Abstract
TMEPAI (transmembrane prostate androgen–induced protein, also called prostate transmembrane protein, androgen-induced 1 (PMEPA1)) is a type I transmembrane (TM) protein, but its cellular function is largely unknown. Here, studying factors influencing the stability of c-Maf, a critical transcription factor in multiple myeloma (MM), we found that TMEPAI induced c-Maf degradation. We observed that TMEPAI recruited NEDD4 (neural precursor cell expressed, developmentally down-regulated 4), a WW domain–containing ubiquitin ligase, to c-Maf, leading to its degradation through the proteasomal pathway. Further investigation revealed that TMEPAI interacts with NEDD4 via its conserved PY motifs. Alanine substitution or deletion of these motifs abrogated the TMEPAI complex formation with NEDD4, resulting in failed c-Maf degradation. Functionally, TMEPAI suppressed the transcriptional activity of c-Maf. Of note, increased TMEPAI expression was positively associated with the overall survival of MM patients. Moreover, TMEPAI was down-regulated in MM cells, and re-expression of TMEPAI induced MM cell apoptosis. In conclusion, this study highlights that TMEPAI decreases c-Maf stability by recruiting the ubiquitin ligase NEDD4 to c-Maf for proteasomal degradation. Our findings suggest that the restoration of functional TMEPA1 expression may represent a promising complementary therapeutic strategy for treating patients with MM.
Keywords: ubiquitylation (ubiquitination), apoptosis, multiple myeloma, protein-protein interaction, transcription factor, ubiquitin ligase
Introduction
TMEPAI (transmembrane protein, androgen induced), which is also called PMEPA1 (prostate transmembrane protein, androgen-induced), is a single-span transmembrane (TM)4 protein originally identified from a prostate cDNA library (1). In addition to androgen, TMEPAI expression is also stimulated by some growth factors such as epidermal growth factor and transforming growth factor-β (TGF-β) (2). The expression profiling found that TMEPAI is overexpressed in solid cancers including prostate cancer, lung cancer (3, 4), breast cancer (3, 5), ovarian cancer (6), and rental cell carcinoma (7), but it is not detectable in liquid cancers such as leukemia and lymphoma samples (7). The specific expression pattern suggests that TMEPAI might function differently upon specific cancer types. For example, TMEPAI displays as a tumor suppressor in prostate cancer progression because silence of TMEPAI leads to accelerated proliferation of prostate cancer cells (8), whereas expression of TMEPAI inhibits prostate cancer cell growth (9) and prevents its bone metastasis (10). However, TMEPAI acts as an oncogene and promotes proliferation and survival of lung and breast cancers. TMEPAI mediates the degradation of receptor-activated SMAD and activates non-canonical phosphatidylinositol 3-kinase/Akt signaling transduction, thus promoting TGF-β-dependent cell growth and metastasis of triple negative breast cancer (3). In lung cancer cells, TMEPAI not only increases cancer cell proliferation, migration, and invasion (11, 12) but also enhances tumorigenicity and the epithelial–mesenchymal transition of lung cancer cells (4). These actions were associated with TMEPAI-induced TGF-β degradation in lysosomes and TMEPAI-induced reactive oxygen species and insulin receptor substrate-1 (4). In contrast, knockdown of TMEPAI enhances Smad2 phosphorylation and significantly suppressed lung cancer cell proliferation in the presence of TGF-β. All the above results collectively suggest that TMEPAI can promote or suppress cancer cell proliferation and tumor growth depending on the cancer types, but the TGF-β signaling pathway is a key factor.
Hematological malignancies are a collection of blood cancers including leukemia, lymphoma, and multiple myeloma in which TMEPAI is less expressed (7), but the function and pathophysiological significance of TMEPAI in these cancers are not yet known. Multiple myeloma (MM) is an incurable hematological malignancy derived from plasma cells; it is the second highest blood cancer and accounts for 2% overall cancer cases. MM is featured with chromosomal translocations that lead to overexpression of several key genes, including the transcription factor c-Maf (13, 14), which promotes MM cell proliferation and development (15, 16). In the present study, we found that TMEPAI induces the ubiquitination and degradation of c-Maf and induces MM cell apoptosis independent of TGF-β signaling.
Results
TMEPAI induces c-Maf degradation
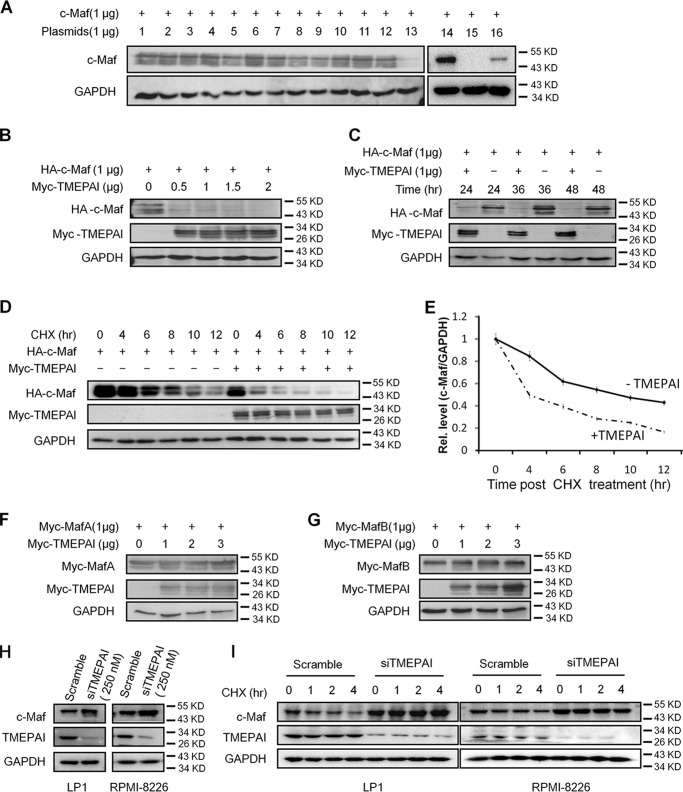
c-Maf is a transcription factor that induces myelomagenesis (15), and it could be degraded via both proteasomes and lysosomes (17). Our previous studies have shown that ubiquitin ligase HERC4 and ubiquitin-conjugating enzyme UBE2O lead to c-Maf degradation (13, 18). To find out whether there were any other enzymes that might mediate c-Maf degradation, a small collection of plasmids (Hospital for Sick Children, Toronto, Canada) that were involved in proteolysis was screened. After these individual plasmids were co-transfected with c-Maf, an immunoblotting screen was performed. The results showed that TMEPAI down-regulated c-Maf protein, similar to the effect from the proven E3 ligase HERC4 (Fig. 1A). To confirm this finding, c-Maf was co-transfected with increased TMEPAI and incubated for various periods, followed by immunoblotting assay. As shown in Fig. 1 (B and C), c-Maf was down-regulated by TMEPAI in a concentration- and time-dependent manner. To further understand the degradation of c-Maf protein by TMEPAI, we applied a cycloheximide (CHX) chase assay. As shown in Fig. 1 (D and E), when c-Maf synthesis was inhibited by CHX, the addition of TMEPAI promoted c-Maf degradation. In the presence of TMEPAI, the half-life of c-Maf was less than 4 h, which was markedly decreased from the control in which c-Maf half-life was more than 8 h (Fig. 1, D and E). Therefore, TMEPAI mediated c-Maf protein degradation.
Figure 1.
TMEPAI induces the degradation of c-Maf but not MafA and MafB. A, the c-Maf plasmid was co-transfected with individual plasmids for 24 h, followed by total protein extract and immunoblotting assay. Lane 1, pcDNA3.1; lane 2, LRSAM1; lane 3, TRIM25; lane 4, RNF31; lane 5, UBE3C; lane 6, TRIM44; lane 7, RNF10; lane 8, RNF32; lane 9, RNF125; lane 10, MKRN2; lane 11, TRIM9; lane 12, TRIM11; lane 13, TMEPAI; lane 14, pcDNA3.1; lane 15, HERC4; lane 16, TMEPAI. B, c-Maf was co-transfected with TMEPAI at the indicated concentrations into HEK293T cells for 24 h, followed by immunoblotting assay. C, c-Maf and TMEPAI plasmids were co-transfected into HEK293T cells with the indicated incubation times followed by immunoblotting assay. D, c-Maf and TMEPAI plasmids were co-transfected into HEK293T cells followed by CHX treatment for indicated periods. All cell lysates were subjected to immunoblotting assay as indicated. E, densitometric analysis of c-Maf in the presence of TMEPAI and CHX treatment from D. F and G, MafA (F) or MafB (G) and TMEPAI plasmids were co-transfected into HEK293T cells for 24 h followed by immunoblotting assay. H, TMEPAI was knocked down in MM cell lines LP1 and RPMI-8226 by siTMEPAI for 48 h, followed by immunoblotting assay. I, LP1 and RPMI-8226 cells were subjected to TMEPAI knockdown by siRNA. Forty-eight hours later, the cells were treated with CHX for indicated periods and immunoblotting assay.
c-Maf belongs to the Maf basic leucine-zipper transcription factor family, which also contains MafA and MafB, another two members that share high sequence similarity with c-Maf. To find out whether TMEPAI also modulated the stability of these two proteins, TMEPAI was co-transfected with MafA or MafB for 24 h, followed by immunoblotting assay. The results showed that neither MafA nor MafB was degraded by TMEPAI (Fig. 1, F and G). This finding was confirmed by the CHX chase assay (data not shown). Therefore, TMEPAI selectively regulates c-Maf protein stability.
To confirm the effects of TMEPAI on c-Maf degradation, we next measured endogenous expression of c-Maf in MM cells by knockdown TMEPAI using TMEPAI-specific siRNA. As shown in Fig. 1H, TMEPAI knockdown led to higher c-Maf protein levels in both LP1 and RPMI-8226, two typical MM cell lines. To further evaluate this result, TMEPAI was knocked down with siTMEPAI followed by CHX treatment and immunoblotting assay. As shown in Fig. 1I, c-Maf was maintained at a high level in 4 h when TMEPAI knockdown; in contrast, c-Maf was steadily decreased following CHX treatment. Therefore, TMEPAI knockdown stabilized c-Maf protein, and TMEPAI induces c-Maf degradation.
TMEPAI induces c-Maf degradation via the proteasomal pathway
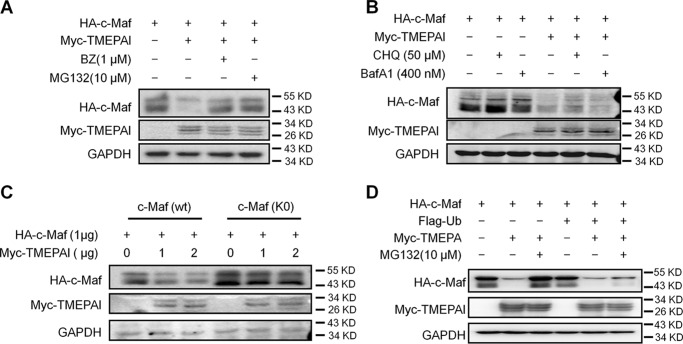
Previous studies have demonstrated that c-Maf could be degraded via both proteasomes and lysosomes (17), and it is interesting that TMEPAI is capable of mediating protein degradation in either proteasomes (2) or lysosomes (11). To find out the pathway in which c-Maf degradation was mediated by TMEPAI, c-Maf stability was evaluated in the presence of TMEPAI and lysosomal or proteasomal inhibitors. As shown in Fig. 2A, TMEPAI mediated c-Maf degradation, but when bortezomib (BZ) or MG132, two typical proteasomal inhibitors, was added, TMEPAI-induced c-Maf degradation was abolished. In contrast, TMEPAI-induced c-Maf degradation was not affected by either chloroquine nor bafilomycin A1, two typical lysosomal inhibitors (Fig. 2B). These findings suggested that TMEPAI mediated c-Maf degradation via the ubiquitin-proteasome pathway. To verify this hypothesis, all lysine residues in c-Maf were replaced with arginine; therefore c-Maf will not be ubiquitinated and could not be degraded in the proteasomes (17). As shown in Fig. 2C, TMEPAI mediated WT c-Maf degradation; however, the protein levels of mutated c-Maf (K0) without lysine residues were not significantly affected. Interestingly, when ubiquitin was overexpressed in cells, TMEPAI-induced c-Maf degradation could not be prevented by proteasomal inhibitors (Fig. 2D). This finding was also found in several subsequent experiments. These results thus demonstrated that TMEPAI mediated c-Maf degradation via the proteasomal pathway and that extra ubiquitin expression markedly increased the degradation rate mediated by TMEPAI.
Figure 2.
TMEPAI induces c-Maf degradation via the proteasomal but not the lysosomal pathway. A, c-Maf and TMEPAI plasmids were co-transfected into HEK293T cells, followed by the treatment of proteasomal inhibitors BZ or MG132. The cell lysates were subjected to immunoblotting assay. B, c-Maf and TMEPAI plasmids were co-transfected into HEK293T cells, followed by the treatment of lysosomal inhibitors chloroquine (CHQ) or bafilomycin A1 (BafA1). The cell lysates were subjected to immunoblotting assay. C, TMEPAI was co-transfected into HEK293T cells with the WT or all-lysine mutant c-Maf (K0) plasmids for 24 h followed by immunoblotting assay. The relative expression of c-Maf protein against GAPDH is shown at the bottom. D, a c-Maf plasmid was co-transfected into HEK293T cells with a TMEPAI plasmid with or without ubiquitin (Ub) plasmid transfection. Twenty-four hours later, the cells were treated with MG132 for 8 h before being applied to an immunoblotting assay for indicated protein expression.
NEDD4 is required for TMEPAI-mediated c-Maf ubiquitination and degradation
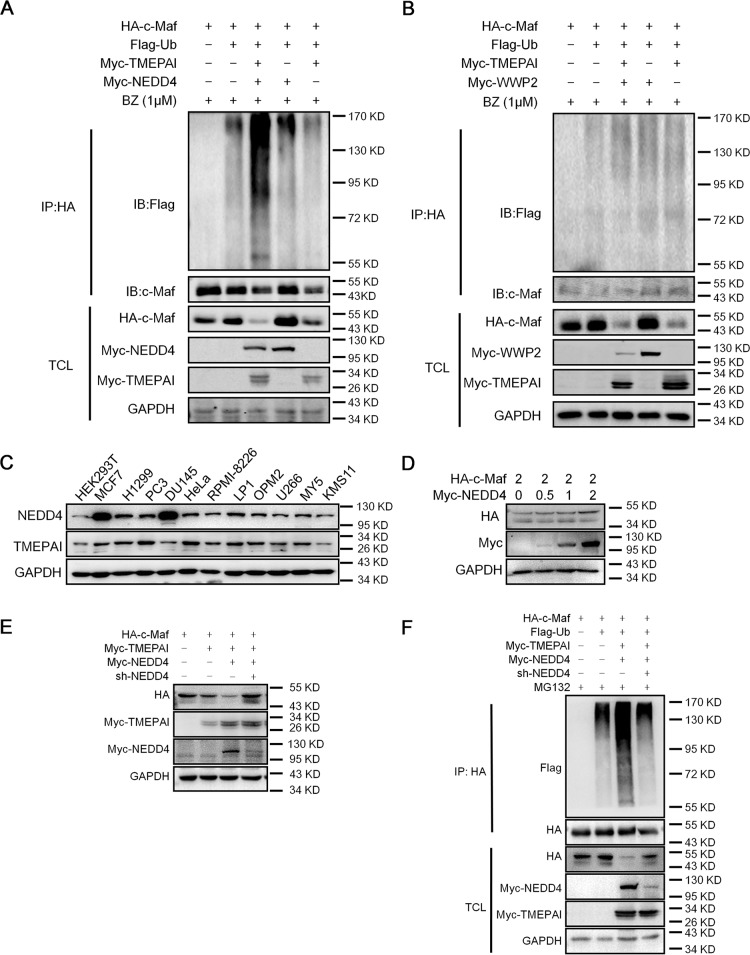
The above results indicated that TMEPAI-induced c-Maf degradation via the ubiquitin proteasomal pathway. However, we found that TMEPAI failed to interact with c-Maf (data not shown). By reviewing the literature, TMEPAI was found to be able to interact with the WW domain of some E3 ubiquitin ligases for protein ubiquitination and degradation (9). To find out the protein that could be the potential partner of TMEPAI in mediating c-Maf stability, we compared c-Maf ubiquitination by TMEPAI in combination with several WW domain-containing E3 ligases, including NEDD4, WWP1, WWP2, and ITCH. The results showed that all these E3s alone failed to induce c-Maf ubiquitination (data not shown); however, NEDD4 markedly increased the polyubiquitination level of c-Maf and enhanced c-Maf degradation in the presence of TMEPAI (Fig. 3A). In contrast, three other ligases such as WWP2, another possible interacting partner, failed to boost such kind of events in c-Maf ubiquitination mediated by TMEPAI (Fig. 3B). Therefore, NEDD4 was required for TMEPAI-induced c-Maf ubiquitination and degradation. However, in the analysis of expression profiles, both TMEPAI and NEDD4 were found in all cell lines tested, including HEK293T and MM cells (Fig. 3C). However, NEDD4 alone could not mediate c-Maf degradation (Fig. 3D). To confirm this finding, NEDD4 was knocked down in HEK293T cells. The immunoblotting assay showed that c-Maf was degraded in the presence of TMEPAI and NEDD4; however, when shNEDD4 was introduced, c-Maf degradation was abolished (Fig. 3E). Consistent with this finding, TMEPAI-mediated c-Maf ubiquitination was also prevented by shNEDD4 (Fig. 3F). All the above studies thus demonstrated that NEDD4 was required for TMEPAI-mediated c-Maf polyubiquitination and degradation.
Figure 3.
NEDD4 is required for TMEPAI-induced c-Maf polyubiquitination and degradation. A and B, a c-Maf plasmid was co-transfected with a TMEPAI plasmid into HEK293T cells along with a NEDD4 (A) or a WWP2 (B) plasmid for 24 h, followed by BZ treatment. Cell lysates were then subjected to the IP and IB assay. C, cell lysates from a panel of cancer cell lines were subjected to evaluation of the expression level of TEMPAI and NEDD4 by immunoblotting assay. D, HEK239T cells were transfected with NEDD4 for 24 h, followed by immunoblotting assay. E, HEK293T cells were infected with shNEDD4 for 48 h, followed by immunoblotting against specific proteins as indicated. F, HEK293T cells were transfected a NEDD4 plasmid for 24 h, followed by infection with shNEDD4. The cells were then incubated for another 48 h before being prepared for immunoprecipitation/immunoblotting assays as indicated. TCL, total cell lysates.
The transmembrane domain is indispensable for TMEPAI-mediated c-Maf degradation
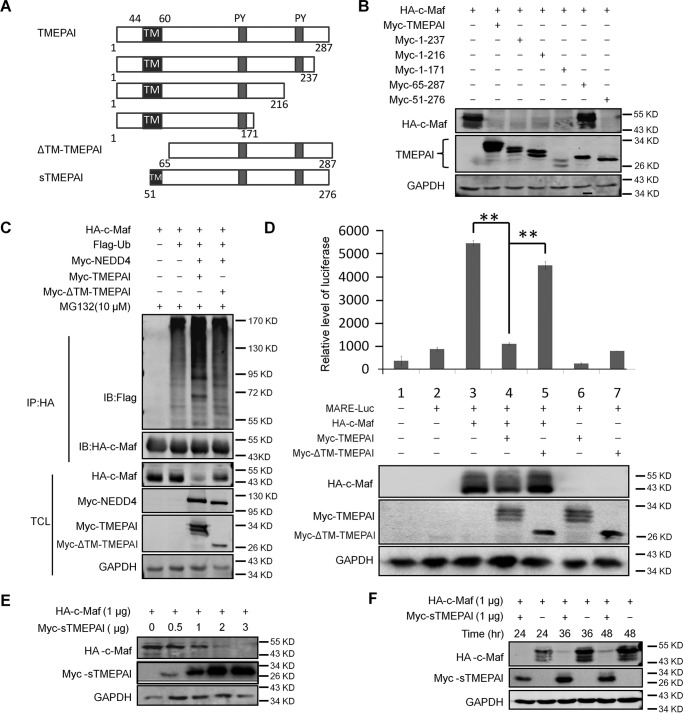
TMEPAI is a type Ib transmembrane protein with the TM domain at the N terminus (Fig. 4A). A previous study found that TMEPAI has three isoforms, of which the cytosolic form TMEPAIc lacks the TM domain and loses its transcriptional activity (10), suggesting the TM domain is critical for TMEPAI activity. To find out whether the TM domain is critical for TMEPAI-mediated c-Maf ubiquitination and stability, we made a series of short truncates of TMEPAI as shown in Fig. 4A. When these TMEPAI truncates were co-transfected with c-Maf, subsequent immunoblotting assays showed that all truncates with the TM domain displayed activity similar to the WT in terms of c-Maf degradation (Fig. 4B). However, TMEPAI lacking the TM domain (ΔTM-TMEPAI) failed to mediate c-Maf degradation (Fig. 4B). To find out whether this TM domain was associated with c-Maf ubiquitination, c-Maf and NEDD4 were co-transfected with a WT or a ΔTM-TMEPAI plasmid into HEK293T cells, followed by the immunoprecipitation and immunoblotting (IP/IB) assay. As shown in Fig. 4C, the wtTMEPAI but not the ΔTM-TMEPAI induced c-Maf ubiquitination, which was consistent with c-Maf degradation (Fig. 4B). Because c-Maf is a transcription factor, we next evaluated whether TMEPAI modulated c-Maf transcriptional activity. To this end, a luciferase construct directed by MARE (Maf recognition element) (19) was co-transfected with wildtype or ΔTM-TMEPAI. The luciferase activity analysis revealed that ΔTM-TMEPAI lost its modularity on the stability and transcriptional activity of c-Maf (Fig. 4D), suggesting that the TM domain was critical for TMEPAI in mediating c-Maf transcriptional activity. To further confirm this finding, a plasmid with TMEPAI containing a part of the TM domain (sTMEPAI retained 10 amino acid residues in the TM domain, as shown in Fig. 4A) was co-transfected with c-Maf for increasing incubation periods or increasing doses, followed by immunoblotting assay. As shown in Fig. 4 (E and F), the sTMEPAI construct with a small part of TM domain was also able to mediate c-Maf degradation in a manner similar to wtTMEPAI. Therefore, the TM domain is indispensable for TMEPAI-mediated c-Maf degradation.
Figure 4.
The TM domain is indispensable for TMEPAI to mediate c-Maf ubiquitination and biological activity. A, the schematic drawing of the TMEPAI truncated constructs. B, c-Maf was co-transfected with full-length or truncated TMEPAI constructs, respectively, into HEK293T cells for 24 h, followed by immunoblotting assay. C, c-Maf was co-transfected with wildtype TMEPAI or TM-deleted TMEPAI into HEK293T cells. 16 h later, the cells were subjected to protein extraction, immunoprecipitation/immunoblotting assays. D, the MARE-driving firefly luciferase and c-Maf plasmids were co-transfected into HEK293T cells along with full-length or TM-deleted TMEPAI constructs for 24 h, followed by luciferase assay and immunoblotting assay. **, p < 0.001. E and F, the c-Maf plasmid was co-transfected into HEK293T cells with the TMEPAI construct containing a part of the TM domain (sTMEPAI) for 24 h at indicated concentration (E) or 1 μg for indicated periods (F), followed by immunoblotting assay. TCL, total cell lysates.
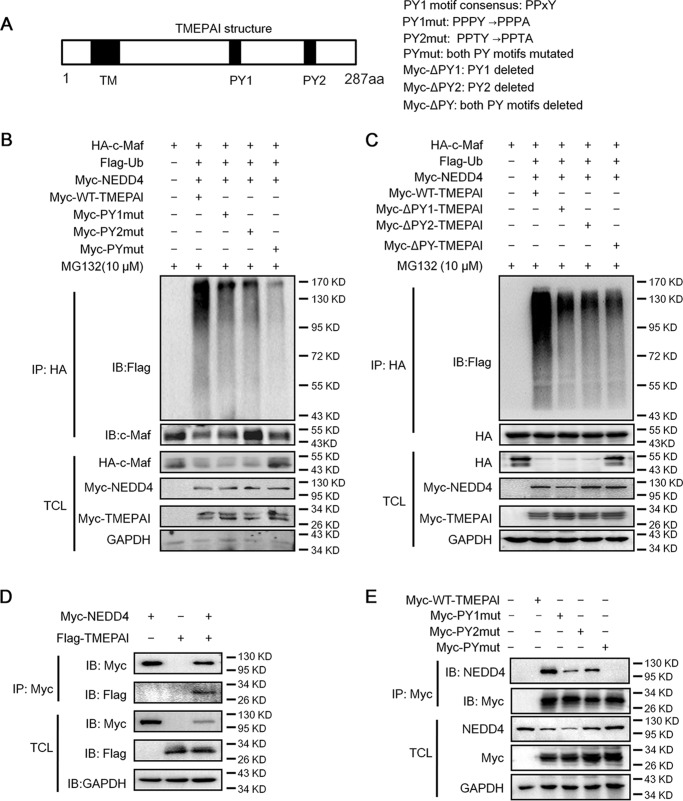
The PY motifs are essential for TMEPAI to mediate c-Maf degradation
TMEPAI is featured with two PY motifs with a consensus sequence PPXY (Fig. 5A) through which TMEPAI interacts with other proteins (9). To find out their importance in c-Maf stability, the PY motifs were mutated by replacing Tyr to Ala or deleted as shown in Fig. 5A, followed by measurement of c-Maf ubiquitination. As shown in Fig. 5B, wtTMEPAI markedly increased c-Maf polyubiquitination, whereas the ubiquitination level of c-Maf was significantly reduced when PY1 or PY2 was mutated. Moreover, when both PY1 and PY2 were mutated, the ubiquitination level of c-Maf was almost completely abolished. To confirm this finding, we next deleted the PY motifs (ΔPY; Fig. 5A), followed by transfection and measurement of c-Maf ubiquitination. As shown in Fig. 5C, when PY motifs were deleted, TMEPAI lost its activity in mediating c-Maf ubiquitination. This result was consistent with the mutation study. Therefore, these results suggested that the PY motifs were indispensable for TMEPAI in mediating c-Maf ubiquitination.
Figure 5.
The PY motifs are essential for TMEPAI-mediated c-Maf ubiquitination. A, the PY motifs in TMEPAI. To make the PY motif mutation, Tyr was mutated into Ala as indicated. B, the wildtype and PY-mutated TMEPAI plasmids were co-transfected into HEK293T cells with c-Maf, respectively. Twenty-four hours later, the cells were treated with MG132, followed by immunoprecipitation and immunoblotting assay. C, the wildtype or PY-deleted TMEPAI plasmids were co-transfected into HEK293T cells with c-Maf, respectively. Twenty-four hours later, the cells were treated with MG132, followed by immunoprecipitation and immunoblotting assay. D, NEDD4 and TMEPAI plasmids were co-transfected into HEK293T cells; 48 h later, cell lysates were prepared for immunoprecipitation and immunoblotting assay. E, TMEPAI and its PY mutants were transfected into HEK293T cells; 48 h later, cell lysates were prepared for immunoprecipitation and immunoblotting assay. TCL, total cell lysates.
The above studies showed that TMEPAI mediated c-Maf ubiquitination in collaboration with NEDD4; therefore, we wondered whether the PY motifs were important for TMEPAI to interact with NEDD4. To this end, TMEPAI was co-transfected with NEDD4 followed by the IP/IB assay. As shown in Fig. 5D, TMEPAI was found in the NEDD4 complexes. To evaluate whether the PY motifs were essential for the interaction between NEDD4 and TMEPAI, HEK293T cells that express endogenous NEDD4 were transfected with the TMEPAI with mutant PY motifs. As expected, the IP/IB assays showed that NEDD4 interacted with wtTMEPAI (Fig. 5E); however, the NEDD4 levels were markedly decreased when a single PY motif was mutated (Fig. 5E). When both PY motifs were mutated, NEDD4 could not be detected in the immunoprecipitates of TMEPAI (Fig. 5E). Therefore, the PY motifs were critical for TMEPAI to interact with NEDD4. These findings further demonstrated that NEDD4 was required for TMEPAI-mediated c-Maf ubiquitination.
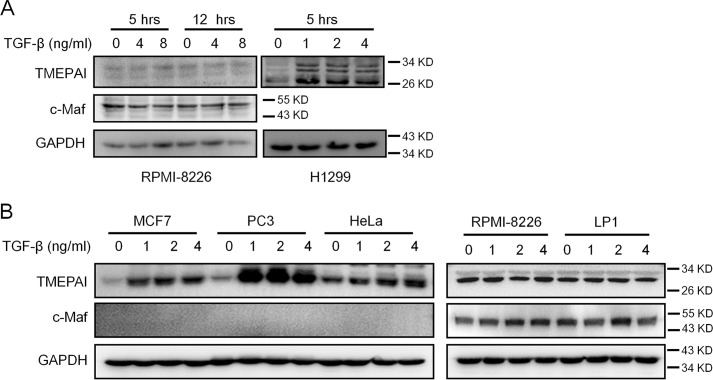
TMEPAI is not induced by TGF-β in myeloma cells
It is known that the expression of TMEPAI is up-regulated by the TGF-β, the key growth factor in the SMAD3 signaling transduction (2, 5). TGF-β–induced TMEPAI in turn promotes the degradation of TGF-β, and SMAD3 therefore contributes to tumorigenesis in lung and breast cancers (3, 5, 11). To find out whether TGF-β could modulate TMEPAI expression in MM cells, the MM cell line RPMI-8226 and lung cancer cell line H1299 were treated with TGF-β (R&D Systems, Shanghai, China). As shown in Fig. 6A, TGF-β induced the expression of TMEPAI in H1299 cells, but it failed to induce TMEPAI expression in RPMI-8226 cells. To confirm this finding, more MM cell lines (RPMI-8226 and LP1) and more solid tumor cell lines including breast cancer cell line MCF7, prostate cancer cell line PC3, and cervical cancer cell line HeLa were examined. Consistent with the above finding, TMEPAI were induced by TGF-β in all solid cancer cells but not in MM cell lines (Fig. 6B). The results thus suggested that TMEPAI activity in MM cells was probably independent of the TGF-β signaling pathway.
Figure 6.
TEMPAI is not modulated by TGF-β in MM cells. A, MM cell line RPMI-8226 and lung cancer cell line H1299 were treated with TGF-β for 5 or 12 h, followed by cell lysates preparation and Western blotting assay. B, MM cell lines (RPMI-8226 and LP1) and solid tumor cell lines (breast cancer cell line MCF7, prostate cancer cell line PC3, and cervical cancer cell line HeLa) were treated with increased TGF-β for 12 h, followed by immunoblotting assay against TMEPAI.
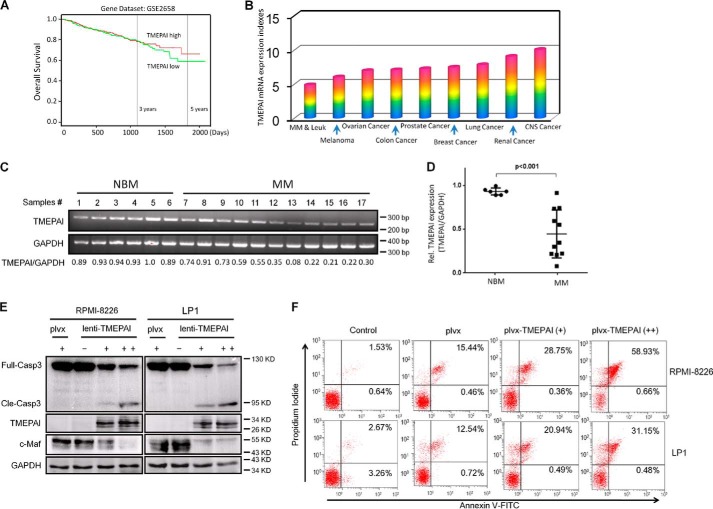
Overexpression of TMEPAI induces MM cell apoptosis
TMEPAI was shown to promote tumorigenesis, especially in lung, renal, and breast cancers (4, 5, 7, 11). In contrast, TMEPAI acts as a tumor suppressor in prostate cancer (8, 9). To find out the effects of TMEPAI on the survival of patients with MM, a fetal hematological malignancy, we analyzed the PROGgene database (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php) (20),5 which curates a large size of reported data on various cancer genes and associated patient survival. There was one confirmative data set (GSE2658) (21) regarding the prognostic value of TMEPAI expression and MM patients. It showed that the expression level of TMEPAI was positively associated with the overall survival periods of MM patients. As shown in Fig. 7A, the higher the TMEPAI expression, the higher the overall survival rate, especially for survival of >1500 days. This result suggested TMEPAI might be important for the survival of MM patients. Subsequently, TMEPAI expression was evaluated in cancer cell lines based on the DNA microarray on the typical 60 cancer cell lines from the National Cancer Institute (NCI 60 cell line) with a Dataset ID of GSE32474. As reported previously (7), it revealed that TMEPAI was high in solid cancers including lung, breast, and renal cancer (Fig. 7B); in contrast, TMEPAI was least expressed in MM and leukemia cell lines among these 60 cancer cell lines (Fig. 7B). To confirm this finding, primary MM cells were applied for RT-PCR assay. As shown in Fig. 7 (C and D), TMEPAI was higher in healthy bone marrow cells but markedly reduced in those cells from MM patients, which suggested that TMEPAI expression was down-regulated in MM cells. Because TMEPAI induces the degradation of c-Maf, a key oncogene in MM, we wondered whether TMEPAI induced MM cell death. To this end, MM cells were infected with lentiviral TMEPAI followed by apoptotic analysis. As shown in Fig. 7E, expression of TMEPAI led to caspase-3 cleavage in RPMI-8226 and LP1, two typical MM cell lines, in association with c-Maf degradation (Fig. 7E). Next we measured the apoptosis of MM cells infected with lentiviral TMEPAI by flow cytometry. As shown in Fig. 7F, the fraction of annexin V-positive cells were markedly increased in MM cells infected with lentiviral TMEPAI. Because annexin V staining is a gold standard for detection of apoptotic cells, all the above data therefore collectively suggested that TMEPAI acts as a tumor suppressor in MM and that re-expression of TMEPAI induced MM cell apoptosis in association with c-Maf degradation.
Figure 7.
TMEPAI overexpression leads to myeloma cell apoptosis in association with c-Maf degradation. A, the TMEPAI expression level was negatively associated with the overall survival of MM patients based on the prognostic gene analysis (PROGgene database, http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php).5 B, TMEPAI is down-regulated in myeloma cells based the DNA microarray analysis on the typical 60 cancer cell lines from National Cancer Institute (NCI 60 cell line). C, TMEPAI expression was analyzed by RT-PCR in bone marrow cells from healthy donors (normal bone marrow (NBM)) and MM patients. D, the densitometric analysis of TMEPAI from C. E, myeloma cell lines RPMI-8226 and LP1 were infected with lentiviral TMEPAI for 96 h, followed by immunoblotting assay against specific antibodies. F, myeloma cell lines were infected with lentiviral TMEPAI for 96 h followed by annexin V/propidium iodide staining and flow cytometric analysis. plvx, the lentivirus without TMEPAI gene.
Discussion
The transcription factor c-Maf is an oncogene in myelomagenesis (15) and an independent factor in the poor prognosis of MM patients. The c-Maf stability was modulated via the ubiquitin-proteasome pathway (17). The present study demonstrated that the transmembrane protein TMEPAI promotes c-Maf ubiquitination and degradation by recruiting the ubiquitin ligase NEDD4. We also found that ectopic expression of TMEPAI leads to MM cell apoptosis.
TMEPAI has been reported to be involved in the degradation of several important proteins, such as TGF-β type I receptor (TβRI) (11) and androgen receptor (AR) (22). However, the specific mechanisms are different. TEMPAI mediates TβRI degradation in lysosomes (11) but mediates AR degradation in proteasomes (22). TMEPAI induces the sequestration of TβRI in lysosomes, which is possibly associated with TMEPAI subcellular location because TMEPAI was found to be localized to the lysosomal membrane and increases lysosomal stability (11, 23). In our present study, TMEPAI-mediated c-Maf degradation is via the ubiquitin-proteasome pathway in a manner similar to AR degradation because TMEPAI mediates c-Maf polyubiquitination, and TMEPAI-induced c-Maf degradation could be abolished by proteasomal inhibitors MG132 and bortezomib but not by lysosomal inhibitors chloroquine and bafilomycin A (Fig. 2). However, TMEPAI is a transmembrane protein, and we found the TM domain is essential for TMEPAI to induce c-Maf ubiquitination and degradation; consistently when the TM domain is deleted, TMEPAI completely loses its activity. This finding suggests that TMEPAI should be localized to the plasma membrane to exert its activity. To our surprise, there is no direct interaction between TMEPAI and c-Maf (dada not shown), suggesting that TMEPAI could recruit another protein (such as ubiquitin ligases) for c-Maf ubiquitination and degradation, which is different from our previous studies in which the ubiquitin-conjugating enzyme UBE2O (18) or the ubiquitin ligase HERC4 (13) could directly mediate c-Maf polyubiquitination and degradation. This is reasonable because TMEPAI is not a ubiquitin enzyme, and it lacks ubiquitination activity, which is different from UBE2O and HERC4. As reported earlier, TMEPAI activity is dependent of its partner NEDD4 because only the presence of NEDD4 significantly enhances TMEPAI-induced c-Maf polyubiquitination (Fig. 3). Compared with the published reports, TMEPAI-induced c-Maf degradation is in a similar manner to AR degradation (10). This could be explained by the specific structure and direct interaction between TMEPAI and NEDD4. It is predicted that TMEPAI interacts with WW domain–containing E3 ligases (11); however, our findings showed that NEDD4 but not other WW-containing proteins such as ITCH, WWP1, or WWP2 enhances TMEPAI activity. Moreover, the interaction between TMEPAI and NEDD4 was also supported by the finding that TMEPAI fails to interact with NEDD4 when the PY motifs are mutated. In accordance with this finding, PY-mutant or PY-deleted TMEPAI loses its activity in c-Maf degradation. This finding is different from a previous report in which all of the TMEPAI mutants lacking the PY motifs (TMEPAIΔPY1, TMEPAIΔPY2, and TMEPAIΔPY) retained their inhibitory ability of TGF-β signaling (11). Therefore, the TMEPAI/NEDD4 forms a complex ubiquitin ligase to induce c-Maf ubiquitination and modulates c-Maf stability via the proteasomal pathway.
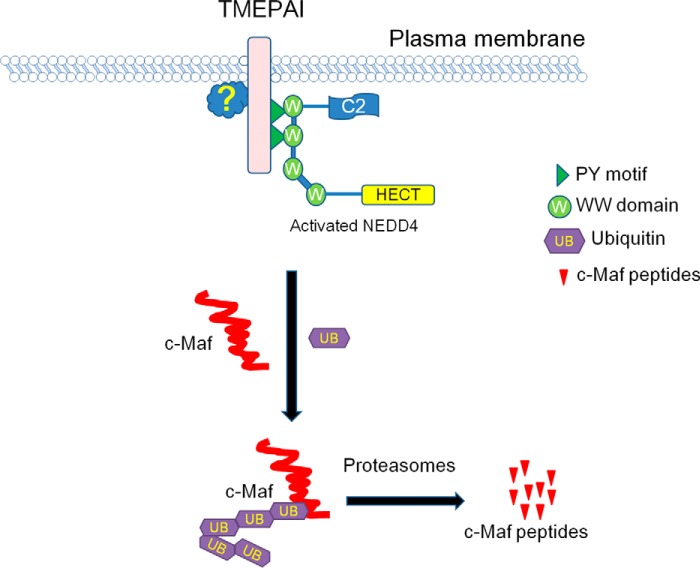
However, why does NEDD4 or TMEPAI alone fail to mediate c-Maf ubiquitination? Why is the TM domain essential for TMEPAI in c-Maf ubiquitination? Why does TMEPAI have to recruit NEDD4 in c-Maf ubiquitination? We may find this answer from a recent study demonstrating that NEDD4 exists as both activated and inactivated forms (24). Persaud et al. (24) found that inactivated NEDD4 is activated after phosphorylation by the kinase c-Src, which is activated by receptor tyrosine kinases such as fibroblast growth factor receptor 1 or epidermal growth factor receptor at plasma membrane. By summarizing our findings in the present study, we proposed an acting model for TMEPAI in mediating c-Maf ubiquitination by recruiting NEDD4 as shown in Fig. 8. Similar to the finding by Persaud et al. (24), to mediate c-Maf ubiquitination, TMEPAI brings NEDD4 to the inner side of the plasma membrane in which NEDD4 is activated by phosphorylation or other as-yet-unknown factors. When NEDD4 is activated, it is able to induce c-Maf ubiquitination and subsequent degradation in the proteasomes. From this model, it is easy to understand that 1) the TM domain is essential for TMEPAI in c-Maf ubiquitination and 2) NEDD4 is required for TMEPAI-mediated c-Maf degradation. Therefore, in the TMEPAI/NEDD4 team, TMEPAI interacts with NEDD4 and leads to NEDD4 activation, and activated NEDD4 is the actual executor in c-Maf ubiquitination. Of course, the hypothesis has to be further demonstrated.
Figure 8.
The proposed model for TMEPAI-induced c-Maf ubiquitination and degradation in partner with NEDD4. TMEPAI on the plasma membrane interacts with the WW domain on NEDD4 via its PY motifs, which leads to NEDD4 activation by phosphorylation or an unknown mechanism in association with the plasma membrane. Activated NEDD4 thus mediates c-Maf ubiquitination and subsequent degradation in the proteasomes.
The above data collectively demonstrated that TMEPAI mediates c-Maf polyubiquitination and promotes its degradation by partnering with NEDD4. Because c-Maf is an oncogenic transcription factor in MM and plays a critical role in regulating MM cell proliferation, cell cycle progress, and MM cell adhesion to bone marrow, therefore rendering MM cell survival (16), TMEPAI is thus considered as a tumor suppressor that neutralizes the oncogenic function of c-Maf. This hypothesis was convinced by overexpression of TMEPAI in MM cells and resultant MM cell apoptosis. Previous studies showed that TMEPAI does not induce apoptosis of prostate cancer cells but inhibits their proliferation (8, 25) and prevents their metastasis (10). This finding is contradicted with its role in some solid cancers, such as lung, ovarian, and breast cancers (5, 6, 11). The discrepancies in TMEPAI cancer biology are probably due to its specific targeted cell signaling pathway. In breast cancer, TMEPAI promotes breast cancer cell proliferation by converting TGF-β from a tumor suppressor to a tumor promoter (5), whereas in lung cancer cells, TMEPAI sequestrates R-SMAD and down-regulates PTEN, thereby enhancing the phosphatidylinositol 3-kinase/Akt signaling pathway (3). In our present study, TMEPAI-induced MM cell apoptosis is probably associated with c-Maf ubiquitination and degradation because c-Maf promotes MM cell survival while silencing c-Maf leads to MM cell apoptosis (19). Moreover, TMEPAI is markedly down-regulated in MM cells but highly expressed in bone marrow cells from healthy donors. Therefore, TMEPAI is a potential tumor suppressor, but it is suppressed in MM. This finding also demonstrates that TMEPAI could exert as an oncogene or tumor suppressor upon the context and cancer types.
In summary, the present study demonstrates that the membranous TMEPAI protein induces MM cell apoptosis by mediating c-Maf polyubiquitination and degradation by recruiting the ubiquitin ligase NEDD4 but independent of the TGF-β signaling. Induction of TMEPAI could be a potential strategy for MM therapy.
Experimental procedures
Cells and cell culture
Human embryonic kidney cells (HEK293T) were cultured in Dulbecco's modified Eagle's medium. MM cell lines (RPMI-8226 and LP1) were generously provided by Dr. Aaron Schimmer (University of Toronto). MM cells were maintained in Iscove's modified Dulbecco's medium. All the media were supplemented with 10% fetal bovine serum, glutamine, and antibiotics. Primary bone marrow species from healthy donors and MM patients were collected from the Department of Hematology of the First Affiliated Hospital of Soochow University. The use of primary bone marrow cells was approved by the Institutional Review Board of Soochow University. Informed consent was obtained in accordance with the Declaration of Helsinki. Mononuclear cells were isolated by Lympholyte® cell separation (Cedarlane, Burlington, Canada) as described previously (13).
Antibodies and plasmids
The antibodies against c-Maf, caspase-3, and TMEPAI were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), Cell Signaling Technology (Danvers, MA), and Abnova (Taiwan, China), respectively. The antibodies against GAPDH, HA, Myc, and FLAG were purchased from MBL (Tokyo, Japan). The c-Maf, MafA, and MafB plasmids were prepared as described previously (13). The TMEPAI gene was cloned from A549 cells and then subcloned into a pcDNA3.1 vector with a FLAG or a Myc tag. Primers for TMEPAI and its mutants were shown in Table 1. TMEPAI with PY motif mutations, in which the Tyr residue was replaced by the Ala residue, were made with Mut ExpresII fast mutagenesis kit V2 (Vazyme Biotech, Nanjing, China). The NEDD4 gene was first cloned from the genomic DNA of MM cell line RPMI-8226, and it was then subcloned into pcDNA3.1 vector with a Myc tag. The luciferase reporter driven by MARE (5′-TGCGAGTGAGGCA-3′) (pGL4-MARE.Luci) was cloned as described previously (19). The PY motif mutants of TMEPAI were cloned by site-directed mutagenesis using the following primers: 5′-CCCCCACCCGCCCAGGGCCCCTGCACCCTCCAGCTT-3′ (forward) and 5′-GCCCTGGGCGGGTGGGGGCTCCTCCCCGTCTGACAG-3′ (reverse) for the PY1 mutant, 5′-CCGCCCACCGCCAGCGAGGTCATCGGCCACTACCCG-3′ (forward) and 5′-ACCTCGCTGGCGGTGGGCGGCGGCCCCTCCATGCG-3′ (reverse) for the PY2 mutant. To mutate both PY motifs, the above primers were used. For PY1 deletion, the forward primer was 5′-TCAGACGGGGAGGAGCAGGGCCCCTGCACCCTC-3′, and the reverse primer was 5′-CTGCTCCTCCCCGTCTGACAGCG-3′. For PY2 deletion, the forward primer was 5′-GAGCGAGGTCATCGGCCAC-3′ and the reverse primer was 5′-TGGCCGATGACCTCGCTCGGCCCCTCCATGCGCCC-3′. These specific primers were designed using the ClonExpress Design software (Vazyme), followed by cloning using the Mut Express II fast mutagenesis kit V2 (Vazyme). All cloning and mutations were confirmed by sequencing (Genewiz, Inc., Suzhou, China). The shNEDD4 sequence was 5′-GTTGGAGAATGTAGCAATA-3′, designed and provided by Genechem Biotech, Inc. (Shanghai, China). siTMEPAI (5′-GGAGCAAAGAGAAGGATAA-3′) was provided by Ribobio Inc. (Guangzhou, China).
Table 1.
Primers for constructs of TMEPAI truncates
F, forward primer; R, reverse primer.
| Amino acid region | Size | Primers (5′ → 3′) |
|---|---|---|
| bp | ||
| 1–287 | 861 | F: CGGGATCCATGCACCGCTTGATGGGGGTC |
| R: CCGCTCGAGCTAGAGAGGGTGTCCTTTCTG | ||
| 1–237 | 711 | F: CGGGATCCATGCACCGCTTGATGGGGGTC |
| R: CCGCTCGAGCTAGCCGATGACCTCGCTGTA | ||
| 1–216 | 648 | F: CGGGATCCATGCACCGCTTGATGGGGGTC |
| R: CCGCTCGAGCTAGGCGCTGATGCCCGAGTTACT | ||
| 1–171 | 513 | F: CGGGATCCATGCACCGCTTGATGGGGGTC |
| R: CCGCTCGAGCTAGTCCCGAAGCTGGAGGGT | ||
| 65–287 | 669 | F: CGGGATCCATGTACAAGCTGTCTGCACGGTCC |
| R: CCGCTCGAGCTAGAGAGGGTGTCCTTTCTG | ||
| 51–276 | 678 | F: CGGGATCCATGATGGTGATGGTGGTGGTG |
| R: CCGCTCGAGCTATTTGCTCCAGATGGCTGCGCT |
CHX chase assay
After transfected with plasmids of interest for 24 h, HEK293T cells were treated with CHX (100 μg/ml, Sigma–Aldrich) for indicated periods as needed. Cell lysates were then prepared for SDS-PAGE and immunoblotting analyses with specific antibodies.
Luciferase assay
To examine the effect of TMEPAI on c-Maf biological function, the luciferase reporter plasmid pGL4-MARE.Luci was co-transfected with c-Maf, full-length TMEPAI, or ΔTM-TMEPAI into HEK293T cells. Twenty-four hours later, the cell lysates were subjected to luciferase analysis using Bright-Glo system (Promega, Madison, WI) as described previously (18). Luciferase activity was normalized to β-gal expression for each sample. All transfection experiments were performed in duplicate. The detailed method was described previously (18).
Immunoblotting assay
Twenty-four hours after transfection with appropriate plasmids, HEK293T cells were lysed on ice in a lysis buffer as described previously (18). After clarifying at a high speed at 4 °C, protein concentrations were determined by the BCA assay (Pierce). Equal amounts of proteins (30 μg) were fractionated by SDS-PAGE, transferred to PVDF membranes (Millipore), and probed with appropriate antibodies.
RT-PCR
Total RNA was extracted from normal bone marrow and multiple myeloma species using TRIzol (TransGen Biotech, Beijing, China). cDNA preparation was accomplished with 2 μg of RNA using a SuperscriptTM-III kit (Invitrogen) according to the manufacturer's instructions. The PCR amplification primers for TMEPAI were 5′-GAGAAGATGCCCTGTCC-3′ and 5′-GCTGATGCCCGAGTTA-3′, and the primers for GAPDH were 5′-CAAGGTCATCCATGACAACTTTG-3′ and 5′-GTCCACCACCCTGTTGCTGTAG-3′. PCR products were visualized by Goldview staining (TransGen Biotech) after electrophoresis on a 2% agarose gel. The optical densities of the genes were quantified using an image analysis system and ImageJ software (National Institutes of Health, Bethesda, MD).
Immunoprecipitation
HEK293T cells were transfected with specific plasmids as needed. Twenty-four hours later, transfection cells were treated with proteasome inhibitors MG132 (10 μm) or BZ (1 μm) for 12 h. The subsequent immunoprecipitation assay was performed as described previously (18).
TMEPAI lentivirus
Primers for TMEPAI cDNA fragment were designed as follows: 5′-CCGCTCGAGATGCACCGCTTGATGGGGGTC-3′ (forward) and 5′-CGGGATCCCAGAGAGGGTGTCCTTTCTG-3′ (reverse). The TMEPAI cDNA fragment was inserted into pLVX-AcGFP lentiviral vector (Clontech) within the XhoI and BamHI sites. To generate TMEPAI lentiviral particles, HEK293T cells at 80% confluence were transfected with 10 μg of pLVX-AcGFP-TMEPAI, 3.5 μg of VSV-G envelope glycoprotein, 2.5 μg of packaging proteins (Rev), and 6.5 μg of packaging proteins (ΔR8.74) using PEI (Sigma) as gene delivery carrier. After 8 h, the cells were washed and refreshed with DMEM. Forty-eight hours later, the lentiviral particle-enriched supernatant was harvested, filtered, and stored for further use. These lentiviral particles were applied to infect RPMI-8226 and LP1 cells. The pLVX-AcGFP lentiviral particles were used as a mock control in the infection assay.
TMEPAI analyses based on the public gene expression databases
The association of TMEPAI expression with the overall survival of MM patients was analyzed using the PROGgene database (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php) (20).5 The TMEPAI expression profile was analyzed using the DNA microarray on the typical 60 cancer cell lines from National Cancer Institute (NCI 60 cell line) with a Dataset ID of GSE32474.
Flow cytometric analysis
To measure MM cell apoptosis induced by TMEPAI, MM cells were infected with lentiviral TMEPAI for 96 h, followed by annexin V and propidium iodide staining and flow cytometric analysis. The specific protocol was described previously (18).
Statistics
Statistical differences between the control and the experimental groups were analyzed by Student's t test.
Author contributions
Y. D. and Z. Z. data curation; Y. D., Z. X., Q. W., and X. M. formal analysis; Y. D., Y. L., and Z. Z. validation; Y. D., Y. L., Y. X., J. J., Z. Z., Z. X., and B. C. investigation; Y. D., Y. L., B. C., and Q. W. methodology; B. C., Y. Z., and X. M. project administration; Q. W. and Y. Z. resources; Y. Z. and X. M. conceptualization; Y. Z. and X. M. writing-review and editing; X. M. supervision; X. M. funding acquisition; X. M. writing-original draft.
Acknowledgments
We thank Dr. Guoqiang Xu (Soochow University, Suzhou, China) and Dr. Michael F. Moran (Hospital for Sick Children, University of Toronto, Toronto, Canada) for critical discussions for this study.
This work was partly supported by National Natural Science Foundation of China Grants 81320108023 (to X. M.), 81600171 (to Z. Z.), and 81770215 (to B. C.), by funds from the Priority Academic Program Development of Jiangsu Higher Education Institutions (to X. M.), and by Jiangsu Key Laboratory for Neuropsychiatric Diseases Grant BK2013003 (to X. M.). This work was also supported in part by Suzhou Key Laboratory for Pediatric Leukemia Grant SZS201615 (to X. M.) and by Suzhou Key Medical Center Grant Szzx201506 (to Y. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TM
- transmembrane
- BZ
- bortezomib
- CHX
- cycloheximide
- MM
- multiple myeloma
- TGF-β
- transforming growth factor-β
- IP
- immunoprecipitation
- IB
- immunoblotting
- TβRI
- TGF-β type I receptor
- AR
- androgen receptor
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Xu L. L., Shanmugam N., Segawa T., Sesterhenn I. A., McLeod D. G., Moul J. W., and Srivastava S. (2000) A novel androgen-regulated gene, PMEPA1, located on chromosome 20q13 exhibits high level expression in prostate. Genomics 66, 257–263 10.1006/geno.2000.6214 [DOI] [PubMed] [Google Scholar]
- 2. Azami S., Vo Nguyen T. T., Watanabe Y., and Kato M. (2015) Cooperative induction of transmembrane prostate androgen induced protein TMEPAI/PMEPA1 by transforming growth factor-β and epidermal growth factor signaling. Biochem. Biophys. Res. Commun. 456, 580–585 10.1016/j.bbrc.2014.11.107 [DOI] [PubMed] [Google Scholar]
- 3. Singha P. K., Pandeswara S., Geng H., Lan R., Venkatachalam M. A., and Saikumar P. (2014) TGF-β induced TMEPAI/PMEPA1 inhibits canonical Smad signaling through R-Smad sequestration and promotes non-canonical PI3K/Akt signaling by reducing PTEN in triple negative breast cancer. Genes Cancer 5, 320–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu Y., He K., Wang D., Yuan X., Liu Y., Ji H., and Song J. (2013) TMEPAI regulates EMT in lung cancer cells by modulating the ROS and IRS-1 signaling pathways. Carcinogenesis 34, 1764–1772 10.1093/carcin/bgt132 [DOI] [PubMed] [Google Scholar]
- 5. Singha P. K., Yeh I. T., Venkatachalam M. A., and Saikumar P. (2010) Transforming growth factor-β (TGF-β)-inducible gene TMEPAI converts TGF-β from a tumor suppressor to a tumor promoter in breast cancer. Cancer Res. 70, 6377–6383 10.1158/0008-5472.CAN-10-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. ÓhAinmhire E., Quartuccio S. M., Cheng W., Ahmed R. A., King S. M., and Burdette J. E. (2014) Mutation or loss of p53 differentially modifies TGFβ action in ovarian cancer. PLoS One 9, e89553 10.1371/journal.pone.0089553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rae F. K., Hooper J. D., Nicol D. L., and Clements J. A. (2001) Characterization of a novel gene, STAG1/PMEPA1, upregulated in renal cell carcinoma and other solid tumors. Mol. Carcinog. 32, 44–53 10.1002/mc.1063 [DOI] [PubMed] [Google Scholar]
- 8. Li H., Mohamed A. A., Sharad S., Umeda E., Song Y., Young D., Petrovics G., McLeod D. G., Sesterhenn I. A., Sreenath T., Dobi A., and Srivastava S. (2015) Silencing of PMEPA1 accelerates the growth of prostate cancer cells through AR, NEDD4 and PTEN. Oncotarget 6, 15137–15149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu L. L., Shi Y., Petrovics G., Sun C., Makarem M., Zhang W., Sesterhenn I. A., McLeod D. G., Sun L., Moul J. W., and Srivastava S. (2003) PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Res. 63, 4299–4304 [PubMed] [Google Scholar]
- 10. Fournier P. G., Juárez P., Jiang G., Clines G. A., Niewolna M., Kim H. S., Walton H. W., Peng X. H., Liu Y., Mohammad K. S., Wells C. D., Chirgwin J. M., and Guise T. A. (2015) The TGF-β signaling regulator PMEPA1 suppresses prostate cancer metastases to bone. Cancer Cell 27, 809–821 10.1016/j.ccell.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bai X., Jing L., Li Y., Luo S., Wang S., Zhou J., Liu Z., and Diao A. (2014) TMEPAI inhibits TGF-β signaling by promoting lysosome degradation of TGF-β receptor and contributes to lung cancer development. Cell Signal. 26, 2030–2039 10.1016/j.cellsig.2014.06.001 [DOI] [PubMed] [Google Scholar]
- 12. Vo Nguyen T. T., Watanabe Y., Shiba A., Noguchi M., Itoh S., and Kato M. (2014) TMEPAI/PMEPA1 enhances tumorigenic activities in lung cancer cells. Cancer Sci. 105, 334–341 10.1111/cas.12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Z., Tong J., Tang X., Juan J., Cao B., Hurren R., Chen G., Taylor P., Xu X., Shi C. X., Du J., Hou J., Wang G., Wu D., Stewart A. K., Schimmer A. D., Moran M. F., and Mao X. (2016) The ubiquitin ligase HERC4 mediates c-Maf ubiquitination and delays the growth of multiple myeloma xenografts in nude mice. Blood 127, 1676–1686 10.1182/blood-2015-07-658203 [DOI] [PubMed] [Google Scholar]
- 14. Chang H., Qi Q., Xu W., and Patterson B. (2007) c-Maf nuclear oncoprotein is frequently expressed in multiple myeloma. Leukemia 21, 1572–1574 10.1038/sj.leu.2404669 [DOI] [PubMed] [Google Scholar]
- 15. Morito N., Yoh K., Maeda A., Nakano T., Fujita A., Kusakabe M., Hamada M., Kudo T., Yamagata K., and Takahashi S. (2011) A novel transgenic mouse model of the human multiple myeloma chromosomal translocation t(14;16)(q32;q23). Cancer Res. 71, 339–348 10.1158/1538-7445.AM2011-339,10.1158/0008-5472.CAN-10-1057 [DOI] [PubMed] [Google Scholar]
- 16. Hurt E. M., Wiestner A., Rosenwald A., Shaffer A. L., Campo E., Grogan T., Bergsagel P. L., Kuehl W. M., and Staudt L. M. (2004) Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 5, 191–199 10.1016/S1535-6108(04)00019-4 [DOI] [PubMed] [Google Scholar]
- 17. Chen G., Xu X., Tong J., Han K., Zhang Z., Tang J., Li S., Yang C., Li J., Cao B., Zhou H., Wu D., Moran M. F., and Mao X. (2014) Ubiquitination of the transcription factor c-MAF is mediated by multiple lysine residues. Int. J. Biochem. Cell Biol. 57, 157–166 10.1016/j.biocel.2014.10.024 [DOI] [PubMed] [Google Scholar]
- 18. Xu Y., Zhang Z., Li J., Tong J., Cao B., Taylor P., Tang X., Wu D., Moran M. F., Zeng Y., and Mao X. (2017) The ubiquitin-conjugating enzyme UBE2O modulates c-Maf stability and induces myeloma cell apoptosis. J. Hematol. Oncol. 10, 132 10.1186/s13045-017-0499-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang S., Juan J., Zhang Z., Du Y., Xu Y., Tong J., Cao B., Moran M. F., Zeng Y., and Mao X. (2017) Inhibition of the deubiquitinase USP5 leads to c-Maf protein degradation and myeloma cell apoptosis. Cell Death Dis. 8, e3058 10.1038/cddis.2017.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goswami C. P., and Nakshatri H. (2013) PROGgene: gene expression based survival analysis web application for multiple cancers. J. Clin. Bioinforma 3, 22 10.1186/2043-9113-3-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhan F., Huang Y., Colla S., Stewart J. P., Hanamura I., Gupta S., Epstein J., Yaccoby S., Sawyer J., Burington B., Anaissie E., Hollmig K., Pineda-Roman M., Tricot G., van Rhee F., Walker R., Zangari M., Crowley J., Barlogie B., and Shaughnessy J. D. Jr. (2006) The molecular classification of multiple myeloma. Blood 108, 2020–2028 10.1182/blood-2005-11-013458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H., Xu L. L., Masuda K., Raymundo E., McLeod D. G., Dobi A., and Srivastava S. (2008) A feedback loop between the androgen receptor and a NEDD4-binding protein, PMEPA1, in prostate cancer cells. J. Biol. Chem. 283, 28988–28995 10.1074/jbc.M710528200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo S., Yang M., Lv D., Jing L., Li Y., Liu Z., and Diao A. (2016) TMEPAI increases lysosome stability and promotes autophagy. Int. J. Biochem. Cell Biol. 76, 98–106 10.1016/j.biocel.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 24. Persaud A., Alberts P., Mari S., Tong J., Murchie R., Maspero E., Safi F., Moran M. F., Polo S., and Rotin D. (2014) Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci. Signal. 7, ra95 10.1126/scisignal.2005290 [DOI] [PubMed] [Google Scholar]
- 25. Liu R., Zhou Z., Huang J., and Chen C. (2011) PMEPA1 promotes androgen receptor-negative prostate cell proliferation through suppressing the Smad3/4-c-Myc-p21 Cip1 signaling pathway. J. Pathol. 223, 683–694 10.1002/path.2834 [DOI] [PubMed] [Google Scholar]