyses (p = 0.0097) (Fig. 5B and C). After 14 days of FMT, the relative abundance of microbes in the genus of Erysipelotrichia, Erysipelotrichaceaeand Erysipelotrichales was significantly elevated, whereas that of Lactobacillaceae, Lactobacillus, Lactobacillales and Bacilli was significantly reduced (Fig. 5D). In addition, mice receiving the faecal microbiota transplantation (FMT 14D group) carried bacteria of the Bacteroides, Parabacteroides and Alloprevotella genus at a lower relative abundance, but harbored Blautia at a higher relative abundance (Fig. 5E). Overall, the transplantation of faecal microbes from alcohol-consumed mice elicited a major structural change in the gut microbiota composition in the recipients.
Fig. 5. FMT altered gut microbiota structure of the recipients.
(A) Principal component analysis was performed in mice before (FMT 0D) and after (FMT 14D) alcohol-addicted mice’s stool supernatant treatment by 16S RNA sequencing, n = 5 per group. (B, C) The β diversity of intestinal bacteria was compared by the weighted and unweighted Unifrac analysis between FMT 0D and FMT 14D group. The top and bottom boundaries of each box indicate the 75th and 25th quartile values, respectively, and lines within each box represent the 50th quartile (median) values. Ends of whiskers mark the lowest and highest diversity values in each instance. (D, E) Linear discriminant analysis (LDA) effect size (LEfSe) results showed bacteria were significantly different in abundance between FMT 0D and FMT 14D, and they indicated the effect size of each differentially abundant bacterial taxon in the small intestine (n = 5). Statistically significant differences are indicated: Student’s t-test.
3.5. Transplantation of gut microbiota from alcohol-exposed mice facilitated depressive behavior in the recipients
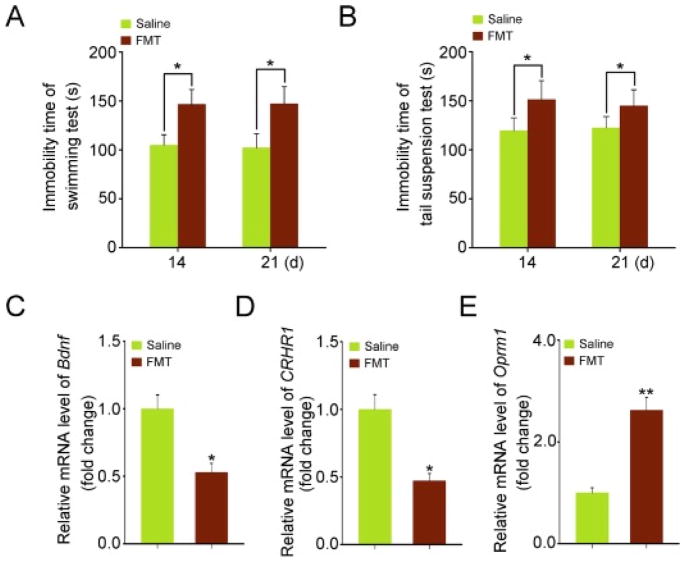
On the basis of the aforementioned observations, we further examined the impact of FMT from alcohol-exposed mice on the possible alcohol withdrawal-induced anxiety in otherwise healthy control mice. Both forced swimming test and tail suspension test were performed to evaluate the impact of FMT on the neurological/cognitive functions. As shown in Fig. 6A and B, mice subjecting to FMT developed behavioral signs of alcohol withdrawal-induced anxiety, as reflected by the significant increase in the immobility time of the forced swimming and tail suspension tests within 14 days of FMT. It suggested a possibility that gut microbiota might contribute to the alcohol withdrawal-induced anxiety. To further verify this hypothesis, qRT-PCR was employed to assess the mRNA expression levels of several alcoholism-relating genes such as Bdnf, CRHR1, and Oprm1 in the hippocampus, which were divergently altered by FMT from the alcohol-exposed mice (Fig. 6C–E). Collectively, these findings suggest that gut microbiota might be involved in the alcohol withdrawal-induced anxiety at least in mice.
Fig. 6. Transplantation of the gut microbiota from alcohol-fed mice facilitated the development of depressive behavior in the recipients.
(A, B) The effects of transplanting the gut microbiota of alcohol-fed mice on the cognitive functions. The forced swimming test (A) and tail suspension test (B) at day 14 and 21. (C–E) The expression levels of Bdnf (C), CRHR1 (D) and Oprm1 (E) were assessed at day 14 in hippocampus of mice from FMT and control group by qRT-PCR. Statistically significant differences are indicated: *P < 0.05; **P < 0.01; Student’s t-test.
4. Discussion
A large body of evidence suggests that excessive alcohol consumption is a worldwide major health risk factor for disability, sickness and even death. Currently, only three drugs have been approved for alcoholism treatment in the United States: disulfiram(Antabuse), acamprosate (N-acetyl homotaurine) and naltrexone, a nonselective opioid receptor antagonist (Mitchell et al., 2012). In addition, there is an increased awareness about the genetic predisposition to alcoholism and long-lasting changes in neurotransmission systems associated with different phases of addiction (Addolorato et al., 2012; Hernández-Nazará et al., 2008; Ma et al., 2015). Even so, the intricate mechanism underlying the pathogenesis of alcoholism remains poorly understood. To this end, we analyzed the effects of alcohol consumption on the possible changes of gut microbiota. Intriguingly, we found that excessive alcohol infusion was able to shape the intestinal bacterial composition. Transplantation of the gut microbiota from alcohol-exposed mice induced depressive behavior resembling alcohol-mediated neurological/cognitive dysfunction, suggesting that gut microbiota might contribute to the development of alcoholism. These findings not only provide novel insights into the underlying mechanism of alcoholism, but also suggest gut microbiota as a novel therapeutic target to ameliorate alcohol withdrawal syndrome in pre-clinical settings.
The gastrointestinal tract is inhabited by an intensive population of organized and highly specialized microbial flora that is a key determinant of health and disease (Petrof and Khoruts, 2014). Compositional and functional changes in commensal microbiota are thought to be involved in the pathogenesis of many diseases (Lozupone et al., 2012). Although the notion that these microbial partners modulating human health is not a recent concept, the extent to which the microbiota controls all physiological systems including nervous system has only recently begun to be appreciated (Ridaura and Belkaid, 2015). For instance, Lactobacillus has anti-Alzheimer properties to fight against D-Galactose-induced Alzheimer’s disease (Nimgampalle and Kuna, 2017), and alleviates the burden of respiratory tract infections (Belkacem et al., 2017). In this study, our observations revealed that alcohol consumption reduced the relative abundance of Lactobacillus, indicating that alcohol consumption has potential to promote Alzheimer’s disease and respiratory tract infections. The intestinal microbiota is an important factor in the prevention and treatment of metabolic dysregulation (Nieuwdorp et al., 2014) and immune system disorder (Hooper et al., 2012). Emerging evidence underpins the link between gut microbiota and Hashimoto’s thyroiditis. Clinically, Hashimoto’s thyroiditis patients harbor lower relative abundance of Bacteroides (Zhao et al., 2018). In this sense, alcohol consumption reduced relative abundance of Bacteroides might promote pathogenesis of Hashimoto’s thyroiditis. In addition, gut microbiota could also directly facilitate pathological by causing gastroenteritis (Jalankatuovinen et al., 2014) and neurodegenerative diseases (Lin and Flint, 2006). Our previous studies revealed that aberrant circadian rhythm shaped gut microbiota community resulting in impairing the radioresistance of hosts (Cui et al., 2016), and transplantation of faecal microbes from healthy donor was able to alleviate toxicity of irradiated recipients (Cui et al., 2017b). Recently, others also showed that mice chronically exposed to ethanol vapors exhibited marked changes in microbiota (Peterson et al., 2017). In this study, we further exploited the pathogenesis of alcoholism, and found that excessive alcohol exposure did not alter the abundance of gut bacteria, but markedly changed the gut microbiota structure. Moreover, we generated evidence for the possible involvement of gut microbiota in the pathogenesis of alcoholism through faecal transplantation, reinforcing the notion that enteric microbes influence the complex function of distant organs.
In conclusion, our work demonstrated that alcohol consumption educated gut bacterial composition leading to behavioral signs of alcohol withdrawal-induced anxiety. Transplantation of faecal microbiota from alcohol-exposed mice elicited depressive behaviors in the otherwise normal healthy recipients via shaping their gut bacterial composition. Thus, our findings provide new insights into the mechanism of alcohol withdrawal-induced anxiety, support that gut microbiota might be a potential therapeutic target to mitigate alcohol withdrawal syndrome in pre-clinical settings.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81502664, 81572969 and 81402541), CAMS Innovation Fund for Medical Sciences (CIFMS, 2016-I2M-1-017), Fundamental Research Funds for CAMS/PUMC(2016ZX310200), the PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (No. 33320140187, 3332016099 and 3332016143), the IRM-CAMS Research Fund (Nos. 1547 and 1522), the Technology and Development and Research Projects for Research Institutes, Ministry of Science and Technology (2014EG150134), the Tianjin Science and Technology Support Plan Project (TJKJZC, 14ZCZDSY00001). H.W. is supported by the U.S. National Center of Complementary and Alternative Medicine (NCCAM, R01AT005076) and the National Institute of General Medical Sciences (NIGMS, R01GM063075).
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
M.C. elaborated the study design. H.W.X., C.G., G.X.F., D.L., Y.L., H.L. and J.L.D. collected the data. M.C. and H.W.X. contributed to data analysis and interpretation. M.C., H.W.X., H.C.W. and S.J.F drafted the article. All authors critically reviewed the content and approved the final version for publication.
References
- Addolorato G, Leggio L, Hopf FW, Diana M, Bonci A. Novel therapeutic strategies for alcohol and drug addiction: focus on GABA, ion channels and transcranial magnetic stimulation. Neuropsychopharmacology. 2012;37:163–177. doi: 10.1038/npp.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentsen T, Raith H, Qian Y, Forssberg H, Heijtz RD. Host microbiota modulates development of social preference in mice. Microb Ecol Health Dis. 2015;26:29719. doi: 10.3402/mehd.v26.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Clément K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol. 2015;12:169–181. doi: 10.1038/nrneph.2015.191. [DOI] [PubMed] [Google Scholar]
- Belkacem N, Serafini N, Wheeler R. Lactobacillus paracasei feeding improves immune control of influenza infection in mice. PLoS One. 2017;12:e0184976. doi: 10.1371/journal.pone.0184976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ. Genetic vulnerability and susceptibility to substance dependence. Neuron. 2011;69:618–627. doi: 10.1016/j.neuron.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Mombereau C, Vassout A. The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neurosci Biobehav Rev. 2005;29:571–625. doi: 10.1016/j.neubiorev.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Cui M, Xiao H, Luo D, Zhang X, Zhao S, Zheng Q, Li Y, Zhao Y, Dong J, Li H. Circadian rhythm shapes the gut microbiota affecting host radiosensitivity. Int J Mol Sci. 2016;17:E1786. doi: 10.3390/ijms17111786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Xiao H, Li Y, Dong J, Luo D, Li H, Feng G, Wang H, Fan S. Total abdominal irradiation exposure impairs cognitive function involving miR-34a-5p/BDNF axis. Biochim Biophys Acta. 2017a;1863:2333–2341. doi: 10.1016/j.bbadis.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Xiao H, Li Y, Zhou L, Zhao S, Luo D, Zheng Q, Dong J, Zhao Y, Zhang X, Zhang J, Lu L, Wang H, Fan S. Faecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol Med. 2017b;9:448–461. doi: 10.15252/emmm.201606932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini G, Adron HR, Dayne MR. Proteomic approaches and identification of novel therapeutic targets for alcoholism. Neuropsychopharmacology. 2014;39:104–130. doi: 10.1038/npp.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Nazará ZH, Ruiz-Madrigal B, Martínez-López E, Roman S, Panduro A. Association of the epsilon 2 allele of APOE gene to hypertriglyceridemia and to early-onset alcoholic cirrhosis. Alcohol Clin Exp Res. 2008;32:559. doi: 10.1111/j.1530-0277.2007.00607.x. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Dan RL, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalankatuovinen J, Salojärvi J, Salonen A, Immonen O, Garsed K, Kelly FM, Zaitoun A, Palva A, Spiller RC, de Vos WM. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2014;63:1737–1745. doi: 10.1136/gutjnl-2013-305994. [DOI] [PubMed] [Google Scholar]
- Kordjazy N, Haj-Mirzaian A, Amiri S, Ostadhadi S, Kordjazy M, Sharifzadeh M, Dehpour AR. Elevated level of nitric oxide mediates the anti-depressant effect of rubidium chloride in mice. Eur J Pharmacol. 2015;762:411. doi: 10.1016/j.ejphar.2015.06.030. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Case NJ, Hulvershorn L, Patel SD, Bowker D, Gupta J, Bell R, Edenberg HJ, Tsuang MT, Kuczenski R. Convergent functional genomic studies of ω-3 fatty acids in stress reactivity, bipolar disorder and alcoholism. Transl Psychiatry. 2011;1:e4. doi: 10.1038/tp.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey DF, Leniculescu H, Frank J, Ayalew M, Jain N, Kirlin B, Learman R, Winiger E, Rodd Z, Shekhar A. Genetic risk prediction and neurobiological understanding of alcoholism. Transl Psychiatry. 2014;4:e391. doi: 10.1038/tp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Yuan W, Jiang X, Cui WY, Li MD. Updated findings of the association and functional studies of DRD2/ANKK1 variants with addictions. Mol Neurobiol. 2015;51:281–299. doi: 10.1007/s12035-014-8826-2. [DOI] [PubMed] [Google Scholar]
- Mitchell JM, O’Neil JP, Janabi M, Marks SM, Jagust WJ, Fields HL. Alcohol consumption induces endogenous opioid release in the human orbitofrontal cortex and nucleus accumbens. Sci Transl Med. 2012;4:116ra116. doi: 10.1126/scitranslmed.3002902. [DOI] [PubMed] [Google Scholar]
- Moreno-Indias I, Cardona F, Tinahones FJ, Queipo-Ortuno MI. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwdorp M, Gilijamse PW, Pai N, Kaplan LM. Role of the microbiome in energy regulation and metabolism. Gastroenterology. 2014;146:1525–1533. doi: 10.1053/j.gastro.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Nimgampalle M, Kuna Y. Anti-alzheimer properties of probiotic, lactobacillus plantarum MTCC 1325 in alzheimer’s disease induced albino rats. J Clin Diagn Res: JCDR. 2017;11:Kc01–kc05. doi: 10.7860/JCDR/2017/26106.10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC. Anxiety and alcohol abuse disorders: a common role for CREB and its target, the neuropeptide Y gene. Trends Pharmacol Sci. 2003;24:456–460. doi: 10.1016/S0165-6147(03)00226-8. [DOI] [PubMed] [Google Scholar]
- Peterson VL, Jury NJ, Cabrera-Rubio R, Draper LA, Crispie F, Cotter PD, Dinan TG, Holmes A, Cryan JF. Drunk bugs: chronic vapour alcohol exposure induces marked changes in the gut microbiome in mice. Behav Brain Res. 2017;323:172–176. doi: 10.1016/j.bbr.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof EO, Khoruts A. From stool transplants to next-generation microbiota therapeutics. Gastroenterology. 2014;146:1573–1582. doi: 10.1053/j.gastro.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V, Belkaid Y. Gut microbiota: the link to your second brain. Cell. 2015;161:193–194. doi: 10.1016/j.cell.2015.03.033. [DOI] [PubMed] [Google Scholar]
- Ron D, Barak S. Molecular mechanisms underlying alcohol-drinking behaviours. Nat Rev Neurosci. 2016;17:576. doi: 10.1038/nrn.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore JE, Aliev F, Edwards AC, Evans DM, Macleod J, Hickman M, Lewis G, Kendler KS, Loukola A, Korhonen T. Polygenic scores predict alcohol problems in an independent sample and show moderation by the environment. Genes. 2014;5:330–346. doi: 10.3390/genes5020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D, Lacadie CM, Tuit K, Hong KI, Constable RT, Sinha R. Disrupted ventromedial prefrontal function, alcohol craving, and subsequent relapse risk. JAMA Psychiatry. 2013;70:1–13. doi: 10.1001/jamapsychiatry.2013.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Kissling C, Frank J, Wiemann S, Dong L, Depner M, Saam C, Lascorz J, Soyka M, Preuss UW, Rujescu D, Skowronek MH, Rietschel M, Spanagel R, Heinz A, Laucht M, Mann K, Schumann G. Genetic association of the human corticotropin releasing hormone receptor 1 (CRHR1) with binge drinking and alcohol intake patterns in two independent samples. Mol Psychiatry. 2006;11:594–602. doi: 10.1038/sj.mp.4001813. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Walther D, Hess J, Naiman D. Polysubstance abuse–-vulnerability genes: genome scans for association, using 1,004 subjects and 1,494 single-nucleotide polymorphisms. Am J Hum Genet. 2001;69:1290. doi: 10.1086/324467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan R, Watts R, Orr CA, Althoff RR, Artiges E, Banaschewski T, Barker GJ, Bokde ALW, Büchel C, Carvalho FM. Neuropsychosocial profiles of current and future adolescent alcohol misusers. Nature. 2014;512:185. doi: 10.1038/nature13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Feng J, Li J, Zhao L, Liu Y, Chen H, Jin Y, Zhu B, Wei Y. Alterations of the gut microbiota in Hashimoto’s thyroiditis patients. Thyroid. 2018 doi: 10.1089/thy.2017.0395. http://dx.doi.org/10.1089/thy.2017.0395. (Epub ahead of print) [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.