Abstract
Recently, a new class of HIV-1 integrase (IN) inhibitors with a dual mode of action, called IN-LEDGF/p75 allosteric inhibitors (INLAIs), was described. Designed to interfere with the IN-LEDGF/p75 interaction during viral integration, unexpectedly, their major impact was on virus maturation. This activity has been linked to induction of aberrant IN multimerization, whereas inhibition of the IN-LEDGF/p75 interaction accounts for weaker antiretroviral effect at integration. Because these dual activities result from INLAI binding to IN at a single binding site, we expected that these activities co-evolved together, driven by the affinity for IN. Using an original INLAI, MUT-A, and its activity on an Ala-125 (A125) IN variant, we found that these two activities on A125-IN can be fully dissociated: MUT-A–induced IN multimerization and the formation of eccentric condensates in viral particles, which are responsible for inhibition of virus maturation, were lost, whereas inhibition of the IN-LEDGF/p75 interaction and consequently integration was fully retained. Hence, the mere binding of INLAI to A125 IN is insufficient to promote the conformational changes of IN required for aberrant multimerization. By analyzing the X-ray structures of MUT-A bound to the IN catalytic core domain (CCD) with or without the Ala-125 polymorphism, we discovered that the loss of IN multimerization is due to stabilization of the A125-IN variant CCD dimer, highlighting the importance of the CCD dimerization energy for IN multimerization. Our study reveals that affinity for the LEDGF/p75-binding pocket is not sufficient to induce INLAI-dependent IN multimerization and the associated inhibition of viral maturation.
Keywords: allosteric regulation, crystal structure, human immunodeficiency virus (HIV), inhibitor, integrase, viral replication, INLAI, LEDGF, multimerization, polymorphism
Introduction
The integrase (IN)9 protein of human immunodeficiency virus type 1 (HIV-1) catalyzes the stable insertion of the viral cDNA genome into the host cell chromatin, a step of the viral life cycle that is required for efficient viral gene expression. Integration occurs via a two-step reaction where IN initially cleaves after a conserved CA dinucleotide at the 3′ end of the viral cDNA genome to free a 3′-OH group (3′-processing), which is next used to carry out a nucleophilic attack on cellular chromosomal DNA (strand transfer).
IN is one of the preferred targets for the development of antiretroviral (ARV) drugs. Inhibitors of IN currently used to treat HIV-1-infected individuals (raltegravir (RAL), elvitegravir (EVG), and dolutegravir (DTG)) bind to the catalytic site and block the strand transfer activity, and they are thus collectively named IN Strand Transfer Inhibitors (INSTIs) (1).
However, given the high genetic variability of HIV-1, IN mutations conferring cross-resistance to the first generation INSTIs, RAL and EVG, were described in patients receiving INSTI-containing regimens (2). The second generation INSTI DTG has a higher genetic barrier and conserves good ARV activity against a number of RAL- and EVG-resistant strains. Recent reports showed that bictegravir, a second generation INSTI still in development from Gilead Sciences, has a resistance profile similar to DTG (3). Nevertheless, DTG and bictegravir are sensitive to the most detrimental INSTI-resistant mutations, albeit at lower levels than first generation INSTIs (4). Therefore, the development of small molecule inhibitors impairing IN functions with distinct mechanisms of action and that conserve full ARV activity on all INSTI-resistant strains is an important objective.
Efficient integration of the HIV-1 genome requires the interaction between IN and LEDGF/p75, a host cell chromatin-associated protein (5, 6), which tethers the viral pre-integration complex at preferred genomic insertion sites (7). Solution of the three-dimensional structure of the LEDGF-binding pocket of IN was recently exploited for the preclinical development of a new class of IN inhibitors. From a chemical point of view, these compounds share a common chain composed of a tert-butyl ether and a carboxylic acid group linked to a wide variety of scaffolds, notably quinoline, naphthalene, benzene, or pyrimidine (8). Many names have been proposed for this new class of IN inhibitors such as LEDGINs, the first inhibitors of this class reported (9), allosteric IN inhibitors (ALLINIs) (10), noncatalytic IN inhibitors (11–13), multimerization IN inhibitors (MINIs) (14), or IN-LEDGF allosteric inhibitors (INLAIs) (15). In the absence of a general consensus name, this latter acronym will be used throughout this article because it has the advantage to recall the dual mode of action of these compounds. Initially designed to prevent the IN-LEDGF/p75 interaction, it was later evidenced that these molecules have an additional biochemical activity based on the induction of allosteric conformational changes of IN that ultimately trigger its aberrant multimerization (10–12, 15–19). This effect is independent of the presence of LEDGF/p75 and the viral DNA and results from the binding of the inhibitors to the IN dimer interface that is also part of the LEDGF-binding pocket.
Given their dual biochemical activities, INLAIs were shown to block HIV-1 replication at two different steps: the inhibition of IN-LEDGF/p75-binding accounts for an “early” block at integration, and the INLAI-promoted IN multimerization results in a “late” effect during virus maturation (11, 12, 17, 18).
Vranckx et al. (20) have shown that LEDGF/p75 depletion hampers HIV-1 reactivation in cell culture, and they demonstrated that LEDGINs relocate and retarget HIV integration, resulting in an HIV reservoir that is refractory to reactivation by different latency-reversing agents.
HIV-1 virions produced in the presence of INLAIs are noninfectious because they are unable to complete reverse transcription upon target cell infection (12, 17, 18). Investigating the molecular bases of the observed infectivity defects, we found that HIV-1 virions produced in the presence of the quinoline INLAI compound BI-D (developed by Boehringer Ingelheim) package normal levels of genomic RNA dimer and harbor a properly placed tRNALys-3 primer that could be extended ex vivo. In addition, reverse transcriptase extracted from these virions is fully active (21).
EM images show that HIV-1 viral particles produced from INLAI-treated cells contain aberrant cores, from which the viral ribonucleoprotein complex is excluded, leading to the formation of “eccentric condensates” with high nucleocapsid content outside the core (22). Their compactness and density distinguish condensates from the other material that typically occupies the space between the core and the viral envelope (22), which are mostly nonpolymerized capsid (23, 24) and host cell proteins (25). Recently, Kessl et al. (26) reported that IN binds the viral RNA genome inside virions and that INLAIs preclude this interaction required for proper viral particle morphogenesis. Madison et al. (27) showed that upon infection with aberrant eccentric virions, IN and genomic RNA that are not protected in the capsid core undergo rapid degradation, which likely accounts for the reverse transcription defect.
Here, we characterize a new type of INLAI, MUT-A. MUT-A shares with all previously described INLAIs a key chain composed of a tert-butyl ether moiety linked to a carboxylic acid group, but it is based on an original scaffold, a 5-membered thiophene ring (Table 1). We studied the molecular mechanism of action of MUT-A on the inhibition of IN-LEDGF/p75 interaction and induction of IN multimerization, and we characterized its ARV activity. MUT-A also influences the appearance of eccentric condensates in the viral particles as visualized by cryo-EM.
Table 1.
Studied compounds, their in vitro biochemical and antiviral activities, and cytotoxicity
Means of at least three independent experiments ± S.D. IBD is Integrase-binding domain of LEDGF/p75.
We also studied the influence of the IN hot spot polymorphism at amino acid residues 124/125 on the activity of MUT-A and other INLAIs. Our findings reveal the importance of this polymorphism in INLAI-induced IN multimerization in correlation with the ARV activity of these compounds.
Results
Biochemical and antiretroviral activities of MUT-A, compared with reference INLAI compounds
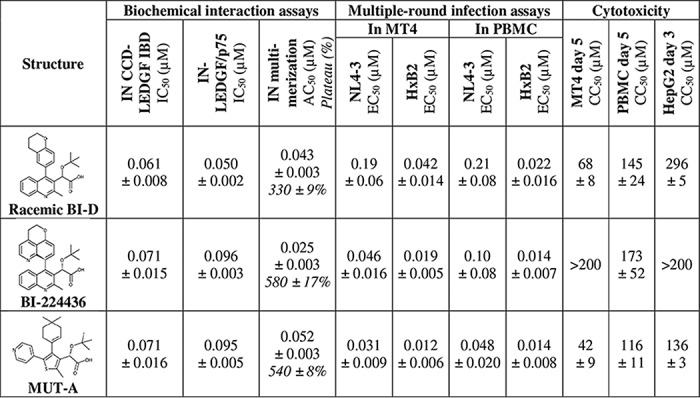
Optimization by medicinal chemistry of the Mut101 series (15) led to the identification of a novel family of potent INLAIs with low EC50 values of ARV activity. These compounds harbor a tert-butoxy acetic acid chain, as all previously described INLAIs, which is crucial for the ARV potency. Rescaffolding of the described INLAIs led to the discovery of an original thienyl series that allows the exploration of a new chemical space. We identified MUT-A lead compound, substituted by a methyl, a gem-dimethylcyclohexenyl, and a 4-pyridinyl at positions 2, 4, and 5, respectively, on the thienyl core (Table 1). MUT-A shows potent in vitro biochemical inhibition of IN-LEDGF/p75 interaction with an IC50 of 95 nm and induces IN multimerization with an activation concentration (AC50) of 52 nm, as determined by homogeneous time-resolved fluorescence (HTRF) assays. These biochemical activities were comparable with those of previously described INLAI compounds BI-D and BI-224436 (Table 1). We also checked by cryo-electron microscopy (cryo-EM) that MUT-A treatment during production of HIV-1 induced the formation of virus particles containing aberrant cores, from which the viral ribonucleoprotein complex is excluded, leading to the formation of eccentric condensates (28). In multiple-round antiviral assays on MT4 cells infected with the HIV-1 NL4-3 strain, MUT-A has a strong ARV activity with a 31 nm EC50, slightly more potent than BI-224436, and is roughly 6-fold more efficient than the BI-D racemate (EC50 = 0.19 μm). MUT-A ARV activity on MT4 cells infected with HIV-1 HxB2 strain was even more potent than that found in NL4-3 infection with an EC50 of 12 nm. Comparable ARV activities were measured upon infection of activated primary peripheral blood mononuclear cells (PBMC) with NL4-3 or HxB2. MUT-A showed low cellular toxicity with CC50 values of 42 or 116 μm on MT4 cells or PBMC, respectively, and high CC50/EC50 selectivity indexes of ≥1355 (for NL4-3) or ≥3500 (for HxB2) (Table 1).
MUT-A ARV activity is strongly affected by an alanine residue at position 125 in the IN sequence
IN is a highly polymorphic protein particularly at positions 124 and 125, which are hot spots of IN polymorphism (29–31). As shown in Table 2, for all clades the AA combination at position 124/125 is the most frequent (46%), followed by TA and AT that occur in about 15% of the sequences. The TT combination has a frequency below 5%. In contrast, analysis of IN sequences of clade B strains revealed that the TT and AA combinations have a frequency of 32 and 9%, respectively (Table 2). Residues 124 and 125 are located on the edge of the INLAI-binding pocket and interact with the part opposite to the carboxylic acid side chain of these compounds, including MUT-A (see Fig. 4). Thus, we investigated the potential impact of these major IN polymorphic sites on the ARV activity of MUT-A and the other INLAI compounds shown in Table 1. Point mutations encoding Ala-124–Ala-125 (AA), Ala-124–Thr-125 (AT), Thr-124–Ala-125 (TA), Asn-124–Thr-125 (NT), and Asn-124–Ala-125 (NA) polymorphisms were introduced in the NL4-3 molecular clone, and the corresponding viral stocks were used to challenge MT4 cells in multiple-round infection assays. We observed that the EC50 of BI-224436 was not significantly affected by the nature of the 124/125 residues (Table 3A), whereas BI-D racemate was 2–3 times more potent on viruses with an Ala than a Thr residue at position 124 (compare EC50 values of 83 nm for AT versus 0.19 μm for TT and 61 nm for AA versus 0.10 μm for TA). MUT-A more efficiently inhibited viruses bearing an Ala or an Asn residue at position 124 (compare EC50 values of 14 or 27 nm for NL4-3 AT or NT, respectively, versus 31 nm for TT). The slight preference of MUT-A for an Ala residue at position 124 explains the more potent ARV activity of MUT-A on HxB2, which harbors an Ala at position 124, whereas NL4-3 bears a Thr residue at this location. More importantly, MUT-A ARV activity was strongly affected by the presence of an Ala residue at position 125 in the IN sequence, with a 30–100-fold decrease in potency (compare EC50 of 1.5 μm for AA versus 14 nm for AT or 1.6 μm for TA versus 31 nm for TT) (Table 3A). The negative impact of A125-polymorphism on MUT-A ARV activity was confirmed by studying several HIV-1 primary isolates (Table 3B). Yet the EC50 fold-changes between NL4-3 and the primary isolates in these assays on PBMCs were ∼5 times lower than in MT4 infection assays with the NL4-3 strains bearing the corresponding IN 124/125 polymorphisms.
Table 2.
| 124–125 | Frequency |
|---|---|
| % | |
| All clades | |
| AA | 46.0 |
| TA | 15.3 |
| AT | 14.5 |
| NA | 8.0 |
| TT | 4.8 |
| NT | 2.5 |
| Other | 8.9 |
| Clade B | |
| TT | 32.0 |
| AT | 17.0 |
| TA | 15.0 |
| NT | 9.0 |
| AA | 8.1 |
| NA | 4.0 |
| Other | 14.9 |
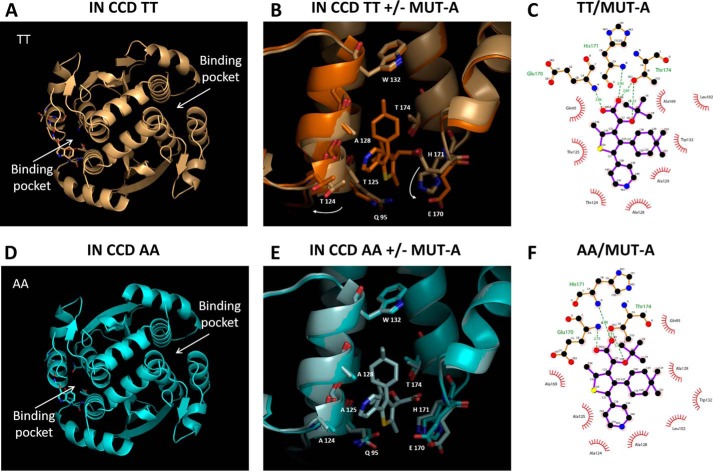
Figure 4.
Structures of IN CCD TT and AA ± MUT-A. A, global view of the IN CCD TT structure. B, superposition of the IN CCD TT ± MUT-A structures, close view of the binding pocket (gold = + ligand; light gold = − ligand). Large displacement of Thr-124 and Glu-170 between + and − MUT-A. C, 2D view of MUT-A interactions with IN CCD TT. D, global view of the IN-CCD AA structure. E, superposition of the IN-CCD AA ± MUT-A structures, close view of the binding pocket (light blue = + ligand, blue = − ligand). F, 2D view of MUT-A interactions with IN-CCD AA.
Table 3.
Antiretroviral activity of INLAIs on polymorphic viruses (A) in MT4 infected by NL4–3 harboring 124–125 polymorphisms or (B) in activated PBMCs infected with HIV-1 laboratory strains or primary isolates harboring 124–125 polymorphisms
Means are of at least three independent experiments ± S.D.
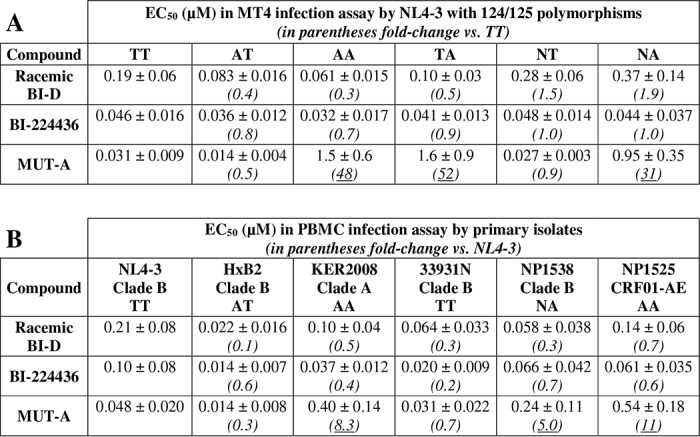
Negative impact of A125-IN polymorphism on MUT-A ARV activity correlates with impairment of IN multimerization but not inhibition of the IN-LEDGF/p75 interaction
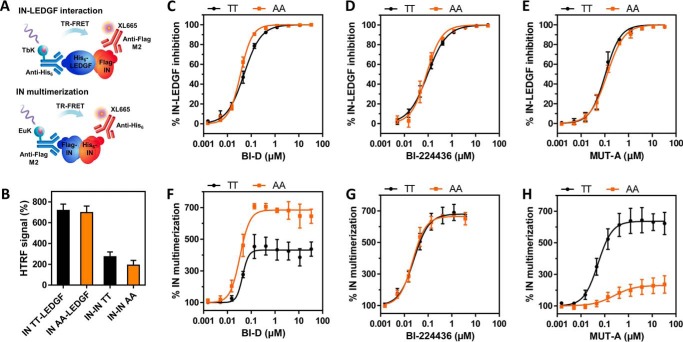
MUT-A like all INLAIs has dual biochemical activity, consisting of the inhibition of the IN-LEDGF/p75 interaction and the promotion of IN multimerization. To establish whether the residue at position 125 affects either activity, we purified recombinant NL4-3 IN proteins harboring TT (corresponding to WT NL4-3 IN sequence) or AA at positions 124/125 and tagged with His6 or FLAG epitopes. Next, the IN-LEDGF/p75 interaction and IN multimerization were assessed by HTRF assays in the presence or absence of MUT-A or reference INLAIs (Fig. 1A). Both IN variants had similar ability to interact with LEDGF/p75 and multimerize in the absence of inhibitor (Fig. 1B). As shown in Fig. 1, C–E, MUT-A, BI-D, and BI-224436 inhibited the binding of both IN variants with LEDGF/p75 to a similar extent. However, the nature of the 124/125 residues strongly affected the extent of MUT-A–induced IN multimerization. MUT-A efficiently promoted IN variant TT oligomerization. In contrast, multimerization of the IN variant AA was inefficient, displaying both a higher AC50 constant and a much lower plateau of maximum multimerization level reached at saturating concentrations of MUT-A (Fig. 1H). These results clearly demonstrate that the weak ARV activity of MUT-A on HIV-1 harboring IN AA 124/125 correlates with strong impairment of IN multimerization and not with the extent of IN-LEDGF/p75 interaction inhibition. Confirming this correlation further, we found that the more potent ARV activity of BI-D on HIV-1 harboring the IN 124/125 AA variant correlated with the ability of this compound to promote a higher level of IN multimerization on this variant. As observed for MUT-A, BI-D equally inhibited the interaction between LEDGF/p75 and either IN variant (Fig. 1F). The increased potency of BI-D to trigger IN multimerization on the IN 124/125 AA variant was not due to a change in the AC50 constant but rather to an ∼2-fold increase in the maximum plateau level reached at saturating concentrations of the compound. As expected, BI-224436 had similar ARV activity on viruses harboring either IN variants and triggered the multimerization of both IN proteins to a similar extent (Fig. 1G).
Figure 1.
Effect of IN 124–125 TT/AA polymorphism on biochemical activities of indicated INLAIs. A, setup of IN-LEDGF/p75 interaction and IN multimerization HTRF assays. B, HTRF signal of interactions with IN TT (WT NL4-3) or AA variant in the absence of inhibitor. C–H, dose-response curves of IN-LEDGF/p75 inhibition (C–E) or IN multimerization (F–H) with IN TT (in black) or IN AA variant (in orange) for the studied compounds. Results shown as average ± S.D. of at least three independent experiments performed in duplicate.
Relationships between the dual biochemical properties of INLAIs and their dual ARV activities
It is believed that the strong ARV effect of INLAIs on virus maturation is mostly linked to the induction of IN multimerization and that the weaker effect at integration is caused by the inhibition of the IN-LEDGF/p75 interaction. We sought to determine whether the differential activities of MUT-A on IN 124/125 AA and TT variants corroborate this theory. To this aim, we extended the analysis to the ARV activity at integration, measured in single-round infection using nonreplicative HIV-1 NL4-3Δenv virions as described under “Experimental procedures.” As shown in Table 4, the ARV activity of MUT-A at integration was similar for both IN TT and AA polymorphs, with an EC50 ratio of 1.4 between the two variants consistent with the IC50 ratio of 1.1 found on the IN-LEDGF/p75 interaction. This result is in sharp contrast with the fold-change of 48 between the ARV activities at late stage estimated by multiple-round infection assays on these variants, which conversely matches with a 14-fold loss in IN multimerization assays (Table 4). These observations further confirm that the ARV activity of INLAIs at integration correlates with their activity on the IN-LEDGF/p75 interaction, whereas their ARV activity at a late stage of HIV replication relates to their ability to promote IN multimerization. In the case of BI-D or BI-224436, the modest gains in ARV activities on the viruses bearing IN AA (fold-changes down to 0.32) are also consistent with the similarly enhanced biochemical activities on this IN variant.
Table 4.
Correlations between biochemical activities and dual antiretroviral activities of compounds
For infection assays, the inhibitor was added to the target MT4 cells and not to the producer 293T cells. Means are of at least three independent experiments ± S.D.
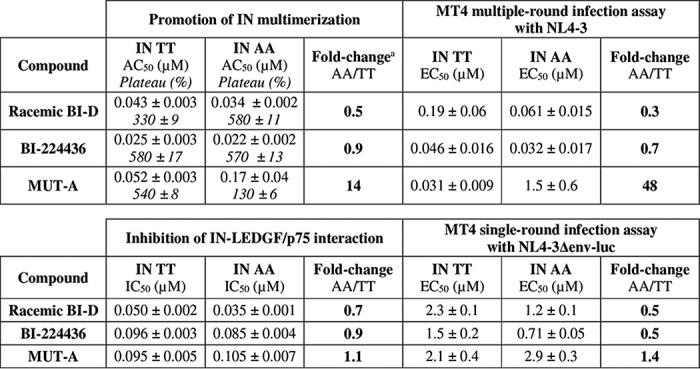
a For IN multimerization, the fold-change takes the AC50 and plateau shifts into account, according to Equation 1.
Lack of MUT-A–induced IN AA 124/125 multimerization correlates with an absence of assembly alterations in the IN AA 124/125 virus variant
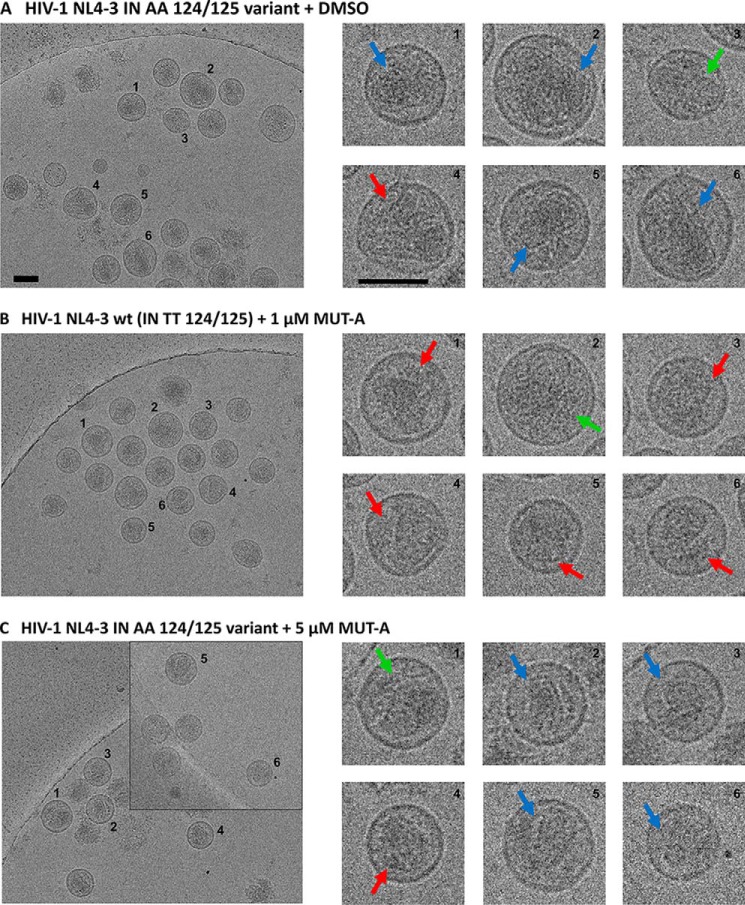
A hallmark of the class of INLAI antiretroviral compounds is the alterations in the virus assembly that are related to IN multimerization. To demonstrate that the lack of MUT-A–induced IN multimerization observed with the AA 124/125 polymorphism at the IN protein level leads to an absence of assembly defects at the virus level, we produced both WT NL4-3 and IN AA 124/125 viruses in the presence or absence of MUT-A. Because the EC50 of MUT-A antiretroviral activity for the AA 124/125 polymorphic virus was 1.5 μm, much higher than that for the WT NL4-3 virus of 31 nm, we produced the IN AA 124/125 virus variant in the presence of 5 μm MUT-A, compared with WT NL4-3 virus produced with 1 μm MUT-A. Although the infectivity of the NL4-3 WT virus was severely impaired by 1 μm MUT-A treatment as shown previously, the virus variant NL4-3 AA 124/125 was only marginally affected by 5 μm MUT-A treatment. Imaging of viral particles by cryo-EM (Fig. 2) reveals that even a concentration of MUT-A of 5 μm was not sufficient to promote in the IN AA 124/125 polymorphic virus (Fig. 2C) a transition to the virus assembly defects observed with WT virus in the presence of 1 μm MUT-A (Fig. 2A), such as the increased occurrence of eccentric condensates outside the viral core (Fig. 2B). Core morphology of the IN AA 124/125 virus variant treated with 5 μm MUT-A (Fig. 2C) was similar to that of the control IN AA 124/125 virus variant produced in the absence of MUT-A (Fig. 2A). Hence, the occurrence of MUT-A–promoted alterations in virus assembly correlates at the protein level with the induction of IN multimerization of WT virus. When MUT-A is unable to promote IN multimerization of IN AA 124/125 (Fig. 1H), assembly alterations at the level of the NL4-3 AA 124/125 virus variant are concomitantly lost.
Figure 2.
Cryo-EM images of HIV-1 NL4-3 WT (IN TT 124/125) and HIV-1 NL4-3 polymorphic IN AA 124/125 virus particles treated in the presence or absence of MUT-A. Red arrows indicate the formation of eccentric condensates; blue arrows indicate normal conical cores; and green arrows show nonconical cores. A, NL4-3 IN AA 124/125 polymorphic virus produced from 293T cells, transfected with pNL4-3 IN AA 124/125, in the presence of DMSO and in the absence of MUT-A (negative control). B, NL4-3 WT (IN TT 124/125) virus produced in the presence of 1 μm MUT-A (positive control showing eccentric condensate). C, NL4-3 IN AA 124/125 polymorphic virus produced in the presence of 5 μm MUT-A. Scale bars, 100 nm.
Impact of modifications at position 5 on the thienyl core of MUT-A on its ARV activity
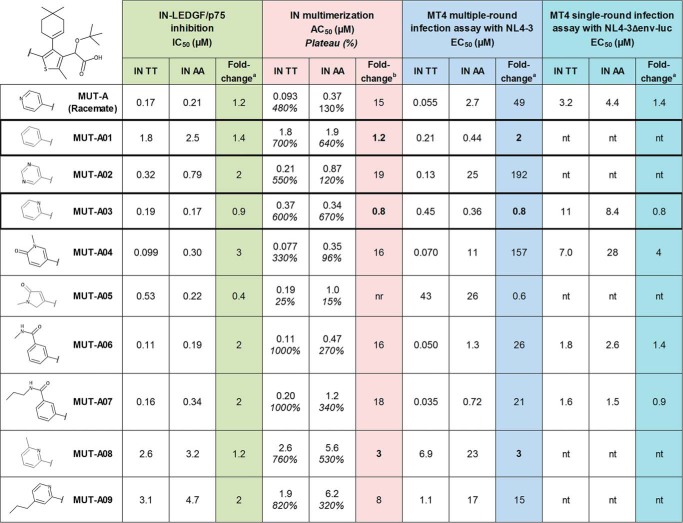
To identify the structural components of MUT-A responsible for its sensitivity to the A125 polymorphism, and eventually to improve the ARV activity of analog compounds on IN polymorphic variants at position 124/125, we explored the impact of the substitution at position 5 on the thienyl core of MUT-A. We considered replacing the 4-pyridyl moiety by regioisomers, other heterocycles, or a phenyl, and adding various substituents. All compounds including MUT-A were synthesized as racemic mixtures. As shown in Fig. 3, substitutions by a 1-methyl-5-oxo-2H-pyrrol-3-yl (MUT-A05), a 6-methyl-2-pyridyl (MUT-A08), or a 4-propyl-2-pyridyl (MUT-A09) had a strong negative effect on the ARV potency in multiple-round antiviral assay on both AA and TT IN variant viruses, which likely results from a 5- to 30-fold decrease in affinity as evidenced by the IN-LEDGF/p75 IC50 values and the IN multimerization AC50 values of these compounds. In addition, MUT-A05 was almost unable to induce IN multimerization (with plateau at saturation below 25%), and thus had the highest EC50 values in multiple-round assay (up to 43 μm). Substitutions by a pyrimidin-5-yl (MUT-A02) or a 1-methyl-6-oxo-3-pyridyl (MUT-A04) significantly affected the ARV activity only on the IN AA variant virus, with a minor effect on the TT polymorph. These molecules had a high IN multimerization fold-change on IN AA variant (19 and 16, respectively). Conversely, compounds substituted by a phenyl (MUT-A01), 2-pyridyl (MUT-A03), a 3-(methylcarbamoyl)phenyl (MUT-A06), or 3-(propylcarbamoyl)phenyl (MUT-A07) had increased ARV activity on the AA variant, from 2.7 μm for MUT-A to 0.44, 0.36, 1.3, and 0.72 μm, respectively. Such improvement in EC50 on the AA variant correlated with an enhanced multimerization of this IN variant, from 130% for the IN multimerization plateau of MUT-A to 640, 670, 270, and 340% for MUT-A01, MUT-A03, MUT-A06, and MUT-A07, respectively. MUT-A01 and MUT-A03 are the most efficient compounds, with equal potency on the multimerization of both IN variants. They also have the best ratio between the AA and the TT IN polymorph with a fold-change in EC50 of 2 and 0.8 for MUT-A01 and MUT-A03, respectively. Interestingly, the improvement in the EC50 of MUT-A03 on the virus bearing IN AA variant was observed only in multiple-round but not in single-round infection assay, demonstrating further that IN multimerization correlates with the potency of the late ARV activity on virus maturation. These findings are also supported by the fact that MUT-A derivatives did not display a significant relative variation in the IC50 on IN-LEDGF/p75 interaction with IN AA or TT variant. Unfortunately, the improved ARV activity on viruses harboring the AA IN variant was accompanied by a variable but significant decrease in the ARV potency on viruses harboring the TT IN variant. In particular, MUT-A03, which has a MUT-A–like affinity for IN (as measured by the IN-LEDGF/p75 IC50), has a 10-fold decrease in ARV potency on the virus bearing IN TT variant. This defect is presumably linked to a lower ability of MUT-A03 to promote multimerization of IN TT protein (with a three times higher AC50).
Figure 3.
Structure-activity relationship study of the substituent at position 5 of the thienyl core in MUT-A series. Shown are activities of racemic MUT-A and analogs with various modifications on the chemical group at position 5 of the thiophene core on IN T124/T125 and A124/A125 variant in biochemical interaction assays and multiple-round and single-round infection assays. Means are from at least three experiments (standard deviation omitted for clarity). The two compounds with better susceptibility of IN AA (fold-change closest to 1) are boxed. a, IC50 or EC50 fold-change AA/TT. b, for IN multimerization, the fold-change takes the AC50 and plateau shifts into account, according to Equation 1. nt, not tested; nr, not reliable (marginal effect).
Structure–function analysis
The results obtained with the chemical series studied in Fig. 3 confirm that the difference in the potency of MUT-A ARV activity on the IN polymorphic variants at positions 124/125 is linked to the nature of the chemical moiety found at position 5 on MUT-A core. To further explore this phenomenon, X-ray structures of IN CCD were solved for the TT and AA variants in the presence or absence of MUT-A or MUT-A03. At first sight, the overall structures were similar, but detailed analysis of the ligand-binding pocket, the dimeric interface, and the predicted ligand-binding free energy explained the effects of MUT-A and MUT-A03 on viral replication and set the structural basis for the design of more efficient INLAIs.
Structure description
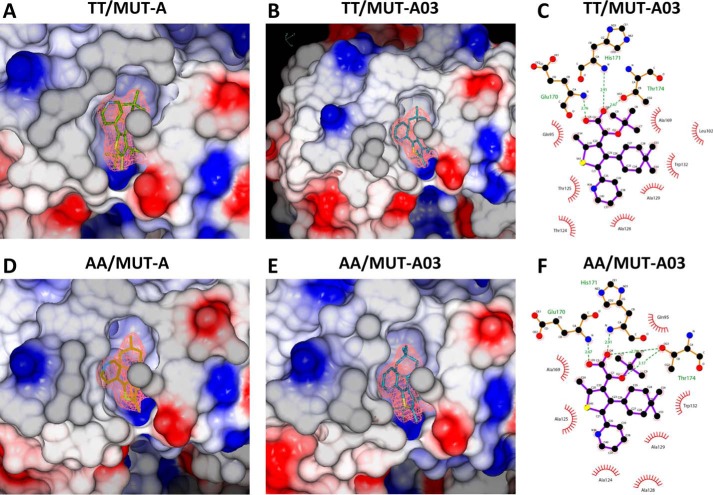
The structure of unliganded IN CCD AA variant was similar to that of the TT variant, including the general topology of the LEDGF/p75-binding site (Fig. 4, A and D). However, the two amino acid substitutions resulted in a more open conformation of the AA site, with the side chains of Gln-95 and Glu-170 flipped outwards, although they are folded in the LEDGF/p75-binding pocket of the TT variant (Fig. 4, B and E). Therefore, whereas large displacements of Thr-124 and Glu-170 are required for the TT variant to accommodate MUT-A (Fig. 4B), there are no major changes in the AA variant (Fig. 4E). Both structures superimpose nicely (Fig. 4E), except for a rotation of the Glu-95 side chain. MUT-A is anchored in the pocket for IN CCD AA and TT through H-bonds with Glu-170, His-171, and Thr-174 (Fig. 4, C and F). Interestingly, the same findings apply to MUT-A03 binding. In particular, there are structural similarities between MUT-A03- and MUT-A-bound IN CCD AA, as well as versus the unliganded protein. However, the pyridine nitrogen of MUT-A is headed toward the solvent (Fig. 4B), whereas for MUT-A03 it is buried in the site (Fig. 5B). The surface of the IN CCD is mainly hydrophobic with basic and acidic patches distributed on the surface (Fig. 5, A and D). The ligand-binding pocket shows an overall positive potential (Fig. 5, A, B, D, and E). As for MUT-A, MUT-A03 is anchored in the pocket for IN CCD AA and TT through H-bonds with Glu-170, His-171, and Thr-174 (Fig. 5, C and F). In the case of IN CCD TT + MUT-A03, there is one missing H-bond (Fig. 5C) present in all other structures (two oxygen atoms of the ligand, which share an H-bond with Thr-174) (Fig. 4, C and F, 5F).
Figure 5.
Surface potential of the ligand-binding pockets of IN TT and AA variants with MUT-A and MUT-A03. A, close view in the binding pocket of the IN CCD TT/MUT-A structure. Negative potential is in red, and positive potential is in blue. B, close view in the IN CCD TT MUT-A03-binding pocket. C, 2D view of MUT-A03 interactions with IN CCD TT. D, close view in the binding pocket of the IN CCD AA/MUT-A structure. E, close view in the IN CCD AA MUT-A03-binding pocket. F, 2D view of MUT-A03 interactions with IN CCD TT. All the ligands are contoured with the experimental map at 1.5 σ.
Interface and ligand-binding analysis
The specificity and the strength of the CCD dimer interface of the TT and AA variants in the presence or absence of MUT-A or MUT-A03 were analyzed by protein interfaces, surfaces, and assemblies (PISA), which estimates the dimer stability based on the binding energy of the interface and the entropy change due to complex formation (32). Low p values are indicative of a specific interface. The predicted binding free energy for MUT-A and MUT-A03 was calculated using the BAPPL server (33) and PDB 2PQR for the ligand net charge (34). The net charges for MUT-A and MUT-A03 were found to be 0 and −1, respectively. The results of this analysis together with biochemical and virological data are summarized in Table S2.
TT versus AA comparison
In the absence of ligand, the reduction of the number of salt bridges in the dimer of the AA variant (−6 versus TT), as well as the lower gain in solvation energy on dimerization together with the increase of the p value, indicate a less stable CCD dimer for the AA IN variant.
TT + MUT-A
In the presence of MUT-A, there is a decrease in the number of H-bonds and salt bridges (−2 and −8), as well as a smaller gain on complex formation, indicating a weaker interface. Correlation between these structural changes and the efficiency of MUT-A to promote IN TT multimerization could be explained by the destabilization of the IN CCD dimer leading presumably to aberrant multimerization of IN through its N- and C-terminal domains.
AA + MUT-A
In this complex, the same numbers of H-bonds and salt bridges as with the IN TT are observed (10 and 4, respectively). However, the loss upon MUT-A binding is smaller for the AA variant (−2 and −2 only) than for TT variant (−2 and −8). It is combined with a reverse effect on the energy gain on dimerization and a low p value. Altogether, these data are indicative of the formation of a strong and specific dimeric interface. Also, this AA/MUT-A pair gives the best ligand-binding free energy indicating a tight interaction. Correlation between these changes and the poor efficiency of MUT-A in promoting IN AA multimerization can be interpreted as the stabilization of the IN CCD dimer leading to the correct positioning of the CTD and NTD domains in the context of the full-length protein.
TT + MUT-A03
Again, we observed a loss in the number of H-bonds and salt bridges (−4 and −2) in the presence of MUT-A03, together with a small increase in solvation energy gain on dimerization, indicating a slightly decreased dimerization propensity with MUT-A03.
AA + MUT-A03
In the presence of MUT-A, there is a more important loss of H-bonds and salt bridges (−6 and −4), with a gain of solvation energy on dimerization relatively higher than for the complex with TT variant. This leads to an equivalent destabilization of the IN CCD dimer explaining the maintained ARV activity of the inhibitor on the AA variant.
Discussion
INLAIs are small molecule inhibitors of HIV-1 IN with a peculiar dual mode of action. Their ARV activity results from an early weak inhibitory effect at integration and a late strong activity during production of mature virus particle. INLAIs also display two distinct biochemical properties, inhibition of IN-LEDGF/p75 interaction and induction of allosteric conformational change of IN leading to aberrant multimerization of the protein. The fact that INLAIs bind to a unique site on IN, the LEDGF-binding pocket that lies at the IN dimer interface, raises the question whether the two distinct biochemical and ARV activities of INLAIs are linked or can be dissociated. Previous studies on point mutants at amino acid residues critical for INLAI binding to IN that dramatically decrease IN affinity for INLAIs showed that both ARV activities were either lost concomitantly in the T174I mutant or both were strongly affected, although at variable levels in the case of the H171T mutant (12, 35). These mutants, the T174I in particular, demonstrate that the two activities of INLAIs are linked to their binding to the LEDGF-binding pocket. However, if this binding is sufficient to inhibit IN-LEDGF/p75 interaction, it is not sufficient, even if fully conserved, to promote the second activity on IN, aberrant multimerization, and consequently the late effect on virus maturation. Some other effects, additional to the binding to the LEDGF-binding pocket, are required to promote IN multimerization, presumably conformational changes of allosteric nature between CCD–CTD interactions, as described previously (36, 37). In this report, we demonstrate that the two ARV activities of INLAIs can indeed be fully dissociated for several compounds such as MUT-A and some of its derivatives. These molecules have lost the ability to induce the multimerization of the A125-polymorphic IN while retaining their affinity for IN and full inhibitory activity on the IN-LEDGF/p75 interaction. Reciprocally, Sharma et al. (14) reported the discovery of MINIs, INLAIs that potently induce IN multimerization, while having a 50-fold lower activity of the inhibition of IN-LEDGF/p75 interaction. Interestingly, the IN allosteric inhibitor family can be quite diverse, with compounds displaying various levels of their dual ARV activities.
Studies from Kvaratskhelia and co-workers (36) to determine the biochemical mechanisms underlying the dual ARV activity of INLAIs were performed with the A128T IN mutant conferring “moderate” INLAI resistance. For the ALLINI-2 compound, they measured a mere 2.6-fold loss in IN-LEDGF/p75 inhibition potency, but a roughly 10-fold increase in IN multimerization AC50 together with a 3-fold reduction in the plateau at saturation (for a 30-fold multimerization loss as we would calculate it here). Reduced IN multimerization correlates with a 19-fold decrease in inhibition of HIV-1 multiple-round infection and 33-fold loss in impairment of virus infectivity in producer cells. The close correlation that we observed between the level of IN multimerization and the potency of INLAI ARV activity in multiple-round infection assays provides further evidence that aberrant IN multimerization is the primary biochemical mechanism for the of INLAI-dependent viral maturation defect. It was also reported that modulating LEDGF/p75 levels in target cells (by knockout or overexpression) determines INLAI BI-D potency at integration (37). In our experiments, the relative inhibition of LEDGF/p75 interaction with IN polymorphs strikingly matches the corresponding ARV activity at integration of INLAIs. This finding is also in agreement with the previous reports showing that competition by LEDGF/p75 controls the INLAI-mediated inhibition of integration and weakens its magnitude compared with the ARV potency at late stage that does not suffer from such competition (15, 37–39).
Using cryo-EM imaging of the NL4-3 IN AA 124/125 variant virus produced in the presence of 5 μm MUT-A, we could confirm that the inability of MUT-A to induce IN AA 124/125 multimerization correlates with an absence of assembly alterations in the IN AA 124/125 virus variant (Fig. 2). Interestingly, despite the lack of late stage alteration of virus assembly and infectivity, MUT-A conserved a weak but non-negligible ARV activity on this variant virus, evaluated in a classical dose-response experiment at 1.5 μm EC50. Such weak ARV activity of MUT-A can be attributed only to the inhibition of IN-LEDGF/p75 interaction because the activity of IN multimerization is lacking with this variant. Such assumption is strengthened by the fact that this ARV activity corresponds essentially to an inhibitory activity at integration with an EC50 in single-cycle infection of similar order of magnitude estimated at 2.9 μm (see Table 4). Overall, these observations consolidate our conclusion that the two ARV activities of INLAIs can indeed be fully dissociated for some INLAIs in certain conditions.
Analysis of the crystallographic structures of IN CCD TT and AA variants in the presence and absence of MUT-A or MUT-A03 pointed out the importance of the interaction energy of the IN CCD dimer interface. Although the results of PISA analysis indicate that MUT-A binding destabilizes the CCD TT dimer but has a stabilizing effect on the AA dimer, and on the contrary MUT-A03 equally destabilizes both IN-CCD dimers, it is unlikely that this phenomenon alone could explain the defect of MUT-A in the induction of the multimerization of the IN A125 variant. Indeed, it has been demonstrated by Shkriabai et al. (40) and others that the C-terminal domain of IN plays a critical role for the induction of IN aberrant multimerization by allosteric inhibitors of IN. More recently, Gupta et al. (41) elucidated the structure of the complex of IN bound to the ALLINIs GSK1264 or GSK002 and showed that aberrant multimerization of IN induced interactions between the catalytic core domain and the CTD. Importantly, by probing the mechanism of resistance and the structural basis for polymorphism-induced resistance to these ALLINIs, they found that substitutions at residues 124 and 125 could affect ALLINI binding or the CCD–CTD interaction. More specifically, they found that substitutions at residues 124 and 125 in the CCD would create a steric clash with the polypeptide backbone of the IN CTD, disrupting the inhibitor-mediated interaction between these domains. So, it is more likely that the polymorphic substitutions at position 125 affect MUT-A–induced CCD–CTD interactions rather than simply the stability of the CCD–CCD dimer observed here in PISA analysis.
Our studies highlight the significant impact of IN polymorphic residues 124/125 for INLAI ARV activity. Consistently, INLAI resistance mutations at these positions were described following serial passage experiments, including known polymorphisms (Asn-124, Thr-124, and Ser-125) or novel substitutions (Asp-124, Ser-125, and Lys-125) (14, 41, 43).10,11 Taking into account the high variation frequency at these positions, it is critical to verify the sensitivity of all INLAIs to such polymorphisms. One of the reference compounds we used in this study, BI-224436, was optimized on A125-IN polymorph (45). Lately, the optimization of a pyridine series on N124-IN variant virus was also reported (43). Because MUT-A is strongly affected by the presence of an Ala residue at IN position 125, its development toward further preclinical studies was stopped. By introducing chemical modifications at position 5 on the thienyl core of MUT-A, we could correct its sensitivity to the A125-polymorphism, confirming further the link between INLAI-induced IN multimerization and late stage ARV activity of these small-molecule inhibitors. However, up to now, all these chemical changes resulted in an overall lower ARV activity. If MUT-A could not be advanced in development, it was a highly valuable reagent to better understand the molecular basis of the biochemical properties of INLAIs, some of the structural changes leading to IN multimerization, and the links between the biochemical properties and the ARV activities of this class of compounds.
Experimental procedures
Chemical compound synthesis
References BI-224436 and racemic BI-D were synthesized according to experimental procedures described in patent application WO2009/062285A1. Chemical synthesis of MUT-A and analogs was developed at Mutabilis and described in detail in patent application WO2014/053666A1 (46). Enantiomerically pure MUT-A corresponds to example 11. Racemates of MUT-A and MUT-A01 to MUT-A09 refer to examples 5, 44, 25, 43, 30, 69, 41, 42, 73, 71, respectively.
Cell culture
MT4, TZM-bl, and HeLa-LAV cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health. MT4 cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS) and 100 IU/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) to obtain RPMI 1640 complete medium. HeLa-LAV, TZM-bl, and 293T cells (ATCC, CRL-11268) were grown in Dulbecco's modified Eagle's medium supplemented with 10% FCS and antibiotics. TZM-bl cells are a HeLa-modified cell line containing separately integrated copies of the luciferase and β-gal genes under control of the HIV-1 promoter. HepG2 cells (ATCC, HB-8065) were cultured in Eagle's minimum essential medium supplemented with 10% FCS and antibiotics.
Virus strains and recombinant HIV-1 molecular clones
HIV-1 NL4-3 and NL4-3Δenv-luc molecular clones were obtained from the AIDS Research and Reference Reagent Program, National Institutes of Health. The SpeI-SalI fragment from pNL4-3 containing the full pol gene was cloned into the pUC18 plasmid. In vitro mutagenesis was performed with the PfuTurbo (Agilent) and specific sets of primers to engineer the mutants. The mutated fragment was validated by sequencing (Eurofins) and cloned back into pNL4-3 to generate an HIV-1 mutant molecular clone.
Viral stocks
293T (2.2 × 106 cells) were transfected with 6 μg of pNL4-3 proviral plasmids (WT or drug-resistant) using X-tremeGENE 9 reagent (Roche Applied Science). Cells were washed 24 h later, and cell supernatants were collected 48 h post-transfection and stored at −80 °C. When indicated, viral stocks were prepared in the presence of various concentrations of MUT-A. Single-round viral stocks were produced by co-transfecting pNL4-3Δenv with stomatitis virus envelope glycoprotein (VSV-G) expression vector. Supernatants were collected 2 days after transfection. All viral stocks were quantified for p24 antigen using the Alliance HIV-1 p24 Antigen ELISA (PerkinElmer Life Sciences) and titrated to measure the quantity of infectious particles per ml by infecting TZM-bl indicator cells.
Multiple-round antiviral assay in MT4 cells
MT4 cells growing exponentially at the density of 106/ml were infected with HIV-1 strain NL4-3 at a multiplicity of infection (m.o.i.) of 0.001 for 2 h. The cells were washed with PBS, resuspended in fresh complete RPMI 1640 medium, and distributed into 96-well white plates (Corning) in the presence of different concentrations of compounds in a final volume of 100 μl per well. The effective concentration of compound required to inhibit 50% (EC50) of HIV-1 replication was determined after 5 days using the CellTiter-Glo® luminescent reagent (Promega) to quantify cell viability.
Single-round HIV assay
MT4 cells (growing exponentially at the density of 106/ml) were infected with VSV-G–pseudotyped NL4-3Δenv-luc at an m.o.i. of 0.0001 for 90 min. The cells were washed with PBS, resuspended in fresh complete RPMI 1640 medium, and distributed into 96-well white plates (Corning) in the presence of different concentrations of compounds in a final volume of 100 μl per well. Luciferase expression was quantified after 2 days using the One-GloTM luciferase assay (Promega) for the determination of compound EC50.
Isolation, activation, and culture of human PBMCs
Human PBMCs were isolated from healthy blood donor buffy coats by centrifugation on Ficoll-Hypaque (GE Healthcare) and then washed in PBS. The PBMCs were resuspended at 107 cells/ml in complete RPMI 1640 medium supplemented with 2 μg/ml phytohemagglutinin P and were incubated for 48–72 h at 37 °C. After stimulation, the PBMCs were centrifuged and resuspended in complete RPMI 1640 medium with 20 units/ml recombinant human interleukin-2 (IL-2). The PBMCs were maintained in this medium at a concentration of 0.8 × 106 cells/ml with medium changes every 2 or 3 days until they were used in the assay protocol.
Primary HIV-1 isolates
Primary isolates were obtained from the AIDS Research Reference Reagent Program, National Institutes of Health (http://aidsreagent.org).12 See supporting information for references (Table S3) and IN sequences (Table S4 and Fig. S1). Viral stocks were prepared by infection of human PBMCs and recovery of supernatant when a sufficient p24 titer was reached.
Antiviral assay with human PBMCs
PBMCs were infected with HIV-1 strain or primary isolate at an m.o.i. of 0.001 for 2 h. The cells were washed with PBS and aliquoted, using 100 μl of fresh complete RPMI 1640 medium with 20 units/ml IL-2, into 96-well white plates (Corning) in the presence of different concentrations of compounds. The effective concentration of compound required to inhibit 50% of HIV-1 replication (EC50) was determined after 5 days by ELISA quantification of p24 antigen in the supernatant (PerkinElmer Life Sciences).
Cytotoxicity assays
Growth inhibition was monitored in proliferating cell cultures with different concentrations of compounds. ATP levels were quantified using the CellTiter-Glo® luminescent reagent (Promega) to measure the ability of a compound to inhibit cell growth, an indication of the compound's cytotoxicity. Cytotoxicity was evaluated at day 5 on MT4 and PBMCs or at day 3 on HepG2 cells.
Cryo-electron microscopy of HIV-1 virions
Cell culture supernatants containing HIV particles were fixed in 8% paraformaldehyde (EM grade) solubilized in a 4% cacodylate buffer. Virus particles were pelleted by ultracentrifugation in an SW32Ti rotor (Beckman) for 3 h at 17,000 rpm together with 10 μl of colloidal gold particles (Aurion, GaR 10) to better localize the pellet. Virus pellets were suspended in 50 μl of 50 mm Tris, pH 8, and 2.5 μl was applied to Quantifoil R2/2 holey carbon grids, which were then plunged frozen into liquid ethane using a Vitrobot instrument (Thermo Fisher Scientific). For data acquisition, grids were transferred to the Polara electron microscope equipped with a Falcon I camera, and images were acquired at 300-kV acceleration voltage under low dose conditions at −6 μm defocus.
Construction of epitope-tagged proteins
The His6-LEDGF plasmid has been previously described (47). The plasmid encoding GST-FLAG-IBD/LEDGF was constructed by cloning the LEDGF DNA sequence (encoding residues 342–507) in fusion with the FLAG epitope into pGEX-2T (GE Healthcare). His6-IN plasmid corresponds to pINSD.His and has been previously described (48). The IN mutants were generated by site-directed mutagenesis from pINSD.His. The full-length FLAG-tagged integrase sequence from WT or mutated NL4-3 was PCR-amplified and cloned between the BamHI and XhoI restriction sites of a pGEX-6P1 vector (GE Healthcare) to generate the expression plasmid GST-FLAG-IN. His6-CCD was obtained by cloning the integrase region (residues 50–202, encoding the catalytic core domain) from pINSD.His.Sol (49) into pET15b. CCD contains the F185K mutation, which greatly improves the solubility of the recombinant protein.
Purification of recombinant proteins
Recombinant tagged proteins have been purified as described earlier (15).
HTRF®-based interaction assays
All HTRF®-conjugated monoclonal antibodies were purchased from Cisbio Bioassays. The assays were performed in 384-well low-volume black polystyrene plates (Corning) and incubated at room temperature before reading the time-resolved fluorescence in a PHERAstar Plus with HTRF module (excitation at 337 nm, dual emission at 620 and 667 nm).
CCD–IBD interaction assay
IN-CCD/LEDGF-IBD HTRF® assay was performed in CCD–IBD assay buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 0.4 m KF, 0.1% BSA, 1 mm DTT). 2 μl of 3-fold serial dilutions of inhibitory compound in 25% DMSO were preincubated for 30 min at room temperature with 8 μl of IN-CCD mixture (75 nm His6-IN-CCD, 17 nm XL665-conjugated anti-His6 mAb). Then, 10 μl of LEDGF-IBD mixture (20 nm GST-FLAG-LEDGF-IBD, 1.8 nm europium cryptate-labeled anti-GST mAb) were added, and the plate was incubated for 2.5 h before reading. The HTRF ratio was converted to % inhibition and analyzed by fitting with a sigmoidal dose-response equation with Hill slope to determine the compound IC50.
IN-LEDGF/p75 interaction assays
IN-LEDGF HTRF® assays were done in IN-LEDGF assay buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 0.4 m KF, 0.1% Igepal CA-630, 0.1% BSA, 1 mm DTT). 2 μl of 3-fold serial dilutions of inhibitory compound in 25% DMSO were preincubated for 30 min at room temperature with 8 μl of IN mixture (50 nm FLAG-tagged IN T124–T125 or A124–A125 variant, 17 nm XL665-conjugated anti-FLAG M2 mAb). 10 μl of LEDGF mixture (60 nm His6-tagged LEDGF/p75, 1.5 nm terbium cryptate-labeled anti-His6 monoclonal) were added, and the plate was incubated for 2.5 h before reading. The HTRF ratio was converted to % inhibition and analyzed by fitting a sigmoidal dose-response equation with Hill slope to determine the IC50 of the compound.
IN multimerization assays
2 μl of 3-fold serial dilutions of inhibitory compound in 25% DMSO were preincubated for 30 min at room temperature with 4 μl of 125 nm FLAG-IN TT or AA variant dilution. 4 μl of 125 nm His6-IN TT or AA variant were added, and the plate was incubated for 3 h at room temperature to allow IN subunit exchange and multimerization. This step was performed in IN2 buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 0.005% Tween 20, 0.1% BSA, 1 mm DTT). 10 μl of revelation mixture (1.1 nm europium cryptate-labeled monoclonal anti-FLAG M2 antibody and 13 nm XL665-labeled anti-His6 mAb in IN2 buffer supplemented with 0.8 m KF) were added, and the plate was incubated for 2 h before reading. The HTRF ratio was converted to % baseline interaction signal, and the dose-response curves were analyzed by fitting to a sigmoidal dose-response equation with Hill slope to determine the maximum level of multimerization at saturating compound concentration (the plateau) and the concentration of compound giving half-maximal activation (AC50) of the compound. To rationalize the structure activities of INLAIs, a fold-change index between WT IN and IN T124A/T125A, taking into account both the change in AC50 and plateau was calculated according to Equation 1,
| (1) |
Expression and purification of His6-tagged IN(50–212) TT or AA variant used for crystallization
The CCD of HIV-1 IN WT or AA variant was expressed and purified as described previously (15). Briefly, bacterial expression constructs were transformed into Escherichia coli BL21(DE3). Cells were grown at 37 °C in LB medium containing 100 μg/ml ampicillin until the OD reached 0.5. IN expression was induced by adding 0.5 mm isopropyl β-d-thiogalactoside. Cells were further grown for 3 h at 37 °C and harvested by centrifugation at 4 °C at 4,000 × g. During purification, protein purity was analyzed by SDS-PAGE, and protein concentrations were measured by UV absorption at 280 nm. The cells were homogenized in 25 mm HEPES, pH 7.5, 500 mm NaCl, 2 mm MgCl2, 2 mm β-mercaptoethanol (affinity binding buffer) with a ratio of 10 ml of buffer for 1 g of cells and lysed by pulse sonication. The extract was centrifuged at 100,000 × g for 1 h. The crude extract was loaded onto a 5-ml nickel affinity column (Hitrap Chelating). Nonspecifically bound proteins were removed by 10 column volumes wash in affinity binding buffer supplemented with 100 mm imidazole. Elution was performed in a 10-column volume imidazole gradient from 0.1 to 0.5 m (affinity elution buffer). The fractions of interest were pooled and concentrated on Centriprep (MWCO 10 kDa). The protein was further purified on a gel-filtration Superdex 200 column equilibrated in 50 mm MES, pH 5.5, 50 mm NaCl, 5 mm DTT (gel filtration buffer). Peak fractions corresponding to the complex were pooled and concentrated by ultrafiltration. With this procedure, one could obtain 17 mg of complex from a 1-liter culture.
Crystallization, data collection, and structure refinement
Crystallization was performed by the hanging-drop vapor diffusion method at 24 °C in 24-well plates. Each hanging drop consisted of 3 μl of protein solution (2–5 mg/ml) and 3 μl of reservoir solution (1.26 m ammonium sulfate, 50 mm sodium cacodylate-HCl, pH 6.5), with 500 μl of reservoir solution in the well. The crystals were soaked with the ligands for 24 h before data collection by adding 10 eq of MUT-A or MUT-A03 to the drop. The crystals were plunged in oil (FOMBLIN Y LVAC 14/6 from Aldrich) for a few seconds and cryo-cooled in a stream of liquid nitrogen at −173 °C. All data were collected at a temperature of −173 °C and processed with XDS (50). Diffraction data for IN CCD AA + MUT-A were collected at the home X-ray source (Rigaku FR-X rotating anode equipped with a Pilatus 300K detector). All other datasets were collected using a Pilatus 2M detector on beamline X06DA (PXIII) at the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland. The structures were solved by molecular replacement using the MOLREP program (51) in the CCP4 program suite (52). The structure was refined with REFMAC (53). The ligands were placed in the structure using ARP/wARP (54). Data collection and refinement statistics are summarized in Table S1. Structure superpositions were performed in Coot (55). All structure drawings were performed with PyMOL (42) and Coot (55). 2D view of ligand interactions have been generated with LigPlot (44). Structures and structure factors have been deposited in the PDB database with codes 4LH4 (IN CCD TT) (15), 5OI3 (IN CCD AA), 5OI2 (IN CCD TT + MUT-A), 5OI5 (IN CCD TT + MUT-A03), 5OI8 (IN CCD AA + MUT-A), and 5OIA (IN CCD AA + MUT-A03).
Structure analysis
The specificity and strength of the CCD dimeric interface were analyzed by PISA (European Bioinformatics Institute. http://www.ebi.ac.uk/pdbe/prot_int/pistart.html)12 (32). The predicted ligand-binding free energy was calculated using the BAPPL server (33). The net charge of the ligand was calculated using PDB 2PQR server (http://nbcr-222.ucsd.edu/pdb2pqr_2.1.1/)12 (34).
Author contributions
J. Brias and J. Barbion synthesized the compounds. F. C., S. C., and B. L. designed medicinal chemistry strategy. D. B., E. L. R., C. A., J. M. B., S. Eiler, I. O., D. S., B. P. K., designed, performed the experiments and analyzed the results; F. M., A. S., S. Emiliani, M. R., A. Z., R. B., B. P. K., were involved in designing the experiments, interpretation of data and revising the manuscript critically; R. B., D. B., M. R., and A. Z. drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank Stéphane Huguet for support; Juliette Nguyen, Roxane Beauvoir, and Elodie Drocourt for assistance in virology; Frédéric Le Strat for fruitful discussions; and the National Institutes of Health AIDS research repository for primary isolates. We thank Nicolas Lévy and Vincent Olieric for help in X-ray data collection on beamline X06DA (PXIII) at the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland. We thank Alastair McEwen for help in X-ray data collection at the home source.
This work was supported in part by Biodim Mutabilis under Authorization Number DUO 2145, assigned by the French Ministry of Research for work with genetically modified organisms, by Grants EU FP7 (to Biodim, S. Emiliani, A. S., and A. Z.) under the HIVINNOV Consortium, Grant Agreement 305137, and by Eurostars Grant ResistAids (to Biodim), Grant Agreement E!10239. D. B., E. L. R., C. A., J. -M. B., J. Brias, J. Barbion, F. C., F. L. S., S. C., B. L., F. M., R. B. are or were employees of Biodim Mutabilis at the time of this study. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1, Tables S1–S4, and supporting Ref. 1.
The atomic coordinates and structure factors (codes 5OI2, 5OI3, 5OI5, 5OI8, and 5OIA) have been deposited in the Protein Data Bank (http://wwpdb.org/).
C. W. Fenwick, S. Tremblay, E. Wardrop, R. Bethell, R. Coulombe, R. Elston, A.-M. Faucher, S. Mason, B. Simoneau, Y. Tsantrizos, and C. Yoakim, Resistance studies with HIV-1 non-catalytic site integrase inhibitors. Poster presented at the International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies, Los Cabos, Mexico (June 7–11, 2011).
M. Mitchell, M. Balakrishnan, G. Brizgys, R. Cai, E. Lansdon, A. Mulato, M. Osier, J. Wang, H. Yu, and R. Sakowicz. Novel non-catalytic site integrase inhibitor with improved resistance profile. Poster 434 presented at the Conference on Retroviruses and Opportunistic Infections, Seattle, Washington (February 13–16, 2017).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- IN
- integrase
- INLAI
- IN-LEDGF/p75 allosteric inhibitor
- CCD
- catalytic core domain
- ARV
- antiretroviral
- CTD
- C-terminal domain
- NTD
- N-terminal domain
- ALLINI
- allosteric IN inhibitor
- m.o.i.
- multiplicity of infection
- RAL
- raltegravir
- EVG
- elvitegravir
- DTG
- dolutegravir
- INSTI
- IN strand transfer inhibitor
- MINI
- multimerization IN inhibitor
- PBMC
- peripheral blood mononuclear cell
- FCS
- fetal calf serum
- PDB
- Protein Data Bank
- PISA
- protein interfaces, surfaces, and assemblies
- VSV-G
- vesicular stomatitis virus envelope glycoprotein
- HTRF
- homogeneous time-resolved fluorescence.
References
- 1. Hazuda D. J. (2012) HIV integrase as a target for antiretroviral therapy. Curr. Opin. HIV AIDS 7, 383–389 10.1097/COH.0b013e3283567309 [DOI] [PubMed] [Google Scholar]
- 2. Métifiot M., Vandegraaff N., Maddali K., Naumova A., Zhang X., Rhodes D., Marchand C., and Pommier Y. (2011) Elvitegravir overcomes resistance to raltegravir induced by integrase mutation Y143. AIDS 25, 1175–1178 10.1097/QAD.0b013e3283473599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsiang M., Jones G. S., Goldsmith J., Mulato A., Hansen D., Kan E., Tsai L., Bam R. A., Stepan G., Stray K. M., Niedziela-Majka A., Yant S. R., Yu H., Kukolj G., Cihlar T., et al. (2016) Antiviral activity of Bictegravir (GS-9883), a novel potent HIV-1 integrase strand transfer inhibitor with an improved resistance profile. Antimicrob. Agents Chemother. 60, 7086–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wares M., Mesplède T., Quashie P. K., Osman N., Han Y., and Wainberg M. A. (2014) The M50I polymorphic substitution in association with the R263K mutation in HIV-1 subtype B integrase increases drug resistance but does not restore viral replicative fitness. Retrovirology 11, 7 10.1186/1742-4690-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Engelman A., and Cherepanov P. (2008) The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 4, e1000046 10.1371/journal.ppat.1000046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., and De Clercq E., and Debyser Z. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 10.1074/jbc.M209278200 [DOI] [PubMed] [Google Scholar]
- 7. Emiliani S., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempé D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Christ F., Rain J.-C., Dargemont C., Debyser Z., and Benarous R. (2005) Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 280, 25517–25523 10.1074/jbc.M501378200 [DOI] [PubMed] [Google Scholar]
- 8. Demeulemeester J., Chaltin P., Marchand A., De Maeyer M., Debyser Z., and Christ F. (2014) LEDGINs, non-catalytic site inhibitors of HIV-1 integrase: a patent review (2006–2014). Expert Opin. Ther. Pat. 24, 609–632 10.1517/13543776.2014.898753 [DOI] [PubMed] [Google Scholar]
- 9. Christ F., Voet A., Marchand A., Nicolet S., Desimmie B. A., Marchand D., Bardiot D., Van der Veken N. J., Van Remoortel B., Strelkov S. V., De Maeyer M., Chaltin P., and Debyser Z. (2010) Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 6, 442–448 10.1038/nchembio.370 [DOI] [PubMed] [Google Scholar]
- 10. Kessl J. J., Jena N., Koh Y., Taskent-Sezgin H., Slaughter A., Feng L., de Silva S., Wu L., Le Grice S. F., Engelman A., Fuchs J. R., and Kvaratskhelia M. (2012) Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 287, 16801–16811 10.1074/jbc.M112.354373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsiang M., Jones G. S., Niedziela-Majka A., Kan E., Lansdon E. B., Huang W., Hung M., Samuel D., Novikov N., Xu Y., Mitchell M., Guo H., Babaoglu K., Liu X., Geleziunas R., and Sakowicz R. (2012) New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J. Biol. Chem. 287, 21189–21203 10.1074/jbc.M112.347534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balakrishnan M., Yant S. R., Tsai L., O'Sullivan C., Bam R. A., Tsai A., Niedziela-Majka A., Stray K. M., Sakowicz R., and Cihlar T. (2013) Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS ONE. 8, e74163 10.1371/journal.pone.0074163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fenwick C., Amad M., Bailey M. D., Bethell R., Bös M., Bonneau P., Cordingley M., Coulombe R., Duan J., Edwards P., Fader L. D., Faucher A. M., Garneau M., Jakalian A., Kawai S., et al. (2014) Preclinical profile of BI 224436, a novel hiv-1 non-catalytic-site integrase inhibitor. Antimicrob. Agents Chemother. 58, 3233–3244 10.1128/AAC.02719-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sharma A., Slaughter A., Jena N., Feng L., Kessl J. J., Fadel H. J., Malani N., Male F., Wu L., Poeschla E., Bushman F. D., Fuchs J. R., and Kvaratskhelia M. (2014) A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog. 10, e1004171 10.1371/journal.ppat.1004171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Rouzic E., Bonnard D., Chasset S., Bruneau J.-M., Chevreuil F., Le Strat F., Nguyen J., Beauvoir R., Amadori C., Brias J., Vomscheid S., Eiler S., Lévy N., Delelis O., Deprez E., et al. (2013) Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology 10, 144 10.1186/1742-4690-10-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christ F., Shaw S., Demeulemeester J., Desimmie B. A., Marchand A., Butler S., Smets W., Chaltin P., Westby M., Debyser Z., and Pickford C. (2012) Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrob. Agents Chemother. 56, 4365–4374 10.1128/AAC.00717-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desimmie B. A., Schrijvers R., Demeulemeester J., Borrenberghs D., Weydert C., Thys W., Vets S., Van Remoortel B., Hofkens J., De Rijck J., Hendrix J., Bannert N., Gijsbers R., Christ F., and Debyser Z. (2013) LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 10, 57 10.1186/1742-4690-10-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jurado K. A., Wang H., Slaughter A., Feng L., Kessl J. J., Koh Y., Wang W., Ballandras-Colas A., Patel P. A., Fuchs J. R., Kvaratskhelia M., and Engelman A. (2013) Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc. Natl. Acad. Sci. U.S.A. 110, 8690–8695 10.1073/pnas.1300703110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gupta K., Brady T., Dyer B. M., Malani N., Hwang Y., Male F., Nolte R. T., Wang L., Velthuisen E., Jeffrey J., Van Duyne G. D., and Bushman F. D. (2014) Allosteric inhibition of human immunodeficiency virus integrase: late block during viral replication and abnormal multimerization involving specific protein domains. J. Biol. Chem. 289, 20477–20488 10.1074/jbc.M114.551119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vranckx L. S., Demeulemeester J., Saleh S., Boll A., Vansant G., Schrijvers R., Weydert C., Battivelli E., Verdin E., Cereseto A., Christ F., Gijsbers R., and Debyser Z. (2016) LEDGIN-mediated inhibition of integrase–LEDGF/p75 interaction reduces reactivation of residual latent HIV. EBioMedicine 8, 248–264 10.1016/j.ebiom.2016.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Bel N., van der Velden Y., Bonnard D., Le Rouzic E., Das A. T., Benarous R., and Berkhout B. (2014) The allosteric HIV-1 integrase inhibitor BI-D affects virion maturation but does not influence packaging of a functional RNA genome. PLoS ONE 9, e103552 10.1371/journal.pone.0103552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fontana J., Jurado K. A., Cheng N., Ly N. L., Fuchs J. R., Gorelick R. J., Engelman A. N., and Steven A. C. (2015) Distribution and redistribution of HIV-1 nucleocapsid protein in immature, mature, and integrase-inhibited virions: a role for integrase in maturation. J. Virol. 89, 9765–9780 10.1128/JVI.01522-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Briggs J. A., Simon M. N., Gross I., Kräusslich H.-G., Fuller S. D., Vogt V. M., and Johnson M. C. (2004) The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11, 672–675 10.1038/nsmb785 [DOI] [PubMed] [Google Scholar]
- 24. Lanman J., Lam T. T., Emmett M. R., Marshall A. G., Sakalian M., and Prevelige P. E. (2004) Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 11, 676–677 10.1038/nsmb790 [DOI] [PubMed] [Google Scholar]
- 25. Chertova E., Chertov O., Coren L. V., Roser J. D., Trubey C. M., Bess J. W. Jr., Sowder R. C. 2nd., Barsov E., Hood B. L., Fisher R. J., Nagashima K., Conrads T. P., Veenstra T. D., Lifson J. D., and Ott D. E. (2006) Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80, 9039–9052 10.1128/JVI.01013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kessl J. J., Kutluay S. B., Townsend D., Rebensburg S., Slaughter A., Larue R. C., Shkriabai N., Bakouche N., Fuchs J. R., Bieniasz P. D., and Kvaratskhelia M. (2016) HIV-1 integrase binds the viral RNA genome and is essential during virion morphogenesis. Cell 166, 1257–1268.e12 10.1016/j.cell.2016.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Madison M. K., Lawson D. Q., Elliott J., Ozantürk A. N., Koneru P. C., Townsend D., Errando M., Kvaratskhelia M., and Kutluay S. B. (2017) Allosteric HIV-1 integrase inhibitors lead to premature degradation of the viral RNA genome and integrase in target cells. J. Virol. 91, e00821–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amadori C., van der Velden Y. U., Bonnard D., Orlov I., van Bel N., Le Rouzic E., Miralles L., Brias J., Chevreuil F., Spehner D., Chasset S., Ledoussal B., Mayr L., Moreau F., García F., et al. (2017) The HIV-1 integrase-LEDGF allosteric inhibitor MUT-A: resistance profile, impairment of virus maturation and infectivity but without influence on RNA packaging or virus immunoreactivity. Retrovirology 14, 50 10.1186/s12977-017-0373-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rhee S.-Y., Liu T. F., Kiuchi M., Zioni R., Gifford R. J., Holmes S. P., and Shafer R. W. (2008) Natural variation of HIV-1 group M integrase: Implications for a new class of antiretroviral inhibitors. Retrovirology 5, 74 10.1186/1742-4690-5-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lataillade M., Chiarella J., and Kozal M. J. (2007) Natural polymorphism of the HIV-1 integrase gene and mutations associated with integrase inhibitor resistance. Antivir. Ther. 12, 563–570 [PubMed] [Google Scholar]
- 31. Ceccherini-Silberstein F., Malet I., D'Arrigo R., Antinori A., Marcelin A.-G., and Perno C.-F. (2009) Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 11, 17–29 [PubMed] [Google Scholar]
- 32. Krissinel E. (2015) Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 43, W314–W319 10.1093/nar/gkv314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jain T., and Jayaram B. (2005) An all atom energy based computational protocol for predicting binding affinities of protein-ligand complexes. FEBS Lett. 579, 6659–6666 10.1016/j.febslet.2005.10.031 [DOI] [PubMed] [Google Scholar]
- 34. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., and Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Slaughter A., Jurado K. A., Deng N., Feng L., Kessl J. J., Shkriabai N., Larue R. C., Fadel H. J., Patel P. A., Jena N., Fuchs J. R., Poeschla E., Levy R. M., Engelman A., and Kvaratskhelia M. (2014) The mechanism of H171T resistance reveals the importance of Nδ-protonated His171 for the binding of allosteric inhibitor BI-D to HIV-1 integrase. Retrovirology 11, 100 10.1186/s12977-014-0100-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng L., Sharma A., Slaughter A., Jena N., Koh Y., Shkriabai N., Larue R. C., Patel P. A., Mitsuya H., Kessl J. J., Engelman A., Fuchs J. R., and Kvaratskhelia M. (2013) The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 288, 15813–15820 10.1074/jbc.M112.443390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feng L., Dharmarajan V., Serrao E., Hoyte A., Larue R. C., Slaughter A., Sharma A., Plumb M. R., Kessl J. J., Fuchs J. R., Bushman F. D., Engelman A. N., Griffin P. R., and Kvaratskhelia M. (2016) The competitive interplay between allosteric HIV-1 integrase inhibitor BI/D and LEDGF/p75 during the early stage of HIV-1 replication adversely affects inhibitor potency. ACS Chem. Biol. 11, 1313–1321 10.1021/acschembio.6b00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kvaratskhelia M., Sharma A., Larue R. C., Serrao E., and Engelman A. (2014) Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 42, 10209–10225 10.1093/nar/gku769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Jurado K. A., Wu X., Shun M. C., Li X., Ferris A. L., Smith S. J., Patel P. A., Fuchs J. R., Cherepanov P., Kvaratskhelia M., Hughes S. H., and Engelman A. (2012) HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 40, 11518–11530 10.1093/nar/gks913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shkriabai N., Dharmarajan V., Slaughter A., Kessl J. J., Larue R. C., Feng L., Fuchs J. R., Griffin P. R., and Kvaratskhelia M. (2014) A critical role of the C-terminal segment for allosteric inhibitor-induced aberrant multimerization of HIV-1 integrase. J. Biol. Chem. 289, 26430–26440 10.1074/jbc.M114.589572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gupta K., Turkki V., Sherrill-Mix S., Hwang Y., Eilers G., Taylor L., McDanal C., Wang P., Temelkoff D., Nolte R. T., Velthuisen E., Jeffrey J., Van Duyne G. D., and Bushman F. D. (2016) Structural basis for inhibitor-induced aggregation of HIV integrase. PLoS Biol. 14, e1002584 10.1371/journal.pbio.1002584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. DeLano W. L. (2002) The PyMOL Molecular Graphics System, version 1.8.2, Delano Sci. LLC, Palo Alto, CA [Google Scholar]
- 43. Fader L. D., Bailey M., Beaulieu E., Bilodeau F., Bonneau P., Bousquet Y., Carson R. J., Chabot C., Coulombe R., Duan J., Fenwick C., Garneau M., Halmos T., Jakalian A., James C., et al. (2016) Aligning potency and pharmacokinetic properties for pyridine-based NCINIs. ACS Med. Chem. Lett. 7, 797–801 10.1021/acsmedchemlett.6b00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laskowski R. A., and Swindells M. B. (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- 45. Fader L. D., Malenfant E., Parisien M., Carson R., Bilodeau F., Landry S., Pesant M., Brochu C., Morin S., Chabot C., Halmos T., Bousquet Y., Bailey M. D., Kawai S. H., Coulombe R., et al. (2014) Discovery of BI 224436, a noncatalytic site integrase inhibitor (NCINI) of HIV-1. ACS Med. Chem. Lett. 5, 422–427 10.1021/ml500002n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chasset S., Chevreuil F., Ledoussal B., Le Strat F., and Benarous R. (2014) Inhibitors of viral replication, their process of preparation and their therapeutical uses. Patent WO2014/053666A1 [Google Scholar]
- 47. Michel F., Crucifix C., Granger F., Eiler S., Mouscadet J. F., Korolev S., Agapkina J., Ziganshin R., Gottikh M., Nazabal A., Emiliani S., Benarous R., Moras D., Schultz P., and Ruff M. (2009) Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J. 28, 980–991 10.1038/emboj.2009.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bushman F. D., Engelman A., Palmer I., Wingfield P., and Craigie R. (1993) Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. U.S.A. 90, 3428–3432 10.1073/pnas.90.8.3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jenkins T. M., Hickman A. B., Dyda F., Ghirlando R., Davies D. R., and Craigie R. (1995) Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc. Natl. Acad. Sci. U.S.A. 92, 6057–6061 10.1073/pnas.92.13.6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kabsch W. (2010) XDS Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 10.1107/S0907444909042589 [DOI] [PubMed] [Google Scholar]
- 52. Potterton E., Briggs P., Turkenburg M., and Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 10.1107/S0907444903008126 [DOI] [PubMed] [Google Scholar]
- 53. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 10.1038/nprot.2008.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.