Abstract
The epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that is an integral component of proliferative signaling. EGFRs on the cell surface become activated upon EGF binding and have an increased rate of endocytosis. Once in the cytoplasm, the EGF·EGFR complex is trafficked to the lysosome for degradation, and signaling is terminated. During trafficking, the EGFR kinase domain remains active, and the internalized EGFR can continue signaling to downstream effectors. Although effector activity varies based on the EGFR's endocytic location, it is not clear how this occurs. In an effort to identify proteins that uniquely associate with the internalized, liganded EGFR in the early endosome, we developed an early endosome isolation strategy to analyze their protein composition. Post-nuclear supernatant from HeLa cells stimulated with and without EGF were separated on an isotonic 17% Percoll gradient. The gradient was fractionated, and early endosomal fractions were pooled and immunoisolated with an EEA1 mAb. The isolated endosomes were validated by immunoblot using antibodies against organelle-specific marker proteins and transmission EM. These early endosomes were also subjected to LC–MS/MS for proteomic analysis. Five proteins were detected in endosomes in a ligand-dependent manner: EGFR, RUFY1, STOML2, PTPN23, and CCDC51. Knockdown of RUFY1 or PTPN23 by RNAi indicated that both proteins play a role in EGFR trafficking. These experiments indicate that endocytic trafficking of activated EGFR changes the protein composition, membrane trafficking, and signaling potential of the early endosome.
Keywords: endosome, epidermal growth factor receptor (EGFR), membrane trafficking, proteomics, receptor endocytosis
Introduction
The epidermal growth factor receptor (EGFR)2 is the prototypical receptor-tyrosine kinase (RTK) with established roles in developmental biology, tissue homeostasis, and wound healing (1). In addition, overexpression and activating mutations of EGFR have been associated with many cancers including brain, lung, breast, pancreatic, and colon cancers (2). Although there are Food and Drug Administration–approved chemotherapies that target the EGFR directly, these therapies can be refractory and do not have long-term efficacy (3). A better understanding of EGFR signaling is an important first step for positively and negatively regulating signaling via pharmacological intervention.
The fundamentals of how a ligand, such as EGF, binds to the EGFR, induces receptor dimerization and transphosphorylation, and elicits downstream effector activation is well-established (4, 5). The molecular mechanisms that regulate the magnitude, duration, and specificity of EGFR·effector communication are less understood, but these factors determine receptor specific cell biology such as cell proliferation, migration, and differentiation. Further, alterations in regulatory mechanisms are associated with cell transformation (6) and have been targeted to promote receptor activity to enhance wound healing (7).
One of the most critical regulatory mechanisms is ligand-mediated endocytosis. Concurrent with receptor dimerization and phosphorylation, ligand binding also increases the rate of EGFR internalization (8). At physiological ligand concentrations, the ligand·receptor complex translocates along the plasma membrane and is enriched in clathrin-coated pits that pinch off to give rise to intracellular clathrin-coated vesicles. The clathrin is shed from these vesicles, and the resulting intermediate vesicle fuses with the early endosome. Depending on multiple factors such as the activating ligand (9), cell type, and receptor density, the early endosome then readies the receptor for its ultimate cellular destination, be it the plasma membrane (10), lysosome (11), endoplasmic reticulum (12), or nucleus (13). This process governs how long the ligand·receptor complex is active, the localization of the receptor, and its proximity to downstream effectors.
The EGFR and other RTKs continue to signal once they are inside the cell (14). Several groups have demonstrated that specificity in EGFR·effector communication is conferred through the spatial regulation of the endocytic pathway. In fact, some work has shown that receptor endocytosis is obligatory for certain receptor·effector interactions (11). Many downstream effectors are associated with the early endosome, either as integral membrane proteins, as associated membrane proteins, or through interactions with integral membrane proteins (15). In many cases, however, the identification of associated membrane proteins entailed a bias selection of candidate proteins. We hypothesized that this strategy may have overlooked key regulatory or effector proteins. In an effort to identify novel proteins that associate with the early endosome in an EGF-dependent manner, we sought an unbiased approach. This study combines classic biochemical approaches for endosomal enrichment with MS to identify associated proteins.
We have developed a noninvasive strategy to enrich and purify early endosomes from cells stimulated with or without physiologic levels of EGF and used LC–MS/MS to analyze the proteome of both populations. In addition to several expected early endosomal proteins, we identified four novel proteins that associate with early endosomes in an EGF-dependent manner: RUFY1, STOML2, PTPN23, and CCDC51. Among the roles for these proteins are membrane trafficking and cargo sorting (16, 17). Knockdown of these proteins resulted in altered kinetics of EGF·EGFR endocytic trafficking. The long-term impact of this work is critical, because it is impartial in protein discovery. There are implications in understanding the contribution of the endocytic pathway in EGFR signaling under physiological and pathological conditions, as well as for the signaling of other cell surface receptors.
Results
The overarching goal of these studies was to use an unbiased approach to identify proteins that associate with the early endosome in an EGF-dependent manner. All experiments were performed using HeLa cells, a human adenocarcinoma cell line (18). HeLa cells express physiologic levels of EGFR (∼50,000 EGFRs/cell) (19), the trafficking of which has been extensively characterized; grow rapidly; and are relatively easy to culture (20).
EGF colocalizes with EEA1-positive vesicles
To determine the time point at which the EGFR was maximally localized to the early endosome, we used indirect immunofluorescence probing for the early endosome marker, EEA1 (early endosome autoantigen 1) (21) following Alexa 647–EGF treatment (Fig. 1). The addition of EGF induces a time-dependent redistribution of EGFR into the cytosol and colocalization with EEA1. Fifteen minutes after the addition of EGF, there is a peak accumulation of EGF costaining with EEA1. These kinetics of endocytic trafficking are consistent with previous reports (9, 22). After 30 min of EGF treatment, there is a decrease in EGF and EEA1 costaining, which is consistent with reports that the EGF·EGFR complex is trafficked out of the early endosome 30–60 min after EGF stimulation (23, 24). Subsequent experiments use 15 min of EGF treatment to maximize the receptor association with the early endosome.
Figure 1.
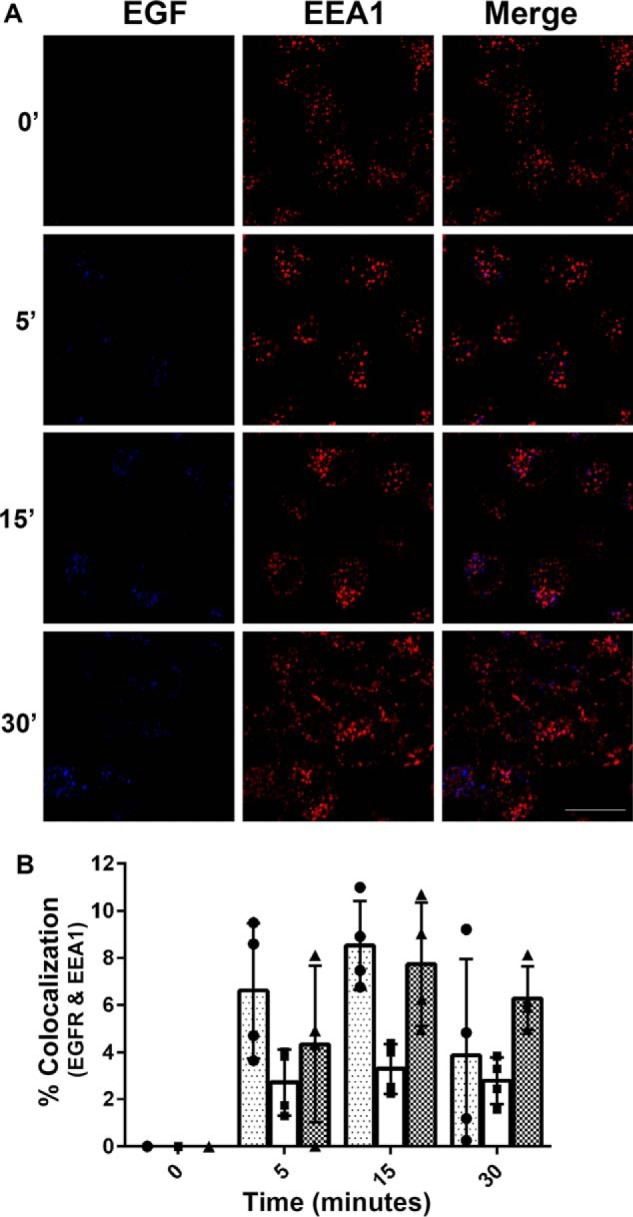
EGF colocalization with EEA1-positive vesicles. Serum-starved HeLa cells were pulse-chased with Alexa 647–EGF (10 ng/ml) for 0, 5, 15, and 30 min. The cells were fixed and processed for indirect immunofluorescence using an EEA1 antibody and fluorescently labeled secondary antibody (goat anti-rabbit Alexa 488). Scale bar, 20 μm. A, images are representative of three independent experiments. B, the extent of colocalization between EGF and EEA1 was measured as described under “Experimental procedures.” The data are plotted as the percentages of colocalization for each time point (four images were taken per time point, i.e. each data point measured one image). Three independent experiments are represented with three distinct bars. Approximately 300 cells total were analyzed per time point per condition, per experiment. Scale bars, 20 μm. The images were quantified using ImageJ software. A Pearson's correlation was calculated for each of the three experiments comparing EGF fluorescence to total EEA1 fluorescence: r = 0.8790, r = 0.9608, and r = 0.9659.
EGFR colocalizes with early endosomal proteins in isotonic Percoll gradient fractions
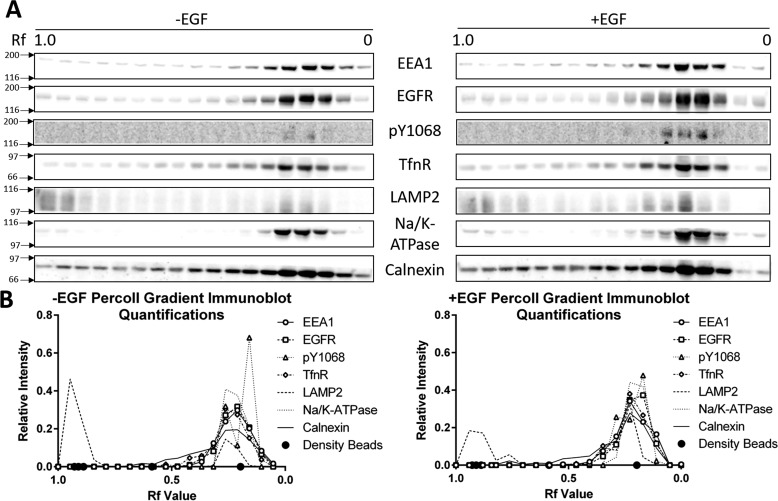
To biochemically enrich the early endosome population, post-nuclear supernatant (PNS) was prepared from HeLa cells treated without and with EGF and separated on a 17% isotonic Percoll gradient (25). Fractions of Percoll gradients were subjected to immunoblot for EEA1, EGFR, phosphorylated EGFR (Tyr(P)-1068), transferrin receptor (TfnR) (early/recycling endosome marker), and LAMP2 (late endosome/lysosome-associated membrane protein 2) (late endosome/lysosome marker) (Fig. 2A). EGFR peaked in the same fractions as EEA1 and TfnR (Rf = ∼0.25–0.10) independent of EGF treatment. The gradient distribution of LAMP2 indicated that there is a distinct, but not complete separation of early and late endosomes/lysosomes within the Percoll gradient, although a lesser amount of LAMP2 is present in the early endosome peak fractions. This less intense peak of LAMP2 increases upon EGF ligand stimulation. Phosphorylated EGFR (at tyrosine residue 1068, i.e. Tyr(P)-1068) was detected to differentiate between active and inactive receptors. There was a low basal level of phosphorylation in the unstimulated fractions, likely reflecting the population of constitutively recycling EGFR. These levels increased upon EGF stimulation and corresponded with EEA1 peak fractions. We also monitored the distribution of Na/K-ATPase (plasma membrane marker) and Calnexin (endoplasmic reticulum marker) in Percoll gradient fractions and found that both markers peaked in the same fractions as EEA1 and EGFR. These immunoblots were quantified and the relative distribution of each protein in the Percoll gradient was plotted (Fig. 2B). We noticed that Na/K-ATPase has peak concentrations in the same fractions as EEA1. This is because the density of plasma membrane is very close to the density of early endosomes (i.e. 1.045 g/ml) (26). Na/K-ATPase has also been shown to undergo endocytosis under certain conditions, and its presence in these fractions may also indicate early endosomal localization (27–29). These data highlighted that despite significant enrichment of the early endosome, there is a need for additional endosome purification.
Figure 2.
Total and phosphorylated EGFR colocalize with early endosomal markers following isotonic Percoll gradient fractionation. A, PNS was prepared from HeLa cells treated with and without EGF (10 ng/ml) for 15 min. PNS was resolved on a 17% isotonic Percoll gradient, fractionated, and resolved by 7.5% SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and immunoblotted for phosphorylated (Tyr(P)-1068, pY1068) and total EGFR as well as the following marker proteins: EEA1 (early endosomes), TfnR (early and recycling endosomes), LAMP2 (late endosomes and lysosomes), Na/K-ATPase (plasma membrane), and Calnexin (endoplasmic reticulum). The immunoblots are representative of three independent experiments. B, relative intensity of the immunoblots in A. Circles on the x axis represent density bead migration (Rf ∼0.93 = 1.109 g/ml, ∼0.91 = 1.070 g/ml, ∼0.89 = 1.057 g/ml, ∼0.59 = 1.049 g/ml, and ∼0.20 = 1.042 g/ml).
EEA1 targeting antibodies purify early endosomes
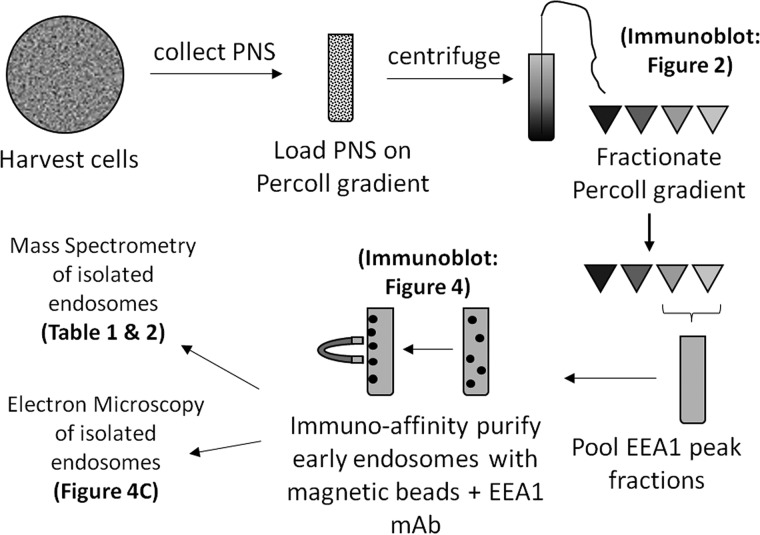
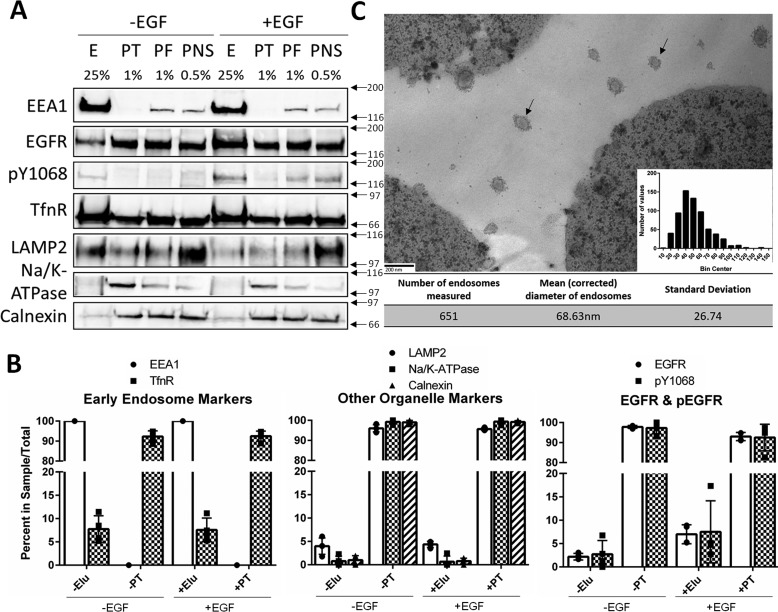
Because of the presence of other contaminating organelles in the EEA1 fractions, an affinity purification strategy was used to further isolate the enriched early endosomes. Percoll gradient fractions with an Rf of ∼0.25–0.10 (∼1.04–1.03 g/ml density) were immunoisolated using an EEA1 mAb (Cell Signaling) and protein G-conjugated to magnetic Dynabeads (Invitrogen). Magnetic beads were selected for the purification strategy to provide a rapid and gentle platform, because the substrate can be quickly and easily precipitated on a magnet. The steps of this newly developed purification strategy are outlined in Fig. 3. Samples from each step of the process were collected and immunoblotted for multiple organelle marker proteins (Fig. 4A). Our strategy yielded 100% pulldown of EEA1 and ∼7% pulldown of the constitutively recycled TfnR in both EGF stimulated and unstimulated samples. Although we expected greater pulldown of TfnR, it is not surprising because the constitutively recycled receptor is also localized to the plasma membrane and recycling endosomes. Alternatively, this low yield of TfnR could indicate that our early endosomes are not remaining intact throughout the isolation procedure.
Figure 3.
Schematic of the Percoll gradient purification protocol.
Figure 4.
Affinity purification of early endosomes from Percoll gradient fractions. A, PNS from HeLa cells treated with or without EGF (10 ng/ml) were separated on a 17% isotonic Percoll gradient. Fractions containing early endosome markers were immunoisolated using an EEA1 antibody as outlined under “Experimental procedures.” Samples were loaded by percentage of total sample volume, and proteins were resolved on a 7.5% SDS-PAGE. E, elution; PT, pass-through; PF, pooled fractions (Percoll gradient fractions with Rf values of ∼0.25–0.10). The percentages of sample total are noted above each lane. Membranes were immunoblotted for EEA1, total EGFR, phospho-EGFR (Tyr(P)-1068, pY1068), TfnR, LAMP2, Na/K-ATPase, and Calnexin. The data are representative of three independent experiments. B, quantifications are shown as percentages of the total IP sample (i.e. elution + pass-through = 100%). The data are plotted ± S.D. C, electron micrograph of immunoisolated early endosomes. A representative micrograph of Dynabeads and early endosomes (30,000×). Scale bar, 200 nm. The diameters of 651 individual endosomes were measured using ImageJ software. Correcting with the Fullman equation, the mean diameter of the endosomes was calculated to be 68.63 nm. A histogram of endosome size is inset in C. Arrows indicate endosomes.
Immunoisolated fractions contained early endosome markers (e.g. EEA1 and TfnR) and were largely devoid of markers of other organelles (LAMP2, Na/K-ATPase, and Calnexin) (Fig. 4B). EGF treatment increased total and phosphorylated EGFR. Together, these data indicate that we are successfully enriching EGFR-containing early endosomes. In both samples, the majority of phosphorylated EGFR was detected in the pass-through and not in the elution. This result, along with the low levels of TfnR in the elution, could also suggest that the early endosomes do not remain intact during isolation. However, EGF stimulation increases the amount of total EGFR precipitated with EEA1 more than 3-fold. Further, our ability to consistently precipitate virtually 100% of EEA1 from enriched gradient fractions indicates that these membrane preparations are highly specific for early endosomes.
The absence of other organelle marker proteins was used as a negative control for early endosome precipitation. The vast majority of LAMP2, Na/K-ATPase, and Calnexin were present in the pass-through of both samples. However, low but detectable amounts were present in the elutions. It is possible that their presence indicates contamination in our preparations. It is not likely that Calnexin would be found in early endosomes under normal conditions; however, we cannot completely rule out this possibility. LAMP2 on the other hand was detected in our Percoll gradient early endosome fractions at low levels (Fig. 2), and because it is involved in trafficking, it is likely that its presence in our sample is representative of hybrid endosomes that are undergoing maturation (30). Very low levels of Na/K-ATPase in the elutions indicate that there is minimal but detectable plasma membrane contamination.
EEA1-purified compartments exhibit early endosome morphology
Although the biochemical data confirmed that early endosome proteins were being purified and other organelles were being excluded, we wanted to determine whether intact vesicles or membrane fragments were being pulled down. Early endosomes were enriched in Percoll gradients and affinity-purified using an EEA1 antibody and magnetic beads. The magnetic bead–endosome complexes were fixed and stained for transmission EM (TEM) as outlined under “Experimental procedures.” The mean (± standard deviation) diameter (corrected using the Fullman equation) of the vesicles was 68.63 ± 26.74 nm (Fig. 4C).
TEM has been used by many investigators to visualize endosomes. Early endosomes appear as round structures with low density staining, whereas late endosomes stain darker and contain intraluminal vesicles. This makes the two endosome types easy to distinguish using TEM. The endosomes we isolated exhibit staining, morphology, and size distribution indistinguishable from reports of early endosomes in the literature (31).
Mass spectrometry reveals early endosome proteins and novel EGF–dependent associations
The affinity-purified endosomes were collected and subjected to LC–MS/MS as outlined under “Experimental procedures.” The resulting data were uploaded into the MassIVE (http://massive.ucsd.edu/) data repository (entry MSV000081692; also shared with the ProteomeXchange (www.proteomexchange.org) under entry PXD008170). Among all three replicates, a total of more than 900 distinct proteins were detected. Here, we report an abridged list of proteins in Table 1. Multiple proteins with known associations and functions with early endosomes and endocytosis were present with and without EGF treatment, including sorting nexins, secretory carrier-associated membrane proteins, EEA1, and TfnR. These data, along with the TEM images, validate our claim of early endosome purification.
Table 1.
Early endosome and membrane trafficking proteins
Proteins were detected in all EGF-treated and untreated samples from three independent LC–MS/MS analyses of EEA1-affinity–purified early endosomes. Early endosomes from HeLa cells treated with ±10 ng/ml EGF for 15 min were enriched in a Percoll gradient and affinity-purified using an EEA1 monoclonal antibody. Purified endosomes were eluted in sample buffer, resolved on a 12% SDS-PAGE, stained with Coomassie, and subjected to LC–MS/MS as outlined under “Experimental procedures.” The data are compiled from three independent experiments and quantified using intensity-based absolute quantification (iBAQ). A complete list of proteins can be found at the MassIVE data repository (entry MSV000081692).
| Protein abbreviation | Protein name | Molecular mass | Molecular function |
|---|---|---|---|
| kDa | |||
| DNAJC13 | DnaJ homolog subfamily C member 13 | 254 | Early endosomal trafficking |
| EEA1 | Early endosome antigen 1 | 162 | Early endosomal trafficking |
| SNX6 | Sorting nexin-6 | 47 | Intracellular trafficking |
| SNX2 | Sorting nexin-2 | 58 | Intracellular trafficking |
| STX7 | Syntaxin-7 (isoform 2) | 27 | Endocytic trafficking |
| MYOF | Myoferlin (isoform 6) | 233 | Endocytic recycling |
| SCAMP1 | Secretory carrier-associated membrane protein 1 | 38 | Intracellular recycling |
| SCAMP2 | Secretory carrier-associated membrane protein 2 | 37 | Intracellular recycling |
| SCAMP3 | Secretory carrier-associated membrane protein 3 | 38 | Intracellular recycling |
| SNX27 | Sorting nexin-27 (Isoform 2) | 60 | Intracellular recycling |
| TBC1D5 | TBC1 domain family member5 (Isoform 2) | 91 | Retrograde transport of cargo from endosomes |
| VP26A | Vacuolar protein sorting-associated protein 26A | 38 | Component of the retromer cargo-selective complex |
| VPS35 | Vacuolar protein sorting-associated protein 35 | 92 | Component of the retromer cargo-selective complex |
| CHMP4B | Charged multivesicular body protein 4b | 25 | Component of ESCRT-III complex |
| CTSZ | Cathepsin Z | 34 | Lysosomal cysteine proteinase |
| STAM2 | Cluster of signal transducing adapter molecule 2 | 58 | Down-regulates RTKs via ESCRT-0 |
We also compared the protein composition of the endosomes collected from cells treated with and without EGF ligand. As expected, both EGF-treated and untreated samples contain EEA1, as well as transport receptors localized to the early endosome (e.g. TfnR, low-density lipoprotein receptor (LDLR), and insulin-like growth factor 2 receptor (IGF2R)) (Table 2). Our methodology was indirectly validated by the presence of proteins involved in endocytosis, intracellular trafficking, membrane recycling, etc. Among the proteins identified, there were five that were specifically present in EGF-treated samples: EGFR, CCDC51, PTPN23, RUFY1, and STOML2 (Table 2). Of these proteins, PTPN23 and RUFY1 play known roles in early endosome trafficking and cargo sorting (16, 17). STOML2 is a mitochondrial protein with well-documented roles in mitochondrial and cardiolipin biogenesis (32, 33). CCDC51 is a recently discovered coiled-coil domain containing protein that has only been described based on its structure. We have used PTPN23, RUFY1, and STOML2 to validate EGF-dependent protein associations with the early endosome.
Table 2.
Receptors detected in early endosomes and proteins that associate with the early endosome in an EGF-dependent manner
Early endosomes from HeLa cells treated with ±10 ng/ml EGF for 15 min were enriched in a Percoll gradient and affinity-purified using an EEA1 monoclonal antibody. Purified endosomes were eluted in sample buffer, resolved on a 12% SDS-PAGE, stained with Coomassie, and subjected to LC–MS/MS as outlined under “Experimental procedures.” The data are compiled from three independent experiments and quantified using intensity-based absolute quantification (iBAQ). The receptors listed (i.e. TFRC, IGF2R, LDLR) and EEA1 were detected in EGF-treated and untreated samples in all three replicates. EGFR and the four novel proteins were detected in only the EGF-treated samples and in at least two of the three replicates.
| Protein abbreviation | Protein name identified | Molecualar mass | Molecular function |
|---|---|---|---|
| kDa | |||
| EEA1 | Early endosome antigen 1 | 162 | Early endosomal trafficking |
| TFRC | Transferrin receptor protein 1 | 85 | Transport of transferrin and iron |
| IGF2R | Cation-independent mannose-6-phosphate receptor | 274 | Transport to lysosome |
| LDLR | Low-density lipoprotein receptor | 105 | Binds and transports low-density lipoprotein |
| EGFR | Cluster of epidermal growth factor receptor | 134 | Receptor tyrosine-kinase |
| STOML2, SLP2 | Cluster of isoform 2 of stomatin-like protein 2 | 33 | Mitochondrial biogenesis |
| CCDC51 | Isoform 2 of coiled-coil domain-containing protein 51 | 34 | Unknown |
| PTPN23, HD-PTP | Tyrosine-protein phosphatase non-receptor type 23 | 179 | Multivesicular body cargo sorting |
| RUFY1, Rabip4′ | RUN and FYVE domain-containing protein 1 | 80 | Early endosomal trafficking |
Analysis of the candidate proteins and other downstream effectors in early endosomes
To determine whether these proteins associate with the early endosome in an EGF-dependent manner, HeLa cells were treated with and without EGF, prepared as a PNS, and separated over an isotonic Percoll gradient. The gradient fractions were immunoblotted for the candidate proteins PTPN23, RUFY1, and STOML2. It was expected that, as primarily early endosome-associated proteins, RUFY1 and PTPN23 would be found in the same fractions in which EEA1 peaks or those with an Rf value of ∼0.25–0.10. There was a peak concentration of both proteins in the EEA1 peak fractions (Fig. 5). STOML2, being a predominately mitochondria-associated protein, was expected to be found in the densest fractions, because mitochondria have a density of ∼1.1g/ml. Interestingly, STOML2 staining was somewhat diffuse throughout the gradient and decreased in the EEA1 peak fractions. This suggests the possibility that STOML2 is not found strictly within mitochondria and may be associated with other organelles of various densities. The concentration of RUFY1 appeared to increase in the EEA1 peak fractions upon EGF stimulation (Fig. 5B). Although RUFY1 (and STOML2) appear in these fractions without EGF stimulation as well, this likely indicates the association of these proteins with other organelles in these fractions, such as plasma membrane. RUFY1 is known to associate with early endosomes regardless of ligand stimulation, and it has also been suggested that RUFY1 may shuttle between the plasma membrane and endosomes (34) and can likely be found localized to both membrane populations.
Figure 5.
Immunoblot validation of candidate proteins from immunoisolated early endosomes. HeLa cells treated with ±10 ng/ml EGF were subjected to Percoll gradient fractionation and EEA1-targeted immunoisolation as described under “Experimental procedures.” A, Percoll gradient fractions were resolved on a 10% SDS-PAGE and immunoblotted for EEA1, PTPN23, RUFY1, or STOML2. B, quantification of the immunoblots in A.
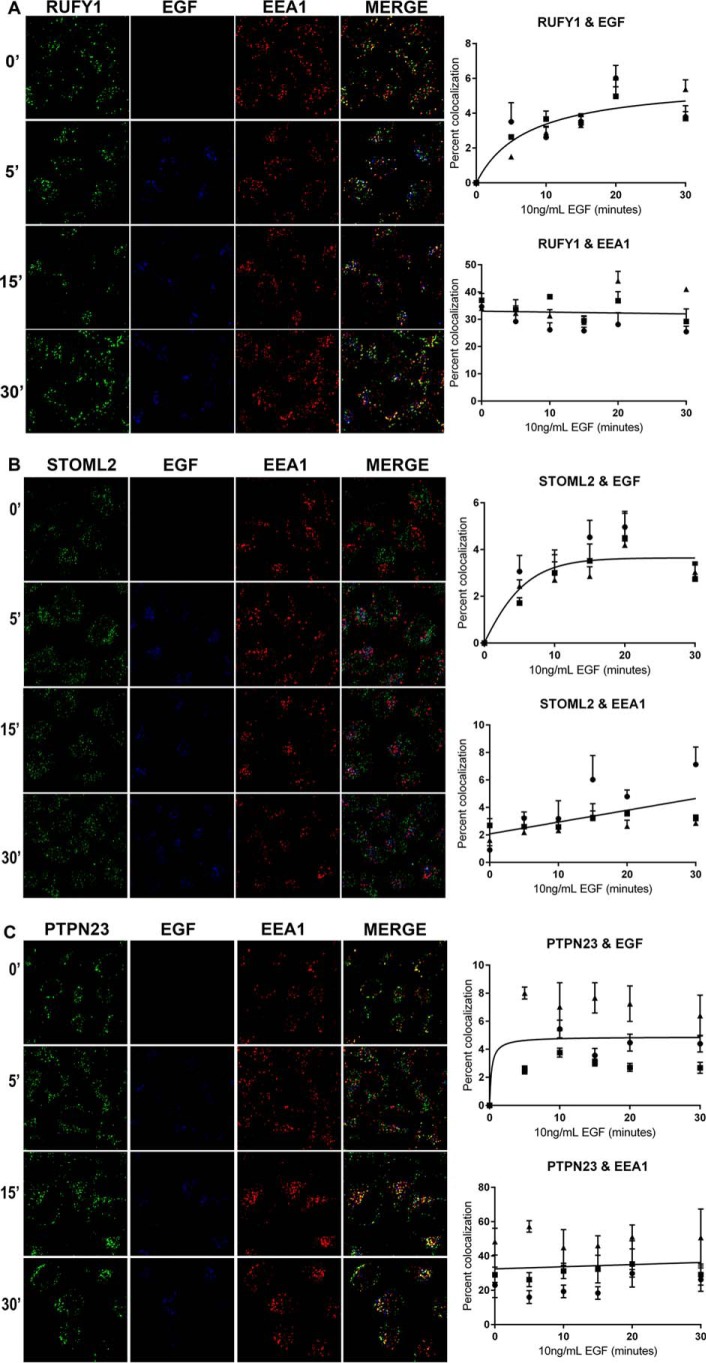
RUFY1, STOML2, and PTPN23 colocalize with EGF and EEA1
We used indirect immunofluorescence to monitor the kinetics of association of our candidate proteins with the early endosome. HeLa cells were pulse-labeled with fluorescently labeled EGF (Alexa 647–EGF) for 0–30 min. The cells were fixed and immunostained for EEA1 and each of the candidate proteins. For all three proteins, there was a time-dependent association with the labeled EGF. This is consistent with the model of the liganded receptor internalizing and trafficking to the early endosome. RUFY1 and PTPN23 colocalized with EEA1 independent of EGF treatment (Fig. 6, A and C), whereas STOML2 had low levels of colocalization with EEA1 that increased with EGF treatment (Fig. 6B).
Figure 6.
RUFY1, STOML2, and PTPN23 colocalize with early endosomes and internalized EGF. A–C, HeLa cells were pulse-labeled with 10 ng/ml Alexa Fluor 647–EGF ligand (Invitrogen) for 0, 5, 10, 15, 20 and 30 min, followed by fixation in 4% paraformaldehyde. The cells were permeabilized and immunostained for EEA1 and either RUFY1, STOML2, or PTPN23 and visualized using either a goat anti-rabbit Alexa 568 or goat anti-mouse Alexa 488, respectively. Images are representative of 0-, 5-, 15-, and 30-min time points from three independent experiments. The extent of colocalization between EGF and each candidate protein or EEA1 and each candidate protein was measured as described under “Experimental procedures.” The data are plotted as the percentages of colocalization for each time point. Approximately 300 cells were analyzed per time point per condition, per experiment. Scale bar, 20 μm. The images were quantified using ImageJ software.
Loss of PTPN23 or RUFY1 changes the kinetics of EGFR endocytic trafficking
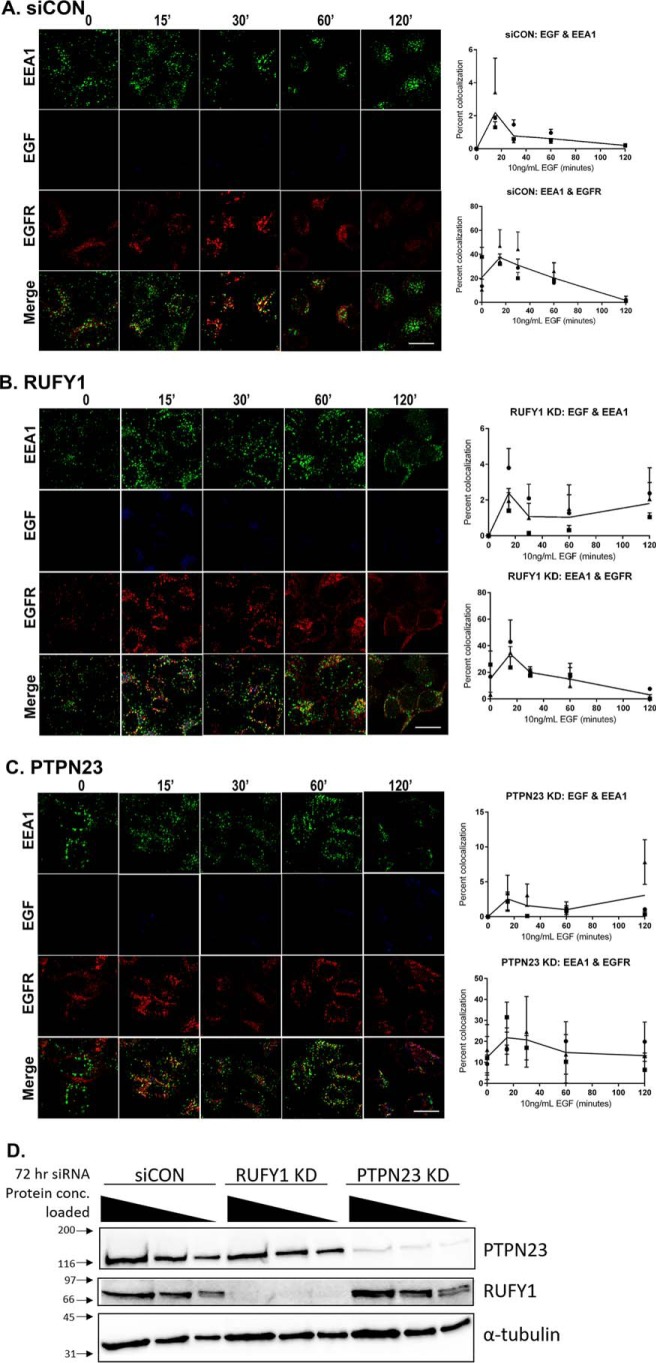
We used RNAi to knock down PTPN23 and RUFY1 in HeLa cells to determine the functional relevance of these proteins in EGFR trafficking. HeLa cells were transfected with 50 nm siRNA or siCON (scramble control) for 72 h, serum-starved, and treated with 10 ng/ml Alexa 647–EGF ligand for 0–120 min as described under “Experimental procedures.” Each experiment was repeated in triplicate, and a knockdown efficiency of more than 90% was achieved for each experiment (Fig. 7D).
Figure 7.
EGF and EGFR colocalization with EEA1 in siCON, RUFY1 KD, and PTPN23 KD cells. A–C, HeLa cells were incubated with siCON, RUFY1, or PTPN23 siRNA for 72 h prior to serum starving. The serum-starved cells were pulse-labeled with 10 ng/ml Alexa Fluor 647–EGF ligand (Invitrogen) for 0, 15, 30, 60, and 120 min, followed by fixation in 4% paraformaldehyde. The cells were permeabilized and immunostained for EEA1 and EGFR and visualized using either a goat anti-rabbit Alexa 488 or goat anti-mouse Alexa 568, respectively. Images are representative of time points from three independent experiments. The extent of colocalization between EGF or EGFR and EEA1 was measured as described under “Experimental procedures.” The data are plotted as the percentages of colocalization for each time point. Approximately 300 cells were analyzed per time point per condition, per experiment. Scale bars, 20 μm. Images were quantified using ImageJ software. D, a representative immunoblot from each knockdown experiment, probing for PTPN23, RUFY1, and α-tubulin. For each knockdown experiment, the samples were loaded in multiple protein concentrations (20, 10, and 5 μg), and the percentage of knockdown was calculated. Only experiments with a knockdown efficiency of >90% were used.
Loss of RUFY1 resulted in sustained activation of EGFR, as evidenced by the prolonged colocalization of EGF ligand with EEA1 at 60- and 120-min when compared with siCON (Fig. 7, A and B). A similar prolonged association of EGF ligand with EEA1 is also seen in the PTPN23 KD samples (Fig. 7C). In addition, the loss of PTPN23 in HeLa cells also yields an increase in the colocalization of total EGFR with EEA1. This suggests that EGFR is sequestered in early endosomes after activation and that PTPN23 plays a role in the endocytic progression (e.g. multivesicular body cargo sorting or endosomal maturation) of the receptor. This is consistent with the results of PTPN23 knockdown in other labs (16, 35, 36).
Discussion
In this study, we developed a novel method for enriching early endosomes. Immunoblotting of Percoll gradient fractions validated the separation of early and late endosomes using EEA1 and LAMP2 as respective marker proteins. In Fig. 2, there is a discreet amount of LAMP2 staining in the EEA1 peak fractions. Although these results could be interpreted as incomplete separation of early and late endosomes, we propose that this lower-density peak of LAMP2 represents a population of early endosomes that are maturing into late endosomes. The literature supports this notion, particularly regarding EGFR internalization driving endosomal maturation (37). This is supported by the increased intensity of the lower density LAMP2 peak after EGF stimulation. Further, the peak of EGFR aligns with the EEA1 peak, independent of EGF treatment. The presence of EGFR in these fractions without ligand stimulation suggests that these receptors could be present in early endosomes, plasma membrane, or endoplasmic reticulum membranes. For this reason, the affinity purification strategy was applied to the gradient enriched early endosomes.
Biochemical and cell biological assays (TEM, immunoblotting, and MS) validated the purification strategy. These assays confirmed that the isolated endosomes were of the proper size and morphology, contained many expected resident proteins, and excluded markers of other organelles. One distinction we noticed is that our TEM images contain vesicles with a halo of extensions around the membrane perimeter. There are several possible explanations for this phenomenon. One is that the EEA1 antibodies conjugated to the endosomes maintained their association with the endosomes. A second is that this is staining of clathrin lattices of clathrin-coated vesicles (38). We favor the idea that the extensions are from the EEA1 antibody, because the endosomes are preincubated with the antibody before conjugation to the beads. Clathrin lattices are unlikely, because clathrin-coated vesicles tend to be distinct compartments from early endosomes. Regardless, the TEM images confirm that the isolated compartments exhibit morphology characteristic of early endosomes.
This combined biochemical/proteomic approach revealed that EGFR, PTPN23, and three previously uncharacterized proteins (i.e. STOML2, RUFY1, and CCDC51) associate with early endosomes in an EGF-dependent manner. Unfortunately, there are no antibodies commercially available for the uncharacterized coiled-coiled domain containing protein CCDC51, so we restricted further studies to RUFY1, STOML2, and PTPN23. The temporal and spatial association with the EGFR-containing endosomes could reflect an important role for these proteins in regulating EGFR signaling.
RUFY1 is a Rab4 effector protein and is commonly referred to by its isoforms rapib4 and rabip4′. It has been previously reported to have a role in receptor tyrosine kinase signaling; loss of RUFY1 has been shown to inhibit PDGF-induced migration of fibroblasts (39). Our data support what other investigators have found—that RUFY1 colocalizes with EEA1 (34, 39, 40) (Fig. 6A). PTPN23 is the only one of the four identified proteins to have been well-studied in the context of EGFR signaling and trafficking. It acts as a coordinator of the ESCRT complex pathway to transport internalized EGFR into multivesicular bodies (35, 36). Our data support the involvement of PTPN23 in EGFR endocytic trafficking (Fig. 6C). The third protein, STOML2, has been studied predominantly in the mitochondria. The literature contains numerous clinical reports of STOML2 being a driver of proliferation in multiple cancer types and is associated with poor patient prognosis (41–44). Interestingly, the mechanism of STOML2 in cancer cell proliferation remains unknown, and the association of STOML2 with EGFR in the early endosomes introduces some interesting possibilities. Together, these findings initiate some important questions: What is the role of these proteins in the early endosome? Are they required for EGFR functions? Do they impact membrane trafficking? Do they affect EGFR signaling?
Biochemical and immunofluorescence analysis indicates that RUFY1 and PTPN23 colocalize with EEA1 independent of EGF treatment (Fig. 6). This observation contradicts the LC–MS/MS data that suggested there was an EGF-dependent association of these proteins with early endosomes. One possible explanation for this discrepancy is that the presence of active EGFR in early endosomes increased the affinity of the candidate proteins with the compartment. As such, the proteins may have dissociated from the endosomes during the biochemical isolation protocol without EGF stimulation; trafficking of the receptor to the endosomes stabilizes these proteins to the vesicles.
In an attempt to further elucidate their functional role in EGFR endocytic trafficking, we used siRNA to knockdown these proteins in HeLa cells. We selected RUFY1 and PTPN23 for RNA silencing, because both RUFY1 and PTPN23, but not STOML2, have previously reported endosomal functions. As such, it is more plausible that their recruitment to the early endosome would affect endocytic trafficking of EGFR.
Loss of either RUFY1 and PTPN23 slowed trafficking of the EGF·EGFR complex and increased its colocalization with EEA1 (Fig. 7). RUFY1 knockdown yielded sustained EGF and EEA1 colocalization. This suggests that RUFY1 loss slows the endocytic trafficking of the EGFR, indicating RUFY1 enhances EGFR trafficking. RUFY1 has been reported to play a role in early endosome transport and recycling (17, 40), and our data support this role. PTPN23 knockdown yielded sustained colocalization of EGFR with EEA1. This suggests that PTPN23 loss resulted in the sequestration of the receptor in early endosomes upon EGF stimulation. This is consistent with PTPN23 regulating EGFR trafficking toward the late endosomes/lysosomal degradation. This aligns with the work of Woodman and co-workers (16, 36), who found that PTPN23/HD-PTP plays a critical role in multivesicular body morphogenesis and (EGFR) cargo sorting.
Of the three proteins, only STOML2 increases its colocalization with EEA1 with EGF treatment using immunofluorescence assays (Fig. 6B). Although this association is modest, it is consistent with a model in which the liganded EGFR recruits STOML2 to the early endosome. This change in subcellular location could permit novel interactions that drive new, previously unknown functions of this protein. This is the first publication testing STOML2 fluorescence using the specified STOML2 antibody, and it is worth noting that the localization of the protein does not mirror what has been published using other STOML2 antibodies (32). This could be due to several possibilities: 1) STOML2 may have an abnormal distribution in HeLa cells, 2) STOML2 may be overexpressed in HeLa cells and a large subgroup of the proteins are mislocalized away from mitochondria and into endosomes, or 3) STOML2 has an undiscovered association with endosomes. Another protein in the STOML2 family, SLP-1/STOML1, has been shown to localize to late endosomes (47), which is consistent with STOML2 having a role in endocytic trafficking.
We were struck by the absence of effector proteins among those associated with the early endosome (MassIVE data repository entry MSV000081692); more specifically, those that had been previously reported to be associated with EGFR containing endosomes (e.g. Shc, Grb2, MEK2, etc.) (10, 45, 46). As such, we wondered why these proteins were not detected in the MS analyses. Despite LC–MS/MS being a more sensitive detection method, it does have limitations. As mentioned above, low-affinity associations with the endosomes may have dissociated during the isolation procedure. Previous work showing an association of these scaffold and effector proteins with early endosomes used immunofluorescence and immunoblotting. An alternative explanation is that the antibodies in those immune-detection methods generate an amplification of a signal, whereas LC–MS/MS quantifies only the total number of peptides present in a sample. As such, it is possible that the quantities of these and other effector proteins in the early endosomes were too low to be detected by LC–MS/MS, but sufficient for detection via immunoblot and immunofluorescence because of signal amplification.
Alternatively, despite our protocol being designed to be rapid and gentle, there is always the possibility that the endosomes did not remain intact during isolation, and some associated proteins were lost. TEM was employed to test for this possibility (Fig. 4C), and the data from those images suggest that the compartments collected were mostly intact.
This method for endosome isolation can be adapted to isolate many other organelles, by selecting an antibody that targets an antigen with high specificity for the organelle of interest. LC–MS/MS provides the sensitivity and unbiased detection in this screen. Although other labs have monitored the subcellular location of other trafficking and signaling proteins, they have relied primarily upon biochemical techniques that require a previous knowledge or predication as to what proteins to monitor. The process developed in this study supersedes this prerequisite. Using this technique, we report an EGF-dependent association of novel proteins with the early endosome. This protocol can be extrapolated to study the spatial regulation of other endocytosed receptors, including the other ErbB family members (i.e. Her2, ErbB3, and ErbB4), and other RTKs such as PDGFR, IGF1-R, and VEGFR.
Experimental procedures
Cell lines
HeLa cells were acquired from American Type Culture Collection. The cells were cultured at 37 °C in 5% CO2 and maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 5% fetal bovine serum (Invitrogen), 100 units/ml streptomycin, 100 units/ml penicillin, and 2 mm glutamine (18).
Post-nuclear supernatant preparation
The cells were grown to confluence in 15-cm dishes, serum-starved for 2 h at 37 °C, and then incubated with or without 10 ng/ml EGF ligand (ProSpec, catalog no. cyt-217-a) for 15 min immediately prior to harvest. The cell lysates were prepared by washing twice with room temperature PBS and equilibrating to 4 °C on ice, followed by equilibrating in ice-cold lysis buffer (TES, 10 mm triethanolamine, 1 mm EDTA, 0.25 m sucrose, pH 7.2). The cells were incubated on ice with TES buffer (supplemented with 2 mm PMSF, 1 mm Na3VO4, 10 μm pepstatin, and 1 μm aprotinin) until cells began to swell, but before bursting (∼5 min), and scraped with a rubber policeman. The collected cells were pipetted up and down 40 times with a P1000 pipetman and centrifuged at 200 × g for 10 min at 4 °C in a tabletop centrifuge to create a PNS, which was subsequently collected. The pellet was resuspended in TES buffer and centrifuged a second time at 200 × g for 10 min at 4 °C. Supernatants were pooled to yield a final PNS (25).
Percoll gradient fractionation
Percoll gradient fractionations were performed as previously described (25, 48).
Affinity purification of early endosomes
Approximately 0.44 μg of EEA1 mAb (Cell Signaling, catalog no. 3288) was preconjugated to ∼2 ml of pooled EEA1 peak fractions (Rf of ∼0.25–0.10) from Percoll gradient samples overnight at 4 °C with rotation. Protein G Dynabeads (Invitrogen) were washed three times in PBS before use, and ∼4.0 × 107 Dynabeads were incubated with each antibody-conjugated sample and rotated at 4 °C for 1 h. Magnetic beads were isolated, and the first supernatant (pass-through) was collected. The beads were then washed three times in ice-cold PBS and eluted in 6× SDS buffer containing 10% β-mercaptoethanol and boiled at 100 °C for 3 min. Remaining samples collected were diluted in 6× SDS buffer with 10% β-mercaptoethanol and boiled. Any samples containing Percoll were centrifuged at 21,000 × g to pellet Percoll.
Immunoblotting
The samples were loaded as a percentage of total sample volume, resolved by SDS-PAGE, and transferred to nitrocellulose. The membranes were probed with the following antibodies according to manufacturer's directions: EGFR (Santa Cruz, catalog no. sc-03), TfnR (BD Biosciences, catalog no. 612124), LAMP2 (University of Iowa Hybridoma Bank, catalog no. H4B4), EEA1 (BD Biosciences, catalog no. 610456), Na/K-ATPase (Sigma, catalog no. A276, only used where indicated), Na/K-ATPase (Cell Signaling, catalog no. 3010), Calnexin (Assay Designs, catalog no. SPA-850), Tyr(P)-1068 (Cell Signaling, catalog no. 2236), PTPN23 (Proteintech, catalog no. A304-883A), RUFY1 (Thermo Fisher Scientific, catalog no. PA5-31400), and STOML2 (Abcam, catalog no. ab191884). Following incubation with the appropriate horseradish peroxidase–conjugated secondary antibody (anti-mouse or anti-rabbit, Thermo Fisher–Pierce), immunoreactive proteins were subjected to Enhanced Chemiluminescence and visualized using a Fotodyne imaging system. Western blotting was quantified using ImageJ software.
Indirect immunofluorescence
HeLa cells were grown to confluency on NaOH-treated, sterile 12-mm round glass coverslips. Serum-starved cells were incubated with EGF (ProSpec, catalog no. cyt-217-a) ligand for the indicated amount of time. Cells were stimulated with 10 ng/ml fluorescent Alexa 647–EGF ligand (Invitrogen) for 10 min and pulse-chased following previously described methods (22). After EGF stimulation, the cells were subjected to indirect immunofluorescence as described previously (49) using EGFR (Ab-1, EMD Millipore, catalog no. GR01), EEA1 (BD Biosciences, catalog no. 610456), PTPN23 (Proteintech, catalog no. A304-883A), RUFY1 (ThermoFisher Scientific, catalog no. PA5-31400), and STOML2 (Abcam, catalog no. ab191884) primary antibodies prepared by manufacturer-recommended dilutions. Immunoreactive proteins were visualized using goat anti-rabbit Alexa 488– and goat anti-mouse Alexa 568–labeled secondary antibodies (Life Technologies), respectively. Coverslips were mounted onto glass slides with Prolong Gold Antifade (Life Technologies) (49). The slides were cured in the dark overnight before imaging. The images were taken in the middle plane of the cells using a 60× oil immersion objective lens on a Nikon A1R confocal microscope.
Colocalization analysis
Colocalization of EGF or EGFR with EEA1 was quantified as described by Lopez-Alcalá et al. (50) and Vanlandingham and Ceresa (22). Briefly, analysis was carried out using ImageJ software and the colocalization plug-in (Pierre Bourdoncle, Institut Jacques Monod, Service Imagerie, Paris) to generate a binary image of colocalized pixels from two separate channels. ImageJ was used to automate channel thresholding, and colocalization was established for pixels whose intensities were higher than threshold and for which the ratio of intensity was greater than 50%. The data were plotted as the ratio of the integrated intensity from the two images. All data represent the averages of three separate experiments, with a total of ∼300 cells measured per experiment.
Coomassie staining
Immunoprecipitated early endosomes were resolved on a 12% SDS-PAGE. The gel was rinsed once in ddH2O and covered with Coomassie (50% MeOH, 0.05% Coomassie Brilliant Blue R (Sigma), 10% acetic acid, 40% ddH2O) and microwaved for 5 s. The gel was incubated with Coomassie at room temperature with gentle rocking for 15 min. The Coomassie was removed, and the gel was rinsed twice in ddH2O. The gel was then covered in destain solution (7% glacial acetic acid, 5% MeOH, 88% ddH2O) and incubated overnight at room temperature with gentle rocking. The gel was rinsed in ddH2O, imaged using a Fotodyne imaging system, and stored in 7% acetic acid/ddH2O at 4 °C.
In-gel protein digestion
This protocol is modified from Jensen et al. (51). A Coomassie-stained SDS-PAGE gel was cut into 1-mm3 plugs and incubated in 100 mm triethylammonium bicarbonate (TEA-BC; Sigma) at room temperature for 15 min. Acetonitrile was added to the TEA-BC solution, and the gel plugs were incubated at room temperature for 15 min with gentle vortexing. The solvent was removed, and the washing process was repeated until the Coomassie Blue stain was no longer visible. Solvent was removed, and the gel plugs were dried in a SpeedVac for 5 min. The dried plugs were incubated in DTT (20 mm DTT [(Bio-Rad), 100 mm TEA-BC) at 56 °C for 45 min, followed by iodoacetamide (55 mm iodoacetamide (Sigma), 100 mm TEA-BC) at room temperature for 30 min protected from light. Iodoacetamide was removed, and gels were washed in 50 mm TEA-BC at room temperature for 15 min, followed by the addition of acetonitrile for 15 min at room temperature with gentle vortexing. The gel plugs were again dried for 5 min in a SpeedVac and incubated in digestion buffer (20 ng/μl modified Trypsin (Promega) in 50 mm TEA-BC) for ∼10 min until the gel plugs swelled. After swelling, 50 mm TEA-BC was added to the plugs, followed by 37 °C overnight incubation in a shaker. Digestion supernatants from the upper and lower half of the gel were combined for each sample.
Extraction of peptides
This protocol is modified from Shevchenko et al. (52). LC–MS grade water was added to the digested gel plugs to give a final concentration of 25 mm TEA-BC. Two volumes of 1:2 5% (v/v) formic acid:acetonitrile was added and incubated at room temperature for 15 min in a shaker (100 rpm in a C25 Incubator Shaker; New Brunswick Scientific). Liquid surrounding the gel pieces was transferred to a clean microtube and dissolved in chromatography buffer A (2% (v/v) acetonitrile, 0.1% (v/v) formic acid). The dissolved sample was filtered through a 0.45-μm regenerated cellulose syringe filter (Thermo catalog no. F2504-7) to remove any remaining gel material. Resolubilized gel band digests were desalted and concentrated using C18 PROTOTM, 300 Å Ultra MicroSpin Column (The Nest Group, Inc., Southborough, MA). The samples were cooled to −80 °C, dried using a SpeedVac and redissolved in chromatography buffer A. Sample absorbance was read at 205 nm using a NanoDrop 2000 spectrophotometer to determine peptide concentration. Sample volumes were adjusted in buffer A to normalize peptide concentrations to 0.1 μg/μl.
Liquid chromatography/tandem mass spectrometry
Gel band digests (0.5 μg) were separated on 12 cm of Aeris Peptide XB-C18, 3.6 μm, 100 Å material (Phenomenex, Torrance, CA) packed into a 360-μm outer diameter - 100-μm inner diameter fused silica tip that was pulled using a model P-2000 micropipette puller (Sutter Instrument Co., Novato, CA). Peptides were eluted from the column using an EASY n-LC UHPLC system (Thermo Fisher Scientific, Waltham, MA) in an 80-min linear gradient using buffer A and buffer B (80% (v/v) acetonitrile, 0.1% (v/v) formic acid) as mobile phases (from 0% buffer B to 50% buffer B). The samples were then separated by a 5-min linear gradient from 50% buffer B to 95% buffer B, followed by a 5-min wash in 95% buffer B. The sample was introduced into the LTQ-Orbitrap Elite (ThermoElectron) mass spectrometer by nanoelectrospray using a Nanospray Flex source (ThermoElectron). The ion transfer capillary temperature was set to 225 °C, and the spray voltage was set to 1.6 kV. An Nth Order Double Play was created in Xcalibur v2.2. Scan event 1 of the methods obtained an FTMS MS1 scan (normal mass range; 240,000 resolution, full scan type, positive polarity, profile data type) for the range 300–2000 m/z. Scan event 2 obtained ITMS MS2 scans (normal mass range, rapid scan rate, centroid data type) of up to 20 peaks that had a minimum signal threshold of 5,000 counts from scan event 1. The lock mass option was enabled (0% lock mass abundance) using the 371.101236 m/z polysiloxane peak as an internal calibrant. Proteome Discoverer v1.4.1.14 (Thermo Fisher Scientific) was used to analyze the mass spectrometer data. MS2 scan data were extracted from the Xcalibur RAW file, CID MS2 scans were searched in Mascot v2.5.1 (Matrix Science, Inc., Boston, MA) and SequestHT, and the results were collected in a single file. The protein database UniprotKB Homosapiens version 3/9/2016 reference proteome canonical and isoform sequences, with cRAP database (thegpm.org) version 1/1/2012 appended to it, were used in the Mascot and SequestHT searches. The resulting files from Proteome Discoverer were loaded into Scaffold Q+S v4.4.5 (Proteome Software, Inc., Portland, OR). The peptide false discovery rate was calculated with Scaffold local false discovery rate algorithm, and protein probabilities were calculated using the Protein Prophet algorithm. The results were annotated with human gene ontology information from the Gene Ontology Annotations Database.
Electron microscopy
Early endosomes were precipitated on protein G Dynabeads as described above. Endosome-Dynabead complexes were pelleted and fixed in 2% paraformaldehyde/2% glutaraldehyde in 0.1 m phosphate buffer (PB) overnight at 4 °C. The pellets were subsequently washed in PB and fixed in 1% OsO4 in PB for 90 min at room temperature. The pellets were washed in PB, dehydrated in an ethanol and propylene oxide series, embedded in Durcupan epoxy resin. Ultrathin sections (80 nm) were cut using a diamond knife and collected on nickel mesh grids. The grids were stained with uranyl acetate and lead citrate. The images were collected using a Hitachi HT7700 transmission electron microscope. Individual vesicles were measured and quantified using ImageJ (National Institutes of Health). The diameters of 651 endosomes were measured, and the corrected mean diameter (D) of all endosomes was calculated using the following Fullman equation: D = (π/2)*N/(1/d1 + 1/d2 … 1/dN), where N represents the total number of compartments, and d represents the diameter of each individual compartment. This equation corrects for the differences in the positioning of the vesicles within the ultrathin sections that were cut.
siRNA knockdown of RUFY1 and PTPN23
RUFY1 siRNA (siGENOME SMARTpool Human RUFY1 catalog no. M-016355-01, lot no. 170711) and PTPN23 siRNA (siGENOME SMARTpool Human PTPN23 catalog no. M-009417-01, lot no. 170711) were obtained from Dharmacon (Lafayette, CO). Each 5-nmol stock siRNA was reconstituted into 20 μm aliquots. Scramble control siRNA (siCON) was acquired from IDTDNA (Coralville, IA). HeLa cells were seeded at 500,000 cells/60-mm dish and transfected with final concentrations of 50 nm siRNA (or 50 nm siCON) with INTERFERin (Polyplus Transfection, Strasbourg, France) according to the manufacturer's instructions. The following day (24 h post-transfection), the cells were split and plated into 24-well dishes with NaOH washed coverslips. Seventy-two hours post-transfection, cells were serum-starved for 2 h and then stimulated for the indicated time points with fluorescent 10 ng/ml Alexa 647–EGF (Invitrogen) ligand, followed by indirect immunofluorescence.
A 6-well dish was plated for each siRNA knockdown experiment. These cells were harvested in radioimmune precipitation assay buffer as previously described (53) 72-hours post-transfection and immunoblotted for RUFY1 and PTPN23 to ensure at least 90% knockdown efficiency.
Statistical analyses
Unpaired Student's t tests were used to determine significance. A p value of less than 0.05 is considered significant and is denoted with a single asterisk (*). A p value of less than 0.001 is denoted with two asterisks (**). A p value of less than 0.0005 is denoted with three asterisks (***).
Author contributions
J. A. G. and D. W. W. data curation; J. A. G., D. W. W., and M. L. M. formal analysis; J. A. G. investigation; J. A. G., D. W. W., M. L. M., and B. P. C. methodology; J. A. G. writing-original draft; J. A. G., M. L. M., and B. P. C. project administration; D. W. W., M. L. M., and B. P. C. writing-review and editing; M. L. M. resources; M. L. M. software; M. L. M. and B. P. C. supervision; B. P. C. conceptualization; B. P. C. funding acquisition.
Acknowledgment
We thank Arkadiusz Slusarczyk for assistance with the preparation of TEM samples.
This work was funded in part by NEI, National Institutes of Health Grant EY027032 (to B. P. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008170.
- EGFR
- EGF receptor
- EGF
- epidermal growth factor
- IGF
- insulin-like growth factor
- IGF
- insulin-like growth factor
- PNS
- post-nuclear supernatant
- RTK
- receptor-tyrosine kinase
- TfnR
- transferrin receptor
- TEM
- transmission EM
- ddH2O
- double-distilled water
- TEA-BC
- triethylammonium bicarbonate
- PB
- phosphate buffer
- siCON
- scramble control siRNA.
References
- 1. Miettinen P. J., Berger J. E., Meneses J., Phung Y., Pedersen R. A., Werb Z., and Derynck R. (1995) Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376, 337–341 10.1038/376337a0 [DOI] [PubMed] [Google Scholar]
- 2. Nicholson R. I., Gee J. M., and Harper M. E. (2001) EGFR and cancer prognosis. Eur. J. Cancer 37, S9–S15 [DOI] [PubMed] [Google Scholar]
- 3. Chong C. R., and Jänne P. A. (2013) The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat. Med. 19, 1389–1400 10.1038/nm.3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H. J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., and Ward C. W. (2002) Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell 110, 763–773 10.1016/S0092-8674(02)00940-6 [DOI] [PubMed] [Google Scholar]
- 5. Hashimoto A., Kurosaki M., Gotoh N., Shibuya M., and Kurosaki T. (1999) Shc regulates epidermal growth factor-induced activation of the JNK signaling pathway. J. Biol. Chem. 274, 20139–20143 10.1074/jbc.274.29.20139 [DOI] [PubMed] [Google Scholar]
- 6. Wells A., Welsh J. B., Lazar C. S., Wiley H. S., Gill G. N., and Rosenfeld M. G. (1990) Ligand-induced transformation by a noninternalizing epidermal growth factor receptor. Science 247, 962–964 10.1126/science.2305263 [DOI] [PubMed] [Google Scholar]
- 7. Rush J. S., Boeving M. A., Berry W. L., and Ceresa B. P. (2014) Antagonizing c-Cbl enhances EGFR-dependent corneal epithelial homeostasis. Invest. Ophthalmol. Vis. Sci. 55, 4691–4699 10.1167/iovs.14-14133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hofman E. G., Bader A. N., Voortman J., van den Heuvel D. J., Sigismund S., Verkleij A. J., Gerritsen H. C., and van Bergen en Henegouwen P. M. (2010) Ligand-induced EGF receptor oligomerization is kinase-dependent and enhances internalization. J. Biol. Chem. 285, 39481–39489 10.1074/jbc.M110.164731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roepstorff K., Grandal M. V., Henriksen L., Knudsen S. L., Lerdrup M., Grøvdal L., Willumsen B. M., and van Deurs B. (2009) Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic 10, 1115–1127 10.1111/j.1600-0854.2009.00943.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Guglielmo G. M., Baass P. C., Ou W. J., Posner B. I., and Bergeron J. J. (1994) Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. EMBO J. 13, 4269–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vieira A. V., Lamaze C., and Schmid S. L. (1996) Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 274, 2086–2089 10.1126/science.274.5295.2086 [DOI] [PubMed] [Google Scholar]
- 12. Stamos J., Sliwkowski M. X., and Eigenbrot C. (2002) Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 277, 46265–46272 10.1074/jbc.M207135200 [DOI] [PubMed] [Google Scholar]
- 13. Liao H. J., and Carpenter G. (2007) Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol. Biol. Cell 18, 1064–1072 10.1091/mbc.E06-09-0802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baass P. C., Di Guglielmo G. M., Authier F., Posner B. I., and Bergeron J. J. (1995) Compartmentalized signal transduction by receptor tyrosine kinases. Trends Cell Biol. 5, 465–470 10.1016/S0962-8924(00)89116-3 [DOI] [PubMed] [Google Scholar]
- 15. Jiang X., and Sorkin A. (2002) Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol. Biol. Cell 13, 1522–1535 10.1091/mbc.01-11-0552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doyotte A., Mironov A., McKenzie E., and Woodman P. (2008) The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 6308–6313 10.1073/pnas.0707601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cormont M., Mari M., Galmiche A., Hofman P., and Le Marchand-Brustel Y. (2001) A FYVE-finger-containing protein, Rabip4, is a Rab4 effector involved in early endosomal traffic. Proc. Natl. Acad. Sci. U.S.A. 98, 1637–1642 10.1073/pnas.98.4.1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Damke H., Gossen M., Freundlieb S., Bujard H., and Schmid S. L. (1995) Tightly regulated and inducible expression of dominant interfering dynamin mutant in stably transformed HeLa cells. Methods Enzymol. 257, 209–220 10.1016/S0076-6879(95)57026-8 [DOI] [PubMed] [Google Scholar]
- 19. Berkers J. A., van Bergen en Henegouwen P. M., and Boonstra J. (1991) Three classes of epidermal growth factor receptors on HeLa cells. J. Biol. Chem. 266, 922–927 [PubMed] [Google Scholar]
- 20. Landry J. J., Pyl P. T., Rausch T., Zichner T., Tekkedil M. M., Stütz A. M., Jauch A., Aiyar R. S., Pau G., Delhomme N., Gagneur J., Korbel J. O., Huber W., and Steinmetz L. M. (2013) The genomic and transcriptomic landscape of a HeLa cell line. G3 3, 1213–1224 10.1534/g3.113.005777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zlobina M. V., Kamentseva R. S., Kornilova E. S., and Kharchenko M. V. (2014) [Analysis of vesicle subpopulations carrying early endosomal autoantigen EEA1]. Tsitologiia 56, 741–748 [PubMed] [Google Scholar]
- 22. Vanlandingham P. A., and Ceresa B. P. (2009) Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 284, 12110–12124 10.1074/jbc.M809277200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oksvold M. P., Skarpen E., Wierød L., Paulsen R. E., and Huitfeldt H. S. (2001) Re-localization of activated EGF receptor and its signal transducers to multivesicular compartments downstream of early endosomes in response to EGF. Eur J. Cell Biol. 80, 285–294 10.1078/0171-9335-00160 [DOI] [PubMed] [Google Scholar]
- 24. Herbst J. J., Opresko L. K., Walsh B. J., Lauffenburger D. A., and Wiley H. S. (1994) Regulation of postendocytic trafficking of the epidermal growth factor receptor through endosomal retention. J. Biol. Chem. 269, 12865–12873 [PubMed] [Google Scholar]
- 25. Kornilova E., Sorkina T., Beguinot L., and Sorkin A. (1996) Lysosomal targeting of epidermal growth factor receptors via a kinase-dependent pathway is mediated by the receptor carboxyl-terminal residues 1022–1123. J. Biol. Chem. 271, 30340–30346 10.1074/jbc.271.48.30340 [DOI] [PubMed] [Google Scholar]
- 26. Wallach D. F., Kamat V. B., and Gail M. H. (1966) Physicochemical differences between fragments of plasma membrane and endoplasmic reticulum. J. Cell Biol. 30, 601–621 10.1083/jcb.30.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chibalin A. V., Katz A. I., Berggren P. O., and Bertorello A. M. (1997) Receptor-mediated inhibition of renal Na+-K+-ATPase is associated with endocytosis of its α- and β-subunits. Am. J. Physiol. 273, C1458–C1465 10.1152/ajpcell.1997.273.5.C1458 [DOI] [PubMed] [Google Scholar]
- 28. Khundmiri S. J., Bertorello A. M., Delamere N. A., and Lederer E. D. (2004) Clathrin-mediated endocytosis of Na+,K+-ATPase in response to parathyroid hormone requires ERK-dependent phosphorylation of Ser-11 within the α1-subunit. J. Biol. Chem. 279, 17418–17427 10.1074/jbc.M311715200 [DOI] [PubMed] [Google Scholar]
- 29. Feldmann T., Glukmann V., Medvenev E., Shpolansky U., Galili D., Lichtstein D., and Rosen H. (2007) Role of endosomal Na+-K+-ATPase and cardiac steroids in the regulation of endocytosis. Am. J. Physiol. Cell Physiol. 293, C885–C896 10.1152/ajpcell.00602.2006 [DOI] [PubMed] [Google Scholar]
- 30. Beas A. O., Taupin V., Teodorof C., Nguyen L. T., Garcia-Marcos M., and Farquhar M. G. (2012) Gαs promotes EEA1 endosome maturation and shuts down proliferative signaling through interaction with GIV (Girdin). Mol. Biol. Cell 23, 4623–4634 10.1091/mbc.E12-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammond J. C., McCullumsmith R. E., Funk A. J., Haroutunian V., and Meador-Woodruff J. H. (2010) Evidence for abnormal forward trafficking of AMPA receptors in frontal cortex of elderly patients with schizophrenia. Neuropsychopharmacology 35, 2110–2119 10.1038/npp.2010.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hájek P., Chomyn A., and Attardi G. (2007) Identification of a novel mitochondrial complex containing mitofusin 2 and stomatin-like protein 2. J. Biol. Chem. 282, 5670–5681 10.1074/jbc.M608168200 [DOI] [PubMed] [Google Scholar]
- 33. Da Cruz S., Parone P. A., Gonzalo P., Bienvenut W. V., Tondera D., Jourdain A., Quadroni M., and Martinou J. C. (2008) SLP-2 interacts with prohibitins in the mitochondrial inner membrane and contributes to their stability. Biochim. Biophys. Acta 1783, 904–911 10.1016/j.bbamcr.2008.02.006 [DOI] [PubMed] [Google Scholar]
- 34. Yang J., Kim O., Wu J., and Qiu Y. (2002) Interaction between tyrosine kinase Etk and a RUN domain- and FYVE domain-containing protein RUFY1: a possible role of ETK in regulation of vesicle trafficking. J. Biol. Chem. 277, 30219–30226 10.1074/jbc.M111933200 [DOI] [PubMed] [Google Scholar]
- 35. Miura G. I., Roignant J. Y., Wassef M., and Treisman J. E. (2008) Myopic acts in the endocytic pathway to enhance signaling by the Drosophila EGF receptor. Development 135, 1913–1922 10.1242/dev.017202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ali N., Zhang L., Taylor S., Mironov A., Urbé S., and Woodman P. (2013) Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr. Biol. 23, 453–461 10.1016/j.cub.2013.02.033 [DOI] [PubMed] [Google Scholar]
- 37. Lakadamyali M., Rust M. J., and Zhuang X. (2006) Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell 124, 997–1009 10.1016/j.cell.2005.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vendeville A., Ravallec M., Jousset F. X., Devise M., Mutuel D., López-Ferber M., Fournier P., Dupressoir T., and Ogliastro M. (2009) Densovirus infectious pathway requires clathrin-mediated endocytosis followed by trafficking to the nucleus. J. Virol. 83, 4678–4689 10.1128/JVI.02401-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vukmirica J., Monzo P., Le Marchand-Brustel Y., and Cormont M. (2006) The Rab4A effector protein Rabip4 is involved in migration of NIH 3T3 fibroblasts. J. Biol. Chem. 281, 36360–36368 10.1074/jbc.M602920200 [DOI] [PubMed] [Google Scholar]
- 40. Fouraux M. A., Deneka M., Ivan V., van der Heijden A., Raymackers J., van Suylekom D., van Venrooij W. J., van der Sluijs P., and Pruijn G. J. (2004) Rabip4′ is an effector of rab5 and rab4 and regulates transport through early endosomes. Mol. Biol. Cell 15, 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cao W., Zhang B., Ding F., Zhang W., Sun B., and Liu Z. (2013) Expression of SLP-2 was associated with invasion of esophageal squamous cell carcinoma. PLoS One 8, e63890 10.1371/journal.pone.0063890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cao W., Zhang B., Liu Y., Li H., Zhang S., Fu L., Niu Y., Ning L., Cao X., Liu Z., and Sun B. (2007) High-level SLP-2 expression and HER-2/neu protein expression are associated with decreased breast cancer patient survival. Am. J. Clin. Pathol. 128, 430–436 10.1309/C6X54HRB580EP2NQ [DOI] [PubMed] [Google Scholar]
- 43. Zhang L., Ding F., Cao W., Liu Z., Liu W., Yu Z., Wu Y., Li W., Li Y., and Liu Z. (2006) Stomatin-like protein 2 is overexpressed in cancer and involved in regulating cell growth and cell adhesion in human esophageal squamous cell carcinoma. Clin. Cancer Res. 12, 1639–1646 10.1158/1078-0432.CCR-05-1858 [DOI] [PubMed] [Google Scholar]
- 44. Song L., Liu L., Wu Z., Lin C., Dai T., Yu C., Wang X., Wu J., Li M., and Li J. (2012) Knockdown of stomatin-like protein 2 (STOML2) reduces the invasive ability of glioma cells through inhibition of the NF-κB/MMP-9 pathway. J. Pathol. 226, 534–543 10.1002/path.3008 [DOI] [PubMed] [Google Scholar]
- 45. Mairhofer M., Steiner M., Salzer U., and Prohaska R. (2009) Stomatin-like protein-1 interacts with stomatin and is targeted to late endosomes. J. Biol. Chem. 284, 29218–29229 10.1074/jbc.M109.014993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bergeron J. J., Searle N., Khan M. N., and Posner B. I. (1986) Differential and analytical subfractionation of rat liver components internalizing insulin and prolactin. Biochemistry 25, 1756–1764 10.1021/bi00355a046 [DOI] [PubMed] [Google Scholar]
- 47. Galperin E., and Sorkin A. (2008) Endosomal targeting of MEK2 requires RAF, MEK kinase activity and clathrin-dependent endocytosis. Traffic 9, 1776–1790 10.1111/j.1600-0854.2008.00788.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gosney J. A., and Ceresa B. P. (2017) Using Percoll gradient fractionation to study the endocytic trafficking of the EGFR. Methods Mol. Biol. 1652, 145–158 10.1007/978-1-4939-7219-7_11 [DOI] [PubMed] [Google Scholar]
- 49. Dinneen J. L., and Ceresa B. P. (2004) Continual expression of Rab5(Q79L) causes a ligand-independent EGFR internalization and diminishes EGFR activity. Traffic 5, 606–615 10.1111/j.1398-9219.2004.00204.x [DOI] [PubMed] [Google Scholar]
- 50. Lopez-Alcalá C., Alvarez-Moya B., Villalonga P., Calvo M., Bachs O., and Agell N. (2008) Identification of essential interacting elements in K-Ras/calmodulin binding and its role in K-Ras localization. J. Biol. Chem. 283, 10621–10631 10.1074/jbc.M706238200 [DOI] [PubMed] [Google Scholar]
- 51. Jensen O. N., Wilm M., Shevchenko A., and Mann M. (1999) Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol. 112, 513–530 [DOI] [PubMed] [Google Scholar]
- 52. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 53. Jackson N. M., and Ceresa B. P. (2016) Protein kinase G facilitates EGFR-mediated cell death in MDA-MB-468 cells. Exp. Cell Res. 346, 224–232 10.1016/j.yexcr.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]