SUMMARY
Teneurins (TENs) are cell-surface adhesion proteins with critical roles in tissue development and axon guidance. Here we report the 3.1-Å electron cryo-microscopy structure of the human TEN2 extracellular region (ECR), revealing a striking similarity to bacterial Tc-toxins. The ECR includes a large β-barrel that partially encapsulates a C-terminal domain, which emerges to the solvent through an opening in the mid-barrel region. An immunoglobulin (Ig)-like domain seals the bottom of the barrel while a β-propeller is attached in a perpendicular orientation. We further show that an alternatively spliced region within the β-propeller acts as a switch to regulate trans-cellular adhesion of TEN2 to latrophilin (LPHN), a transmembrane receptor known to mediate critical functions in the central nervous system. One splice variant activates trans-cellular signaling in a LPHN-dependent manner, whereas the other induces inhibitory postsynaptic differentiation. These results highlight the unusual structural organization of TENs giving rise to their multifarious functions.
Keywords: Adhesion GPCR, embryogenesis, Cryo-EM, ADGRL1, teneurin
TOC Blurb
An adehsion protein with an unexpected, toxin-like fold shows how alternative splicing can regulate synaptic connections
INTRODUCTION
Teneurins (TENs) are evolutionarily conserved cell-adhesion molecules that play a central role in embryogenesis, axon guidance, and synapse formation (Leamey and Sawatari, 2014; Tucker and Chiquet-Ehrismann, 2006). Mammals contain four TEN genes that are broadly expressed during embryonic development, and most abundantly synthesized in the brain at later developmental and adult stages. TENs are unusual because they are large type-II transmembrane proteins (>2000 amino-acids) that lack the requisite domains generally observed in cell-adhesion molecules, such as classical Ig-, cadherin-, LNS- or integrin domains. Even though these proteins are highly conserved with a homolog observed even in unicellular choanoflagellates (Tucker et al., 2012), sequence analyses of TENs have been relatively uninformative because most of their ECRs do not exhibit readily identifiable domains.
Consistent with an ancient origin, TENs have multiple essential functions in animals. During embryogenesis, TENs are expressed in distinct overlapping patterns in different developing tissues. Detailed genetic analysis revealed that in mice, TEN4 is essential for gastrulation and the epithelial to mesenchymal transition (Lossie et al., 2005; Nakamura et al., 2013). Although similar analyses are lacking for other TENs, it is likely that they also have an important role in embryogenesis given their widespread expression (Ben-Zur et al., 2000; Zhou et al., 2003). In C. elegans, the single TEN homolog Ten-1 is essential for the correct formation of basement membranes in a variety of tissues, including the gonads (Trzebiatowska et al., 2008). In human patients, heterozygous TEN mutations predispose to an array of developmental impairments, again illustrating their broad range of functions (Aldahmesh et al., 2012; Alkelai et al., 2016; Hor et al., 2015).
The roles of TENs in the brain have been studied more extensively than their roles in embryonic development. Overwhelming evidence suggests that during brain development, TENs perform an essential function in guiding axons to the correct targets both in C. elegans (Drabikowski et al., 2005) and in mice (Dharmaratne et al., 2012; Leamey et al., 2007; Young et al., 2013). Furthermore, TENs have been implicated in synapse formation, a role that has received the most attention despite the indirect nature of the evidence. Arguably, the best support for a role of TENs in synaptogenesis derives from its strong binding to latrophilins (LPHNs), adhesion G-protein coupled receptors that are localized in synapses (Anderson et al., 2017; Boucard et al., 2014; Silva et al., 2011). Given that TENs are also localized to synapses and at least LPHN2 has been shown to be essential for hippocampal synapse formation (Anderson et al., 2017), a role for the heterophilic TEN-LPHN complex in synapse formation is plausible. Overexpression experiments in Drosophila support such a role (Hong et al., 2012; Mosca et al., 2012), although in those studies TENs were proposed to act as homophilic adhesion molecules. Given their co-expression patterns, it is possible that the interaction of TENs with LPHNs also mediates their respective functions during embryonic development, but the role of this interaction in embryonic development has not been studied. Interestingly, TENs are processed by multiple proteolytic events that may release multiple soluble fragments, including a short C-terminal fragment that stimulates neurons by an unknown mechanism (Vysokov et al., 2016; Wang et al., 2005). These proteolytic events may convert TEN cell-adhesion molecules into diffusible signals, which could act during axonal pathfinding.
Despite their central importance in multiple physiological roles, the lack of information on the structure of TENs is one of the limiting factors in delineating their mechanisms of action. Here we report the 3.1-Å cryo-electron microscopy structure of the large ECR of human TEN2 (Figure 1). The ECR has an unusual architecture whereby a large cylindrical β-barrel with clear similarity to bacterial Tc-toxins partially encapsulates a C-terminal toxin-like domain that emerges from the barrel and is tethered to its outer surface. We demonstrate that the tethered C-terminal domain mediates interactions with LPHN, and that these interactions activate trans signaling by controlling intracellular cAMP levels. In addition, we show that a splice variant of TEN2, which involves a seven-amino-acid extension in a β-propeller domain region, is incapable of mediating interactions with LPHN and instead induces inhibitory postsynaptic specifications through interactions with alternative synaptic partners. These results reveal that TEN2, and by extension other TENs, form highly unusual structures consistent with biological activities spanning a range of function.
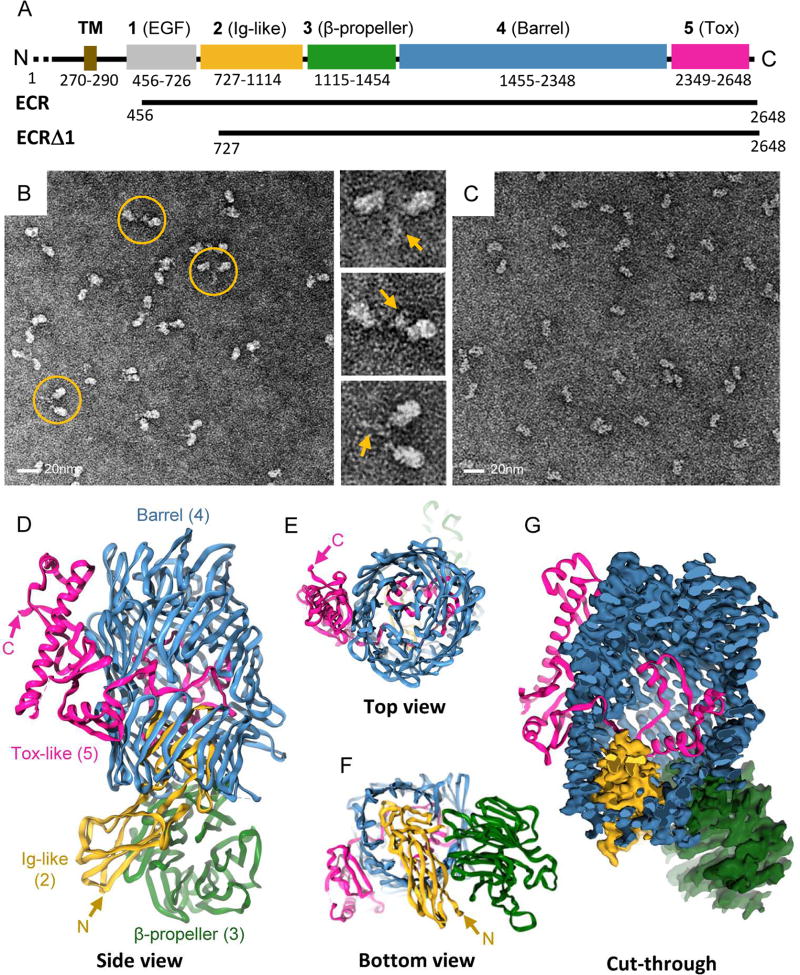
Figure 1. The structure of TEN2.
A, Domain composition of TEN2 and constructs used in the present study. Extracellular domains are colored grey, yellow, green, blue and magenta for domains 1–5, respectively; transmembrane region (TM) in gold. Domain numbers and descriptions are indicated in black above scheme. Domain borders are indicated below. B-C, Representative negative stain EM micrographs of TEN2 ECR (B) and ECRΔ1 (C) indicating dimerization through the ECR domain 1 (B). Scale bar is 20 nm. Close up views of selected dimers (1–3) are indicated to the right with domain 1 dimerization pointed with yellow arrow. C, ECRΔ1 is of monomeric nature. D, Model overview of monomeric TEN2 as obtained from cryo-EM analysis of ECRΔ1. The structure reveals that the TEN ECR is folded as a barrel-shaped container (domain4, YD-repeats, blue) assembled of β-hairpins which is sealed in both ends (E-F). A spiral domain is sealing the barrel top (E). An Ig-like domain (domain2, yellow) seals the barrel bottom (F). A beta-propeller domain (domain 3, green) decorates the bottom of the barrel in a perpendicular orientation (D, F). G, A toxin like domain (domain 5, magenta) is partially encapsulated within the barrel, and further stretches outside the barrel through a side opening. An exploded view of domains architecture is given in Figure S4.
RESULTS
TENs have striking similarity to bacterial toxins
TENs form constitutive cis-dimers via highly conserved disulfide bonds formed between their ECRs in close proximity to the transmembrane pass (Feng et al., 2002; Oohashi et al., 1999; Vysokov et al., 2016). Examination of the purified human TEN2 ECR (residues 456-2648, Figure 1A) by negative stain electron microscopy showed cis-dimerizaton with two large globular domains extended by thin flexible linkers that are tied together through a stalk-like region at their end (Figure 1B, and Figure S1A). Deletion of the stalk-like region from this construct yielded a soluble monomeric protein with a molecular mass of 215 kDa that was suitable for high-resolution structural studies (TEN2 ECRΔ1, encoding residues 727-2648, Figure 1A,C and Figure S1).
We obtained a three-dimensional map of monomeric TEN2 ECRΔ1 by single-particle cryo-EM at a nominal resolution of 3.1 Å, thereby facilitating a near atomic resolution model of the protein (Figure 1D–G; Figures S1–S3 and Table S1). The TEN2 ECR is assembled as a large cylindrical barrel composed of a spiral of β-hairpins and sealed at both ends (domain 4, 1455-2348; Figure 1D–F and Figure S4). The bottom of the barrel is capped with an Ig-like domain (domain 2, 727-1114; Figure 1D,F) that extends to the inner barrel cavity (Figure S4), while a flexible six-bladed β-propeller domain is attached perpendicularly to the barrel bottom (domain 3, 1115-1454; Figure 1D,F, Figure S2 and Figure S4). The C-terminal part of the protein is partially encapsulated within the barrel and emerges to the solvent through an opening in the mid-barrel region (domain 5, 2349-2648; Figure 1G and Figure S4). The external part of the C-terminal region adopts a stable fold and is anchored to the barrel outer surface via electrostatic interactions (Figure 1D,G and Figure S5A).
The fold of TEN2 described here has not been encountered in any eukaryotic protein. However, despite a limited protein sequence identity (12%), the TEN2 β-barrel has a striking structural similarity with the BC-components of bacterial Tc-toxins (Busby et al., 2013; Meusch et al., 2014) (Figure 2 and Table S2). Tc-toxins are tripartite complexes composed of A, B and C protein components that are individually secreted from bacteria and assemble into a megadalton sized complex outside the producing cell (Busby et al., 2013; Landsberg et al., 2011; Meusch et al., 2014). The B and C components of Tc-toxins compose a large macromolecular barrel that encapsulates the covalently attached C-terminal toxic domain (Figure 2A), thereby serving as an immunity cap that shields the producing strain from being exposed to the toxin during production. As opposed to bacteria, the C-terminal region of TEN2 is only partially encapsulated within the barrel (Figure 1G and Figure S4). This difference might explain the larger dimensions of bacterial toxins designed to fully encapsulate C-terminal domains of ~200 residues, compared to TEN2 that only accommodates 86 out of ~300 residues of the C-terminal region (Figures 1G and 2A). The C-component of Tc-toxins acts as an aspartyl protease and is capable of autoproteolytic processing of the toxic domain that will be later released into the host cell. Sequence alignment of TENs with the catalytic pocket region of bacterial toxins revealed a high level of conservation (Figure 2C), with all catalytic residues present in all TENs apart from the present TEN2 variant, which carries a single aspartic acid to glycine alteration (D2345G) within catalytic residues (v1, Figure 2C). It thus appears that TEN2 holds a dormant catalytic pocket that may be active in other TENs. Structural comparison between the dormant pocket in TEN2 and the active catalytic pocket in bacterial toxins (Figure 2D–F) further indicates that two conserved residues that were shown to mediate the catalytic activity in toxins (R2302 and D2315) are similarly oriented in both structures. However, the glycine residue in TEN2 is pulled away from the catalytic pocket, mainly due to several contacts of the encapsulated domain with the barrel inner cavity as well as with the Ig-like domain extension penetrating into the barrel (Figures 2E–F and S5B–C). These observations suggest that, even in the presence of a full aspartic acid protease dyad, a major conformational change would have to take place in TENs in order to stimulate catalytic activity.
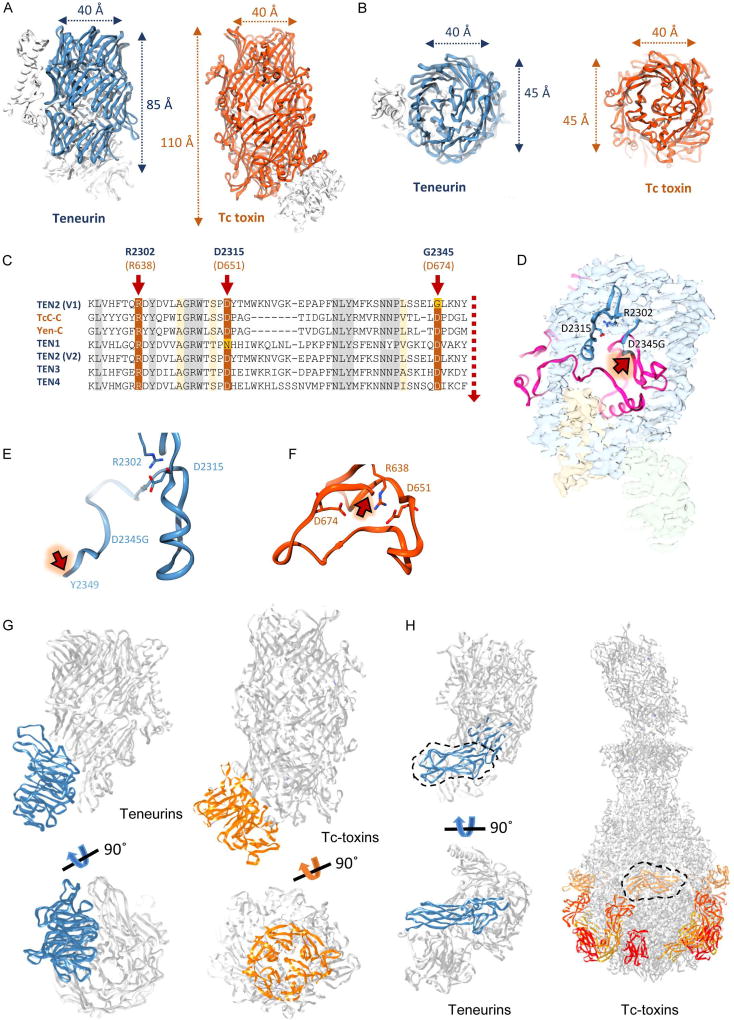
Figure 2. The TEN2 central domain resembles bacterial toxins and contains an ancient domain with dormant autoproteolytic activity.
The TEN2 domain 4 (blue) assembles into a hollow barrel that resembles the BC components of bacterial Tc toxins (TcC-toxin, orange, PDB ID 4O9X). Top (A) and side (B) views indicate similar radial dimensions with a significantly enlarged volume in the bacterial toxin barrels compared to TEN. C, Multiple sequence alignment of TENs with bacterial Tc toxins reveals similarity with an aspartate protease domain localized within the barrel. A D2345G mutation in one of the conserved catalytic pocket residues of aspartate protease domain results in inactivation of the proteolytic activity of TEN2 variant used in this study. Conserved residues that were shown to mediate catalytic activity in bacteria are highlighted by arrows. Putative cleavage site is shown in red dashed line. Corresponding residue numbers in TEN2 and in TcC are shown in blue and orange, respectively. D, Three-dimensional representation of TEN residues with suspected catalytic activity. The cleavage site is indicated by arrow. E-F, Snapshots of the catalytic pockets of TEN2 (E, pre-cleavage) and Tc-toxin (F, post-cleavage) with catalytic residues represented as sticks and putative cleavage site indicated by red arrow. The two conserved residues (R2302/R638 and D2315/D651 in TEN/Tc-toxin) are similarly oriented in the catalytic pocket, whereas the non-conserved TEN2 G2345 and the residue with putative cleavage activity (Y2349) are remote to the catalytic pocket compared to the bacterial toxin. G, The TEN β-propeller domain resembles the TcA binding domain in Tc-toxins. TEN is presented to the left, Tc-toxin to the right. The β-propeller is highlighted in blue in TEN and orange in the toxin. H, The Ig-like domain in TEN plugs the bottom of the barrel. In bacterial toxins, multiple copies of domains with a similar Ig-like fold are attached to the A-component and serve as anchoring points to cell surface receptors. The Ig-like domain is highlighted with a dashed black line.
In addition to the barrel, the six-bladed β-propeller is similar to the TcA binding domain in bacterial component B, while the Ig-like domain is structurally similar to the receptor binding domains that compose the Tc-toxin A-component (Figure 2G–H and Table S2). In the toxin system, the TcA binding domain (β-propeller) plugs the bottom of the barrel and is proposed to serve as a pH-triggered gate that opens to allow the fully processed toxin through a syringe-like component into the host cell (Figure 2G). In TEN2, the β-propeller is attached to the barrel in a perpendicular orientation and instead the barrel bottom is sealed with an Ig-like domain (Figure 2G–H). The receptor-binding domains in TcA are Ig-like domains that decorate the A-component and are thought to mediate the binding of the fully assembled complex to cell surface receptors on host cells (Figure 2H). Considering the role of these components in mediating binding of bacterial toxins to cell surface receptors, it is likely that these domains play similar roles in TENs and mediate interactions with partner receptors.
The TEN2 toxin-like region resembles hormone and neuropeptide precursors
The structural homology of TEN2 with bacterial toxins implies that, through evolution, eukaryotes adopted toxin structural elements to fulfill new roles in these organisms. One of the most peculiar features in TENs is the partially encapsulated C-terminal region that emerges out of the barrel, and is thus partially exposed to the extracellular environment. Comparison of this domain with common bacterial toxins did not indicate any significant sequence homology. One exception is the Geobacillus virus E2 endonuclease precursor GVE2-HNH (PDB ID 5h0m, 15% sequence identity) (Zhang et al., 2012), which encodes a protein with DNase activity (Figure S5 and Table S2) and is structurally similar to the C-terminal region that lies outside the TEN barrel (Figure S5). Sequence alignment with enzymes with known DNase activity shows that TEN2 lacks two out of the three conserved HNH catalytic residues that are considered to mediate DNA cleavage (Figure S5). Thus, the TEN C-terminal region is not expected to serve as a DNase, even though the possibility that it binds DNA cannot be excluded at this point.
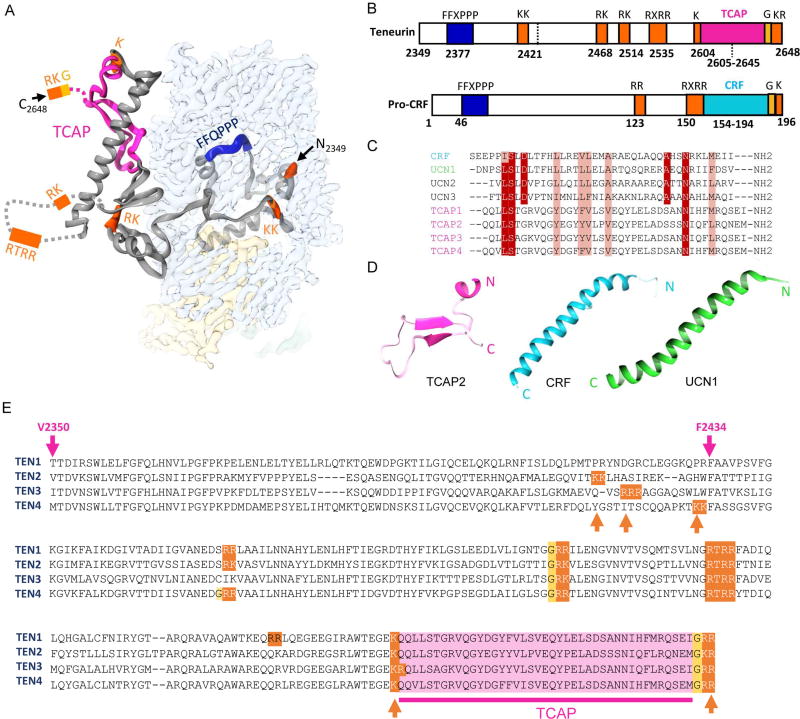
A 41-amino-acid C-terminally amidated peptide that corresponds to the TEN1 C-terminal sequence in mice brain, termed ‘teneurin C-terminal associated peptide 1’ (TCAP1), has been shown to share conserved elements with neuropeptides and corticotrophin releasing factor (CRF) (Figure 3) (Lovejoy et al., 2006; Lovejoy and de Lannoy, 2013). CRF, also a 41-amino-acid long peptide that is amidated at the C-terminus, is produced from a hormone precursor through sequential cleavage by protein proteases. Comparison of the CRF precursor and the C-terminal toxin-like region of TEN2 (Figure 3) reveals the presence of elements that are highly conserved in pre-pro-hormones and neuropeptide precursors; these include sequential pairs of basic residues (KK, KR, RR or RK) known to be sensitive to proteolytic cleavage by peptidases, a highly conserved RXRR furin cleavage site, and two C-terminal basic residues that are prone to processing by carboxy-peptidases. In addition, similar to CRF, the TCAP sequence is flanked by a cleavable basic residue at the N-terminus and an amidation motif at the carboxy terminus. Similar motifs are also found in the TCAP of other TENs (Figure 3E). These shared elements imply that, like CRF, the C-terminus of TENs may be post-translationally processed into TCAP as a putative bioactive neuromodulator. In support of this role, TCAP1 injection in mice was found to induce a relatively broad spectrum of neuromodulatory effects that were mostly related to behavior and stress (Al Chawaf et al., 2007; Erb et al., 2014; Lovejoy et al., 2006; Lovejoy and de Lannoy, 2013; Tan et al., 2011; Wang et al., 2005). Our results, however, show that the non-cleaved TCAP structure diverges structurally from CRF and other related neuropeptides (Figure 3D), raising the possibility that upon cleavage and processing TCAP may undergo major structural transitions.
Figure 3. The TEN2 toxin like domain shares sequence motifs with hormone and neuropeptide precursors.
The TEN2 toxin like domain shares similarity with pre-pro-hormones bearing multiple basic residues that are sensitive to post translational cleavage events by proteases. 3D (A) and 2D (B) representations of structural motifs with resemblance to pre-pro-hormones and neuropeptides. B, Comparison of structural motifs with Pre-Pro-CRF. C, Multiple alignment of fully processed CRF and TCAP1-4 sequences reveal conservation. D, In contrast to CRF (PDB ID 1GOE) and UCN1 (PDB ID 2RMF), the unprocessed TCAP does not assume an alpha helical fold. E, The potentially cleaved product in TENs shares hormone/neuropeptide precursor properties and may result in the post-translational processing of TCAP peptides. Basic residues sensitive to protease and furin cleavage are highlighted in orange, glycines that could be the target of alpha-amidating enzymes in yellow, and TCAP in pink. V2345 in TEN2 is predicated to be cleaved by the aspartic acid protease embedded in domain 4. F2434 is the exit point of the encapsulated domain of the barrel.
The toxin-like region of TEN2 binds to LPHNs and regulates cAMP in neighboring cells
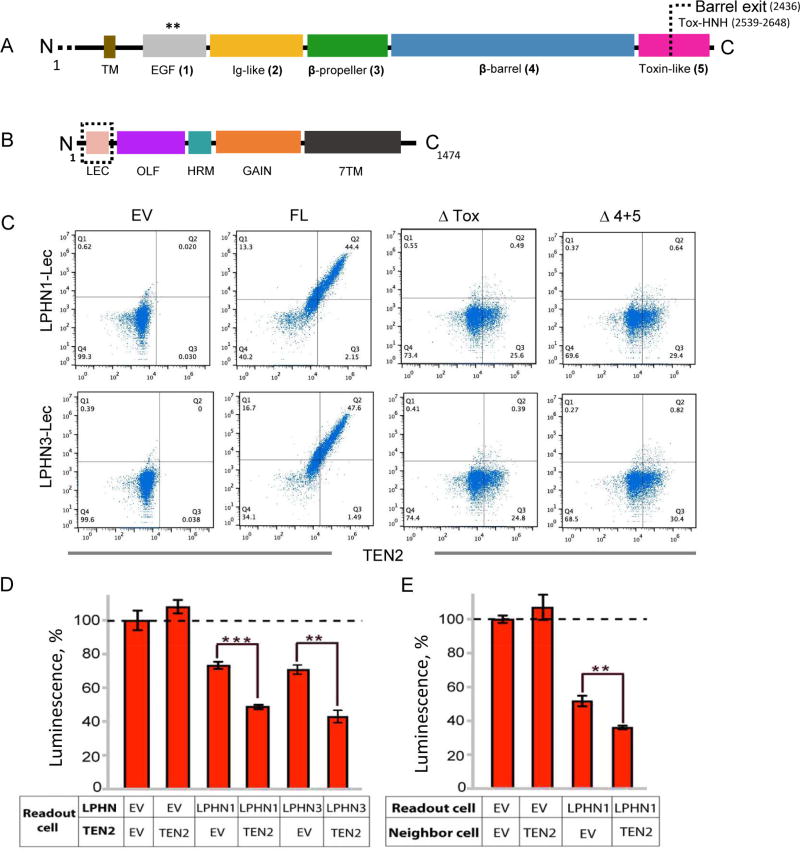
Recent studies have shown that TENs are involved in heterotypic interactions with different protein partners to mediate multifaceted physiological roles (Leamey and Sawatari, 2014; Sudhof, 2017; Woelfle et al., 2016) One of the main interaction partners are LPHNs (LPHN1-3), a family of adhesion GPCRs that form high affinity trans-cellular adhesion complexes with TENs and have roles in embryogenesis, tissue polarity and synapse development (Anderson et al., 2017; Boucard et al., 2014; Hamann et al., 2015; Silva et al., 2011). The TEN/LPHN interaction is mediated by the lectin (LEC) and olfactomedin (OLF) domains of the LPHN ECR (Figure 4A–B), with the LEC domain contributing most of the binding affinity (Boucard et al., 2014; O’Sullivan et al., 2014). To characterize the TEN domains involved in LEC interactions, we assessed the binding of the soluble biotinylated LEC domain of LPHN1 or LPHN-3 to mammalian cells overexpressing TEN2 variants by flow cytometry (Figure 4C, and Figure S6). As expected, cells overexpressing full-length (FL)-TEN2 showed strong binding to biotinylated LEC, whereas control cells that were transfected with an empty vector (EV) did not. Two truncated TEN2 constructs where either the C-terminus of the toxin-like region or both the toxin-like region and the β-barrel were removed (ΔTox and Δ4,5, respectively), did not bind LEC (Figure 4C). In contrast, the binding affinity was not affected by mutating the unpaired cysteine residues in the EGF repeats to eliminate cis-dimer formation (Cis, Figure S6). These results suggest that the soluble LEC domain interacts with the TEN2 toxin-like region regardless of TEN2 cis-dimerization. The findings are also supported by an earlier study showing that a soluble form of the TEN2 C-terminal region that fused to an N-terminal GST tag was able to bind to cells expressing LPHN1 (Silva et al., 2011).
Figure 4. The Toxin-like domain of TEN2 mediates LPHN-dependent trans-cellular signaling.
A, Domain architecture of TEN2 with ECR domains corresponding to the EGF-repeat (gray), Ig-like (yellow), β-propeller (green), β-barrel (blue) and toxin-like (magenta) domains. The barrel exit point, splitting the C-terminal domain to two segments, is indicated. ECR domain numbers are indicated in parenthesis. Residue numbers are indicated. B, Domain architecture of LPHNs with large extracellular N-terminal region containing four main structural domains (lectin-binding domain-LEC, olfactomedin-OLF, hormone-binding domain - HRM and GAIN domain) with the typical GPCR transmembrane domain (7TM). C, Flow cytometry analysis of purified biotinylated lectin domain of LPHN1 (top) or LPHN3 (bottom) binding to TEN2 constructs expressed on HEK293T cells. Shown are representative dot plots demonstrating the correlation between TEN2 expression and LPHN binding in TEN2-transfected cells and un-transfected control. TEN2 construct variants correspond to the domain description in A. EV stands for empty vector that served as control, indicating the lack of binding. The effect of TEN2 on downstream G-protein signaling of LPHN as measured by the decrease in intracellular cAMP level. The cAMP-based signaling assay was performed in cis (D) and trans (E) setups: D, HEK293 cells were co-transfected with TEN2 and LPHN to monitor the effect of TEN2 on LPHN activity where both proteins are presented on the same cell. E, A population of cells expressing TEN2 was mixed with LPHN expressing cells. Intracellular cAMP level of the LPHN-expressing “readout cells” was measured. Overexpression of TEN2 in the neighboring cell further decreased cAMP level in the LPHN-transfected readout cells.
Although TENs and LPHNs are co-expressed and have key roles during embryogenesis, the role of the TEN/LPHN interaction in embryonic development has not been studied. LPHNs were reported to mediate embryogenesis in C. elegans by modulating intracellular cAMP levels (Winkler and Promel, 2016), prompting us to test whether human TEN binding to LPHN also induces trans-cellular cAMP signaling. We established a cAMP-based signaling assay for LPHN1 or LPHN-3 in mammalian cells (Figure 4D–E and Figure S7A–F), and used it as a readout to monitor receptor activity. The experiments were performed in two experimental setups, “cis” and “trans” (Figure S7): In the “cis” setup, the same cell population was co-transfected with both full-length TEN2 and full-length LPHN1 or LPHN-3 in order to monitor the effect of both proteins present on the same cell (Figure 4D, Figure S7). Comparison of the cAMP levels in cells overexpressing LPHN to cells overexpressing both LPHN and TEN2 revealed that the presence of TEN2 causes a further decrease in cAMP levels in a LPHN-dependent manner, suggesting activation of LPHN by TEN. To mimic trans-cellular signaling, we designed the “trans” setup whereby a population of cells expressing TEN2 was mixed with a different population of cells that were expressing LPHN (Figure 4E, Figure S7). We then assessed the effect of TEN2 on LPHN activity in neighboring cells by monitoring the cAMP levels only from the LPHN-expressing cells. Comparison of the cAMP levels in LPHN-expressing cells mixed with empty vector-expressing or TEN2-expressing cells showed that TEN2 enhanced the LPHN-dependent cAMP decrease in neighbor cells (Figure 4E). These results suggest that TEN2 activates LPHN and induces trans-cellular signaling by modulating cAMP levels. Synthetic TCAP has also been observed to reduce cAMP levels in immortalized neuron-like cells, but in these experiments its dependence on LPHN was not explored (Wang et al., 2005). However, although these experiments show that TEN-binding to LPHNs stimulates trans-cellular signaling, the reduced system employed (transfected HEK293 cells) differs from a physiological environment and does not allow for conclusions about the physiological signaling mode of LPHNs.
Alternative splicing regulates trans-cellular ligand interaction and synapse formation
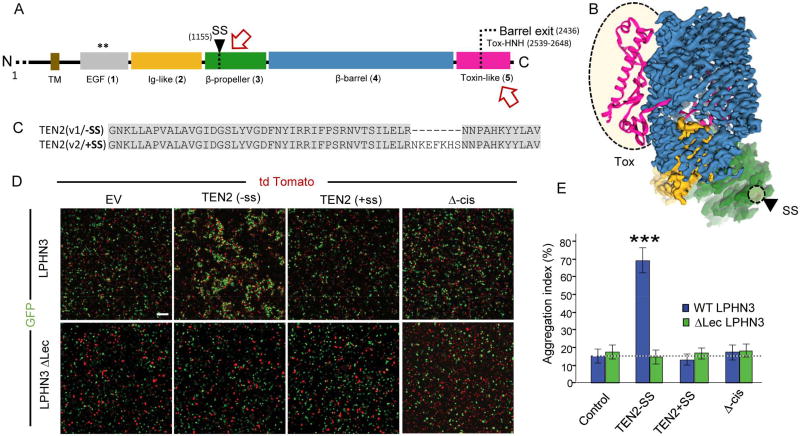
TENs induce multiple distinct physiological activities that may not be solely mediated through their interactions with LPHNs. Accordingly, several studies have suggested that TENs engage additional ligands, including other TENs, integrins and dystroglycans (Chand et al., 2013; Trzebiatowska et al., 2008), even though the evidence for these associations is mostly indirect. With regard to the mechanism regulating such a diverse spectrum of activities, one suggestion is that both TENs and their matching partners are differentially expressed in tissues. Indeed, vertebrate TEN2 expression was shown to be temporally regulated (Otaki and Firestein, 1999a, b), while different TEN species are expressed in divergent CNS regions. Another mechanism that receives increasing experimental support is the utilization of alternate splice variants. In LPHNs, alternatively splicing of a five-residue insert located in the linker connecting the LEC and OLF domains regulates binding to TEN2 and TEN4 (Boucard et al., 2014). TEN is also alternatively spliced with two splice variants of the TEN2 ECR identified in humans: One variant carries a nine-amino-acid insertion in the EGF region responsible for dimerization (domain 1), while the other splice variant carries instead a seven-amino-acid insertion in the β-propeller domain (domain 3; Figure 5A–C).
Figure 5. Alternative splicing in the β-propeller domain regulates LPHN binding to TEN2.
2D (A) and 3D (B) schematic representation of TEN2, highlighting relevant regions that were shown to influence the interaction with LPHN (influenced regions are indicated by arrows on the 2D scheme and circled in the 3D model). C, Comparison of the two TEN2 splice variants within their β-propeller region. V1 is the construct used for structural studies and has a seven-amino-acid deletion. D, Examination of trans-cellular adhesion mediated by TEN2 and LPHN3 via cell aggregation assays. Freestyle HEK293 cells were co-transfected with the indicated TEN2 or LPHN3 and either tdTomato or EGFP, respectively. LPHN3 lacking the Lectin domain (ΔLec LPHN3), which is the domain responsible for binding to TENs, was used as a control. Scale bar denotes 50mm. E, Quantification of aggregation index (%) in indicated conditions (n = 3 independent replicates with quantifications from 10 randomly selected imaging fields per replicate; *** p<0.001 by ANOVA).
To probe the effect of alternative splicing in the TEN β-propeller on TEN/ligand interactions, we conducted cellular assays with non-adherent HEK293 cells in which the full-length proteins are expressed on different cell populations, and the cells are then mixed to monitor cell aggregation (Figure 5, and Figure S7G). In these experiments, we examined which TEN domains are required for trans-cellular LPHN interactions and/or homotypic TEN interactions, and if these interactions are regulated by the alternative splice variants within the β-propeller (domain 3, Figure 5C–E and Figure S7). Full-length TEN2 lacking the β-propeller splice insert robustly induced trans-cellular aggregation with full-length LPHN3, while truncation of the LEC domain of LPHN3 abolished trans-cellular adhesion. Intriguingly, inclusion of the seven-amino-acid splice insert in the β-propeller eliminated trans-cellular adhesion with LPHN3 for all TEN2 forms tested, even though this splice site is distant from the C-terminal domain responsible for LEC binding. Furthermore, trans-cellular aggregation with LPHN3 is prevented when TEN2 includes the β-propeller splice insert or carries point mutations that abolish cis-dimerization (Figure 5B). We also note that none of the TEN2 forms tested exhibited homotypic trans-cellular interactions using the assay that demonstrated heterotypic cell aggregation (Figure S7G). These observations contrast those of the flow cytometry experiments in which the affinity of TEN2 for the soluble LEC domain was not affected by inserting the seven-residue segment or by eliminating dimerization (Figure S6). We thus postulate that trans-cellular TEN2/LPHN3 interactions are regulated by β-propeller alternative splicing likely due to conformational restrains in the context of full-length proteins.
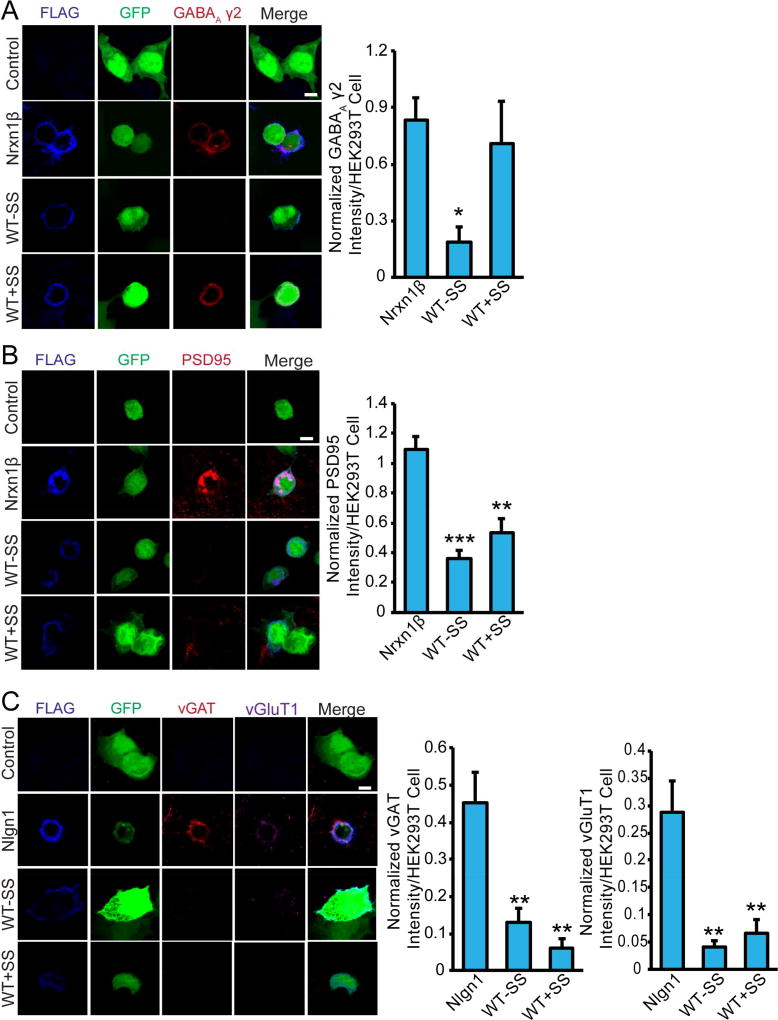
We subsequently examined the ability of TEN2 +/− splice insertion (+/− SS) in the β-propeller to induce artificial synapse formation. HEK293 cells expressing these TEN variants were co-cultured with primary hippocampal neurons, using HEK293 cells expressing neurexin-1 that is known to induce postsynaptic specializations as a positive control (Figure 6). These experiments revealed that while TEN2 -SS failed to recruit synaptic markers, TEN2 +SS potently induced postsynaptic GABAergic specializations similar to neurexins (Figure 6). TEN2 +SS selectively recruited postsynaptic but not presynaptic markers, consistent with TENs being localized to the presynaptic compartment. Together with our cell adhesion assay results, these findings suggest that TEN alternative splicing regulates binding to different interaction partners and has functional impacts on specific synapse subtypes (Figure 7).
Figure 6. TEN alternative splicing regulates inhibitory synapse formation.
Artificial synapse formation assays testing the effect of TEN splice forms. (A) TEN2 WT+SS but not WT-SS induced inhibitory post-synaptic specialization. Left, Mouse cortical neurons were co-cultured with HEK293 cells expressing GFP and either no other protein (Control), Flag-tagged Nrxn1β, WT-SS, or WT+SS at DIV16. 24-hrs later, immunocytochemistry for Flag (blue) and GABAA γ2 (red) was performed. Right, Bar graphs, Fluorescent intensity of GABAA γ2 was normalized with that of Flag and was quantified (n = 2 independent replicates and 12 cells per replicate; * p<0.05 by ANOVA; scale bar denotes 10µm). (B) Neither WT-SS nor WT+SS induced excitatory post-synaptic specialization. Artificial synapse formation assay was performed as in A, except that immunocytochemistry for Flag (blue) and PSD95 (red) was performed (n = 2 independent replicates with 15 cells per replicate** p<0.01; *** p<0.001 by ANOVA; scale bar denotes 10µm). (C) Neither WT-SS nor WT+SS induced pre-synaptic specialization. Artificial synapse formation assay was performed similar to A, with immunocytochemistry for Flag (blue), vGlut1 (purple), and vGAT (red). Nlgn1 was used as a positive control (n = 2 independent replicates with 10 cells per replicate** p<0.01 by ANOVA; scale bar denotes 10µm). Significance between experimental groups is marked with asterisks, * p≤0.05; **p≤0.01; ***p≤0.001.
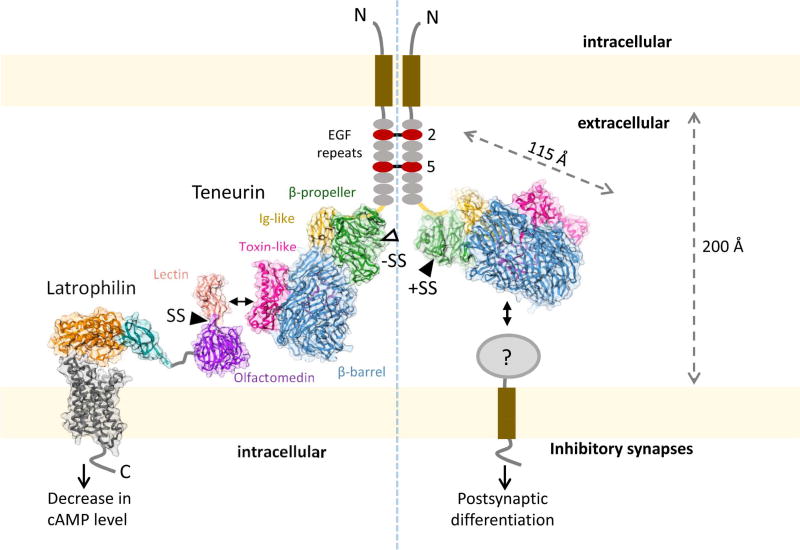
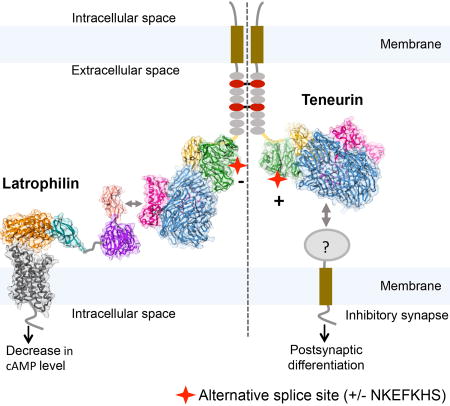
Figure 7. Model for alternative-splice dependent functions of TEN2.
The model depicts how alternative splicing acts as a molecular switch to determine which adhesion partner TEN2 binds to, and, accordingly, which cellular functions TEN2 mediates. On the left, the TEN2 variant lacking the β-propeller splice insert is able to mediate trans-cellular interaction with LPHN and modulate cAMP levels in the neighbor cell. On the right, TEN2 variant including the splice insert is unable to interact with LPHN, and instead induces postsynaptic differentiation in inhibitory synapses by interacting with an unknown partner. The left and right sides of the TEN2 dimer represent various cell-cell junctions and inhibitory synapses, respectively. Model drawn to scale. The LPHN structure is based on the GAIN and HormR domain structure (4DLQ, colored orange and teal, respectively), Lectin and Olfactomedin domain structure (5AFB, colored light pink and violet, respectively) and the modeled TM domain using the corticotrophin-releasing hormone receptor structure (4K5Y, dark grey).
DISCUSSION
TENs are membrane attached proteins found in all multicellular organisms. In mammals, TENs are broadly expressed during embryonic development and are essential for mesoderm induction (Lossie et al., 2005; Nakamura et al., 2013; Tucker and Chiquet-Ehrismann, 2006). By contrast, TENs in adults are primarily expressed in the brain where they mediate multiple functions in tissue polarity, axon guidance, and synapse formation (Leamey and Sawatari, 2014; Sudhof, 2017; Woelfle et al., 2016). Here we show that the large ECR of TENs has an unusual architecture that shares an unexpected structural homology with bacterial Tc-toxins (Busby et al., 2013; Meusch et al., 2014; Minet and Chiquet-Ehrismann, 2000; Young and Leamey, 2009). Interestingly, no similar motif has been reported in any other eukaryotic protein, raising questions about the evolutionary mechanism that transferred the Tc toxin elements from prokaryotes to eukaryotes and about the physiological role of this unusual cell-adhesion protein.
Recent evidence suggests that the evolution of TEN-like homologs from bacteria to humans might have originated from the incorporation of bacterial polymorphic toxin genes into the genome of choanoflagellates which were later adopted by higher eukaryotes (Tucker et al., 2012). Choanoflagellates are eukaryotic unicellular organisms that are often referred to as the origin of multicellularity because their genome contains genes that mediate cell-cell adhesion in Metazoans and because several choanoflagellate species form highly ordered multi-cellular colonies (King, 2004). In addition to TENs, choanoflagellate genomes also include LPHN-like adhesion GPCR homologs (King et al., 2003), hinting that the TEN-LPHN trans-cellular interaction may have occurred early in evolution and have contributed to the generation of multicellularity. In this context, it is possible that TEN function diversified through evolution from toxin encapsulation and release to diverse roles in tissue development, embryogenesis, axon guidance and synapse formation. One notable example to support this notion can be found in the partially encapsulated C-terminal TEN region, which we show here to mediate interactions with LPHNs through their LEC N-terminal domain (Figures 4 and S6). In contrast to Tc toxins, the C-terminal region in TEN has to emerge from the barrel in order to be fully accessible for mediating the interactions with its binding partners. This observation, however, does not explain the functional role of the TEN barrel itself, which in bacterial toxins serves as a chamber to protect the producing cell from being exposed to the toxic cargo and also functions as an autoproteolysis machinery that cleaves and activates the encapsulated toxin (Busby et al., 2013; Meusch et al., 2014). The possibility that the barrel of TEN species encompasses an active auto-protease machinery is exciting because of the similarity with the presumably cleaved domain with HNH-DNases, pre-pro-hormones and neuropeptide precursors (Figure 3), and because their mature product, TCAP, is detected in brain lysates and was shown to mediate multiple functions in the brain (Al Chawaf et al., 2007; Erb et al., 2014; Lovejoy et al., 2006). Moreover, such an autoproteolysis event may release the toxin-like region into the extracellular space and convert TEN into a diffusible ligand that may act during axonal pathfinding. This might give the TEN barrel a possible catalytic role in modulating the integrity of the encapsulated aspartate protease, but also suggests a role as a shield, protecting the protease-sensitive cargo from the protease enriched extracellular environment.
Alternative splicing in proteins is an important mechanism to greatly expand the functional capacity of metazoan genomes, and its regulatory role in brain function has been repeatedly demonstrated. For instance, DSCAMs, protocadherins, calcium channels, neurexins and neuroligins have been shown to use alternative splicing for diversifying their functions (Aoto et al., 2013; Fuccillo et al., 2015; Irimia et al., 2014; Thalhammer et al., 2017). Similarly, the divergence of TENs from their bacterial toxin ancestors is associated with the emergence of spliced isoforms. TENs are alternatively spliced at two canonical sites that are localized to the ECR and include nine- and seven-amino-acid insertions at the EGF repeats and the β-propeller regions, respectively (Tucker et al., 2001). Here we show that alternative splicing within the β-propeller domain acts as a switch to regulate selective binding of TEN2 to adhesion partners, thus controlling the cellular functions TENs perform. The TEN variant that includes the insertion was unable to mediate trans-cellular adhesion with full-length LPHN presented on the neighboring cell, whereas the variant without the insertion interacted with LPHN with high affinity. The alternatively spliced site, however, localizes at least 8 nm away from the LEC binding site (Figures 4 and S6) and is thus unlikely to participate in any interactions with the LPHN lectin domain. These observations suggest that alternative splicing may facilitate a conformational change of the entire ECR that renders the LPHN binding site inaccessible to the full-length LPHN on the neighboring cell (Figure 7). It is also possible that additional domains in LPHN, localized remotely from the binding interface, contribute to their binding to TENs while mediating their cell-cell adhesion properties.
As TENs play key roles in numerous functions, from tissue development and embryogenesis to axon guidance and synapse formation, it is likely that different splice variants have different roles during development (Figure 7). Here we found that the TEN2 variant interacting with LPHNs can modulate cAMP levels in cells expressing LPHN, suggesting that TEN activates LPHN in a trans-cellular manner (Figure 4D). Similar to TENs, LPHNs are also conserved between vertebrates and invertebrates, have broad expression patterns, and are involved in embryogenesis (Langenhan et al., 2009; Muller et al., 2015; Scholz et al., 2015). In both C. elegans and Drosophila, LPHN was suggested to act in development by modulating cAMP levels (Scholz et al., 2017; Winkler and Promel, 2016). Considering that cAMP regulates diverse cellular functions including migration, adhesion and differentiation, our results suggest a role for the TEN-LPHN interaction and the subsequent modulation of cAMP levels in various contexts, including development.
Unexpectedly, we found that the TEN2 splice variant that is incapable of interacting with LPHN is a potent inducer of inhibitory synapses, indicating that it has functional impact on specific synapse subtypes through interactions with alternative binding partners that remain to be discovered (Figures 6, 7). Specifically, the TEN2 variant including the β-propeller splice site induced inhibitory postsynaptic differentiation, suggesting a LPHN-independent role for TEN in synapse formation. We did not detect induction of excitatory presynaptic specialization by splice isoforms of TEN, possibly because unknown co-factors may be required for excitatory but not inhibitory synapse formation. Although it has been suggested that homotypic binding of TENs in trans may may contribute to synapse formation and/or axonal pathfinding (Hong et al., 2012; Mosca et al., 2012), our cell aggregation assays did not detect robust homotypic trans-cellular interactions between various TEN2 splice variants (Figure S7G) (Boucard et al., 2014). At present we cannot exclude potential low-affinity homotypic interactions between TENs, but it is difficult to envision how low-affinity homotypic interactions between TENs could physiologically compete with the high-affinity TEN-LPHN interaction, given the universal expression of these protein partners in all neurons (although in distinct developmental and isoform-specific patterns). A heterotypic role for TENs is also suggested by the fact that they only induced post-but not pre-synaptic specializations in the synapse formation assay (Figure 6). Overall, our results reveal that alternative splicing of TENs may serve as a general principle to regulate their diverse interactions with multiple adhesion partners, giving rise to the multifaceted roles they play. Future studies investigating developmental and spatial patterns of alternative splicing, as well as regulation by extrinsic factors such as neuronal activity, will generate important insights into the regulation of synapse formation and specification as well as embryogenesis and tissue development by TENs. As these investigations intensify, the structure of TEN2 described here will form the structural framework to start delineating the diverse mechanisms of TEN function.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed and will be fulfilled by the Lead Contact Demet Araç (arac@uchicago.edu).
Experimental Model and Subject Details
Cell Culture
High-Five insect cells (Trichoplusia ni, female, ovarian) were cultured in Insect-Xpress medium (Lonza) supplemented with 10 µg/mL gentamicin at 27°C and were used for productio n of recombinant proteins. HEK293/HEK293T mammalian cells (Homo sapiens, female, embryonic kidney) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% FBS (Sigma) at 37°C in 5% CO 2 and were used for cell-surface expression assays, G protein signaling assays and flow cytometry binding assays.
Method Details
Cloning and expression in insect cells
Various ECR constructs of human TEN2 (TEN) variant Lasso (including no splice site in propeller or EGF repeats) (UniProt: Q9NT68-2) and LPHN (LPHN1, UniProt: O88917; LPHN3, UniProt: Q9HAR2) were cloned into a pAcGP67a vector and expressed in High-Five insect cells using the baculovirus expression system as previously described (Arac et al., 2012). For the structural studies, TEN ECR (residues N456-R2648) and TEN ECRΔ1 (residues T727-R2648) were cloned with N-terminal 8XHis-DesG-tags (Skinner et al., 2014). 72 hr after viral infection of High-Five cells, the medium containing secreted glycosylated proteins was collected and centrifuged at 900 g for 15 min at room temperature. After centrifugation, the supernatant was transferred into a beaker. The following were added to the supernatant (to final concentrations): 50 mM Tris pH 8.0, 5 mM CaCl2 and 1 mM NiCl2 and stirred for 30 min. After centrifugation at 8,000 g for 30 min, the supernatant was collected and incubated with nickel-nitrilotriacetic agarose resin (Qiagen) for 3 hr at room temperature. The resin was transferred to a glass Buchner funnel and rinsed with HBS buffer containing 20 mM imidazole, then transferred to a poly-prep chromatography column (Bio-rad). Target protein was eluted with HBS buffer containing 200 mM imidazole and then applied to size-exclusion chromatography (Superdex 200 10/300 GL; GE Healthcare), purified in a final buffer comprised of 10 mM Tris pH 8.5, 150 mM NaCl. The peak fractions were pooled for further analysis. For the flow cytometry assays, LPHN1 lectin (residues S26-Y131) and LPHN3 lectin (residues S21-Y126) were cloned with carboxyl-terminal 6XHis-AVI-tags and purified as described above.
Cloning and expression in mammalian cells
Full-length TEN (residues M1-R2648), ECRΔ5 (residues M1-E2384) and ECRΔ4,5 (residues M1-F1454) constructs and other TEN mutants bearing HA-tag (inserted between K405/E406) and FLAG-tag at the carboxyl-terminus were cloned into a pcDNA3.1 vector for cell-surface expression assays and flow cytometry binding assays in HEK293T cells. TEN mutants were generated with a standard two-step PCR-based strategy.
On-column biotinylation
His-AVI-tagged lectin was captured on nickel-nitrilotriacetic resin as described above. Following a 20 mM imidazole HBS buffer wash, final concentrations of 50 mM Bicine pH 8.3, 10 mM Mg-acetate, 100 mM NaCl, 10 mM ATP, 0.5 mM biotin and 5 µM BirA were added to the resin, which was then rotated at 27°C for 2 hr. Following incubation, the column was washed with 20 mM imidazole HBS buffer to remove residual BirA and ATP, and then biotinylated lectin was eluted with HBS buffer containing 200 mM imidazole. Purified protein was applied to size-exclusion chromatography as described above. The efficiency of biotinylation was assessed using streptavidin beads.
Flow cytometry
HEK293T cells were cultured in 6-well plates in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% FBS (Sigma), at 37°C i n 5% CO2. Transfection was performed with cells at 50–60% confluence. Cells were transiently transfected as follows: Dilute 2 µg cDNA in 50 µl serum-free DMEM. At the same time, dilute 3 µl LipoD293 transfection reagent with 47 µl serum-free DMEM. Add diluted LipoD293 to diluted cDNA and incubate the mixture for approximately 10 min. Then, gently add the transfection complex mixture dropwise to each well. After 48 hr incubation, the cells were detached using citric saline solution (135 mM KCl, 50 mM sodium citrate) and washed with PBS + 2% BSA. To test cell-surface expression of FLAG/HA-tagged TEN, cells were stained with a primary antibody mixture: mouse anti-FLAG M2 1:1000 and rabbit anti-HA 1:1000 for 30 min at room temperature. After wash with PBS + 2% BSA, cells were incubated with a secondary antibody mixture: donkey anti-mouse Alexa Fluor 488 1:3000 and goat anti-rabbit Alexa Fluor 647 1:3000 for 30 min. After washing, cell pellets were resuspended in PBS + 2% BSA immediately before flow cytometry acquisition using an Accuri C6 flow cytometer (BD). Acquired data was analyzed using the FlowJo analysis software (FlowJo LLC).
For the binding assays, 100 nM biotinylated lectin was tetramerized and fluorescently labeled through incubation with NeutrAvidin DyLight 650 on ice for 20 min. Cultured cells expressing FLAG-tagged TEN were detached using citric saline solution and then washed as described above. Next, the cells were stained with mouse anti-FLAG M2 1:1000 antibody and, following two wash cycles, stained with donkey anti-mouse Alexa Fluor 488 antibody in the presence of the labeled lectin mixture. Similarly, rabbit anti-HA, goat anti-rabbit Alexa Fluor 647, and NAV488-lectin were used to test the expression of HA-tagged TEN in HEK293T cells and its binding with purified lectin.
G protein signaling assay
HEK293 cells were grown to 50–60% confluence in 6-well plates in DMEM + 10% FBS. In-cis signaling assay: cells were co-transfected with 0.35 µg LPHN1/LPHN3 or pCMV5 empty vector; 0.35 µg luciferase reporter plasmid 22F (E2301, Promega; a generous gift from R. Lefkowitz lab); 0.009 µg β2AR, and 0.35µg TEN or pcDNA3.1 empty vector, using a ratio of 1:4 DNA (µg): Fugene6 (µl) according to the manufacturer’s instructions. Following 24 hr incubation in 5% CO2 incubator, cells were detached and transferred to a 96-well white opaque plate (3917; Corning) at a density of 50,000 cells per well. Cell media was changed to pre-warmed CO2-independent Opti-MEM I Reduced-Serum Medium after 24 hr of incubation. The cells were then incubated for another 30 min in CO2 incubator before equilibrating with Glosensor substrate at 1% final concentration. Basal cAMP signal was measured by detecting luminescence using a Synergy HTX plate reader (BioTek) at 25°C, GAIN 200. β2AR was activated by adding isoproterenol at a final concentration of 50 nM. Kinetic measurements of luminescence were done at 5 min intervals immediately after activation. In-trans signaling assay: HEK293 cells were similarly prepared while a second set of cells were transfected with 0.35 µg TEN or pcDNA3.1 empty vector. Following 24 hr incubation, the two sets of cells were detached, mixed at a 1:1 ratio, and then transferred to a 96-well white opaque plate. After an additional 24 hr incubation, the signal was recorded similarly to the in-cis method.
Cell aggregation assays
FreeStyle HEK293 cells (Life Technologies) grown to a density of 1×106 cells/mL in a 30mL volume were co-transfected with 30µg of either pCMV-Emerald or pCMV-tdTomato and 30µg of the indicated construct using a FreeStyle Max reagent (Life Technologies). All cDNAs were encoded in the pCMV5 vector. FreeStyle HEK293 cells were grown at 37°C/8%CO2 with shaking at 125rpms. Transfected cells were mixed in non-coated 12 well plates at a 1:1 ratio two days post-transfection and subsequently incubated for an additional 2 hours. Live cells were imaged in 12 well plates using a Nikon A1 confocal microscope. Aggregation index was calculated as previously described (Boucard et al., 2014).
Artificial synapse formation assays
Expression vectors were transfected into HEK293 cells. 24 hrs later, HEK293T cells were co-cultured with cultured cortical neurons (DIV16) from neonatal mice at P0. After 24 hrs, cells were fixed with 4% PFA and immunostained with mouse or rabbit anti-Flag (Sigma; 1:1000 both) together with mouse anti-PSD95 (Sysy; 1:500), mouse anti-GABAA γ2 (Sysy; 1:500), or guinea pig anti-vGlut1 (Millipore; 1:500) and mouse anti-vGAT (Sysy; 1:500) respectively. Species-specific AlexaFluor-405-, −546-, and −633-conjugated antibodies (Invitrogen) were used as secondary antibodies. Images were collected with a Nikon A1 confocal microscope using a 60x objective. The quantification of the artificial synapse formation assay results was performed as follows. The fluorescent intensities of the Flag-tagged proteins expressed in the HEK293T cells (e.g. TENs, Nlg1, Nrn1β) and that of the synaptic markers that were recruited to the surface of the HEK293T cells (e.g. GABAγ2, PSD95, vGlut, vGAT) were quantified using Image J. Normalized values equal the fluorescent intensity of the synaptic marker that was examined (GABAγ2/PSD95/vGlut1/vGAT) / the fluorescent intensity of the Flag-tagged protein expressed in the HEK cells (TENs/Nlg1/Nrn1β).
Negative stain-EM
Purified TEN2-ECR samples (0.01 mg ml-1) were prepared for negative stain EM using uranyl formate (Peisley and Skiniotis, 2015). The negative stained samples were imaged at room temperature with a Morgagni electron microscope (FEI) operated at 80 kV. Images were recorded at ×25,000 magnification and a defocus value of −0.5 µm on an Orius CCD camera (Gatan, Inc.).
Cryo-EM data acquisition
3.5µl purified TEN2-ECR-2–5 sample (0.15 mg ml-1) was applied on glow-discharged holey carbon grids (Quantifoil R2/2, 300 mesh, EMS), and vitrified using a Vitrobot Mark IV (FEI Company). The specimen was visualized using a Titan Krios electron microscope (FEI) operating at 300 kV and equipped with a K2 Summit direct electron detector (Gatan, Inc.). Images were recorded with a nominal magnification of x29,000 in counting mode, corresponding to a pixel size of 1.0 Å on the specimen level. In total, 1,625 images with defocus values in the range of −1.5 to −2.5 µm were recorded using a dose rate of ~1.1 electrons per Å2 per second. The total exposure time was set to 8s with frames recorded every 0.2s, resulting in an accumulated dose of about 44 electrons per Å2 and a total of 40 frames per movie stack.
Image processing and 3D reconstructions
Stack images were subjected to beam-induced motion correction using MotionCor2 (Li et al., 2013). CTF parameters for each micrograph were determined by CTFFIND4 (Mindell and Grigorieff, 2003). Particle selection, two- and three- dimensional classifications were performed on a binned dataset with a pixel size of 2 Å using Relion2.1 (Fernandez-Leiro and Scheres, 2017). In total, 742,520 particle projections were selected using semi-automated procedures and subjected to reference-free two-dimensional classification to discard false-positive particles or particles categorized in poorly defined classes, resulting in 537,785 particle projections for further processing. An ab initio 3D map generated with VIPER (Moriya et al., 2017) was used as initial reference model for maximum-likelihood-based 3D classification in Relion2.1. Two particle subgroups accounting for 268,146 and 157,961 projections showed detailed features for ECR-domains-2-4-5 and ECR-domains-2-3-4-5, respectively, and were subjected to 3D refinement resulting in maps with global nominal resolutions of 3.3 Å and 3.4 Å, respectively. A final 3.1 Å map for ECR-domains-2-4-5 was calculated after combining the two classes (426,107 particles) and masking out the relatively flexible domain 3.
Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) using the 0.143 criterion and were in agreement with both Relion 2.1 and M-triage as implemented in Phenix (Adams et al., 2010). All density maps were corrected for the modulation transfer function (MTF) of the K2 summit direct detector and then sharpened by applying a temperature factor that was estimated using post-processing in RELION. Local resolution was determined using ResMap (Kucukelbir et al., 2014) with half-reconstructions as input maps.
Model building and refinement
An initial template of TEN-main domain (domain 4) was derived from a homology-based model calculated by SWISS model builder (Biasini et al., 2014). The model was docked into the EM density map using Chimera (Pettersen et al., 2004) and then manually adjusted and real-space refined to fit the density using COOT (Emsley et al., 2010). The Ig-like domain (ECR2) and ECR5 were manually traced. The final model was subjected to global refinement and minimization in real space using the phenix_real_space_refine module in Phenix (Adams et al., 2010). FSC curves were calculated between the resulting model and the half map used for refinement as well as between the resulting model and the other half map for cross-validation using Phenix M-triage (Figure S2). The final refinement statistics are provided in Table S1.
Quantification and Statistical Analysis
Error bars in Figure 4, 5, 6 and Figure S7 represent means ± SEM. Each measurement was repeated at least three times independently. Data was analyzed using software GraphPad Prism and ImageJ.
See “Methods Details” for details on cryo-EM analysis.
Data and Software Availability
Data Resources
The cryo-EM density map has been deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) under accession code EMD-7526 and the model coordinates have been deposited in the Protein Data Bank (http://www.rcsb.org) under accession number 6CMX.
Supplementary Material
(A) Gel filtration chromatography (left) and SDS-PAGE analysis (right) for the corresponding fractions of ECR (magenta) and ECRΔ1 (green) indicates that soluble ECR is an oligomer whereas ECRΔ1 is a monomer. (B) Cryo-EM micrographs of TEN2. Scale bar is 20 nm. (C), Representative reference-free two-dimensional cryo-EM averages of TEN2. The diameter of the circular windows is 26 nm. (D) Flow chart of cryo-EM data processing of TEN2 extracellular region, including particle selection, classification, and 3D density map reconstruction. Details are provided in the methods section.
(A) Resolution estimation of the EM map reconstructed from 426,107 particle projections. Gold standard Fourier shell correlation (FSC) curves, indicating an overall nominal resolution of 3.1 Å at 0.143 FSC. (B) FSC curve of the final refined model versus the cryo-EM map. FSC graphs were calculated with M-triage, by Phenix. (C) Three-dimensional density map colored according to local resolution indicating a resolution range of 2.5–3.5 Å in most map regions. Local resolution was calculated by ResMap. (D) Local resolution calculations for map resulting of 157,961 particles with a stable β-propeller indicating lower resolution for ECR 3 (β-propeller indicated by arrow) due to local flexibility. Color scheme for resolution values is as presented in panel C.
(A) Snapshots of map vs. model agreement by domain. Map is contoured at σ=3.0 and each region is highlighted by a dashed black box. Snapshots include the representative regions of domain 5 (top left), domain 2 (top right) and domain 4 (bottom left). Snapshot of one representative sugar modification in the cryo-EM density is presented at the right bottom panel. (B) Domain 3 homology model fitting to the low-resolution map area. The protein backbone is clearly delineated in the map. Visible side chains were used as anchoring points for the docking of the homology model (template model PDB ID 3fvz). The model was real-space refined in the map using Coot and Phenix_real_space_refine.
Two- (A) and three-dimensional (B) representations of TEN2, with domains color coded as in Figure 1. Residue numbers are indicated. (C) An exploded view of each TEN2 domain. Domain borders are numbered, N- and C- termini of each domain are highlighted by spheres. The Ig-like region in ECR2 is indicated by a dashed line. Alternative splicing variant region of ECR3 is indicated by SS. Dormant aspartate dyad in ECR4 is highlighted in yellow. Barrel exit point and furin cleavage site in ECR5 are pointed by arrows, Tox-HNH is highlighted in yellow (domain 5) and TCAP is circled by a dashed line (domain 5). Domain borders and highlighted regions are indicated in black.
(A) The C-terminal of ECR5 emerges from the barrel and remains attached to the outer barrel region by a network of hydrogen bonds. ECR5 is in magenta. H-bonds are indicated by dashed lines. (B) The encapsulated domain (ECR5, magenta) interacts with domain 2 (yellow) that penetrates into the barrel and plugs its bottom. Contact points are highlighted by black arrows. (C) Two notable lysins of the encapsulated domain (K2421, K2422) mediate interactions of the barrel bottom with Y2349, the residue presumed to be cleaved by the aspartic acid protease. (D) The C-terminal of ECR5 (residues 2539-2648) shares structural similarity with HNH DNases. (E) Superposition of TEN2 Tox-HNH like region (magenta) with Geobacillus virus E2 (GVE2, blue) and Bacillus virus SPO1 (I-HmuI, yellow) HNH endonucleases (PDB IDs 5h0m and 1u3e, respectively) indicates great similarity in ternary structure arrangement of the catalytic pocket. The TEN2 α-helical region corresponding to residues 2560-2590 is also similar to the DNA binding region (DB region I) of GVE2, but lacks the second DNA binding region (DB region II). (F-H) Comparison of the TEN2 (F) and the two DNases (G-H) catalytic regions indicates that despite the similar fold, TEN2 lacks that HNH catalytic pocket residues which were found to mediate the DNase activity. (I) Sequence alignment of TENs 1-4 with GVE2 and I-HmuI indicates high secondary structure conservation with low sequence similarity (helices are indicated as magenta boxes, β-sheets as yellow arrows). HNH motif residues are highlighted in blue with residues conserved in TENs and DNases marked by **, and in DNases only by *. A furin cleavage site is present upstream to the nuclease region in both GVE2 and TENs (RTRR motive, orange). In the phage nuclease furin cleavage is necessary to activate the catalytic activity.
(A) Domain architecture schematics and function-metrics of mammalian expression constructs for TEN2. (B) Constructs that did not express TEN2. (C) Flow cytometry analysis of cell-surface expression of full length (FL)-TEN2 on non-permeabilized HEK293T mammalian cells, compared to un-transfected cells (UT), cells that were transfected with an empty vector (EV) and mutant TEN2-transfected cells. (D) Representative flow cytometry plots demonstrating binding of purified biotinylated lectin domain of LPHN3 (top row) or LPHN1 (bottom row) to TEN2 constructs expressed on HEK293T cells. TEN2 expression was determined by Flag tag fluorescence (x-axis) while purified Lectin binding to TEN2-expressing cells was quantified by fluorescence of DyLight attached to neutravidin (y-axis). Dot plots show correlation between TEN2 expression and LPHN binding in TEN2-transfected cells. Black cross on the plot indicates the “high TEN2 expression and high LPHN binding” gate. (E) Co-expression of TEN2 and lectin domains of LPHN3 (top row) and LPHN1 (bottom row) assessed through HA fluorescence of the TEN2 constructs.
(A) Diagram illustrating the experimental setup used for monitoring LPHN activity in mammalian cells. (B) cAMP-based signaling assay for detecting constitutive activity of rat LPHN1 and human LPHN3 in transfected HEK293 cells. LPHN inhibits ISO-induced cAMP elevation by the β2-adrenergic receptor. cAMP levels were measured by the Glosensor assay. Signaling data were obtained from three independent experiments performed in triplicates and are presented as means±SEM (***p≤0.001). EV indicates an empty vector. (C) The cell-surface expression of LPHN1 on non-permeabilized HEK293 cells was quantified by flow cytometry using an N-terminal FLAG tag and presented as a function of DNA concentration. (D) Transiently transfected HEK293 cells display DNA amount-dependent cAMP signaling of LPHN1 determined by β2-adrenergic receptor cAMP assay. Diagrams showing cis (E) and trans (F) setups for monitoring the effect of TEN on LPHN activity using a cAMP-based assay in mammalian cells. (G) Cell aggregation assays testing homotypic TEN2 trans-cellular adhesion using TEN2 splice variants and mutations. Scale bar indicates 50µm. Quantification of aggregation index (%) testing homotypic TEN2 interactions. LPHN3/TEN2 (WT-SS) were used as a positive control (*** p<0.001).
Highlights.
The structure of human teneurin2 has striking homology with bacterial Tc-toxins
Teneurin2 toxin-like domain resembles pre-pro-hormones and neuropeptide precursors
Teneurin2 regulates cAMP signaling in trans by interacting with latrophilins
Alternative splicing in teneurin regulates ligand interaction and synapse formation
Acknowledgments
We thank Tobin R. Sosnick for generously providing DesG tag construct to improve behavior of the proteins, Engin Ozkan for the use of flow cytometer and providing feedback on the manuscript, Chuan He lab for the use of their luminescence plate reader, Gabriel. S. Salzman for technical assistance on flow cytometry binding assays, Adam. M. Zmyslowski for discussions on the β-barrel, and Haim Rozenberg for advice on model refinement. This work was supported by NIH grants R01-GM120322 (to D.A.), R01 DK090165 (to G.S.), R37 MH052804-22 (to T.C.S) and T32GM007183.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.L. cloned, expressed and purified proteins, carried out TEN2-related biochemical characterizations and specimen screening by negative-stain EM, designed and performed mutagenesis, HEK cell-surface expression assays, flow cytometry binding assays and signaling assays. M.S.B. performed negative-stain EM, cryo-EM data collection and map calculation, and model building and refinement. M.S.B. and D.A. did structural analysis with assistance from J.L. M.S.B. prepared the structural figures. R.S., X.J. and J.W. designed and performed the cell aggregation assays and synapse formation assay. O.N. performed cAMP-based signaling assay. A.K and C.K participated in TEN2 EGF and propeller related biochemical characterizations. K.L assisted in specimen screening by negative-stain EM. Y.C.L designed and cloned EGF and propeller domain constructs. D.A., G.S., J.L., and M.S.B. designed experiments. D.A, G.S, T.C.S and M.S.B. wrote the paper with assistance from J.L. D.A. and G.S. supervised the project.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Chawaf A, St Amant K, Belsham D, Lovejoy DA. Regulation of neurite growth in immortalized mouse hypothalamic neurons and rat hippocampal primary cultures by teneurin C-terminal-associated peptide-1. Neuroscience. 2007;144:1241–1254. doi: 10.1016/j.neuroscience.2006.09.062. [DOI] [PubMed] [Google Scholar]
- Aldahmesh MA, Mohammed JY, Al-Hazzaa S, Alkuraya FS. Homozygous null mutation in ODZ3 causes microphthalmia in humans. Genet Med. 2012;14:900–904. doi: 10.1038/gim.2012.71. [DOI] [PubMed] [Google Scholar]
- Alkelai A, Olender T, Haffner-Krausz R, Tsoory MM, Boyko V, Tatarskyy P, Gross-Isseroff R, Milgrom R, Shushan S, Blau I, et al. A role for TENM1 mutations in congenital general anosmia. Clin Genet. 2016;90:211–219. doi: 10.1111/cge.12782. [DOI] [PubMed] [Google Scholar]
- Anderson GR, Maxeiner S, Sando R, Tsetsenis T, Malenka RC, Sudhof TC. Postsynaptic adhesion GPCR latrophilin-2 mediates target recognition in entorhinal-hippocampal synapse assembly. J Cell Biol. 2017;216:3831–3846. doi: 10.1083/jcb.201703042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Sudhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154:75–88. doi: 10.1016/j.cell.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arac D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Sudhof TC, Brunger AT. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. The EMBO journal. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zur T, Feige E, Motro B, Wides R. The mammalian Odz gene family: homologs of a Drosophila pair-rule gene with expression implying distinct yet overlapping developmental roles. Dev Biol. 2000;217:107–120. doi: 10.1006/dbio.1999.9532. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Maxeiner S, Sudhof TC. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. The Journal of biological chemistry. 2014;289:387–402. doi: 10.1074/jbc.M113.504779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby JN, Panjikar S, Landsberg MJ, Hurst MR, Lott JS. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature. 2013;501:547–550. doi: 10.1038/nature12465. [DOI] [PubMed] [Google Scholar]
- Chand D, Casatti CA, de Lannoy L, Song L, Kollara A, Barsyte-Lovejoy D, Brown TJ, Lovejoy DA. C-terminal processing of the teneurin proteins: independent actions of a teneurin C-terminal associated peptide in hippocampal cells. Mol Cell Neurosci. 2013;52:38–50. doi: 10.1016/j.mcn.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Dharmaratne N, Glendining KA, Young TR, Tran H, Sawatari A, Leamey CA. Ten-m3 is required for the development of topography in the ipsilateral retinocollicular pathway. PLoS One. 2012;7:e43083. doi: 10.1371/journal.pone.0043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabikowski K, Trzebiatowska A, Chiquet-Ehrismann R. ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev Biol. 2005;282:27–38. doi: 10.1016/j.ydbio.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb S, McPhee M, Brown ZJ, Kupferschmidt DA, Song L, Lovejoy DA. Repeated intravenous administrations of teneurin-C terminal associated peptide (TCAP)-1 attenuates reinstatement of cocaine seeking by corticotropin-releasing factor (CRF) in rats. Behav Brain Res. 2014;269:1–5. doi: 10.1016/j.bbr.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Feng K, Zhou XH, Oohashi T, Morgelin M, Lustig A, Hirakawa S, Ninomiya Y, Engel J, Rauch U, Fassler R. All four members of the Ten-m/Odz family of transmembrane proteins form dimers. The Journal of biological chemistry. 2002;277:26128–26135. doi: 10.1074/jbc.M203722200. [DOI] [PubMed] [Google Scholar]
- Fernandez-Leiro R, Scheres SHW. A pipeline approach to single-particle processing in RELION. Acta Crystallogr D Struct Biol. 2017;73:496–502. doi: 10.1107/S2059798316019276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo MV, Foldy C, Gokce O, Rothwell PE, Sun GL, Malenka RC, Sudhof TC. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann J, Aust G, Arac D, Engel FB, Formstone C, Fredriksson R, Hall RA, Harty BL, Kirchhoff C, Knapp B, et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol Rev. 2015;67:338–367. doi: 10.1124/pr.114.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Mosca TJ, Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012;484:201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor H, Francescatto L, Bartesaghi L, Ortega-Cubero S, Kousi M, Lorenzo-Betancor O, Jimenez-Jimenez FJ, Gironell A, Clarimon J, Drechsel O, et al. Missense mutations in TENM4, a regulator of axon guidance and central myelination, cause essential tremor. Hum Mol Genet. 2015;24:5677–5686. doi: 10.1093/hmg/ddv281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallieres M, Tapial J, Raj B, O’Hanlon D, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N. The unicellular ancestry of animal development. Dev Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg MJ, Jones SA, Rothnagel R, Busby JN, Marshall SD, Simpson RM, Lott JS, Hankamer B, Hurst MR. 3D structure of the Yersinia entomophaga toxin complex and implications for insecticidal activity. Proc Natl Acad Sci U S A. 2011;108:20544–20549. doi: 10.1073/pnas.1111155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhan T, Promel S, Mestek L, Esmaeili B, Waller-Evans H, Hennig C, Kohara Y, Avery L, Vakonakis I, Schnabel R, et al. Latrophilin signaling links anterior-posterior tissue polarity and oriented cell divisions in the C. elegans embryo. Developmental cell. 2009;17:494–504. doi: 10.1016/j.devcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamey CA, Merlin S, Lattouf P, Sawatari A, Zhou X, Demel N, Glendining KA, Oohashi T, Sur M, Fassler R. Ten_m3 regulates eye-specific patterning in the mammalian visual pathway and is required for binocular vision. PLoS Biol. 2007;5:e241. doi: 10.1371/journal.pbio.0050241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamey CA, Sawatari A. The teneurins: new players in the generation of visual topography. Semin Cell Dev Biol. 2014;35:173–179. doi: 10.1016/j.semcdb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossie AC, Nakamura H, Thomas SE, Justice MJ. Mutation of l7Rn3 shows that Odz4 is required for mouse gastrulation. Genetics. 2005;169:285–299. doi: 10.1534/genetics.104.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy DA, Al Chawaf A, Cadinouche MZ. Teneurin C-terminal associated peptides: an enigmatic family of neuropeptides with structural similarity to the corticotropin-releasing factor and calcitonin families of peptides. Gen Comp Endocrinol. 2006;148:299–305. doi: 10.1016/j.ygcen.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Lovejoy DA, de Lannoy L. Evolution and phylogeny of the corticotropin-releasing factor (CRF) family of peptides: expansion and specialization in the vertebrates. J Chem Neuroanat. 2013;54:50–56. doi: 10.1016/j.jchemneu.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Meusch D, Gatsogiannis C, Efremov RG, Lang AE, Hofnagel O, Vetter IR, Aktories K, Raunser S. Mechanism of Tc toxin action revealed in molecular detail. Nature. 2014;508:61–65. doi: 10.1038/nature13015. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Minet AD, Chiquet-Ehrismann R. Phylogenetic analysis of teneurin genes and comparison to the rearrangement hot spot elements of E. coli. Gene. 2000;257:87–97. doi: 10.1016/s0378-1119(00)00388-7. [DOI] [PubMed] [Google Scholar]
- Moriya T, Saur M, Stabrin M, Merino F, Voicu H, Huang Z, Penczek PA, Raunser S, Gatsogiannis C. High-resolution Single Particle Analysis from Electron Cryo-microscopy Images Using SPHIRE. J Vis Exp. 2017 doi: 10.3791/55448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca TJ, Hong W, Dani VS, Favaloro V, Luo L. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature. 2012;484:237–241. doi: 10.1038/nature10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Winkler J, Fiedler F, Sastradihardja T, Binder C, Schnabel R, Kungel J, Rothemund S, Hennig C, Schoneberg T, et al. Oriented Cell Division in the C. elegans Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1. PLoS genetics. 2015;11:e1005624. doi: 10.1371/journal.pgen.1005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Cook RN, Justice MJ. Mouse Tenm4 is required for mesoderm induction. BMC Dev Biol. 2013;13:9. doi: 10.1186/1471-213X-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan ML, Martini F, von Daake S, Comoletti D, Ghosh A. LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural Dev. 2014;9:7. doi: 10.1186/1749-8104-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oohashi T, Zhou XH, Feng K, Richter B, Morgelin M, Perez MT, Su WD, Chiquet-Ehrismann R, Rauch U, Fassler R. Mouse ten-m/Odz is a new family of dimeric type II transmembrane proteins expressed in many tissues. J Cell Biol. 1999;145:563–577. doi: 10.1083/jcb.145.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaki JM, Firestein S. Neurestin: putative transmembrane molecule implicated in neuronal development. Dev Biol. 1999a;212:165–181. doi: 10.1006/dbio.1999.9310. [DOI] [PubMed] [Google Scholar]
- Otaki JM, Firestein S. Segregated expression of neurestin in the developing olfactory bulb. Neuroreport. 1999b;10:2677–2680. doi: 10.1097/00001756-199908200-00044. [DOI] [PubMed] [Google Scholar]
- Peisley A, Skiniotis G. 2D Projection Analysis of GPCR Complexes by Negative Stain Electron Microscopy. Methods Mol Biol. 2015;1335:29–38. doi: 10.1007/978-1-4939-2914-6_3. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Scholz N, Gehring J, Guan C, Ljaschenko D, Fischer R, Lakshmanan V, Kittel RJ, Langenhan T. The Adhesion GPCR Latrophilin/CIRL Shapes Mechanosensation. Cell reports. 2015;11:866–874. doi: 10.1016/j.celrep.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Scholz N, Guan C, Nieberler M, Grotemeyer A, Maiellaro I, Gao S, Beck S, Pawlak M, Sauer M, Asan E, et al. Mechano-dependent signaling by Latrophilin/CIRL quenches cAMP in proprioceptive neurons. Elife. 2017;6 doi: 10.7554/eLife.28360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JP, Lelianova VG, Ermolyuk YS, Vysokov N, Hitchen PG, Berninghausen O, Rahman MA, Zangrandi A, Fidalgo S, Tonevitsky AG, et al. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc Natl Acad Sci U S A. 2011;108:12113–12118. doi: 10.1073/pnas.1019434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JJ, Yu W, Gichana EK, Baxa MC, Hinshaw JR, Freed KF, Sosnick TR. Benchmarking all-atom simulations using hydrogen exchange. Proc Natl Acad Sci U S A. 2014;111:15975–15980. doi: 10.1073/pnas.1404213111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LA, Al Chawaf A, Vaccarino FJ, Boutros PC, Lovejoy DA. Teneurin C-terminal associated peptide (TCAP)-1 modulates dendritic morphology in hippocampal neurons and decreases anxiety-like behaviors in rats. Physiol Behav. 2011;104:199–204. doi: 10.1016/j.physbeh.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Thalhammer A, Contestabile A, Ermolyuk YS, Ng T, Volynski KE, Soong TW, Goda Y, Cingolani LA. Alternative Splicing of P/Q-Type Ca(2+) Channels Shapes Presynaptic Plasticity. Cell reports. 2017;20:333–343. doi: 10.1016/j.celrep.2017.06.055. [DOI] [PubMed] [Google Scholar]
- Trzebiatowska A, Topf U, Sauder U, Drabikowski K, Chiquet-Ehrismann R. Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol Biol Cell. 2008;19:3898–3908. doi: 10.1091/mbc.E08-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Beckmann J, Leachman NT, Scholer J, Chiquet-Ehrismann R. Phylogenetic analysis of the teneurins: conserved features and premetazoan ancestry. Mol Biol Evol. 2012;29:1019–1029. doi: 10.1093/molbev/msr271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R. Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development. Dev Biol. 2006;290:237–245. doi: 10.1016/j.ydbio.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R, Chevron MP, Martin D, Hall RJ, Rubin BP. Teneurin-2 is expressed in tissues that regulate limb and somite pattern formation and is induced in vitro and in situ by FGF8. Dev Dyn. 2001;220:27–39. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1084>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Vysokov NV, Silva JP, Lelianova VG, Ho C, Djamgoz MB, Tonevitsky AG, Ushkaryov YA. The Mechanism of Regulated Release of Lasso/Teneurin-2. Front Mol Neurosci. 2016;9:59. doi: 10.3389/fnmol.2016.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Rotzinger S, Al Chawaf A, Elias CF, Barsyte-Lovejoy D, Qian X, Wang NC, De Cristofaro A, Belsham D, Bittencourt JC, et al. Teneurin proteins possess a carboxy terminal sequence with neuromodulatory activity. Brain Res Mol Brain Res. 2005;133:253–265. doi: 10.1016/j.molbrainres.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Winkler J, Promel S. The adhesion GPCR latrophilin - a novel signaling cascade in oriented cell division and anterior-posterior polarity. Worm. 2016;5:e1170274. doi: 10.1080/21624054.2016.1170274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle R, D’Aquila AL, Lovejoy DA. Teneurins, TCAP, and latrophilins: roles in the etiology of mood disorders. Transl Neurosci. 2016;7:17–23. doi: 10.1515/tnsci-2016-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TR, Bourke M, Zhou X, Oohashi T, Sawatari A, Fassler R, Leamey CA. Ten-m2 is required for the generation of binocular visual circuits. J Neurosci. 2013;33:12490–12509. doi: 10.1523/JNEUROSCI.4708-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TR, Leamey CA. Teneurins: important regulators of neural circuitry. Int J Biochem Cell Biol. 2009;41:990–993. doi: 10.1016/j.biocel.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XH, Brandau O, Feng K, Oohashi T, Ninomiya Y, Rauch U, Fassler R. The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr Patterns. 2003;3:397–405. doi: 10.1016/s1567-133x(03)00087-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Gel filtration chromatography (left) and SDS-PAGE analysis (right) for the corresponding fractions of ECR (magenta) and ECRΔ1 (green) indicates that soluble ECR is an oligomer whereas ECRΔ1 is a monomer. (B) Cryo-EM micrographs of TEN2. Scale bar is 20 nm. (C), Representative reference-free two-dimensional cryo-EM averages of TEN2. The diameter of the circular windows is 26 nm. (D) Flow chart of cryo-EM data processing of TEN2 extracellular region, including particle selection, classification, and 3D density map reconstruction. Details are provided in the methods section.
(A) Resolution estimation of the EM map reconstructed from 426,107 particle projections. Gold standard Fourier shell correlation (FSC) curves, indicating an overall nominal resolution of 3.1 Å at 0.143 FSC. (B) FSC curve of the final refined model versus the cryo-EM map. FSC graphs were calculated with M-triage, by Phenix. (C) Three-dimensional density map colored according to local resolution indicating a resolution range of 2.5–3.5 Å in most map regions. Local resolution was calculated by ResMap. (D) Local resolution calculations for map resulting of 157,961 particles with a stable β-propeller indicating lower resolution for ECR 3 (β-propeller indicated by arrow) due to local flexibility. Color scheme for resolution values is as presented in panel C.
(A) Snapshots of map vs. model agreement by domain. Map is contoured at σ=3.0 and each region is highlighted by a dashed black box. Snapshots include the representative regions of domain 5 (top left), domain 2 (top right) and domain 4 (bottom left). Snapshot of one representative sugar modification in the cryo-EM density is presented at the right bottom panel. (B) Domain 3 homology model fitting to the low-resolution map area. The protein backbone is clearly delineated in the map. Visible side chains were used as anchoring points for the docking of the homology model (template model PDB ID 3fvz). The model was real-space refined in the map using Coot and Phenix_real_space_refine.
Two- (A) and three-dimensional (B) representations of TEN2, with domains color coded as in Figure 1. Residue numbers are indicated. (C) An exploded view of each TEN2 domain. Domain borders are numbered, N- and C- termini of each domain are highlighted by spheres. The Ig-like region in ECR2 is indicated by a dashed line. Alternative splicing variant region of ECR3 is indicated by SS. Dormant aspartate dyad in ECR4 is highlighted in yellow. Barrel exit point and furin cleavage site in ECR5 are pointed by arrows, Tox-HNH is highlighted in yellow (domain 5) and TCAP is circled by a dashed line (domain 5). Domain borders and highlighted regions are indicated in black.
(A) The C-terminal of ECR5 emerges from the barrel and remains attached to the outer barrel region by a network of hydrogen bonds. ECR5 is in magenta. H-bonds are indicated by dashed lines. (B) The encapsulated domain (ECR5, magenta) interacts with domain 2 (yellow) that penetrates into the barrel and plugs its bottom. Contact points are highlighted by black arrows. (C) Two notable lysins of the encapsulated domain (K2421, K2422) mediate interactions of the barrel bottom with Y2349, the residue presumed to be cleaved by the aspartic acid protease. (D) The C-terminal of ECR5 (residues 2539-2648) shares structural similarity with HNH DNases. (E) Superposition of TEN2 Tox-HNH like region (magenta) with Geobacillus virus E2 (GVE2, blue) and Bacillus virus SPO1 (I-HmuI, yellow) HNH endonucleases (PDB IDs 5h0m and 1u3e, respectively) indicates great similarity in ternary structure arrangement of the catalytic pocket. The TEN2 α-helical region corresponding to residues 2560-2590 is also similar to the DNA binding region (DB region I) of GVE2, but lacks the second DNA binding region (DB region II). (F-H) Comparison of the TEN2 (F) and the two DNases (G-H) catalytic regions indicates that despite the similar fold, TEN2 lacks that HNH catalytic pocket residues which were found to mediate the DNase activity. (I) Sequence alignment of TENs 1-4 with GVE2 and I-HmuI indicates high secondary structure conservation with low sequence similarity (helices are indicated as magenta boxes, β-sheets as yellow arrows). HNH motif residues are highlighted in blue with residues conserved in TENs and DNases marked by **, and in DNases only by *. A furin cleavage site is present upstream to the nuclease region in both GVE2 and TENs (RTRR motive, orange). In the phage nuclease furin cleavage is necessary to activate the catalytic activity.
(A) Domain architecture schematics and function-metrics of mammalian expression constructs for TEN2. (B) Constructs that did not express TEN2. (C) Flow cytometry analysis of cell-surface expression of full length (FL)-TEN2 on non-permeabilized HEK293T mammalian cells, compared to un-transfected cells (UT), cells that were transfected with an empty vector (EV) and mutant TEN2-transfected cells. (D) Representative flow cytometry plots demonstrating binding of purified biotinylated lectin domain of LPHN3 (top row) or LPHN1 (bottom row) to TEN2 constructs expressed on HEK293T cells. TEN2 expression was determined by Flag tag fluorescence (x-axis) while purified Lectin binding to TEN2-expressing cells was quantified by fluorescence of DyLight attached to neutravidin (y-axis). Dot plots show correlation between TEN2 expression and LPHN binding in TEN2-transfected cells. Black cross on the plot indicates the “high TEN2 expression and high LPHN binding” gate. (E) Co-expression of TEN2 and lectin domains of LPHN3 (top row) and LPHN1 (bottom row) assessed through HA fluorescence of the TEN2 constructs.
(A) Diagram illustrating the experimental setup used for monitoring LPHN activity in mammalian cells. (B) cAMP-based signaling assay for detecting constitutive activity of rat LPHN1 and human LPHN3 in transfected HEK293 cells. LPHN inhibits ISO-induced cAMP elevation by the β2-adrenergic receptor. cAMP levels were measured by the Glosensor assay. Signaling data were obtained from three independent experiments performed in triplicates and are presented as means±SEM (***p≤0.001). EV indicates an empty vector. (C) The cell-surface expression of LPHN1 on non-permeabilized HEK293 cells was quantified by flow cytometry using an N-terminal FLAG tag and presented as a function of DNA concentration. (D) Transiently transfected HEK293 cells display DNA amount-dependent cAMP signaling of LPHN1 determined by β2-adrenergic receptor cAMP assay. Diagrams showing cis (E) and trans (F) setups for monitoring the effect of TEN on LPHN activity using a cAMP-based assay in mammalian cells. (G) Cell aggregation assays testing homotypic TEN2 trans-cellular adhesion using TEN2 splice variants and mutations. Scale bar indicates 50µm. Quantification of aggregation index (%) testing homotypic TEN2 interactions. LPHN3/TEN2 (WT-SS) were used as a positive control (*** p<0.001).