Abstract
Muscles together with tendons and the skeleton enable animals including humans to move their body parts. Muscle morphogenesis is highly conserved from animals to humans. Therefore, the powerful Drosophila model system can be used to study concepts of muscle-tendon development that can also be applied to human muscle biology. Here, we describe in detail how morphogenesis of the adult muscle-tendon system can be easily imaged in living, developing Drosophila pupae. Hence, the method allows investigating proteins, cells and tissues in their physiological environment. In addition to a step-by-step protocol with helpful tips, we provide a comprehensive overview of fluorescently tagged marker proteins that are suitable for studying the muscle-tendon system. To highlight the versatile applications of the protocol, we show example movies ranging from visualization of long-term morphogenetic events – occurring on the time scale of hours and days – to visualization of short-term dynamic processes like muscle twitching occurring on time scale of seconds. Taken together, this protocol should enable the reader to design and perform live-imaging experiments for investigating muscle-tendon morphogenesis in the intact organism.
Keywords: Developmental Biology, Issue 132, Live imaging, Drosophila, pupal development, muscle, tendon, sarcomere, myofibril, attachment, self-organization
Introduction
The muscle-tendon apparatus allows animals including humans to move their body parts. The molecular building blocks of the muscle-tendon system are highly conserved. Therefore, concepts of muscle-tendon development relevant for human muscle biology, for example muscle morphogenesis, muscle-tendon attachment and myofibril self-organization, can be studied using Drosophila melanogaster as an easily accessible model system. The Drosophila pupal system has several experimental advantages. First, at the pupal stage – when the adult muscles are formed – the organism is sessile and therefore easy to image on a microscope over a period of hours or even days. Second, many muscles form close enough underneath the pupal surface so that they can be imaged inside the intact, partially translucent organism. Third, the muscles can be investigated in their natural environment, where they are connected to the forming exoskeleton via tendon cells and tissue tension is built up. This is not possible in muscle cell culture systems. And finally, a plethora of genetic tools is available in Drosophila. Among these are many fluorescently tagged markers that allow labeling of specific cell types or subcellular structures for imaging in vivo.
Table 1 summarizes the most important markers used for studying muscle-tendon morphogenesis. It includes markers overexpressed using the GAL4-UAS-system1 and endogenously tagged protein markers2,3,4. The advantage of the GAL4-UAS-system is that the markers are generally expressed at high levels, resulting in a strong signal that can easily be imaged in whole-mount pupae. In addition, tissue specificity can be achieved by choosing GAL4 drivers carefully. The advantage of fusion proteins expressed under endogenous control is that the dynamics of the respective proteins can be studied in vivo, while they can also be used as markers for different cell types or specific subcellular structures, for example, βPS-Integrin-GFP for muscle attachment sites. Together, these markers provide high flexibility in experimental design and choice of research problems that can be solved now and in the future.
| Labeled structure | Marker | Expression and localization | Class | Stock number | Comment | Ref. |
| Muscle | Mef2-GAL4 | all myoblasts and all muscles at all stages | GAL4 line | BL 27390 | 5 | |
| 1151-GAL4 | adult muscle precursors and early myotubes until ≈24 h APF | GAL4 line, enhancer trap | - | 6 | ||
| Act79B-GAL4 | jump muscle upon differentiation | GAL4 line | - | 7 | ||
| Act88F-GAL4 | indirect flight muscles starting ≈14 h APF | GAL4 line | - | 7 | ||
| Act88F-Cameleon 3.1 | indirect flight muscles starting ≈14 h APF | Act88F enhancer/ promoter driving Cameleon 3.1 | - | Ca2+ indicator | 8 | |
| Act88F-GFP | indirect flight muscles starting ≈14 h APF | GFP-fusion (fly TransgeneOme line) | fTRG78 and fTRG10028 | 4 | ||
| Him-nls-GFP | adult muscle precursors, nuclear, until ≈24 h APF in indirect flight muscles | enhancer/promoter with nls-GFP reporter | - | 1.5 kb enhancer fragment | 9 | |
| Mhc-Tau-GFP | microtubules in DLM templates and in differentiating muscles | enhancer/promoter with Tau-GFP reporter | BL 53739 | 10 | ||
| βTub60D-GFP | microtubules in myotubes (e.g. in indirect flight muscles from ≈14 h AFP, strongly decreasing after ≈48 h APF) | GFP-fusion (fly TransgeneOme line) | fTRG958 | 4 | ||
| Mhc-GFP (weeP26) | sarcomeres (thick filament) in all body muscles (e.g. in indirect flight muscles starting from ≈30 h APF) | GFP-trap | - | use heterozygous, labels an isoform subset | 11 | |
| Sls-GFP | sarcomeres (Z-disc) in all body muscles (e.g. in indirect flight muscles starting from ≈30 h APF) | GFP-trap (FlyTrap line) | - | G53, use heterozygous | 2 | |
| Zasp66-GFP | Z-disc in all body muscles | GFP-trap (FlyTrap line) | BL 6824 | ZCL0663 | 2 12 | |
| Zasp52-GFP | Z-disc in all body muscles | GFP-trap (FlyTrap line) | BL 6838 | G00189 | 2 12 | |
| Hts-GFP | actin binding; expressed in epithelium, myoblasts and myotubes | GFP-fusion (fly TransgeneOme line) | fTRG585 | 4 | ||
| Dlg1-GFP | epithelial cell junctions, myoblasts and membranes in muscles at all stages | GFP-fusion (fly TransgeneOme line) | fTRG502 | 4 | ||
| Muscle attachment site | βPS-Integrin-GFP | muscle attachment sites (e.g., starting ≈18 h AFP in indirect flight muscles) | GFP-knock-in | - | 13 | |
| Talin-GFP and -mCherry | muscle attachment sites (e.g., starting ≈18 h AFP in indirect flight muscles) | GFP-trap (MiMIC line) | - | 3 | ||
| Talin-GFP | muscle attachment sites (e.g., starting ≈18 h AFP in indirect flight muscles) | GFP-fusion (fly TransgeneOme line) | fTRG587 | 4 | ||
| Ilk-GFP | muscle attachment sites (e.g., starting ≈18 h AFP in indirect flight muscles) | GFP-trap (FlyTrap line) | Kyoto 110951 (ZCL3111) | ZCL3111, ZCL3192 | 2 | |
| Vinc-GFP and -RFP | muscle attachment sites (e.g., starting ≈18 h AFP in indirect flight muscles) | GFP-fusion (transgene) | - | 13 | ||
| Tendon | sr-GAL4 | thorax tendon cells, throughout pupal stage | GAL4 line, enhancer trap | BL 26663 | homozygous lethal | 14 |
| Muscle and Tendon | Duf-GAL4 | muscles and epithelia, early onset | GAL4 line | BL 66682 | kirre-rP298, founder cell marker | 15 |
| UAS-reporters | UAS-GFP-Gma | actin binding | UAS line | BL 31776 | actin binding domain of Moesin fused to GFP | 16 |
| UAS-mCherry-Gma | actin binding | UAS line | - | Gma fused to mCherry | 17 | |
| UAS-Lifeact-GFP | actin binding | UAS line | BL 35544 | 18 | ||
| UAS-Lifeact-Ruby | actin binding | UAS line | BL 35545 | 18 | ||
| UAS-CD8-GFP | membrane binding | UAS line | various stocks, e.g.: BL 32184 | 19 | ||
| UAS-CD8-mCherry | membrane binding | UAS line | BL 27391 and 27392 | 20 | ||
| UAS-palm-mCherry | membrane binding through palmitoylation | UAS line | BL 34514 | UAS-brainbow | 21 |
Table 1: Fluorescently tagged protein markers suitable for studying muscle-tendon morphogenesis in vivo.
Here, we describe in detail how imaging of muscle-tendon morphogenesis in living pupae can be performed easily and successfully (Figure 1). Alternatively, pupae can be fixed, dissected and immunostained, which allows using antibodies to also label proteins for which no live markers are available22. In this case, the imaging quality is generally higher because there is no movement and the structure of interest can be placed in close proximity to the coverslip. However, dissection and fixation can lead to damage and molecular or tissue dynamics, for example, muscle twitching, can only be studied in the living organism.
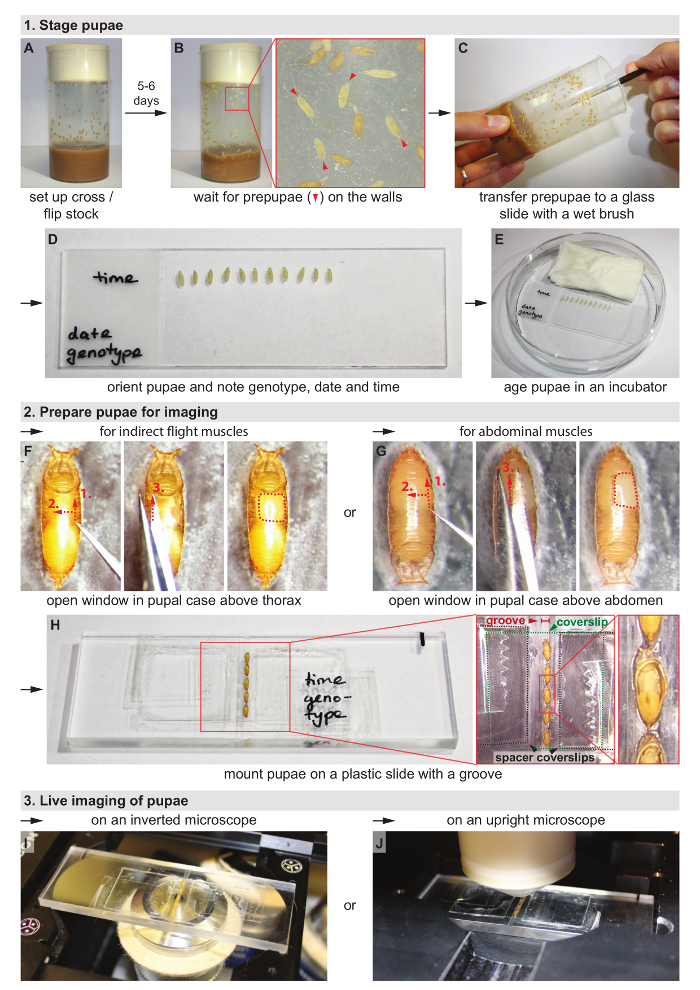
Protocol
1. Stage Pupae
Set up a cross with 20 - 40 virgin females and about 20 males per bottle of fly food (Figure 1A). Alternatively, if stocks will be used, flip each stock into new bottles. To get enough pupae, consider setting up two bottles per genotype.
Incubate the bottles at 25 °C (or 27 °C according to the experimental design) for five to six days (Figure 1B). NOTE: Keep flipping the flies every two to three days to avoid overcrowding and to ensure a continuous supply of pupae.
Use a wet brush to collect white prepupae from the walls of the bottles and transfer them to the glass slides (Figure 1C). Time the staging such that the pupae reach the desired age when starting live imaging under the microscope. NOTE: These prepupae look like older pupae in terms of their shape, but they are still white like larvae. The onset of the prepupal stage is defined as 0 h after puparium formation (0 h APF).
Remove clumps of fly food sticking to the pupae with a wet brush and orient the ventral side of the pupae towards the glass slide using a stereomicroscope (Figure 1D). Discard pupae that are still moving (too young) or that have started to turn brown (too old). This ensures that all pupae have the same age (0 - 1 h APF).
Label the slides with genotype, date and time of collection. Transfer the glass slides to one petri dish each and add a wet tissue to prevent the pupae from drying out (Figure 1E).
Store the pupae in an incubator at 25 °C or 27 °C until they reach the desired age. Continue with the next steps 1 h before imaging, so that the pupae have the desired age when starting live imaging (step 3), for example at 13 h APF for the movie shown in Figure 2A-D. NOTE: The earliest feasible starting time point is after head-eversion of the pupae occurring at 8 - 10 h APF at 27 °C.
2. Prepare Pupae for Imaging
Ensure that the pupae now stick to the glass slide just like they naturally stick to the walls of the bottles because of residual fly food. No extra glue is required. If the pupae do not stick well enough because of high humidity, open the petri dish for a few min to let them dry. Alternatively, transfer them to double-sided tape on a glass slide, but usually, this is not necessary.
- Open window in the pupal case for imaging of indirect flight muscles (Figure 1F; Skip to section 2.3 for imaging of abdominal muscles.)
- Orient the pupae sticking to the glass slide with the anterior facing away from the researcher under a stereomicroscope. Adjust the zoom to 2X.
- Using one end of biology grade #5 forceps, gently poke a hole into the pupal case dorsal to the wing where the abdomen ends, and the thorax begins.
- To slice open the pupal case, gently move the forceps to the anterior end of the thorax along the wing but avoid damaging the vulnerable wing tissue. NOTE: If fluid leaks from the pupa, it is damaged; discard it and start over with a different pupa.
- Lift up the section of the pupal case above the thorax with the forceps and then cut this section off with fine, sharp scissors on the opposite side of the dorsal midline.
- Open window in pupal case for imaging of abdominal muscles (Figure 1G)
- Orient the pupae sticking to the glass slide with the anterior facing towards the researcher under a stereomicroscope. Adjust the zoom to 2X.
- Using one end of biology grade #5 forceps, gently poke a hole into the pupal case dorsal to the wing where the thorax ends, and the abdomen begins.
- To slice open the pupal case, gently move the forceps towards the posterior end of the abdomen. NOTE: If fluid leaks from the pupa, it is damaged; discard it and start over with a different pupa.
- Lift up the section of the pupal case above the abdomen with the forceps and then cut this section off with fine, sharp scissors on the opposite side of the dorsal midline.
- Mount Pupae (Figure 1H)
- Use a wet brush to transfer up to five pupae to a plastic slide with a groove (custom-built, reusable, see discussion and list of materials). Add one or two spacer coverslips to each side depending on the depth of the groove and the thickness of the pupae. Put a small drop of water underneath each spacer coverslip to make it stick to the slide and leave some space between the groove and the spacer coverslips on each side. NOTE: The spacer coverslips can also be permanently attached to the slide with super glue, in advance.
- Orient the pupae such that the opening in the pupal case faces upwards using the brush. Ensure careful positioning of the pupae at the correct angle for good imaging quality. Optimize the angle for each tissue and developmental time point. NOTE: A small amount of water in the groove makes positioning of the pupae easier. Drain excess water with the brush afterward to avoid drowning.
- Hold a coverslip (18 x 18 mm) above the pupae without touching them. Place small droplets (about 0.5 µL) of 50% glycerol solution above each pupal case opening onto the coverslip using a 20 µL pipette. NOTE: This procedure ensures that the droplets have the same spacing as the pupae.
- Turn the coverslip over and position it above the pupae by hand or using forceps (standard #5) while resting one side of the coverslip on the spacer coverslips next to the pupae. Next, gently drop the coverslip onto the pupae. Ensure that the pupal case openings are properly covered with 50% glycerol for good imaging quality and to prevent the pupae from drying out.
- Fold over one end of a piece of adhesive tape and grab it with forceps (standard #5) at that end. Gently place the tape such that it covers one edge of the top coverslip, the spacer coverslip(s), and the plastic slide on one side. Do not press down the tape, yet. First, turn the slide around and repeat for the other side.
- Use both index fingers to press down the tape simultaneously on both sides. This procedure ensures that the pupae are not displaced.
- Check whether the pupae are well in contact with the coverslip. For optimal imaging quality the pupae should be gently squeezed underneath the coverslip. Adjust the number of spacer coverslips if necessary.
- Label the slide by writing on the sticky tape.
3. Live Imaging of Pupae
The mounting method allows imaging of the pupae both on inverted microscopes (Figure 1I) and on upright microscopes (Figure 1J). Especially suitable for live imaging are scanning confocal microscopes, two-photon microscopes and spinning disc confocal microscopes irrespective of whether they are inverted or upright. For long-term movies, use a temperature-controlled stage if available.
Using the ocular and a UV lamp or transmission light, locate a pupa and focus inside the pupa.
Using the camera, find the desired structure and adjust the zoom level.
For long-term movies, define a z-stack that encompasses the structure of interest well (for example, the indirect flight muscles, their attachment sites, the tendon cells or a combination the aforementioned) and choose a time interval. Consider doing averaging for better image quality. For high-speed, short-term movies, choose a single z-plane to achieve a high frame rate.
Adjust the laser power to give the best possible signal but little saturation. However, too high laser power can damage the pupa over time.
Start imaging and return from time to time to check the movie and readjust the z-stack positioning if necessary.
Figure 1: Workflow for live imaging of muscle-tendon morphogenesis in Drosophila pupae. See protocol for details. Please click here to view a larger version of this figure.
Representative Results
Various tissues can be imaged in vivo in developing fly pupae, making them an ideal model system to study morphogenesis of adult organs. Among these are the indirect flight muscles, the thorax epithelium including the tendon cells, the wing epithelium, abdominal muscles and the heart22,23,24,25,26,27. Here, we focus on live imaging of muscle and tendon morphogenesis. For a detailed description of indirect flight muscle and abdominal muscle morphogenesis and additional methods for studying muscle biology in Drosophila, we refer the reader to Weitkunat and Schnorrer 201422.
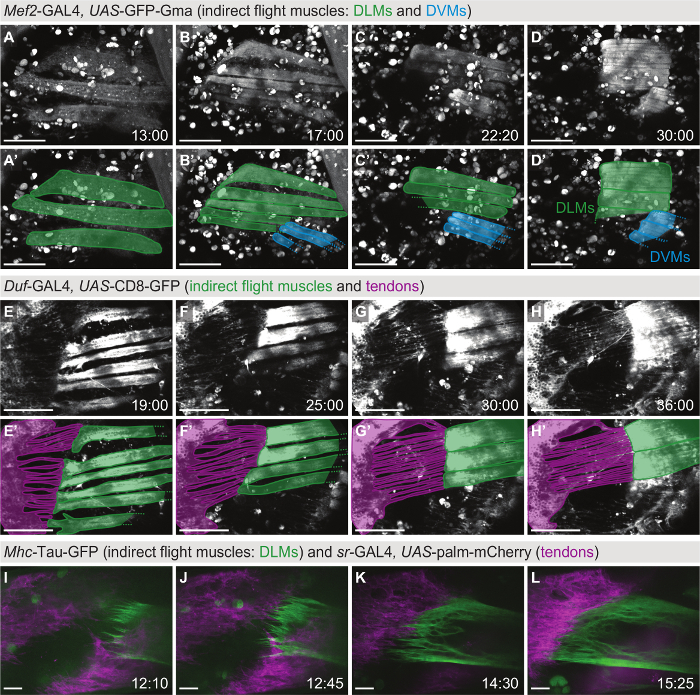
Live imaging of indirect flight muscles
For studying the long-term development of the indirect flight muscles consisting of the dorsolongitudinal muscles (DLMs) and the dorsoventral muscles (DVMs), globular Moesin actin-binding domain tagged with GFP (UAS-GFP-Gma) was expressed in all muscles using the Myocyte enhancer factor 2 (Mef2)-GAL4 driver (Figure 2A-D, Movie S1). Prior to imaging, a window in the pupal case was opened above the thorax as shown in Figure 1F. On a two-photon microscope, z-stacks were acquired every 20 min for 21 h starting at ≈11 h after puparium formation (11 h APF). In this time frame, the DLMs (green overlays in Figure 2A'-D') first initiate attachment to the tendon cells (Figure 2A) and then split while the muscle attachments mature (Figure 2B). Next, the myotubes shorten (Figure 2C) until they finally reach the maximally compacted stage at 30 h APF (Figure 2D). Taken together, this movie highlights the dramatic changes in muscle morphology that occur on the time scale of hours.
Figure 2: Live imaging of indirect flight muscle and tendon morphogenesis.(A-D) Time points from a two-photon movie (Movie S1) using Mef2-GAL4, UAS-GFP-Gma as a marker for actin in the indirect flight muscles consisting of the dorsolongitudinal muscles (DLMs, highlighted in green in A'-D') and the dorsoventral muscles (DVMs, highlighted in blue in A'-D'). Scale bars are 100 µm. (E-H) Time points from a two-photon movie (Movie S2) using Duf-GAL4, UAS-CD8-GFP as a marker for indirect flight muscles and the thorax epithelium including tendon cells. Panels E'-H' show an overlay with a model of the muscle-tendon system at each time point, highlighting the long cellular extensions formed by the tendon epithelium (magenta) in contact with the muscles (green). Scale bars are 100 µm (estimated from size of imaged structures). (I-L) Time points from a two-color, spinning disc confocal movie (Movie S3) focusing on muscle-tendon attachment initiation. The tendon cells are labeled with sr-GAL4, UAS-palm-mCherry (magenta) and the dorsolongitudinal indirect flight muscles with Mhc-Tau-GFP (green). Scale bars are 10 µm. Time is indicated as hh:mm after puparium formation (APF). Note that not all movies were acquired on a temperature-controlled stage, therefore, developmental timing may diverge. Please click here to view a larger version of this figure.
For studying tendon and muscle morphogenesis at the same time, membrane-bound GFP (UAS-CD8-GFP) was expressed using Dumbfounded (Duf)-GAL4 as a driver, which is expressed both in the tendon cells and the indirect flight muscles (Figure 2E-H, Movie S2). A z-stack was taken on a two-photon microscope every 20 min starting at 16 h APF for 20 h. While the myotubes compact (green overlay in Figure 2E'-H'), the tendon cells form long cellular extensions that elongate with time (magenta overlay in Figure 2E'-H'). Taken together, this movie highlights the close interplay between the tendon and muscle cells in vivo.
To investigate muscle-tendon interaction in more detail, two-color, high-magnification imaging was performed. Pupae with myosin heavy chain (Mhc)-Tau-GFP in the DLMs and UAS-palmitoylated-mCherry (UAS-palm-mCherry) driven by stripe (sr)-GAL4 in the tendons were imaged every 5 min starting at 12 h APF for 4.5 h on a spinning disc confocal microscope (Figure 2I-L, Movie S3). At 12 h APF, the myotubes migrate towards their tendon target cells while forming long filopodia at their tips (Figure 2I). Subsequently, the muscle and tendon tissues interdigitate (Figure 2J, K) to form a stable attachment. As the attachment matures, fewer filopodia form and the muscle-tendon interface smoothens (Figure 2L). Thus, two-color, high-magnification imaging can be used to reveal cellular dynamics in detail in the living organism.
Live imaging of abdominal muscles
For live imaging of the abdominal muscles (Figure 3, Movie S4), Mef2-GAL4>UAS-CD8-GFP was used as a marker and a window was opened above the abdomen in the pupal case as detailed in Figure 1G. Similar to the indirect flight muscle movie represented in Figure 2A-D, the formation and growth of the abdominal muscles can be followed over many hours of development (55 h in Movie S4). During this time, the myoblasts fuse to form growing myotubes (Figure 3A). The myotube tips migrate to their tendon targets, and after attaching to the tendon cells, the contractile units of the muscles, the sarcomeres, are formed (Figure 3B-D).
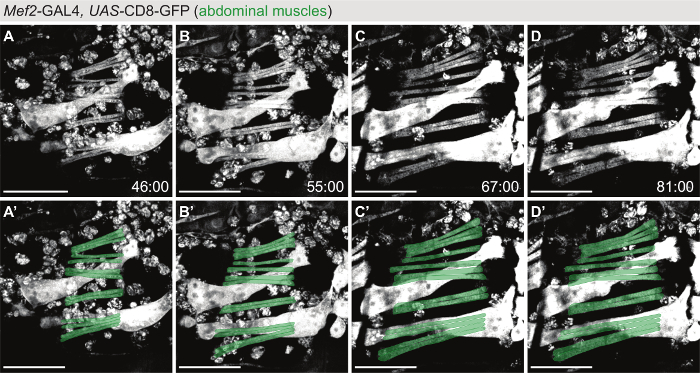
Figure 3: Live imaging of abdominal muscle morphogenesis.(A-D) Time points from a movie (Movie S4) using Mef2-GAL4, UAS-CD8-GFP as a marker to follow abdominal muscle morphogenesis. Panels A'-E' show overlays with models of an abdominal muscle set (green) that forms de novo during the pupal stage. Scale bars are 100 µm. Time is indicated as hh:mm APF. The movie was acquired at room temperature. Please click here to view a larger version of this figure.
Live imaging of twitching muscles
In contrast to Figure 2 and Figure 3, Figure 4 shows muscle dynamics occurring on the time scale of seconds: the live recording of muscle contractions. Pupae expressing βPS-Integrin-GFP under endogenous control were imaged with a time resolution of 0.65 s in a single z-plane on a confocal microscope (Movie S5). In the example displayed in Figure 4, the attachment sites of three DVMs (Figure 4A) were imaged for 10 min starting at 42 h APF. During this time, five twitch events were observed (Figure 4B-F), showing that the sarcomeres are already assembled well enough to support coordinated contractions at this time point in development. Hence, imaging of muscle twitching can be used as a functional read-out for sarcomerogenesis already during indirect flight muscle development28, as opposed to for example flight tests, which can only be performed after eclosion.
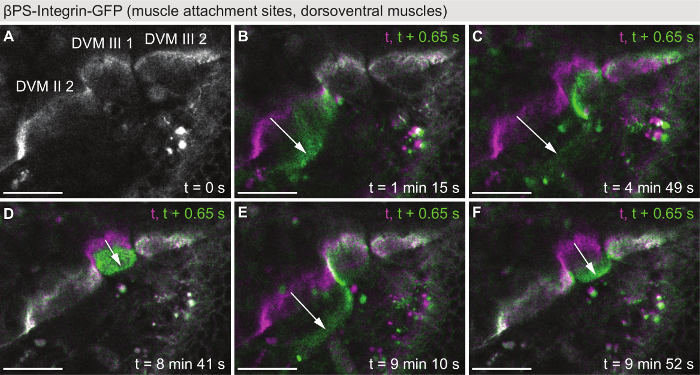
Figure 4: Live imaging of twitching muscles. (A) First time-point from a movie (Movie S5) showing twitching of dorsoventral indirect flight muscles at 42 h APF using βPS-Integrin-GFP as a marker. In the field of view are the muscle attachment sites of DVM II 2, DVM III 1 and DVM III 2. (B-F) Color overlays of five individual twitch events, each showing the frame before the twitch (magenta) and the first frame of the twitch event (green). Note that the individual muscle fibers twitch independently from each other. Arrows highlight the twitching motion. The time resolution of the movie is 0.65 s. Scale bars are 25 µm. Please click here to view a larger version of this figure.
Live imaging of endogenously tagged proteins
Similar to βPS-Integrin-GFP, a large collection of fusion proteins expressed in Drosophila under endogenous control has been generated2,3,4. These fly lines can be used to study for example the subcellular localization or the expression profiles of proteins of interest. The collection is especially helpful if specific antibodies are not available or protein dynamics need to be investigated in vivo without fixation. Figure 5 shows three examples of endogenously expressed fusion proteins from the fly TransgeneOme (fTRG) library, Hu li tai shao (Hts)-GFP (Figure 5A, B), Talin-GFP (Figure 5C, D) and βTubulin60D-GFP (Figure 5E, F). The expression of these proteins can be studied in all tissues that express them naturally. Here, we show the thorax epithelium (Figure 5A, C, E) and the indirect flight muscles or their attachment sites (Figure 5B, D, F) as examples.
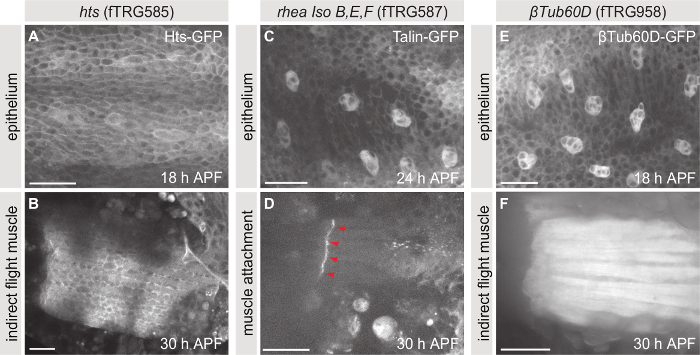
Figure 5: Live imaging of endogenously tagged proteins. (A, B) Maximum projections of z-stacks taken of pupae expressing Hts-GFP. (C, D) Maximum projections of z-stacks taken of pupae expressing Talin-GFP. (E, F) Maximum projections of z-stacks taken of pupae expressing βTubulin60D-GFP. Panels A, C and E show the thorax epithelium at 18, 24 and 18 h APF, respectively, and panels B, D and F show the indirect flight muscles or their attachment sites at 30 h APF. Scale bars are 25 µm. Please click here to view a larger version of this figure.
Discussion
The presented protocol describes how to image muscle-tendon morphogenesis in living Drosophila pupae using a variety of fluorescently tagged proteins. This in vivo imaging strategy can be used to study developmental processes in their natural environment of the entire organism.
It is crucial for a successful experiment to find the correct developmental time point to analyze. For example, dorsolongitudinal indirect flight muscles initiate attachment to their tendon targets at ≈16 h APF23 while abdominal muscles develop later and attach on both ends only between 30 and 40 h APF26. Consequently, previously published literature should be used to find the right time points of development to analyze or, if the tissue or structure of interest has not been studied in detail before, the overall development has to be characterized first.
For mounting pupae successfully on the custom-built plastic slides, it is important that the grooves have suitable dimensions: The grooves need to be 1.0 - 1.5 mm wide and 0.3 - 0.4 mm deep. This depth allows adjusting the precise distance to the top coverslip with spacer coverslips as needed. However, at least one spacer coverslip should be used to avoid draining the 50% glycerol away from the sample by capillary forces. The correct positioning of the pupae in the groove requires some experience and should be optimized such that the structure of interest is as close as possible to the coverslip.
If a large number of pupae is supposed to be imaged in one microscope session, they can all be mounted beforehand and then stored in an incubator until imaging to ensure proper developmental timing. The pupae should survive the entire procedure and also at least try to eclose if kept on the slide after imaging. The survival rate can be used as a readout to check whether the imaging conditions harm the pupae.
The imaging settings should be chosen carefully according to the experimental requirements. For short-term movies, a high frame rate versus a high signal-to-noise ratio needs to be balanced, while relatively high laser power can be used without damaging the pupae too much. However, for long-term movies, the laser power has to be kept at a moderate level and the pupae should not be imaged continuously but rather at certain time points, for example, every 20 min. To ensure that the structure of interest does not move out of the field of view, it might be necessary to readjust the positioning of the z-stack between time points. To our knowledge, the opening of the pupal case per se does not affect developmental timing. However, a temperature-controlled stage should be used for long-term movies to ensure proper developmental timing. Keeping these considerations in mind, highly informative movies can be acquired.
The presented protocol can be used to visualize not only muscle-tendon morphogenesis but also other developing tissues, for example, the wing epithelium29. Only three modifications to this protocol are required: (1) opening of the pupal case above the wing instead of the thorax or abdomen, (2) positioning of pupae with the wing towards the top coverslip, and (3) the use of different fluorescent marker proteins. With the advancement of the CRISPR/Cas9-technology, more and more endogenously tagged fluorescent proteins will be available, because it has become more straightforward to target endogenous loci in Drosophila30,31,32. In the future, this will allow elucidating the dynamics of numerous proteins, cells and entire tissues in their physiological environment in detail.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Manuela Weitkunat for the acquisition of Movie S3. We are grateful to Reinhard Fässler for generous support. This work was supported by the EMBO Young Investigator Program (F.S.), the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC Grant 310939 (F.S.), the Max Planck Society (S.B.L., F.S.), the Centre National de la Recherche Scientifique (CNRS) (F.S.), the excellence initiative Aix-Marseille University AMIDEX (F.S.), the LabEX-INFORM (F.S.) and the Boehringer Ingelheim Fonds (S.B.L.).
References
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. P. Natl. Acad. Sci. USA. 2001;98(26):15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Meth. 2011;8(9):737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M, et al. A genome-wide resource for the analysis of protein localisation in Drosophila. eLife. 2016;5:e12068. doi: 10.7554/eLife.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganayakulu G, Schulz RA, Olson EN. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev. Biol. 1996;176(1):143–148. doi: 10.1006/dbio.1996.9987. [DOI] [PubMed] [Google Scholar]
- Roy S, Raghavan KV. Development. 17. Vol. 124. Cambridge, England: 1997. Homeotic genes and the regulation of myoblast migration, fusion, and fibre-specific gene expression during adult myogenesis in Drosophila; pp. 3333–3341. [DOI] [PubMed] [Google Scholar]
- Bryantsev AL, Baker PW, Lovato TL, Jaramillo MS, Cripps RM. Differential requirements for Myocyte Enhancer Factor-2 during adult myogenesis in Drosophila. Dev. Biol. 2012;361(2):191–207. doi: 10.1016/j.ydbio.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Dickinson MH. Role of calcium in the regulation of mechanical power in insect flight. P. Natl. Acad. Sci. USA. 2006;103(11):4311–4315. doi: 10.1073/pnas.0510109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, et al. Genome-Wide Screens for In Vivo Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures. PLoS Genet. 2013;9(1):e1003195. doi: 10.1371/journal.pgen.1003195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev. Cell. 2001;1(5):705–715. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Brotman JS, Sweeney ST, Davis G. Green Fluorescent Protein Tagging Drosophila Proteins at Their Native Genomic Loci With Small P Elements. Genetics. 2003;165(3):1433–1441. doi: 10.1093/genetics/165.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzemich A, Liao KA, Czerniecki S, Schöck F. Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly. PLoS Genet. 2013;9(3):e1003342. doi: 10.1371/journal.pgen.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz B, et al. Alternative mechanisms for talin to mediate integrin function. Curr. Biol. 2015;25(7):847–857. doi: 10.1016/j.cub.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F, Kalchhauser I, Dickson BJ. The transmembrane protein Kon-tiki couples to Dgrip to mediate myotube targeting in Drosophila. Dev. Cell. 2007;12(5):751–766. doi: 10.1016/j.devcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Menon SD, Chia W. Drosophila rolling pebbles: a multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev. Cell. 2001;1(5):691–703. doi: 10.1016/s1534-5807(01)00075-2. [DOI] [PubMed] [Google Scholar]
- Bloor JW, Kiehart DP. zipper Nonmuscle myosin-II functions downstream of PS2 integrin in Drosophila myogenesis and is necessary for myofibril formation. Dev. Biol. 2001;239(2):215–228. doi: 10.1006/dbio.2001.0452. [DOI] [PubMed] [Google Scholar]
- Millard TH, Martin P. Development. 4. Vol. 135. Cambridge, England: 2008. Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure; pp. 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatan M, Shinder V, Israeli D, Schnorrer F, Volk T. The Drosophila blood brain barrier is maintained by GPCR-dependent dynamic actin structures. J. Cell Biol. 2011;192(2):307–319. doi: 10.1083/jcb.201007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic Analysis with a Repressible Cell Marker for Studies of Gene Function in Neuronal Morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Roy S, Huang H, Liu S, Kornberg TB. Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science. 2014;343(6173):1244624. doi: 10.1126/science.1244624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster D, Luschnig S. Src42A-dependent polarized cell shape changes mediate epithelial tube elongation in Drosophila. Nat. Cell Biol. 2012;14(5):526–534. doi: 10.1038/ncb2456. [DOI] [PubMed] [Google Scholar]
- Weitkunat M, Schnorrer F. A guide to study Drosophila muscle biology. Methods. 2014;68(1):2–14. doi: 10.1016/j.ymeth.2014.02.037. [DOI] [PubMed] [Google Scholar]
- Weitkunat M, Kaya-Çopur A, Grill SW, Schnorrer F. Tension and force-resistant attachment are essential for myofibrillogenesis in Drosophila flight muscle. Curr. Biol. 2014;24(7):705–716. doi: 10.1016/j.cub.2014.02.032. [DOI] [PubMed] [Google Scholar]
- Guirao B, et al. Unified quantitative characterization of epithelial tissue development. eLife. 2015;4:e08519. doi: 10.7554/eLife.08519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etournay R, et al. Interplay of cell dynamics and epithelial tension during morphogenesis of the Drosophila pupal wing. eLife. 2015;4:e07090. doi: 10.7554/eLife.07090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitkunat M, Brasse M, Bausch AR, Schnorrer F. Development. 7. Vol. 144. Cambridge, England: 2017. Mechanical tension and spontaneous muscle twitching precede the formation of cross-striated muscle in vivo; pp. 1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin J, Albrecht S, Kölsch V, Paululat A. Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expr. Patterns. 2006;6(4):360–375. doi: 10.1016/j.modgep.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Lemke SB, Schnorrer F. Mechanical forces during muscle development. Mech. Develop. 2017;144(Pt A):92–101. doi: 10.1016/j.mod.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Classen A-K, Aigouy B, Giangrande A, Eaton S. Imaging Drosophila pupal wing morphogenesis. Methods Mol. Biol. 2008;420:265–275. doi: 10.1007/978-1-59745-583-1_16. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Koolhaas WH, Schnorrer F. G3. 12. Vol. 4. Bethesda: 2014. A versatile two-step CRISPR- and RMCE-based strategy for efficient genome engineering in Drosophila; pp. 2409–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen H-M, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. P. Natl. Acad. Sci. USA. 2014;111(29):E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]


