Abstract
Objectives
To determine whether Tumor Necrosis Factor alpha (TNFα) –308 G/A polymorphism is associated with oral lichen planus (OLP).
Material and Methods
A systematic electronic search of the literature was conducted to identify all published studies on the association between TNFα –308 G/A polymorphism and OLP. All case-control studies evaluating the TNFα –308 G/A polymorphisms in OLP were selected. A meta-analysis of the studies that fulfilled the inclusion criteria was performed. Odds ratios (OR) with 95% confidence intervals (CI) were also calculated.
Results
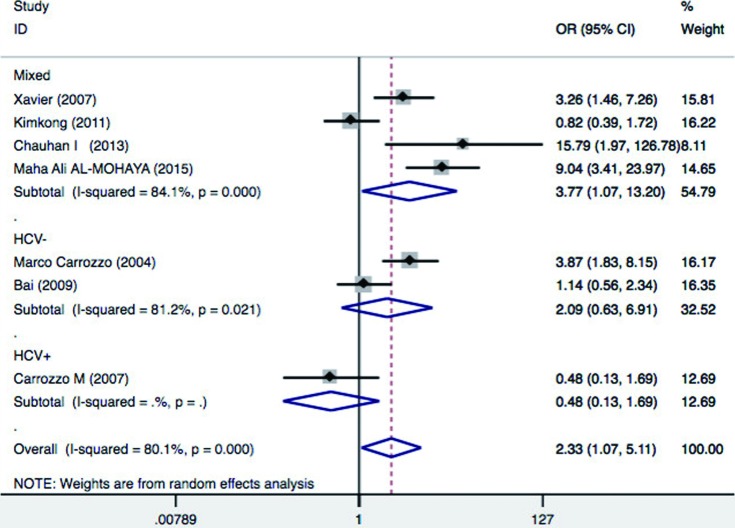
Seven studies comprising 450 OLP cases and 867 controls were included in the meta-analysis. In the pooled analysis, TNFα –308 G/A polymorphism was associated with OLP with random effects and OR of 2.33 (95%CI=1.07-5.11; p=0.03), assuming a dominant mode of inheritance (AA+GA vs. GG). In the subgroup analysis by ethnicity, TNFα –308 G/A was associated with a significantly increased odds ratio of OLP in mixed ethnicity (OR=5.22; 95%CI=1.93-14.15; p=0.001), but not in Asians (OR=1.57; 95%CI=0.54-4.54; p=0.41) or Caucasians (OR=1.45; 95%CI=0.19-11.22; p=0.72). For subgroup analysis based on HCV (hepatitis C virus) infection status, significant increased risk of OLP was found among patients with mixed HCV infection status (OR=3.77; 95%CI=1.07-13.2; p=0.038), but not in patients without HCV infection (OR=2.09; 95%CI=0.63-6.91; p=0.22) and patients with HCV infection (OR=0.48; 95%CI=0.13-1.69; p=0.25).
Conclusion
Our results suggest that –308 G/A polymorphism in TNFα is a potential genetic marker for OLP.
Keywords: Oral lichen planus, Tumor necrosis factors, Genetic polymorphism
Introduction
Oral lichen planus (OLP) is a chronic inflammatory oral condition characterized by oral lichenoid lesions. The etiopathogenesis of OLP involves an interaction between genetic, environmental, and lifestyle factors. OLP lesions can be induced by drugs or dental materials, and are often called idiopathic, involving abnormal immune responses 16 . Previous studies have suggested the involvement of hepatitis C virus (HCV) infection in the etiology of oral lichen planus 14 . However, the etiopathogenesis of OLP is still unclear.
Cytokines play an important role in the pathogenesis of OLP, and a bulk of evidence suggests that OLP is a T-cell-mediated disease 12 , 16 . TNFα is a potent immunomodulator and proinflammatory cytokine that has been found to play roles in the pathogenesis of multiple inflammatory or autoimmune diseases 2 . A higher level of TNFα has been found in serum from OLP patients compared to controls 15 . The subepithelial T cells of OLP contain TNFα mRNA and express TNFα cytokine 6 , 17 . TNFα production may be partially determined at the genetic level. The TNFα gene is a member of the TNF superfamily located on chromosome 6q21, within the major histocompatibility complex (MHC) class III region 3 . Several polymorphisms and mutations have been identified within TNFα, among which a genetic variation at position –308 of the TNFα gene that results in two allelic forms, including a guanine (G) that represents the common variant and an adenine (A) that defines the less common one. G/A polymorphism at position –308 of the TNFα gene promoter is reported to increase TNFα transcription and produce higher levels of this cytokine 11 , 18 . The possible association between TNFα –308 (rs1800629) G/A polymorphism and OLP has been studied by several investigators 1 , 2 , 5 – 7 , 10 , 19 . However, the results have been inconclusive or even contradictory. Differences in ethnicity, lack of statistical power, and involvement of HCV infections are potential factors underlying the discrepant results of these studies. Meta-analysis is a widely used tool to overcome the problem of the small sample sizes and inadequate statistical power of genetic studies of complex traits 13 . Jin, et al. 9 (2012) conducted a meta-analysis on TNFα –308 G/A polymorphism and lichen planus and did not find significant association between them. The studies available at that time were limited. Herein, we conducted a meta-analysis to assess the association between genetic variants of TNFα –308 G/A polymorphism and risk of OLP with the most updated literature on this association.
Material and methods
Study selection and data extraction
A comprehensive literature search was conducted on Medline, PubMed, Embase, and Science Citation Index for all articles that were published by April 17, 2017 on the association between TNFα –308 G/A polymorphism and OLP risk. The search terms were used as follows: (“oral lichen planus” or “OLP”) and (“tumor necrosis factor” or “TNF” or “tumor necrosis factor-α” or “TNF-α” or “TNF-α” or “tumor necrosis factor-a”) and (“Polymorphism” or “mutation” or “variant”). Selection criteria of an eligible study were: (a) investigation of TNFα gene polymorphism and OLP risk; (b) case-control studies; (c) sufficient genotype distributions for cases and controls so that an OR with 95%CI could be assessed. Two investigators collected the information from all eligible publications based on the abovementioned criteria. Data including publication year, first author, country, ethnicity, HCV infection status, case number and control number, and genotyping method were extracted independently by these two investigators and reached conformity on all items by consultation.
Statistical study
Odds ratios with 95% confidence intervals were used to estimate the association between the TNFα –308 G/A polymorphism and OLP risk. The pooled ORs were performed assuming a dominant genetic model (AA+AG vs. GG). The statistical significance of the ORs was analyzed by Z-test, and p<0.05 was considered statistically significant. In addition to the comparison among all subjects, we also performed subgroup analyses by HCV infection and ethnicity.
Heterogeneity was evaluated by a chi-square-based Q statistic, and statistical significance was assumed for a p-value lower than 0.05. A fixed-effect model was used when p heterogeneity <0.05, otherwise a random-effects model was used. Sensitivity analysis was performed to verify the robustness of the results by sequentially deleting single study and check the stability of the result. The underlying publication bias was visually examined by Begg's funnel plot and the degree of asymmetry was evaluated by Egger's test.
All the statistical tests were performed by STATA 14.2.
Results
Overall analysis
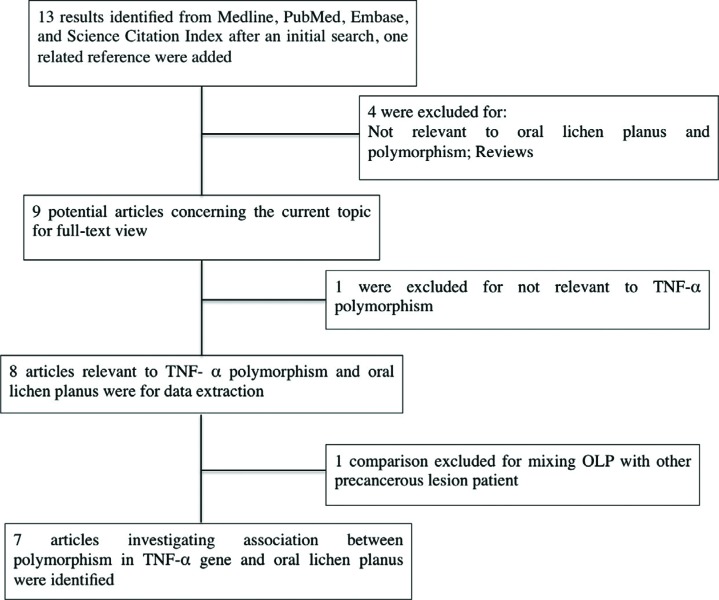
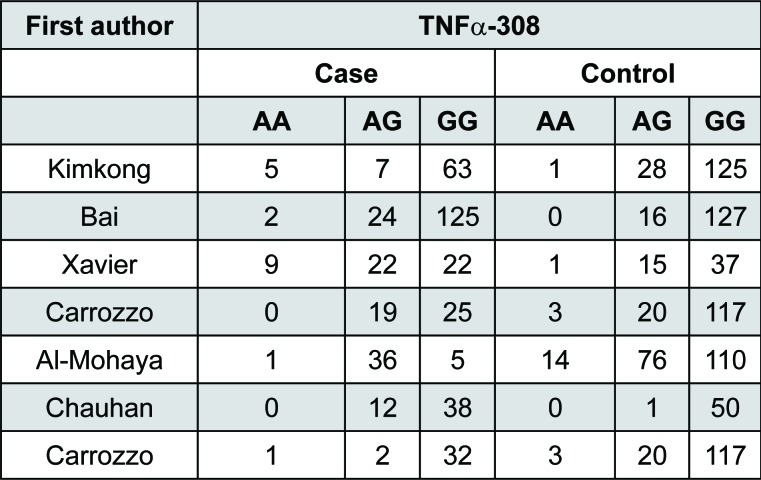
The combined search yielded seven case-control studies that provided information on the association of OLP with TNFα –308 G/A polymorphism (Figure 1). In each studies the clinical diagnosis was confirmed histologically based on the WHO diagnostic criteria of oral lichen planus 19 . Three studies were from Asia 2 , 7 , 10 . Two were conducted in Italy 5 , 6 , one in Saudi Arabia 1 and one in Brazil 19 . One study was specifically performed in patients with HCV positive infection 5 , two with negative HCV infection 2 , 6 , and four with mixed (studied population with or without HCV infection or HCV information not mentioned) HCV infection status 1 , 7 , 10 , 19 . We evaluated 450 patients and 867 controls. In each study, we tested for HWE using the χ2 test. The genotype distribution of the TNFα –308 G/A polymorphism among cases and controls in each study did not deviate from the expected HWE, except for the study conducted in Saudi Arabia 1 . Summary results of comparisons are listed in Figure 2. Distribution of genotypes are listed in Figure 3.
Figure 1. Flow chart for the process of selecting associated publications.
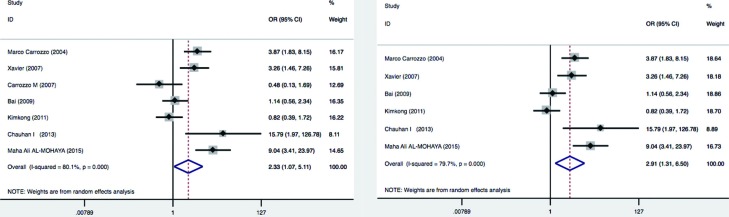
Figure 2. Summary of meta-analysis results. Left panel: Meta-analysis with a random-effects model for the association between OLP risk and the TNFα –308 G/A polymorphism (AA+AG vs. GG). Total random effects p= 0.03. Right panel: Meta-analysis excluding the HCV+ population study. Total random effects p=0.009. For each study, the estimate of OR and its 95%CI is plotted with a box and a horizontal line.
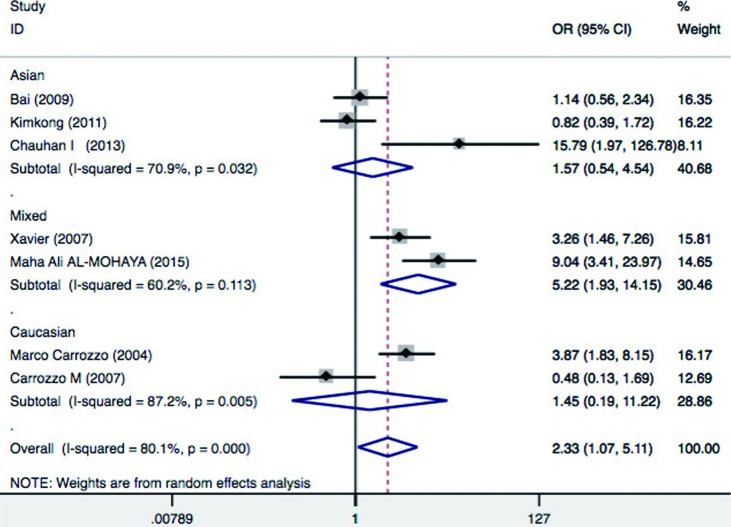
Figure 3. Forest plot of oral lichen planus (OLP) risk associated with TNF-α–308 G/A stratified by ethnicity. For each study, the estimate of OR and its 95%CI is plotted with a box and a horizontal line.
The heterogeneity for the seven studies included was analyzed. The TNFα –308 G/A polymorphism showed significant evidence of genetic heterogeneity between studies (I 2=80.1%, p = 0.03). Random-effects models should be used in the prescience of heterogeneity. Thus, the pooled effect was calculated with a random-effects model. In the pooled analysis, the TNFα –308 G/A polymorphism appeared to be a genetic risk factor for susceptibility to OLP with a random effects and odds ratio (OR) of 2.33 [95% confidence interval (CI): 1.07–5.11; p = 0.03 for AA+AG vs. GG model] (Figure 4, left panel). The results suggest that the AG heterozygote and AA homozygote had an increased risk of OLP compared with those individuals with the GG homozygote genotype. It appears that the effect of TNFα –308 G/A polymorphism may be confounded by HCV infection status in OLP 5 . Therefore, we considered HCV infection status and excluded one study that only studied Hepatitis C virus (HCV) infected OLP patients, and the meta-analysis yielded a higher significant association with a OR of 2.91 [95% confidence interval (CI): 1.31–6.5; p=0.009] (Figure 4, right panel).
Figure 4. Forest plot of oral lichen planus (OLP) risk associated with TNF-α–308 G/A stratified by hepatitis C virus (HCV) infection status. For each study, the estimate of OR and its 95%CI is plotted with a box and a horizontal line.
Subgroup analysis
The prevalence of OLP varies among different ethnic populations mainly due to variations in genetic background. Therefore, to determine whether the effects were due to ethnic differences, subgroup analysis by ethnicity was performed. As for ethnicity subgroup, our meta-analysis revealed that TNFα –308 G/A was associated with a significantly increased risk of OLP in mixed ethnicity (OR: 5.22; 95% CI = 1.93, 14.15 p=0.001), but not in Asians (OR=1.57; 95% CI = 0.54-4.54; p=0.41) and Caucasians (OR=1.45, 95%CI = 0.19-11.22, p = 0.72) for AA+GA vs. GG comparison (Figure 5). These results indicate that A allele carriers (AA+AG) had significantly higher risk of OLP in mixed ethnic population, but not in Asians and Caucasians.
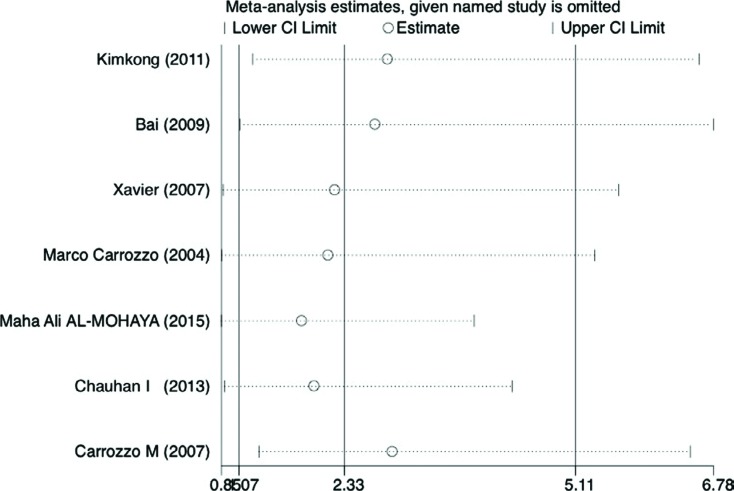
Figure 5. Sensitivity analyses. Effect of individual studies on the pooled OR under allele comparison for TNFα –308G/A polymorphism in OLP patients (random-effects model). Pooled odds ratios and 95% confidence intervals for excluding each data set in the meta-analysis. The vertical axis at 2.33 indicates the overall OR, and the two vertical axes at 1.07 and 5.11 indicate the 95% CI. Every hollow round indicates the pooled OR when the left study was omitted in a meta-analysis with a random model.
Additionally, we performed subgroup analysis based on HCV infection status. Significant increased risk of OLP was found among patients with mixed HCV infection status (OR=3.77, 95%CI = 1.07–13.2, p=0.04), but not in patients without HCV infection (OR=2.09, 95%CI=0.63-6.91, p=0.225) and patients with HCV infection (OR=0.48, 95%CI = 0.13-1.69, p=0.25) (Figure 6). This suggest that A allele carriers (AA+AG) have significantly increased risk of OLP in patients with mixed HCV infection status, but not in HCV positive and/or negative patients. HCV infection also induce a heightened TNFα production, and thus may be a confounding factor in association studies between OLP risk and cytokine gene polymorphisms, underlying the need for future studies to investigate the HCV status of OLP patients. Aggregated ORs for the association are shown in Table 1 and the forest plots of OLP associated with TNFα –308 G/A (AA+AG vs. GG) in Figures 3, 4, 5 and 6.
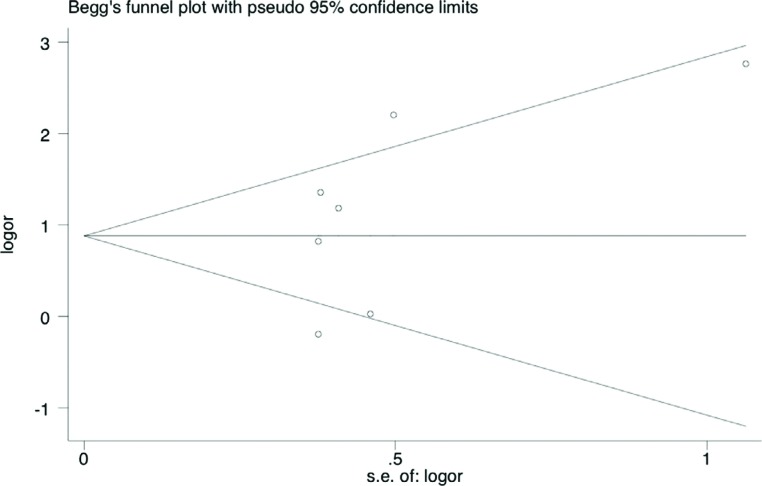
Figure 6. Begg's funnel plot for publication bias in selection of studies on the TNFα –308 G/A polymorphism. The horizontal line represents the meta-analysis summary estimate, and the diagonal linespseudo-95%CI limits about the effect estimate. In the absence of publication bias, studies will be distributed symmetrically above and below the horizontal line. Asymmetry on the bottom of the graph indicates evidence of publication bias towards studies. logOR, natural logarithm of the OR; s.e. of logOR, standard error of the logOR.
Table 1. Associations between TNFα –308 G/A polymorphism and oral lichen planus (OLP).
| Variables | na | OR(95%CI) | Pb |
|---|---|---|---|
| Overall | 7 | 2.33 (1.07- 5.11) c | 0.03 |
| Overall excluding pure HCV+ population | 6 | 2.91(1.30- 6.49) c | 0.009 |
| Subgroup by ethnicity | |||
| Asian | 3 | 1.57(0.54- 4.54) c | 0.41 |
| Caucasian | 2 | 1.44(0.18-11.22) c | 0.72 |
| Mixed | 2 | 5.22(1.92-14.15) c | 0.001 |
| Subgroup by HCV infection | |||
| HCV+ | 1 | 0.47 (0.13- 1.69) c | 0.25 |
| HCV- | 2 | 2.09(0.63-6.90) c | 0.23 |
| Mixed | 4 | 3.77(1.07-13.19) c | 0.04 |
Number of comparisons.
p value for Z-test.
Random-effects model was used for all analysis
Sensitivity analysis and publication bias
In addition, sensitivity analysis was performed by removing one data set at a time. Statistically excluding any individual study did not resolve the genetic heterogeneity. The pooled OR was recalculated in the absence of each study, and OR was not significantly changed (Figure 7 and Table 2).
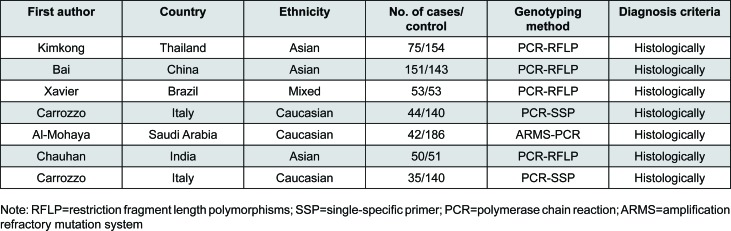
Figure 7. Characteristics of the seven case-control studies included. RFLP: restriction fragment length polymorphisms; SSP: single-specific primer, PCR: polymerase chain reaction; ARMS: Amplification Refractory Mutation System.
Table 2. Summary results of the sensitivity analysis.
| First author | Year | Country | OR | 95% CI | p-value | I2 (%) |
|---|---|---|---|---|---|---|
| Omitting Kimkong, et al. | 2011 | Thailand | 2,852 | [1.2318 6.6029] | 0.014 | 0.778 |
| Omitting Bai, et al. | 2009 | China | 2,7051 | [1.0791 6.7810] | 0.034 | 0.813 |
| Omitting Xavier, et al. | 2007 | Brazil | 2,2216 | [0.8755 5.6376] | 0.093 | 0.826 |
| Omitting Carrozzo, et al. | 2004 | Italy | 2,1411 | [0.8577 5.3451] | 0.103 | 0.814 |
| Omitting Al-Mohaya, et al. | 2015 | Saudi Arabia | 1,8232 | [0.8529 3.8974] | 0.001 | 0.755 |
| Omitting Chauhan, et al. | 2013 | India | 1,9743 | [0.8949 4.3557] | 0.092 | 0.811 |
| Omitting Carrozzo, et al. | 2007 | Italy | 2,9121 | [1.3055 6.4961] | 0.009 | 0.797 |
The shape of the funnel plots did not reveal any evidence of obvious asymmetry (Figure 8), suggesting that there was no obvious publication bias. Egger's test showed no significant publication bias in this meta-analysis (t=0.65, p=0.54 for AA+AG vs. GG).
Figure 8. Distribution of TNF-α –308 genotype.
Discussion
OLP is a complex disorder influenced by multiple genes and environmental factors. Genetic factors that influence the host immune response have been suggested to contribute to OLP risk 4 , 9 . The gene polymorphisms of TNFα has been reported to affect the susceptibility and the progression of OLP 2 , 10 . In this study, we focused on a functional polymorphism of TNFα –308 G/A. Based on seven case-control studies, our meta-analysis revealed that TNFα –308 G/A polymorphism was associated with a significant increase in the risk of OLP (OR=2.33; 95%CI:1.07-5.11; p=0.03). Hepatitis C virus has been proposed to be involved in the etiopathogenesis of OLP. Significant increased risk of OLP was found among patients with mixed HCV infection status (OR=3.77, 95%CI = 1.07-13.2, p=0.04), but not in patients without HCV infection (OR=2.09, 95%CI = 0.63-6.91, p=0.22) and patients with HCV infection (OR=0.47, 95%CI = 0.13-1.69, p=0.25). HCV infection induces an abnormal immune response in the host, enhancing TNFα production, and potentially influences the pathogenesis of OLP lesions. Thus, distinct pathogenetic mechanisms are involved in OLP patients with HCV infection and without HCV infection. The HCV status may be a confounding factor in association studies between OLP risk and TNFα gene polymorphisms, underlying the need for future studies to investigate the HCV status of OLP patients. When we excluded one research that only studied HCV infected OLP patients, the overall meta-analysis yielded higher significance of association with random effects OR of 2.91 [95% confidence interval (CI)=1.31-6.5; p=0.009].
Subgroup analysis based on ethnicity revealed that A allele carriers (AA+AG) had significantly higher risk of OLP in mixed ethnic population, but not in Asians and Caucasians. The two studies with mixed populations are from Brazil and Saudi Arabia respectively. Considering their complex racial genetic makeup, it may be one of the ethnic groups that account for the increased risk. Interestingly, both countries may have mixture of African ethnicity. This also justifies future studies focusing on African populations.
The current meta-analysis has several limitations. First, the sample size for some subgroup analysis was limited. Second, obvious heterogeneity was found in overall analysis. Moreover, one of the studies included in our meta-analysis did not follow HWE. Deviation from HWE reflects potential mistakes, such as laboratory or genotyping errors, population stratification or selection bias in controls. When we excluded one study that deviated from HWE, the pooled ORs was altered and the association became statistically insignificant (p>0.05). Lastly, in this study, we only evaluated a dominant model (AA+GA vs. GG). Other models [i.e. recessive model (AA vs. GG+GA) and homozygote model (AA vs. GG)] could have been conducted. Despite those limitations listed aforementioned, our meta-analysis also has its advantages. First, it contains the latest data on the association between TNFα –308 G/A polymorphism and OLP risk. Also, to our knowledge, it is the first meta-analysis that investigated the association between TNFα –308 G/A polymorphism and oral lichen planus risk.
Conclusion
Our meta-analysis suggests that the substitution of G to A of TNFα –308 G/A polymorphism is a risk factor for OLP. Its effect exists among mixed populations with mixed HCV status. The TNFα –308 G/A polymorphism may be a useful genomic marker for oral lichen planus.
References
- 1.Al-Mohaya MA, Al-Harthi F, Arfin M, Al-Asmari A. TNF-α, TNF-β and IL-10 gene polymorphism and association with oral lichen planus risk in Saudi patients. J Appl Oral Sci. 2015;23(3):295–301. doi: 10.1590/1678-775720150075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai J, Jiang L, Lin M, Zeng X, Wang Z, Chen Q. Association of polymorphisms in the tumor necrosis factor-α and interleukin-10 genes with oral lichen planus: a study in a Chinese cohort with Han ethnicity. J Interferon Cytokine Res. 2009;29(7):381–388. doi: 10.1089/jir.2008.0089. [DOI] [PubMed] [Google Scholar]
- 3.Carroll MC, Katzman P, Alicot EM, Koller BH, Geraghty DE, Orr HT, et al. Linkage map of the human major histocompatibility complex including the tumor necrosis factor genes. Proc Natl Acad Sci U S A. 1987;84(23):8535–8539. doi: 10.1073/pnas.84.23.8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrozzo M. Understanding the pathobiology of oral lichen planus. Curr Oral Health Rep. 2014;1(3):173–179. [Google Scholar]
- 5.Carrozzo M, Dametto E, Fasano ME, Arduino P, Bertolusso G, Uboldi de Capei F, et al. Cytokine gene polymorphisms in hepatitis C virus-related oral lichen planus. Exp Dermatol. 2007;16(9):730–736. doi: 10.1111/j.1600-0625.2007.00577.x. [DOI] [PubMed] [Google Scholar]
- 6.Carrozzo M, Uboldi de Capei M, Dametto E, Fasano ME, Arduino P, Broccoletti R, et al. Tumor necrosis factor-α and interferon-γ polymorphisms contribute to susceptibility to oral lichen planus. J Invest Dermatol. 2004;122(1):87–94. doi: 10.1046/j.0022-202X.2003.22108.x. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan I, Beena VT, Srinivas L, Sathyan S, Banerjee M. Association of cytokine gene polymorphisms with oral lichen planus in Malayalam-speaking ethnicity from South India (Kerala) J Interferon Cytokine Res. 2013;33(8):420–427. doi: 10.1089/jir.2012.0115. [DOI] [PubMed] [Google Scholar]
- 8.Eigler A, Sinha B, Hartmann G, Endres S. Taming TNF: strategies to restrain this proinflammatory cytokine. Immunol Today. 1997;18(10):487–492. doi: 10.1016/s0167-5699(97)01118-3. [DOI] [PubMed] [Google Scholar]
- 9.Jin X, Wang J, Zhu L, Wang L, Dan H, Zeng X, et al. Association between–308 G/A polymorphism in TNF-α gene and lichen planus: a meta-analysis. J Dermatol Sci. 2012;68(3):127–134. doi: 10.1016/j.jdermsci.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Kimkong I, Hirankarn N, Nakkuntod J, Kitkumthorn N. Tumour necrosis factor-alpha gene polymorphisms and susceptibility to oral lichen planus. Oral Dis. 2011;17(2):206–209. doi: 10.1111/j.1601-0825.2010.01722.x. [DOI] [PubMed] [Google Scholar]
- 11.Kroeger KM, Carville KS, Abraham LJ. The –308 tumor necrosis factor-α promoter polymorphism effects transcription. Mil Immunol. 1997;34(5):391–399. doi: 10.1016/s0161-5890(97)00052-7. [DOI] [PubMed] [Google Scholar]
- 12.Lavanya N, Jayanthi P, Rao UK, Ranganathan K. Oral lichen planus: an update on pathogenesis and treatment. J Oral Maxillofacial Pathol. 2011;15(2):127–132. doi: 10.4103/0973-029X.84474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozturk A, Vieira AR. TLR4 as a risk factor for periodontal disease: a reappraisal. J Clin Periodontol. 2009;36(4):279–286. doi: 10.1111/j.1600-051X.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 14.Petti S, Rabiei M, De Luca M, Scully C. The magnitude of the association between hepatitis C virus infection and oral lichen planus: meta-analysis and case control study. Odontology. 2011;99(2):168–178. doi: 10.1007/s10266-011-0008-3. [DOI] [PubMed] [Google Scholar]
- 15.Rhodus NL, Cheng B, Myers S, Bowles W, Ho VU, Ondrey F. A comparison of the pro-inflammatory, NF-κB-dependent cytokines: TNF-alpha, IL-1-alpha, IL-6, and IL-8 in different oral fluids from oral lichen planus patients. Clin Immunol. 2005;114(3):278–283. doi: 10.1016/j.clim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Scully C, Beyli M, Ferreiro MC, Ficarra G, Gill Y, Griffiths M, et al. Update on oral lichen planus: etiopathogenesis and management. Crit Rev Oral Biol Med. 1998;9(1):86–122. doi: 10.1177/10454411980090010501. [DOI] [PubMed] [Google Scholar]
- 17.Simark-Mattsson C, Bergenholtz G, Jontell M, Eklund C, Seymour GJ, Sugerman PB, et al. Distribution of interleukin-2, -4, -10, tumour necrosis factor-α and transforming growth factor-β mRNAs in oral lichen planus. Arch Oral Biol. 1999;44(6):499–507. doi: 10.1016/s0003-9969(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 18.Wilson AG, Di Giovine FS, Blakemore AIF, Duff GW. Single base polymorphism in the human tumour necrosis factor alpha (TNFα) gene detectable by Ncol restriction of PCR product. Hum Mol Genet. 1992;1(5):353–353. doi: 10.1093/hmg/1.5.353. [DOI] [PubMed] [Google Scholar]
- 19.Xavier GM, Sá AR, Guimarães AL, Silva TA, Gomez RS. Investigation of functional gene polymorphisms interleukin-1β, interleukin-6, interleukin-10 and tumor necrosis factor in individuals with oral lichen planus. J Oral Pathol Med. 2007;36(8):476–481. doi: 10.1111/j.1600-0714.2007.00560.x. [DOI] [PubMed] [Google Scholar]