Abstract
The essential histone chaperone FACT (facilitates chromatin transcription) promotes both nucleosome assembly and disassembly. FACT is a heterodimer of Spt16 with either SSRP1 or Pob3, differing primarily by the presence of a high-mobility group B (HMGB) DNA-binding domain furnished only by SSRP1. Yeast FACT lacks the intrinsic HMGB domain found in SSRP1-based homologs such as human FACT, but yeast FACT activity is supported by Nhp6, which is a freestanding, single HMGB-domain protein. The importance of histone binding by FACT domains has been established, but the roles of DNA-binding activity remain poorly understood. Here, we examined these roles by fusing single or multiple HMGB modules to Pob3 to mimic SSRP1 or to test the effects of extended DNA-binding capacity. Human FACT and a yeast mimic both required Nhp6 to support nucleosome reorganization in vitro, indicating that a single intrinsic DNA-binding HMGB module is insufficient for full FACT activity. Three fused HMGB modules supported activity without Nhp6 assistance, but this FACT variant did not efficiently release from nucleosomes and was toxic in vivo. Notably, intrinsic DNA-binding HMGB modules reduced the DNA accessibility and histone H2A–H2B dimer loss normally associated with nucleosome reorganization. We propose that DNA bending by HMGB domains promotes nucleosome destabilization and reorganization by exposing FACT's histone-binding sites, but DNA bending also produces DNA curvature needed to accommodate nucleosome assembly. Intrinsic DNA-bending activity therefore favors nucleosome assembly by FACT over nucleosome reorganization, but excessive activity impairs FACT release, suggesting a quality control checkpoint during nucleosome assembly.
Keywords: histone chaperone, chromatin structure, nucleosome, DNA-binding protein, chromatin regulation, chromatin dynamics, FACT, HMGB domain, nucleosome reorganization
Introduction
FACT2 is a highly conserved histone chaperone capable of either partially disassembling or assembling nucleosomes (1–3). It promotes increased accessibility of nucleosomal DNA to nucleases and chemical probes (4–6), enhances transcription of chromatin templates (7–9), and alters nucleosomal structure in a single-particle Förster resonance energy transfer (spFRET) assay (10, 11). These effects are reversible, can occur without changing the composition of the nucleosome or the position of the histone octamer relative to DNA, and do not require ATP hydrolysis, so this constellation of activities has been called nucleosome reorganization to distinguish FACT from ATP-dependent remodelers (1). Consistent with the reversibility of reorganization, FACT can also assemble nucleosomes from mixtures of histones and DNA (9), and it promotes chromatin duplication in vivo and in vitro (12, 13). Alterations of FACT activity in vivo result in a loss of transcriptional repression (14–17) and elevated histone turnover rates (18), further indicating a key role in chromatin maintenance. FACT therefore contributes to various aspects of transcription, replication, centromere function, and repair and is essential for the viability of eukaryotic cells (1, 2, 7, 19, 20).
FACT is a heterodimer of the Spt16 and SSRP1/Pob3 family members (8, 14, 21). The SSRP1 and Pob3 versions of the small subunit are distinguished primarily by the presence of a DNA-binding domain in SSRP1 (the subunit found in most eukaryotes) that is lacking in Pob3 (found in yeasts and fungi (Fig. 1)). The DNA-binding domain is a single copy of an HMGB domain, a motif typically found in paired tandem repeats in the highly abundant high-mobility group B family of proteins (22–24) (Fig. 1). HMGB proteins bind the minor groove of DNA without sequence specificity, resulting in a bend of nearly 90° (24, 25). They can therefore act as “DNA chaperones” that promote transitions between normal B-form DNA and curved forms that promote looping or assembly of nucleosomes (26, 27). DNA bending by HMGB modules could therefore destabilize nucleosomes by disrupting DNA–histone contacts, but it could also stimulate nucleosome assembly by overcoming the inherent stiffness of DNA to provide the curvature needed.
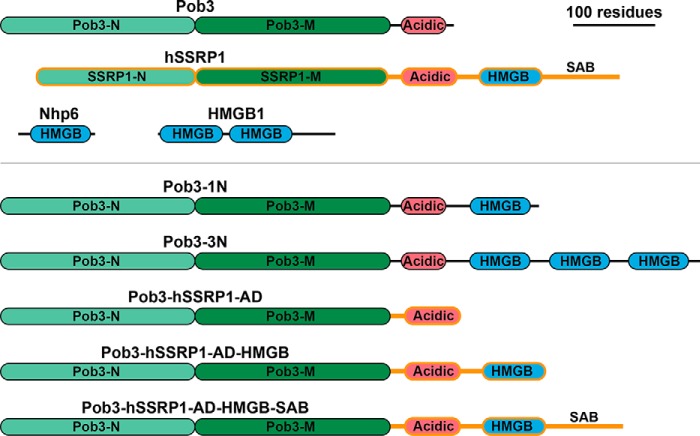
Figure 1.
Architecture of Pob3/SSRP1 constructs used. Native Pob3 and Nhp6 proteins (S. cerevisiae) are drawn to scale, with the N-terminal (Pob3-N (light green)), middle (Pob3-M (dark green)), and acidic domains (AD (red)) of Pob3 and the HMGB domains (blue) of various proteins indicated as ovals flanked by unstructured linkers. Similar domains found in human SSRP1 are shown with the same color scheme but with orange outlines. SAB indicates the serine-acidic-basic domain of hSSRP1. The native architecture of human HMGB1 is shown to contrast with the single HMGB motif architecture of Nhp6. Various fusion proteins with combinations of these domains used here are shown below the gray line.
Yeast FACT (yFACT) lacks the intrinsic HMGB domain found in SSRP1-based versions like human FACT (hFACT), but previous work shows that yFACT activity is supported in vivo and in vitro by Nhp6, which is essentially a freestanding single HMGB motif (6, 23, 28, 29) (Fig. 1). This has been interpreted to mean that FACT function has a universal requirement for DNA-binding activity, but it can either be supplied in cis (Spt16–SSRP1) or in trans (Nhp6 plus Spt16–Pob3, Ref. 1). Nhp6 is encoded by two similar genes in the yeast Saccharomyces cerevisiae, NHP6A and NHP6B, with the loss of both genes leading to severe growth defects but not inviability (23, 28). In addition to supporting FACT activity, Nhp6 also contributes to the functions of RSC and other ATP-dependent nucleosome remodelers (26, 30, 31), transcription of the spliceosomal RNA encoded by SNR6 (32), and other chromatin-mediated processes (23). This broad range of roles makes it difficult to assign individual physiological functions of Nhp6 or other HMGB domain proteins; as a result, little is known about how they contribute to specific processes or what drives the preference for architectural features, such as inclusion of an intrinsic domain in SSRP1 but not in Pob3, the dual repeats of the motif in HMGB1, or the single motif observed in Nhp6.
yFACT (Spt16–Pob3) does not stably bind to or reorganize nucleosomes in the absence of Nhp6 in vitro, indicating a requirement for a DNA-binding domain to initiate or stabilize this structural change in vitro (4–6, 10, 33). Genetic analysis shows that Nhp6 has an important but not essential role in yFACT function in vivo, but it is not clear whether other DNA-binding factors partially complement the loss of Nhp6. Therefore, it is not known whether some DNA-binding activity is just helpful for FACT function or is essential. Broad conservation of FACT at the sequence level suggests that its activities are also conserved, and the architecture of hFACT implies that the role contributed by Nhp6 in yFACT activity is instead performed by the HMGB domain in SSRP1. However, hFACT alone does not form stable complexes with intact nucleosomes (3, 11) and has not been reported to induce nucleosome reorganization, although this was not tested in the presence of additional HMGB proteins. The role of a DNA-binding/-bending domain in FACT activity and the different architectures of FACT, in different organisms, that are responsible for this activity therefore remain puzzling.
To begin to address questions regarding the functional architecture of DNA-binding domains in FACT activity, we fused HMGB domains to the Pob3 subunit of yeast FACT and investigated the activities of the fusion proteins in vitro and in vivo. This allowed us to probe the effects of providing the DNA-binding activity to FACT in different structural contexts. We also compared the effects of intrinsic and freestanding HMGB domains on FACT activity by adding Nhp6 to yFACT constructs with different numbers of domains fused to Pob3 and to hFACT with its single native domain.
Our results show that yFACT and hFACT have comparable activities, but stable binding to and reorganization of nucleosomes requires more than a single intrinsic HMGB domain in both cases. Notably, intrinsic DNA-binding domains reduced the amount of H2A–H2B dimer loss and the level of accessibility of nucleosomal DNA to nucleases during reorganization. The results therefore support the expected importance of DNA binding and bending by HMGB-domain proteins in initiating reorganization of nucleosomes by FACT, but they also suggest a role for intrinsic domains in promoting the resolution of the reorganized state to assemble nucleosomes. We propose a model in which FACT can form weak complexes with nucleosomes without the assistance of DNA-binding activity; this form is in equilibrium with a stably bound, reorganized structure whose formation requires freestanding HMGB proteins. DNA-bending activity provided by intrinsic HMGB domains skews the equilibrium toward intact nucleosomes, explaining the lower accessibility of the DNA and decreased H2A–H2B dimer loss. This model proposes a role for the known DNA-bending activity of HMGB domains (22, 34, 35) during nucleosome assembly (which requires the introduction of curvature to the DNA), suggests a nucleosome assembly checkpoint function for FACT, and provides a potential basis for the two distinct architectures of FACT observed in nature.
Results
Fusing Nhp6 modules to Pob3
SSRP1 and Pob3 are similar over their first ∼450 residues followed by 60–90 residues with a high proportion of acidic amino acids (Fig. 1). This inherently unstructured acidic region is the C terminus of Pob3 and contains one of the two binding sites for H2A–H2B dimers in FACT, with the other being a similar domain in Spt16 (3, 36). SSRP1 proteins also have this feature, but it is followed by an HMGB DNA-binding domain and then a serine-rich (30% Ser in human SSRP1) and highly charged (20% Asp or Glu and 26% Lys or Arg) C-terminal tail, which we are calling the SAB (serine-acidic-basic) domain here. We fused the NHP6A gene in-frame with the 3′-end of the POB3 ORF, retaining all residues of each protein and adding a 3–6-residue linker between them, depending on the construct (see “Experimental procedures” and Pob3–1N in Fig. 1). Human SSRP1 has 27 residues separating the end of the acidic domain from the HMGB motif, and the Pob3–Nhp6 fusions were designed to mimic the length of this linker. These fusions therefore resembled the architecture of SSRP1 but did not include the serine-rich/charged tail for in vitro studies.
Previous results with human FACT and Spt16–Pob3 suggested that a single HMGB domain would be insufficient to promote stable nucleosome binding or reorganization in vitro (3, 11, 37, 38). We therefore also developed a strategy for inserting multiple Nhp6 modules at the C terminus of Pob3 in a plasmid-based expression construct (see “Experimental procedures”). Plasmids containing intact Pob3 were somewhat toxic to Escherichia coli even without explicit bacterial expression signals; this toxicity increased with each Nhp6 module added (not shown). We were therefore unable to recover constructs with more than three Nhp6 modules fused to Pob3 (Pob3–3N (Fig. 1)).
For purification of FACT variants, we co-expressed the POB3 gene or its fusion derivatives along with normal SPT16, both from inducible GAL1 promoters in yeast cells (Fig. 2A and Ref. 4). A high-copy GAL1p-POB3 plasmid expressed enough Pob3 to complement a deletion of this essential gene even when the GAL1 promoter was repressed with glucose (Fig. 2B, top panel). In contrast, POB3–3N did not complement a deletion, and overexpression of POB3–3N blocked the growth of a strain with a normal POB3 gene (Fig. 2B, bottom panel). This fusion protein was therefore toxic to yeast cells but provided a source for purifying the Spt16–Pob3–3N (SP–3N) heterodimers (Fig. 2A).
Figure 2.
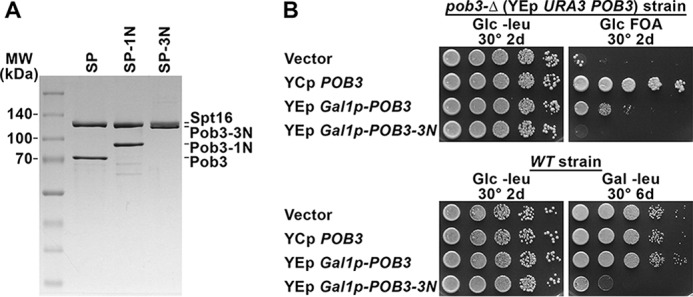
Purification of factors and consequences of their expression in vivo. A, 2.5 μg of purified SP (Spt16–Pob3) or the derivatives with one or three Nhp6 modules fused to Pob3 (-1N, -3N) were separated by SDS-PAGE and stained with Coomassie Blue dye. B, top panel, strain 7789-2-1 pJW4 (Table S1, pob3-Δ YEp URA3 POB3) was transformed with pTF162 (YEp LEU2 vector), pTF139 (YCp LEU2 POB3 (16)), pHX13 (YEp LEU2 GAL1p-POB3) or pHX16 (YEp LEU2 GAL1p-POB3–3N). Cultures were grown in rich medium to permit loss of pJW4, and then 10-fold serial dilutions were placed on synthetic glucose medium lacking leucine (Glc −leu) or containing fluoroorotic acid (Glc FOA) to determine retention of the LEU2 plasmids and loss of the URA3 plasmid, respectively (50). Complementation of pob3-Δ was robust for the YCp POB3 plasmid, weaker for the YEp POB3 plasmid under repressive conditions for the GAL1 promoter, and absent for the YEp POB3–3N plasmid. Expression of POB3 in this configuration was therefore adequate to produce some growth, but Pob3–3N either did not complement pob3-Δ or its toxicity outweighed any benefit. Bottom panel, strain 8127-7-4 (WT, Table S1) was transformed with the same plasmids as above and tested under selection for the plasmids on medium that repressed (Glc) or induced (Gal) the GAL1 promoter. GAL1p-POB3–3N blocked growth only on Gal, demonstrating toxicity of the Pob3–3N protein.
FACT activity in vitro requires multiple HMGB domain modules
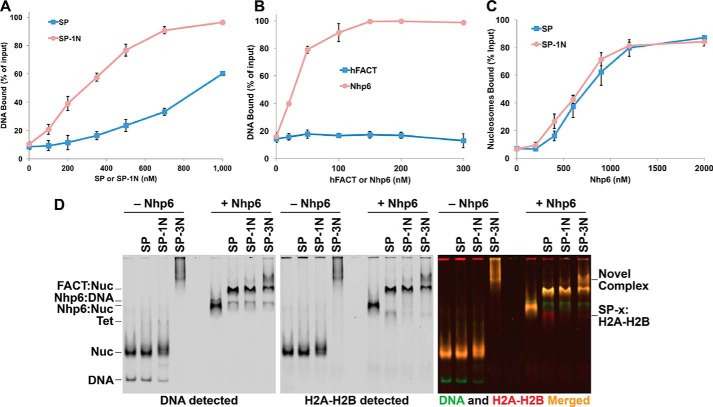
SSRP1 was initially identified as a structure-specific recognition protein because of the affinity of its HMGB domain for distorted DNA molecules (22, 35). However, the middle domains of both Pob3 and SSRP1 have also been reported to bind DNA with low affinity (39). We therefore tested the Spt16–Pob3 complexes with 0, 1, or 3 Nhp6 modules for the ability to bind 147–181-bp dsDNA fragments using an electrophoretic mobility shift assay (EMSA) (Figs. 3A and B, and S1). SP (Spt16–Pob3 heterodimers) alone bound DNA with an apparent affinity of 950 ± 60 nm, and a single fused Nhp6 module (Spt16–Pob3–1N (SP–1N)) increased this affinity, but only moderately, to 310 ± 10 nm. SSRP1 and hFACT were reported previously to bind short 30-bp DNA fragments with affinities near 10 nm (40). We observed little or no stable binding to free DNA at levels up to 300 nm hFACT in our EMSA, whereas free Nhp6 displayed an affinity of 43 ± 14 nm (Figs. 3B and S1). The phosphorylation of residues in hFACT is known to inhibit the DNA-binding activity of the HMGB domain (37), and the reported high affinity for short DNA fragments was observed after phosphatase treatment of hFACT (40). As hFACT is active in other assays without this treatment (Refs. 3, 7, and 11 and see below), we did not test this parameter in our assays but noted that, in contrast, SP–3N bound DNA with affinity of 23 ± 7 nm (Fig. S1). We concluded that HMGB modules are capable of binding DNA with high affinity from within FACT, but the accessibility of nucleosome-sized DNA fragments appears to be limited when only one module is present; the affinity increases with increased numbers or flexibility of the modules, and the DNA-binding affinity of FACT may be regulated by phosphorylation.
Figure 3.
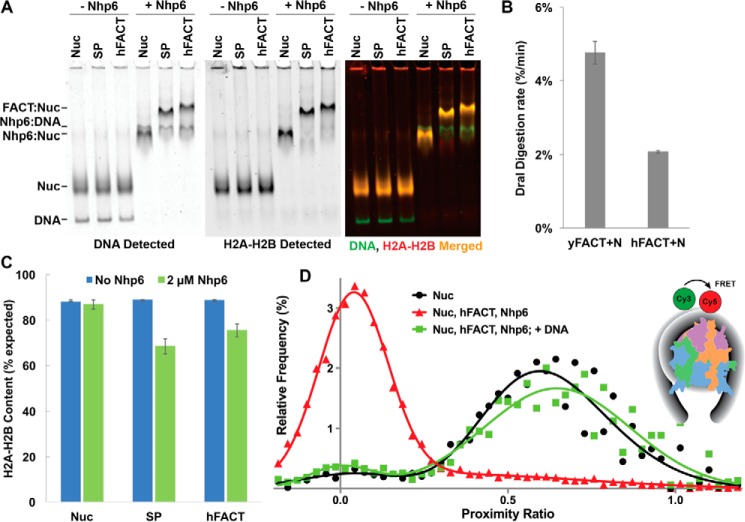
Analysis of complex formation between SP, SP–1N, SP–3N, or hFACT and DNA or nucleosomes. A, Spt16–Pob3 (SP) or the SP–1N derivative were mixed at the indicated concentrations with a 181-bp DNA fragment, and then the fraction of total bound DNA was determined following native PAGE (Fig. S1). The average ± S.D. is shown for four independent repeats. B, same as in A, except Nhp6A and hFACT were tested, and the results from three repeats are shown. Subsequent assays were performed with 200 nm hFACT, a condition that demonstrated little or no binding to free DNA in this assay. C, nucleosomes were mixed with 200 nm SP or SP–1N and the indicated concentrations of Nhp6, and the bound fraction (FACT:Nuc complexes as shown in D) was determined following native PAGE. The average ± S.D. for four independent repeats is shown. D, nucleosomes alone or mixed with 200 nm SP, SP–1N, or SP–3N without or with 2 μm Nhp6 were separated by native PAGE. Gels were scanned to detect the Cy5-labeled DNA (green signal) and the Oregon Green-labeled H2B (red signal) with the merged images on the right. Complexes formed between FACT and displaced dimers are indicated as SP-x:H2A-H2B, and a typically weak band whose prominence increased in proportion to the number of Nhp6 modules fused to Pob3 is indicated as a Novel Complex.
The concentration of Nhp6 needed to support the binding of SP to nucleosomes is about 10-fold higher than the concentration observed to bind DNA (33, 41). This requirement for a molar excess of Nhp6 has been interpreted to mean that multiple Nhp6-binding events are needed within a short window of time to disrupt the histone–DNA contacts sufficiently to allow FACT to compete for binding to the histones to initiate reorganization (1–3, 36). We reasoned that a single intrinsic HMGB domain might provide a high enough local concentration of DNA-binding activity, or an optimal positioning of the module, to decrease the requirement for free Nhp6. Titration of Nhp6 showed that SP–1N required only slightly less Nhp6 than SP (half-maximal complex formation occurred at 690 ± 40 nm Nhp6 with SP and 580 ± 30 nm Nhp6 with SP–1N (Fig. 3C)). A single intrinsic HMGB domain was therefore not sufficient to promote stable nucleosome binding and also did not substitute for a significant amount of free Nhp6 in supporting nucleosome binding. In contrast, similar experiments with SP–3N resulted in binding to nucleosomes in the absence of free Nhp6 (Fig. 3D) with an apparent affinity of 29 ± 3 nm (146-bp DNA, no linker DNA) or 19 ± 4 nm (181-bp DNA, ∼35 bp of linker DNA). Free Nhp6 modules were therefore not needed to produce stable complexes of SP with nucleosomes, but multiple HMGB domains were required.
Native gel analysis showed that the complexes formed with SP–3N alone were poorly behaved, migrating as a smear near the well, but the addition of free Nhp6 resolved these smears into distinct bands (Fig. 3D). This suggests that individual DNA-binding modules within SP–3N can bind to different nucleosomes, forming networks that are disrupted by competition from free Nhp6. If this is correct, it means that at least two of the three HMGB domain modules in this construct are capable of binding DNA independently. When saturating levels of Nhp6 were added, all three FACT variants produced the slow-migrating form shown previously to contain one Spt16–Pob3 heterodimer per nucleosome (FACT:Nuc in Fig. 3D) (4). SP–1N, and especially SP–3N, also produced significant amounts of an even slower-migrating complex (Fig. 3D, Novel Complex).
SP–3N induced nucleosome reorganization without additional Nhp6
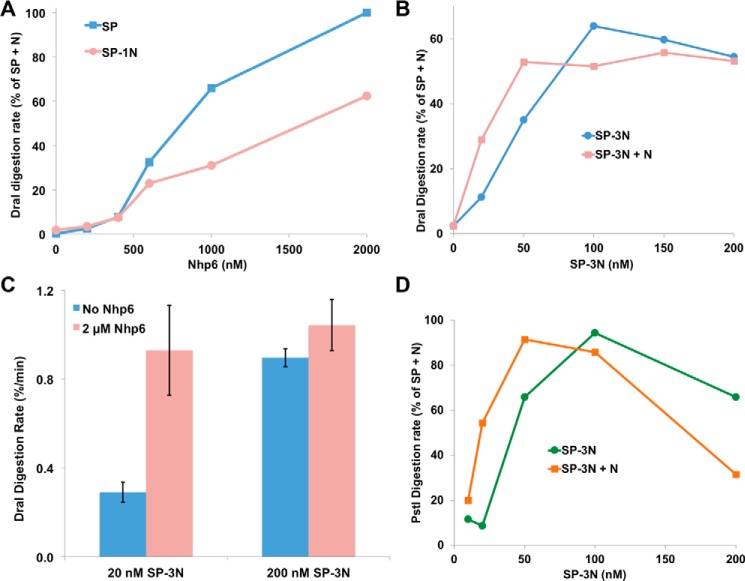
Nucleosome reorganization by FACT is associated with increased accessibility of the DNA to restriction endonuclease digestion (1, 5). Consistent with the binding results above, neither SP nor SP–1N was able to enhance the DraI digestion rate of a 5S rDNA fragment incorporated into nucleosomes, but both were able to produce high rates of digestion when Nhp6 was also added (Fig. 4A). Contrary to our expectation, SP–1N supported lower rates of digestion than SP at all Nhp6 concentrations, indicating that the availability of an intrinsic DNA-binding domain inhibited this activity instead of enhancing it. As discussed below, we interpreted this as support for a model in which the intrinsic DNA-binding module skews the distribution of FACT–nucleosome complexes toward a nuclease-resistant form.
Figure 4.
Effect of fused Nhp6 modules on nuclease sensitivity. A, 200 nm SP or SP–1N was mixed with nucleosomes, and then DraI was added and the amount of digestion determined after 8, 16, and 24 min by denaturing SDS-PAGE analysis. The initial rate of digestion was determined from the time courses and normalized to the value with SP + 2 μm Nhp6 tested in parallel. A similar experiment with three repeats gave no digestion for SP and SP–1N without Nhp6, and the rate for SP–1N + N was 68% ± 5% of the SP + N rate (not shown). B, same as in A, except the concentration of SP–3N was varied with or without 2 μm Nhp6 and the digestion rate was compared with a parallel control reaction with 200 nm SP and 2 μm Nhp6. C, same as in B, except four repeats were performed with 20 nm and 200 nm SP–3N with the average ± S.D. of the absolute rate of digestion (% of total DNA digested/min) for each condition reported without normalization. D, similar to B, except an AT-hook nucleosome positioning sequence was used (4) and the rate of digestion with PstI was measured.
Notably, SP–3N was able to promote nuclease digestion without added Nhp6 (Fig. 4, B–D). As noted above, SP–3N may bind two nucleosomes simultaneously, so at high concentrations this could simply mean that one SP–3N heterodimer provides Nhp6 activity for another heterodimer. However, the rate of digestion was enhanced even at 10–20 nm SP–3N, which is well below the concentration of Nhp6 that supports reorganization by native SP (Refs. 4, 5, and 33 and Fig. 4A). These results therefore suggest that a single FACT heterodimer was able to reorganize a nucleosome in isolation when three locally constrained DNA-binding modules were available. Additional free Nhp6 enhanced the rate of digestion at these low concentrations of SP–3N, so the availability of three localized HMGB domains was not optimal, but it was effective. Activity with SP–3N plateaued at about 60% of the level of SP + Nhp6 with DraI and a 5S rDNA template (Fig. 4B) but was closer to 100% with a different nucleosome positioning sequence probed with PstI (Fig. 4D). Previous results show that different restriction enzymes respond to reorganization by FACT to different extents (4), presumably revealing some structural or kinetic feature of reorganized nucleosomes. The apparent inhibition of activity at high SP–3N concentrations with PstI was not observed previously with free Nhp6 or SP (4, 5, 33) but could be related to the formation of distinct bands detected by EMSA with SP–3N (Fig. 3D) and the interplay between the reorganized structure and the enzyme used as a probe. The ability of SP–3N to induce endonuclease sensitivity indicates that two to three DNA-binding motifs were adequate to support nucleosome reorganization by FACT.
Fusion of Nhp6 to Pob3 reduced H2A–H2B loss during reorganization
An early model proposed that FACT functions by removing one of the two H2A–H2B dimers from a nucleosome to form a hexasome (9), a conclusion based primarily on the significant loss of dimers from immobilized nucleosomes exposed to hFACT. Later assays examined the nucleosome population in solution and found that although some dimers are permanently lost during FACT binding, many nucleosomes contain a full complement of histones after reorganization and reassembly (4). However, it remains unclear whether these intact nucleosomes retain the full complement of histones during FACT exposure or regain H2A–H2B from a pool of displaced molecules during or after dissociation of FACT, and whether the DNA-binding domain has a role in displacement, retention, or reinsertion of H2A-H2B.
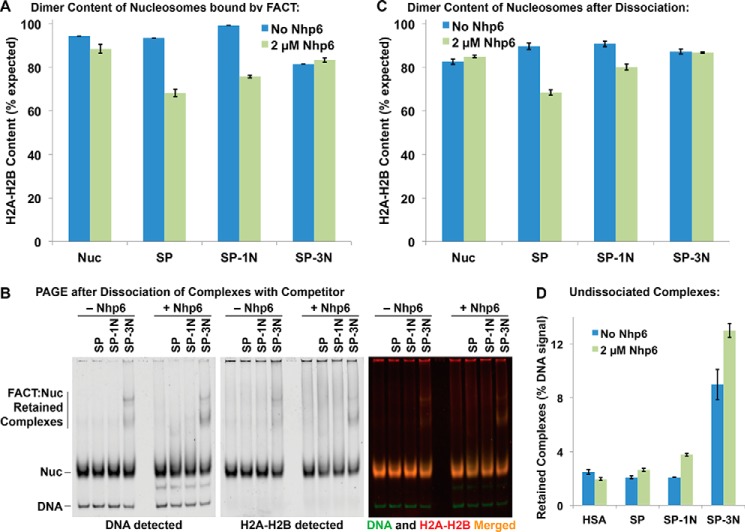
We therefore measured the amount of dimer displacement caused by the formation of complexes with different versions of FACT. Complexes were formed as shown in Fig. 3D with distinct fluors attached to H2A–H2B and DNA; then the ratio of H2A-H2B to DNA was determined for each band and compared with the ratio for intact nucleosomes to calculate the H2A–H2B content (Fig. 5A). Excess unlabeled genomic DNA was then added to bind the HMGB domains, leading to rapid dissociation of the FACT–nucleosome complexes as described previously (4) (Fig. 5B). The amount of H2A–H2B not associated with nucleosomes was then measured, and the composition of the dissociated nucleosomes was also determined from the H2A–H2B/DNA signal ratio, with both methods producing similar estimates of the amount of H2A–H2B loss (Fig. 5C and not shown).
Figure 5.
Fusion of Nhp6 to Pob3 inhibits H2A–H2B displacement from nucleosomes by FACT. A, complexes were formed as described in the legend for Fig. 3D. The H2A–H2B content in the FACT:Nuc band (see Fig. 3D) was determined by comparing the ratio of dimer signal to DNA signal and normalizing it to intact nucleosomes. B, samples were prepared and analyzed as described in the legend for Fig. 3D, except competitor DNA was added prior to electrophoresis. C, the H2A–H2B content of the released nucleosomes prepared as in B above was determined as in A. The average ± S.D. for four independent repeats is shown. D, the fraction of signal remaining in the Retained Complexes region defined in B was calculated for the same samples as described for C.
SP and SP–1N did not affect the H2A–H2B content of nucleosomes in the absence of Nhp6 (Fig. 5). In contrast, SP–3N alone or combinations of Nhp6 with SP, SP–1N, or SP–3N all caused measurable decreases in the H2A–H2B signal in the intact complexes they formed with nucleosomes (Fig. 5A). Nucleosomes alone were prone to some loss of H2A–H2B content when challenged with competitor DNA, but the loss was enhanced by FACT binding (Fig. 5C) as reported previously (4). However, the loss did not reach the 50% level expected for complete hexasome formation. Importantly, fusing Nhp6 to Pob3 reduced the loss of dimers in proportion to the number of modules added, which was true both in intact complexes and after dissociation (Fig. 5, A and C). These results indicate that intact FACT–nucleosome complexes are missing the same fraction of H2A–H2B as nucleosomes released from the complexes, suggesting that dimers are neither lost nor gained during dissociation in response to competitor DNA. They also show unexpectedly that intrinsic HMGB domains reduce the loss of H2A–H2B during reorganization, revealing a mechanistic difference between free and constrained DNA-binding domains.
The dissociation of complexes with nucleosomes after the addition of competitor DNA was essentially complete with SP and SP–1N, but complexes formed with SP–3N were partially resistant to disruption (Fig. 5, B and D). SP–3N did not bind nucleosomes when competitor was added first (not shown); so, this reveals slow or inefficient dissociation of SP–3N·nucleosome complexes. A similar phenomenon was observed with the Spt16–Pob3–Q308K complexes previously (42) and was interpreted as failed resolution of the reorganized state back to canonical nucleosomes. A similar interpretation here would suggest that fusing three Nhp6 modules to Pob3 promotes the formation of complexes but also inhibits the successful reassembly of the nucleosome and that this resolution is necessary for the release of FACT.
Pob3–Nhp6 performs many functions of both Pob3 and Nhp6 in vivo
A Pob3–1N fusion provides some functions of both Pob3 and Nhp6 in vivo (Ref. 43 and Fig. 6A). Pob3 function is not severely impaired by fusion of epitopes or purification tags (16, 44), so the ability of Pob3–1N to perform the functions of Pob3 may just indicate that Nhp6 is tolerated as a nonfunctional addition to Pob3. The Pob3–1N fusion protein had lower activity in vitro (Fig. 4), but this deficiency did not appear to cause a significant problem in vivo.
Figure 6.
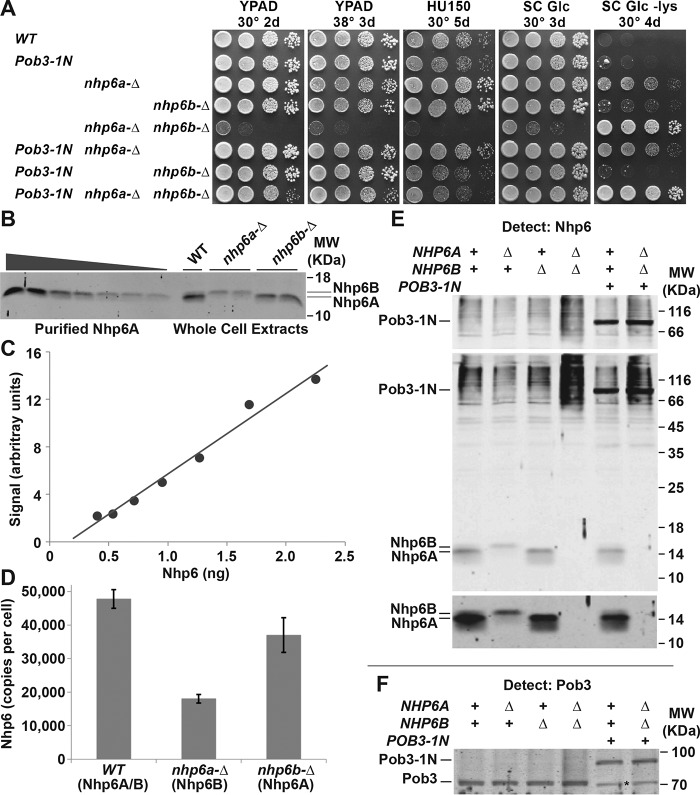
Effects of Pob3–1N in vivo and characterization of Nhp6 levels. A, 10-fold serial dilutions of strains with the genotypes indicated (Table S1) were tested as indicated. YPAD is rich medium (yeast extract, peptone, dextrose, adenine sulfate), HU150 is YPAD with 150 mm hydroxyurea, SC Glc is synthetic complete medium with glucose as the carbon source, and −Lys is the same but lacking lysine. Growth on −Lys reveals the Spt− phenotype in these strains with the lys2–128∂ allele (46). B, purified recombinant Nhp6A or 7 μg of extract from cells with the genotypes shown (Table S1) were separated by SDS-PAGE, transferred to nitrocellulose, and then detected with polyclonal antiserum directed against Nhp6A protein. The signal from the western blotting was quantitated with NIH ImageJ software (51), yielding the standard curve in C. Total amounts of Nhp6 were calculated from the standard curve and converted to copy number/cell (in D, the average ± S.D. for eight independent repeats is shown) using the previously determined relationship between cell number and total protein in these extracts (49). E, same as in B, except strains lacking Nhp6 and with Pob3–Nhp6 were included. The top panel shows a short exposure to optimize detection of Pob3–1N with Nhp6 antiserum, the middle panel shows the entire gel at a medium exposure time, and the bottom panel shows a long exposure time for the region containing free Nhp6 to emphasize the lack of signal in nhp6-ΔΔ strains. F, same as in E, except the gel was probed with antibodies against Pob3. A cross-reacting species observed in all strains but visible as a distinct band only with Pob3–1N fusions is marked by an asterisk.
The ability of Pob3–1N to provide Nhp6 activity is more surprising, as Nhp6 represents only 11 kDa of this 77-kDa fusion protein, and the fusion protein exhibited much lower affinity for DNA or nucleosomes than Nhp6 did in vitro (Fig. 3). Cells lacking a source of Nhp6 (nhp6a-Δ nhp6b-Δ; denoted as nhp6-ΔΔ here) display several phenotypes (6, 16, 31, 43, 45), including a severe growth defect, inability to grow at 38 °C, sensitivity to the replication toxin hydroxyurea, and the Spt− phenotype (reflecting loss of repression of a weak transposon-associated promoter (46)). Although complementation was not complete, POB3-NHP6 significantly reversed most of these defects, with the notable exception of the Spt− phenotype (Fig. 6A). Pob3–1N was therefore able to perform some roles of Nhp6 adequately but could not maintain transcriptional repression.
A trivial explanation for the partial complementation of nhp6-ΔΔ could be that the Pob3–Nhp6 fusion is degraded to release free Nhp6. Western blotting did not reveal any free Nhp6 protein in nhp6-ΔΔ strains, including those with the POB3-NHP6A gene (Fig. 6E). Pob3–Nhp6 was detected at about the same level as Pob3 in a normal cell, and no free Pob3 was detected in Pob3–Nhp6 strains (Fig. 6F). To determine how much Nhp6 was needed to support different functions, we quantified the amount of Nhp6 in various strains (Fig. 6, B–D). Our result of ∼48,000 molecules/cell in a WT strain (∼24 μm in nuclei, about two-thirds of the level of nucleosomes) was in general agreement with a previous estimate of 50,000–70,000 copies/cell (45). NHP6B expression increased somewhat to compensate for loss of NHP6A, so we concluded that ∼36,000 copies of Nhp6A is nearly adequate to promote the repression of the lys2–128∂ reporter of the Spt− phenotype (an nhp6b-Δ strain displayed only slight growth on medium lacking lysine (Fig. 6A)), whereas ∼18,000 copies of Nhp6B was less sufficient, leading to some loss of transcriptional repression but no other phenotypes. We estimated from our titrations that we would detect about ∼2000 copies of free Nhp6/cell, a level unlikely to complement the Spt− phenotype according to these calculations but with unknown effects on other phenotypes. We therefore found no evidence of detectable levels of free Nhp6 in strains expressing Pob3–1N; but different phenotypes displayed different thresholds for Nhp6 activity, and we cannot completely rule out the possibility that small amounts of free Nhp6 contribute to the genetic results.
Nhp6 supports nucleosome reorganization by human FACT
Sequence homology suggests that hFACT and yFACT should have similar activities, but hFACT has not been observed to bind nucleosomes stably or to reorganize them (3, 10, 11). However, the effects of additional free HMGB domain proteins have not been tested. Human cells contain high levels of these factors, so they could support FACT activity in vivo (22, 24) in much the way that Nhp6 supports yFACT function. To address this possibility, we assayed hFACT activity with and without added Nhp6. As reported previously (3), hFACT alone did not form stable complexes with intact nucleosomes, but complexes were observed when Nhp6 was added (Fig. 7A). hFACT also promoted sensitivity to DraI when supplemented with Nhp6 (Fig. 7B) but to a lower extent than yFACT. Notably, hFACT with Nhp6 caused enhanced loss of H2A–H2B (Fig. 7C), and the level of dimer dissociation was lower than that observed with SP + Nhp6 but similar to the result obtained with SP–1N + Nhp6 (Fig. 5). The effect of hFACT on nucleosomes was therefore similar to yFACT, especially the SP–1N version, including the limited extent of dimer displacement and nuclease accessibility relative to native SP.
Figure 7.
Human FACT reversibly reorganizes nucleosomes in the presence of added Nhp6. A, nucleosomes with SP or human FACT with or without 2 μm Nhp6 were separated by native PAGE as described in the legend for Fig. 3. B, complexes were treated with DraI as described in the legend for Fig. 4, except a single 10-min time point was analyzed for three independent samples of 200 nm SP, 2 μm Nhp6 (yFACT + N) or 200 nm hFACT, 2 μm Nhp6 (hFACT + N) with the average ± S.D. shown. The absolute rate of digestion varies with batches of DraI (compare with Fig. 4C). No digestion was observed in parallel experiments with hFACT, SP, or Nhp6 alone (not shown). C, FACT–nucleosome complexes formed as in A were treated with competitor DNA, and the H2A–H2B dimer content in the resulting dissociated nucleosomes was determined as described in the legend for Fig. 5C, with the average ± S.D. of three independent samples shown. D, spFRET analysis of nucleosome N35/112 (Nuc) alone and then with hFACT + Nhp6 before and after the addition of competitor DNA (+DNA) is shown (see Refs. 10 and 11 for details and additional control experiments). Frequency distributions of proximity ratios (EPR) calculated from the FRET efficiencies of individual nucleosomes were based on analysis of 4,315 (Nuc), 16,233 (hFACT + Nhp6), and 5,141 (hFACT + Nhp6 + competitor DNA) single particles. Control experiments published previously show that the Cy3 and Cy5 fluorophores inserted 35 and 112 bp into this 603-nucleosome positioning sequence produced high EPR values only when the DNA was tightly coiled around a histone core (10). Neither hFACT nor Nhp6 alone altered the EPR distribution in this assay (10, 11), but their combination caused a dramatic decrease in EPR values (hFACT + Nhp6). This effect was reversible, as the addition of competitor DNA restored the characteristic EPR distribution for nucleosomes (hFACT + Nhp6 + competitor DNA). Human FACT therefore caused the same disruption of nucleosome structure (uncoiling of DNA that affects about 70% of the DNA in a nucleosome) that was observed for yeast FACT (10). This depended on the further addition of HMGB proteins beyond the single domain available in SSRP1 and was reversible. Inset, an approximate representation of N35/112.
Dramatic uncoiling of nucleosomal DNA was detected previously with yeast FACT using an spFRET assay (10), and hFACT also had this activity but only in the presence of Nhp6 (Fig. 7D). In this assay, 603-nucleosome positioning sequence DNA was used to assemble nucleosomes with donor (Cy3) and acceptor (Cy5) dyes attached to bases that were far apart in the linear DNA fragment but close enough in the context of a nucleosome to produce FRET. The fluorescence intensities of Cy3 and Cy5 were measured when single nucleosomes diffusing freely in solution crossed the laser focus and proximity ratios (EPR) (which are directly proportional to FRET efficiencies) were calculated. Canonical nucleosomes produced the expected high EPR values in this assay, but the distribution was centered near zero in the presence of hFACT with Nhp6, indicating loss of FRET because of uncoiling of the DNA (see the “Discussion” in Refs. 10 and 11), and was restored when unlabeled competitor DNA was added to the sample to bind the Nhp6 (Fig. 7D). Neither hFACT nor Nhp6 alone had this effect on nucleosomal structure in this assay (additional controls and further discussion of the extent of uncoiling inferred are described in Refs. 10 and 11), so the data show that hFACT was able to induce the same dramatic unwrapping of nucleosomal DNA coils observed with yeast SP, with both being dependent on additional Nhp6 and both being reversible upon the sequestration of the free DNA-binding protein. The ability to reversibly reorganize nucleosomes is therefore conserved between yeast and human FACT architectures, as is the dependence of this activity on multiple HMGB domains.
In addition to the HMGB domain, SSRP1 also differs from Pob3 in the presence of the unusual SAB tail (Fig. 1). As an initial test of the effects of this domain on FACT function in vivo, we constructed yeast strains with normal Pob3 N-terminal and middle domains but with the acidic, HMGB, and SAB domains of hSSRP1 (Fig. 1). These constructs had only weakly deleterious effects (Fig. S2) and, unlike the Nhp6 fusions described above (Fig. 6), were unable to perform any of the functions of Nhp6 in vivo. This suggests that the HMGB domain of SSRP1 is not functionally identical to Nhp6, indicating further dissection of these domains will be needed to understand their functions.
Discussion
FACT can either destabilize or promote the assembly of nucleosomes without consuming an energy source (1, 7, 9). It appears to destabilize nucleosomes by recognizing multiple sites that are buried in the intact form and binding to these surfaces as they are transiently exposed, thereby forming a sequential series of intermediates along a pathway to a more open, reorganized form. In the reverse reaction, the same contacts guide loosely associated histones and DNA toward the canonical structure, promoting assembly by maintaining a high local concentration of components in appropriate orientations or conformations compatible with assembly (9, 42). In the currently available structures, histones bound by FACT have the same conformations found in nucleosomes, but FACT clashes with the expected positions of DNA (3, 36). DNA–histone contacts in existing nucleosomes must therefore be disrupted to form stable complexes with FACT, and these contacts must be reestablished to resolve the complexes into canonical nucleosomes. The most obvious candidate for a role in managing the conformation of the DNA during these transitions is the HMGB component, as these factors are known to bind DNA in a bent conformation, thereby stabilizing curved forms (22, 25). This bending could produce the stress that is needed to initiate reorganization, and it could also overcome the inherent stiffness of DNA to promote nucleosome assembly during resolution of the reorganized form. Here, we asked how the HMGB domain contributes mechanistically to FACT activity by comparing the effects of supplying it in different architectural contexts.
FACT heterodimers with single intrinsic HMGB domains (yeast SP–1N or hFACT) were able to reorganize nucleosomes in vitro but only when assisted by free Nhp6 (Figs. 4 and 7). One DNA-binding module was therefore not sufficient to support this activity in either native or artificial constructs. Notably, SP–1N and hFACT were each less active than SP in promoting endonuclease digestion of nucleosomal DNA (Figs. 4 and 7), so supplying a single HMGB domain in cis appeared to inhibit reorganization and not promote it. Cells with Pob3–1N or Pob3–hSSRP1 C-terminal domain fusions as the only source of this essential protein did not display any significant phenotypes (Figs. 6 and S2), so this apparent impairment of activity in vitro was not strongly detrimental in vivo. Although the physiological role of the complete reorganization of mononucleosomes as detected by our in vitro nuclease sensitivity assay remains speculative, these results suggest that providing a DNA-binding domain within FACT does not enhance this activity.
In contrast, an artificial construct with three Nhp6 modules fused to Pob3 was able to reorganize nucleosomes without assistance, supporting the previous conclusion that multiple DNA-binding events are required to initiate reorganization (5, 33) and indicating that two to three such events are adequate, whereas one is not. High concentrations of Nhp6 (Fig. 6) provide this activity in yeast, and the abundant HMGB family of factors presumably plays the same role in other eukaryotes (22, 24).
Reorganization by FACT causes displacement of some H2A–H2B dimers from nucleosomes (9), but even full reorganization of all nucleosomes under the saturating reaction conditions used here leads to an average loss of less than one dimer per nucleosome (Figs. 5 and 7 and Ref. 4). We proposed previously that dimer loss results from accidental dissociation of reorganized nucleosomes, but that interpretation implies that loss should be proportional to the time spent in the reorganized state. Instead, we find that buffer conditions (4) or modifications of FACT architecture (Figs. 5 and 7) drive the amount of displacement that occurs. Intact complexes and the nucleosomes that resulted from their dissociation had similar H2A–H2B contents (Figs. 5 and 7), suggesting that dimer loss did not occur during or after dissociation but was instead a property of the complexes themselves that was in dynamic equilibrium when complex dissociation was triggered. In this view, the observed decrease in H2A–H2B displacement that accompanied increasing numbers of intrinsic HMGB domains suggests that local DNA-binding activity shifts the equilibrium from a form more prone to dimer loss to one less prone to this loss.
Free Nhp6 had a high affinity for DNA in our assays, but fusing this same module to the C terminus of Pob3 reduced this affinity about 10-fold (Fig. 3). The native HMGB domain within hFACT showed little or no DNA-binding activity in this same assay (Fig. 3), although others have reported binding to shorter substrates under different conditions (40). Although the presence of this domain did not support reorganization, it reduced H2A–H2B displacement during reorganization, and the effect was proportional to the number of modules provided (Figs. 5 and 7). Single intrinsic HMGB modules within either hFACT or yFACT therefore seem to have limited access to free DNA, but they alter the properties of FACT–nucleosome complexes, making them less amenable to reorganization and less prone to loss of H2A–H2B dimers.
SP–3N failed to release efficiently from nucleosomes in vitro, and it was toxic in vivo (Figs. 2 and 5). The pob3-Q308K allele also causes multiple defects in vivo (47) and inefficient release from nucleosomes in vitro (42), which was interpreted as failure of a proposed nucleosome assembly checkpoint in which the release of FACT from complexes depends on the successful assembly of a canonical nucleosome. Extending this interpretation, SP–3N may be unable to complete nucleosome assembly effectively because its enhanced DNA-binding capacity makes it too prone to initiate another cycle of reorganization or unable to satisfy some test of the quality of the assembly process. This highlights a conceptual problem associated with histone chaperones in that they must bind to regions of histones and DNA that are buried in the nucleosome to prevent inappropriate interactions prior to assembly, but they must also dissociate at the proper time to allow nucleosomes to form. Release defects like these may therefore reveal checkpoints that monitor the appropriate coordination of multiple steps that occur in parallel during assembly.
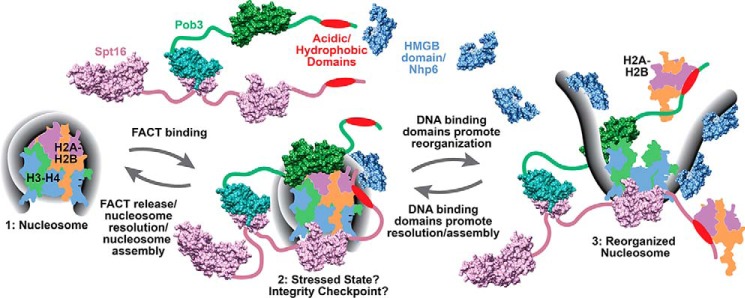
We propose a model (Fig. 8) to explain the effects of intrinsic DNA-binding modules on FACT activity. The new feature introduced here is a proposed unstable complex of FACT with nucleosomes that is not detected by EMSA. The nucleosomes in this proposed state are stressed in some way, making them more permissive to processes like transcription, but the DNA remains tightly coiled and resistant to nucleases. This state could represent multiple tentative steps taken as different domains of FACT interact weakly with individual binding surfaces, sequentially loosening the nucleosomal structure prior to achieving the stable reorganization associated with the observed large-scale uncoiling of the DNA (10, 11). These steps are rapidly reversible if reorganization is not achieved, and the reverse steps are also the normal pathway to nucleosome assembly. If the canonical configuration is not formed, release of FACT is blocked, providing a checkpoint for nucleosomal integrity. This intermediate could therefore explain how FACT affects the properties of nucleosomes without forming stable complexes (3, 7, 11), as well as the blocked release of some mutant versions of FACT from nucleosomes (Fig. 5 and Ref. 42).
Figure 8.
Model for the role of the DNA-binding module in FACT activity. FACT makes multiple weak contacts with a nucleosome leading to a more permissive but unstable “stressed” form. Converting this to a fully reorganized form requires multiple DNA-binding events to sequentially expose higher-affinity binding sites, allowing the formation of a stable complex detectable by EMSA. This activity can be provided by high concentrations of free Nhp6 or by multiple fused HMGB domains on Pob3 but not by the single HMGB domain in SSRP1 or Pob3–1N. Resolution of the reorganized form requires DNA-bending activity to produce curved DNA to enclose the H2A–H2B dimers, and an intrinsic HMGB domain favors this, skewing the equilibrium toward form 2. This wrapping step has similarities with the proposed role of the ATP-dependent remodeler ACF in converting pre-nucleosomes to nucleosomes (48), suggesting potential parallels between active and passive nucleosome assembly processes. The DNA in form 2 is tightly coiled, producing the same spFRET and nuclease sensitivity profiles as canonical nucleosomes (11), but this form is not fully resolved and can promote active processes like transcription or remodeling. Complete resolution requires coordinated assessment of the integrity of the nucleosome, with FACT variants like SP–3N (Fig. 5) and Spt16–Pob3–Q308K failing to release efficiently due to inefficient completion of this checkpoint (42). Factors are drawn approximately to scale using the color scheme as described in Fig. 1 and the following Protein Data Bank structures: 1ID3 (52), 1J5N (25), 4IOY (53), 2GCL (47), 3BIT (54), 4KHB (55), 4WNN, (36) and 4Z2M (3).
We further propose that DNA-binding modules alter the equilibrium between this proposed stressed intermediate and the reorganized state, with multiple DNA-binding events promoting full reorganization and the availability of an intrinsic, dedicated HMGB module favoring resolution to the stressed intermediate. The HMGB module functions through its known ability to stabilize bent DNA in both cases, as this weakens histone–DNA contacts to allow reorganization and provides curvature to the DNA to promote assembly. The stressed intermediate is less prone to H2A–H2B loss and is insensitive to nucleases; therefore, skewing the equilibrium toward this state could explain the effects observed here.
This model provides a mechanistic distinction between the Pob3 and SSRP1 architectures, suggesting that the relative degree of openness of the chromatin is the parameter that drives the evolutionary preference for one or the other. Pob3 lacks an intrinsic HMGB domain and is therefore more likely to promote reorganization, with its more open chromatin status providing enhanced DNA accessibility and H2A–H2B dimer exchange. SSRP1 is less disruptive, leading to more stable chromatin. This model highlights the importance of DNA bending during the nucleosome assembly phase of FACT action, as also proposed for nucleosome assembly by ATP-dependent remodelers (48). Questions remain regarding the nature and functional roles of reorganized nucleosomes, but the refined models proposed herein and the tools developed provide ways to address those questions.
Experimental procedures
Yeast strains
Strains congenic with the A364a genetic background were used, with the genotypes indicated in Table S1. NHP6A was fused to the endogenous POB3 gene as described in the supporting “Methods.” Plasmids were constructed also as described in the supporting “Methods” with oligonucleotides for strain and plasmid constructions listed in Table S2.
Protein purification
All Spt16–Pob3 variants were expressed in yeast and purified by nickel–nitrilotriacetic acid and size exclusion chromatography as described (4). Nhp6A was expressed and purified from bacteria by differential TCA precipitation followed by ion exchange chromatography with Mono-Q matrices (GE Life Sciences) as described (33). Human FACT was expressed in insect cells and purified as described (7, 9).
Nucleosomes
S. cerevisiae histone sequences were expressed in bacteria, purified, and assembled into octamers as described (4). H2A–Q114C or H2B–T119C variants (4) were used for labeling with Oregon Green dye (Invitrogen), and nucleosomes were assembled with Cy5-labeled DNA fragments (147 or 181-bp 5S rDNA sequences or 189-bp AT-hook sequences) and purified by sucrose gradient centrifugation as described (4).
EMSA
Nucleosomes were incubated with various factors, as indicated in the legends for Figs. 3, 5, and 7 for 10 min at 30 °C, and then electrophoresed on prerun 4% polyacrylamide mini-gels in 0.4× Tris borate-EDTA for 1 h at 80 volts. Dyes were detected using a TYPHOON scanner and quantitated using ImageQuant software (GE Life Sciences). Affinities were estimated from the half-maximal binding point, H2A–H2B displacement was measured using the dimer signal not associated with nucleosomes or nucleosomal DNA after the addition of a large excess of unlabeled genomic DNA (4), and H2A–H2B content was measured using the ratio of Oregon Green to Cy5 signal compared with the ratio with an intact nucleosome.
Restriction endonuclease assays
Nuclease sensitivity assays were performed as described previously using DraI (New England Biolabs) with 5S rDNA substrates and PstI (New England Biolabs) with AT-hook substrates (4). The fraction of molecules digested at multiple intervals over a 24-min time course was used to determine the initial rate of digestion, which was normalized to the rate obtained with parallel samples containing nucleosomes with 200 nm Spt16–Pob3 and 2 μm Nhp6 (SP + N) after subtracting the amount of digestion observed with nucleosomes alone. For human FACT, the yield of digested products at a single 10-min time point was used.
Western blotting
Rabbit polyclonal antisera against purified Nhp6 and Pob3 were prepared by Covance and validated using strains lacking Nhp6 or with fusions that altered the size of Pob3 (Ref. 49 and Fig. 6). Whole-cell extracts were prepared using the TCA method and normalized for total protein loaded, and the number of cells represented was calculated as described (49).
Phenotypic analysis
Strains were grown to saturation in rich medium or under selection for the plasmids indicated. Then 10-fold serial dilutions were prepared in sterile water, and aliquots were placed on solid medium and incubated as described in the legends for Figs. 2 and 6 and Fig. S2.
spFRET assay
The spFRET assay was performed as described (10). Nucleosomal DNA was labeled with Cy3 and Cy5 fluorophores at +35 and +112 bp relative to the 603-nucleosome positioning boundary (template N35/112 in Ref. 10). Nucleosomes were examined in a buffer containing 17 mm HEPES, pH 7.6, 2 mm Tris-HCl, pH 7.5, 0.8 mm EDTA, 0.11 mm 2-mercaptoethanol, 11 mm NaCl, 1.1% glycerin, and 12% sucrose.
Author contributions
L. L. M., Z. C., V. M. S., M. E. V., and T. F. data curation; L. L. M., Z. C., H. X., M. E. V., and T. F. formal analysis; L. L. M., Z. C., H. X., and M. E. V. investigation; L. L. M., Z. C., H. X., A. V. F., and M. E. V. methodology; H. X., V. M. S., A. V. F., M. E. V., and T. F. conceptualization; V. M. S., A. V. F., and T. F. supervision; V. M. S., A. V. F., and T. F. writing-review and editing; A. V. F. resources; A. V. F. software; A. V. F. and T. F. funding acquisition; A. V. F. and T. F. project administration; T. F. writing-original draft.
Supplementary Material
Acknowledgments
We thank Fukai Hsieh for providing purified human FACT and David Stillman and his laboratory for providing critical feedback on the experiments and this manuscript. Development and application of spFRET was supported by Russian Science Foundation Grant 14-24-00031.
This work was supported by National Institutes of Health Grants R01 GM064649 (to T. F.) and R01 GM119398 (to V. M. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1 and S2.
- FACT
- facilitates chromatin transcription
- yFACT
- yeast FACT
- hFACT
- human FACT
- spFRET
- single-particle Förster resonance energy transfer
- HMGB
- high-mobility group B
- SAB
- serine-acidic-basic
- SSRP
- structure-specific recognition protein
- EMSA
- electrophoretic mobility shift assay
- SP
- Spt16-Pob3.
References
- 1. Formosa T. (2012) The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819, 247–255 10.1016/j.bbagrm.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hondele M., and Ladurner A. G. (2013) Catch me if you can: How the histone chaperone FACT capitalizes on nucleosome breathing. Nucleus 4, 443–449 10.4161/nucl.27235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsunaka Y., Fujiwara Y., Oyama T., Hirose S., and Morikawa K. (2016) Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686 10.1101/gad.274183.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xin H., Takahata S., Blanksma M., McCullough L., Stillman D. J., and Formosa T. (2009) yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 35, 365–376 10.1016/j.molcel.2009.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rhoades A. R., Ruone S., and Formosa T. (2004) Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 24, 3907–3917 10.1128/MCB.24.9.3907-3917.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., and Stillman D. J. (2001) Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20, 3506–3517 10.1093/emboj/20.13.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsieh F. K., Kulaeva O. I., Patel S. S., Dyer P. N., Luger K., Reinberg D., and Studitsky V. M. (2013) Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 110, 7654–7659 10.1073/pnas.1222198110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orphanides G., Wu W. H., Lane W. S., Hampsey M., and Reinberg D. (1999) The chromatin-specific transcription elongation factor FACT comprises the human SPT16/CDC68 and SSRP1 proteins. Nature 400, 284–288 10.1038/22350 [DOI] [PubMed] [Google Scholar]
- 9. Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., and Reinberg D. (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 10.1126/science.1085703 [DOI] [PubMed] [Google Scholar]
- 10. Valieva M. E., Armeev G. A., Kudryashova K. S., Gerasimova N. S., Shaytan A. K., Kulaeva O. I., McCullough L. L., Formosa T., Georgiev P. G., Kirpichnikov M. P., Studitsky V. M., and Feofanov A. V. (2016) Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat. Struct. Mol. Biol. 23, 1111–1116 10.1038/nsmb.3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valieva M. E., Gerasimova N. S., Kudryashova K. S., Kozlova A. L., Kirpichnikov M. P., Hu Q., Botuyan M. V., Mer G., Feofanov A. V., and Studitsky V. M. (2017) Stabilization of nucleosomes by histone tails and by FACT revealed by spFRET microscopy. Cancers (Basel) 9, E3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurat C. F., Yeeles J. T. P., Patel H., Early A., and Diffley J. F. X. (2017) Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell 65, 117–130 10.1016/j.molcel.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang J., Zhang X., Feng J., Leng H., Li S., Xiao J., Liu S., Xu Z., Xu J., Li D., Wang Z., Wang J., and Li Q. (2016) The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 14, 1128–1141 10.1016/j.celrep.2015.12.096 [DOI] [PubMed] [Google Scholar]
- 14. Evans D. R., Brewster N. K., Xu Q., Rowley A., Altheim B. A., Johnston G. C., and Singer R. A. (1998) The yeast protein complex containing Cdc68 and Pob3 mediates core-promoter repression through the Cdc68 N-terminal domain. Genetics 150, 1393–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malone E. A., Clark C. D., Chiang A., and Winston F. (1991) Mutation in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 5710–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schlesinger M. B., and Formosa T. (2000) POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 155, 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., and Martens J. A. (2011) Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25, 29–40 10.1101/gad.1975011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jamai A., Puglisi A., and Strubin M. (2009) Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell 35, 377–383 10.1016/j.molcel.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 19. Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates JR 3rd, Cleveland D. W. (2006) The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- 20. Prendergast L., Müller S., Liu Y., Huang H., Dingli F., Loew D., Vassias I., Patel D. J., Sullivan K. F., and Almouzni G. (2016) The CENP-T/-W complex is a binding partner of the histone chaperone FACT. Genes Dev. 30, 1313–1326 10.1101/gad.275073.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wittmeyer J., and Formosa T. (1997) The Saccharomyces cerevisiae DNA polymerase α catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17, 4178–4190 10.1128/MCB.17.7.4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bruhn S. L., Pil P. M., Essigmann J. M., Housman D. E., and Lippard S. J. (1992) Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc. Natl. Acad. Sci. U.S.A. 89, 2307–2311 10.1073/pnas.89.6.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stillman D. J. (2010) Nhp6: A small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1799, 175–180 10.1016/j.bbagrm.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas J. O., and Stott K. (2012) H1 and HMGB1: Modulators of chromatin structure. Biochem. Soc. Trans. 40, 341–346 10.1042/BST20120014 [DOI] [PubMed] [Google Scholar]
- 25. Allain F. H., Yen Y. M., Masse J. E., Schultze P., Dieckmann T., Johnson R. C., and Feigon J. (1999) Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 18, 2563–2579 10.1093/emboj/18.9.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonaldi T., Längst G., Strohner R., Becker P. B., and Bianchi M. E. (2002) The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 21, 6865–6873 10.1093/emboj/cdf692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Travers A. A., Ner S. S., and Churchill M. E. (1994) DNA chaperones: A solution to a persistence problem? Cell 77, 167–169 10.1016/0092-8674(94)90306-9 [DOI] [PubMed] [Google Scholar]
- 28. Kolodrubetz D., and Burgum A. (1990) Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J. Biol. Chem. 265, 3234–3239 [PubMed] [Google Scholar]
- 29. Singer R. A., and Johnston G. C. (2004) The FACT chromatin modulator: Genetic and structure/function relationships. Biochem. Cell Biol. 82, 419–427 10.1139/o04-050 [DOI] [PubMed] [Google Scholar]
- 30. Szerlong H., Saha A., and Cairns B. R. (2003) The nuclear actin-related proteins Arp7 and Arp9: A dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 22, 3175–3187 10.1093/emboj/cdg296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu Y., Eriksson P., and Stillman D. J. (2000) Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol. 20, 2350–2357 10.1128/MCB.20.7.2350-2357.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kruppa M., Moir R. D., Kolodrubetz D., and Willis I. M. (2001) Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol. Cell 7, 309–318 10.1016/S1097-2765(01)00179-4 [DOI] [PubMed] [Google Scholar]
- 33. Ruone S., Rhoades A. R., and Formosa T. (2003) Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and reorganize nucleosomes. J. Biol. Chem. 278, 45288–45295 10.1074/jbc.M307291200 [DOI] [PubMed] [Google Scholar]
- 34. Malarkey C. S., and Churchill M. E. (2012) The high mobility group box: The ultimate utility player of a cell. Trends Biochem. Sci. 37, 553–562 10.1016/j.tibs.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yarnell A. T., Oh S., Reinberg D., and Lippard S. J. (2001) Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J. Biol. Chem. 276, 25736–25741 10.1074/jbc.M101208200 [DOI] [PubMed] [Google Scholar]
- 36. Kemble D. J., McCullough L. L., Whitby F. G., Formosa T., and Hill C. P. (2015) FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell 60, 294–306 10.1016/j.molcel.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsunaka Y., Toga J., Yamaguchi H., Tate S., Hirose S., and Morikawa K. (2009) Phosphorylated intrinsically disordered region of FACT masks its nucleosomal DNA binding elements. J. Biol. Chem. 284, 24610–24621 10.1074/jbc.M109.001958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winkler D. D., and Luger K. (2011) The histone chaperone Fact: Structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286, 18369–18374 10.1074/jbc.R110.180778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang W., Zeng F., Liu Y., Shao C., Li S., Lv H., Shi Y., Niu L., Teng M., and Li X. (2015) Crystal structure of human SSRP1 middle domain reveals a role in DNA binding. Sci. Rep. 5, 18688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Winkler D. D., Muthurajan U. M., Hieb A. R., and Luger K. (2011) The histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286, 41883–41892 10.1074/jbc.M111.301465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yen Y. M., Wong B., and Johnson R. C. (1998) Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities: Role of the unique N terminus and putative intercalating methionine. J. Biol. Chem. 273, 4424–4435 10.1074/jbc.273.8.4424 [DOI] [PubMed] [Google Scholar]
- 42. McCullough L., Poe B., Connell Z., Xin H., and Formosa T. (2013) The FACT histone chaperone guides histone H4 into its nucleosomal conformation in Saccharomyces cerevisiae. Genetics 195, 101–113 10.1534/genetics.113.153080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brewster N. K., Johnston G. C., and Singer R. A. (2001) A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21, 3491–3502 10.1128/MCB.21.10.3491-3502.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoffmann C., and Neumann H. (2015) In Vivo mapping of FACT-histone interactions identifies a role of Pob3 C-terminus in H2A-H2B binding. ACS Chem. Biol. 10, 2753–2763 10.1021/acschembio.5b00493 [DOI] [PubMed] [Google Scholar]
- 45. Paull T. T., Carey M., and Johnson R. C. (1996) Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 10, 2769–2781 10.1101/gad.10.21.2769 [DOI] [PubMed] [Google Scholar]
- 46. Simchen G., Winston F., Styles C. A., and Fink G. R. (1984) Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc. Natl. Acad. Sci. U.S.A. 81, 2431–2434 10.1073/pnas.81.8.2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. VanDemark A. P., Blanksma M., Ferris E., Heroux A., Hill C. P., and Formosa T. (2006) The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 22, 363–374 10.1016/j.molcel.2006.03.025 [DOI] [PubMed] [Google Scholar]
- 48. Khuong M. T., Fei J., Ishii H., and Kadonaga J. T. (2015) Prenucleosomes and active chromatin. Cold Spring Harb. Symp. Quant. Biol. 80, 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McCullough L., Connell Z., Petersen C., and Formosa T. (2015) The abundant histone chaperones Spt6 and FACT collaborate to assemble, inspect, and maintain chromatin structure in Saccharomyces cerevisiae. Genetics 201, 1031–1045 10.1534/genetics.115.180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boeke J. D., Trueheart J., Natsoulis G., and Fink G. R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175 10.1016/0076-6879(87)54076-9 [DOI] [PubMed] [Google Scholar]
- 51. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White C. L., Suto R. K., and Luger K. (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20, 5207–5218 10.1093/emboj/20.18.5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kemble D. J., Whitby F. G., Robinson H., McCullough L. L., Formosa T., and Hill C. P. (2013) Structure of the Spt16 middle domain reveals functional features of the histone chaperone FACT. J. Biol. Chem. 288, 10188–10194 10.1074/jbc.C113.451369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. VanDemark A. P., Xin H., McCullough L., Rawlins R., Bentley S., Heroux A., Stillman D. J., Hill C. P., and Formosa T. (2008) Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J. Biol. Chem. 283, 5058–5068 10.1074/jbc.M708682200 [DOI] [PubMed] [Google Scholar]
- 55. Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., Zhang E. T., Nijmeijer B., Kotthoff C., Rybin V., Amlacher S., Hurt E., and Ladurner A. G. (2013) Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature 499, 111–114 10.1038/nature12242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.