Abstract
Plasmodium falciparum (Pf), the causative agent of malaria, has an iron–sulfur cluster–containing class I fumarate hydratase (FH) that catalyzes the interconversion of fumarate to malate, a well-known reaction in the tricarboxylic acid cycle. In humans, the same reaction is catalyzed by class II FH that has no sequence or structural homology with the class I enzyme from Plasmodium. Fumarate is generated in large quantities in the parasite as a by-product of AMP synthesis and is converted to malate by FH and then used in the generation of the key metabolites oxaloacetate, aspartate, and pyruvate. Previous studies have identified the FH reaction as being essential to P. falciparum, but biochemical characterization of PfFH that may provide leads for the development of specific inhibitors is lacking. Here, we report on the kinetic characterization of purified recombinant PfFH, functional complementation of fh deficiency in Escherichia coli, and mitochondrial localization in the parasite. We found that the substrate analog mercaptosuccinic acid is a potent PfFH inhibitor, with a Ki value in the nanomolar range. The fh gene could not be knocked out in Plasmodium berghei when transfectants were introduced into BALB/c mice; however, fh knockout was successful when C57BL/6 mice were used as host, suggesting that the essentiality of the fh gene to the parasite was mouse strain-dependent.
Keywords: enzyme inhibitor, gene knockout, parasitology, Plasmodium, tricarboxylic acid cycle (TCA cycle) (Krebs cycle), class I fumarate hydratase, essentiality of fumarate hydratase, mercaptosuccinic acid
Introduction
Plasmodium falciparum, the causative agent of the most lethal form of malaria, during its intra-erythrocytic asexual stages, derives ATP primarily from glycolysis with low contribution from mitochondrial pathways (1, 2). The bulk of pyruvate formed is converted to lactic acid with a minor amount entering the tricarboxylic acid (TCA)5 cycle, the flux through which is up-regulated in sexual stages (2). Key intermediates that anaplerotically feed into the TCA cycle are α-ketoglutarate derived from glutamate, oxaloacetate (OAA) from phosphoenolpyruvate, and fumarate from AMP synthesis. Synthesis of AMP in the parasite is solely from IMP through a pathway involving the enzymes adenylosuccinate synthetase (ADSS) and adenylosuccinate lyase (ASL). The net reaction of ADSS and ASL involves consumption of GTP and aspartate and generation of GDP, Pi, and fumarate. In the rapidly-dividing parasite with an AT-rich genome and high-energy requirements, leading to a high demand for adenine pools, one would expect a high flux of fumarate generation. The parasite does not secrete fumarate, but instead, the carbon derived from this metabolite can be traced in malate, OAA, aspartate, pyruvate (through phosphoenolpyruvate), and lactate (3). The metabolic significance of this fumarate anaplerosis is still obscure. In this context, fumarate hydratase (FH, fumarase), the key enzyme to metabolize fumarate, becomes an important candidate for further investigation.
Fumarate hydratase (fumarase, EC 4.2.1.2) catalyzes the reversible conversion of fumarate to malate. The stereospecific reaction involves the anti-addition of a water molecule across the carbon–carbon double bond of fumarate resulting in the formation of S-malate (l-malate). The reverse reaction proceeds with the elimination of a molecule of water from malate in an anti-fashion (4–6). FH is found in two biochemically distinct forms: class I FH is a thermolabile, oxygen-sensitive 4Fe-4S cluster-containing enzyme; class II FH is a stable, oxygen-insensitive, and iron-independent enzyme (7). Class I FH is further divided into two types, two-subunit and single-subunit, depending on the number of genes that encode the functional enzyme (8). There is no sequence homology between these two classes of enzymes. Class I fumarases display substrate promiscuity; apart from catalyzing the interconversion of fumarate and malate, these enzymes also interconvert S,S-tartrate and oxaloacetate and mesaconate and S-citramalate with varying catalytic efficiencies (9, 10). The 4Fe-4S cluster is bound to the enzyme by three metal–thiolate bonds formed between three conserved cysteine residues in the protein and three ferrous ions (11). The fourth iron in the cluster, proposed to be held loosely by a hydroxyl ion, is thought to be directly involved in substrate binding and catalysis as seen in the enzyme aconitase (12, 13).
Both classes of FHs are distributed in all three domains of life with class I FH being more prevalent in archaea, prokaryotes, and lower eukaryotes. Many organisms have genes corresponding to both classes, as in Escherichia coli (Ec), which has three FH-encoding genes, viz. fumA, fumB, and fumC. FumA and FumB are 4Fe-4S cluster-containing class I enzymes, and FumC belongs to class II-type FH. Recently, another gene, fumD, has been identified in the E. coli genome to code for a class I fumarase with altered substrate preferences (14).
The structural and biochemical characteristics of class II FH are thoroughly studied from different organisms, viz. human, porcine, yeast, E. coli, and other sources (15–19). However, class I FH is not well-studied because of its thermolabile and oxygen-sensitive nature. All apicomplexans and kinetoplastids possess only class I FH, whereas dinoflagellates have both the classes (20). Biochemical characterization of class I FH from Leishmania major (Lm) and Trypanosoma cruzi, both kinetoplastids (21, 22), and the three-dimensional structure of LmFH II (11) are the only reports of class I FH from eukaryotes. All Plasmodium species have one gene annotated putatively as fumarate hydratase that remains to be characterized. Genetic investigations on the role of TCA cycle enzymes in P. falciparum have revealed the non-essentiality of all genes of the cycle except FH and malate-quinone oxidoreductase to asexual intra-erythrocytic stages (23). Recently, a metabolic network reconstruction of pathways in artemisinin-resistant P. falciparum strains has identified FH reaction as uniquely essential to these parasites (24). Biochemical characterization of PfFH could throw light on unique features of the enzyme and also provide leads for the development of inhibitors.
We report here the kinetic characterization and substrate promiscuity of PfFH, studied using in vitro assays on the recombinant enzyme and E. coli-based functional complementation. dl-Mercaptosuccinic acid (dl-MSA), a malate analog, was found to be a competitive inhibitor of the P. falciparum enzyme. dl-MSA inhibited the growth of the ΔfumACB strain of E. coli-expressing PfFH as well as the asexual intra-erythrocytic stages of P. falciparum in in vitro cultures. Attempts at generating fh-null Plasmodium berghei grown in BALB/c mice yielded drug-resistant clonal populations that had retained the fh gene, implying its essentiality. However, fh gene knockout was obtained when the transfectants were grown in C57BL/6 mice. This suggests mouse-strain–dependent essentiality of the fh gene in P. berghei.
Results
Distribution of class I fumarate hydratase in eukaryotes
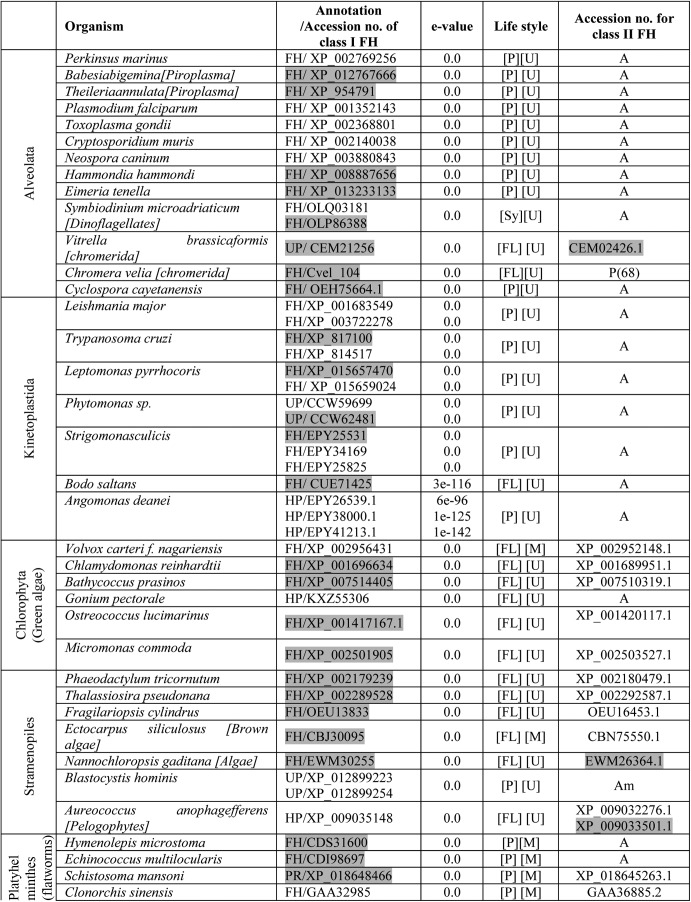
Although both class I and class II FHs catalyze the conversion of fumarate to malate, it is the class II FHs that are widely distributed across eukaryotic organisms. To elicit possible correlations between the presence of class I FH and the nature of the organisms, such as their uni- or multicellularity and parasitic or free-living lifestyle, eukaryotes with class I FH were catalogued (Table 1). In addition, Table 1 reports the presence/absence of class II FH in organisms having class I FH and on FH proteins with mitochondrial targeting sequence (accession number is shaded in gray). Class I FHs are sparsely distributed in both uni- and multicellular eukaryotes and are of the single-subunit type. Although most multicellular eukaryotes with class I FH also have class II FH, Hymenolepis microstoma and Echinococcus multilocularis (flatworms) are the only multicellular eukaryotes that have only the class I fh gene. Eukaryotes, including Entamoeba histolytica, Hymenolepis microstoma, Echinococcus multilocularis, Chrysochromulina sp., and organisms belonging to alveolata and kinetoplastida having only class I FH, are all parasitic in nature with the exception of Chrysochromulina sp., Gonium pectorale, and Bodo saltans that are free-living. In addition, the symbiont Symbiodinium microadriaticum has only class I FH. Most other eukaryotes having class I FH also have the gene for class II FH. Vitrella brassicoformis, a photosynthetic ancestor of apicomplexans (25), has genes for both class I and class II type FH, suggesting the occurrence of a gene loss event with respect to class II FH during the evolution of the apicomplexan lineage as has been noted previously (20). Most organisms belonging to Kinetoplastida have more than one gene for Class I FH, with the mitochondrial targeting sequence predicted to be present only in one. Further, in many organisms listed in Table 1 where both classes of FH are present, it was seen using MitoFates (26) that predominantly Class I FHs were predicted to have the mitochondrial targeting sequence.
Table 1.
Eukaryotic organisms with class I fumarate hydratase
The abbreviations used in table are as follows: FH, fumarate hydratase; HP, hypothetical protein; UP, unknown protein; PR, pol related; [P], parasitic; [FL], free living; [Sy], symbiont; [U], unicellular; [M], multicellular; P, present; A, absent; Am, ambiguous. The common names of the organisms are given in square brackets following the Latin names. The list of organisms was obtained from the output of BLASTP using E. coli class I FH protein sequence as the query. An e-value cut-off of 10−10 and query coverage of 65% were used as criteria for selecting the protein sequences from different organisms. The name of the taxon to which the organism belongs is indicated in the 1st column. The E. coli FumC protein sequence was used as the query to ascertain the presence or absence of class II fumarate hydratase in these organisms and, if present, the accession number of the protein is given in the 6th column. Ambiguity in the presence of class II FH in some cases is due to the annotation of these proteins as adenylosuccinate lyase with which class II FHs share high sequence similarity. Proteins that are predicted by MitoFates (26) to have a mitochondrial targeting sequence are shaded in grey. The gene, XP_001683549, though not predicted by MitoFates to have a mitochondrial targeting sequence, has been shown to localize to the mitochondria (21).
Mitochondrial localization of P. falciparum FH
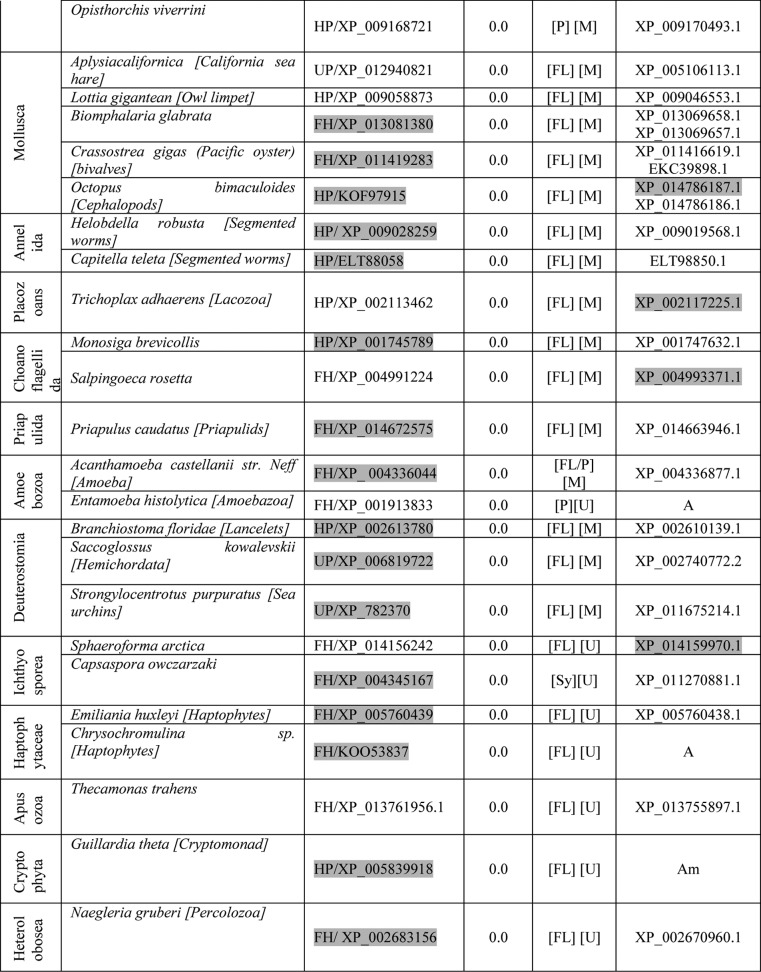
FH in eukaryotes is known to be localized to mitochondria. Although biochemical evidence suggests that FH is mitochondrially localized in P. falciparum (3), microscopic images showing localization to this organelle are not available. To examine the localization of the protein in P. falciparum, the fh gene on chromosome 9 was replaced with DNA encoding FH-regulatable fluorescent affinity tag (comprising green fluorescent protein (GFP), E. coli dihydrofolate reductase (EcDHFR) degradation domain, and a hemagglutinin (HA) tag in tandem) fusion protein by single crossover recombination in PM1KO (27) strain of the parasite (Fig. 1a). The genotype of the strain (Fig. 1b) was validated by PCR using primers P1–P4 (Table S1) and used for live-cell imaging after staining with Hoechst and MitoTracker Red CM-H2XRos. The GFP-positive parasites clearly showed colocalization of GFP signal with MitoTracker Red staining (Fig. 1c) showing mitochondrial localization of fumarate hydratase in P. falciparum. Despite this localization, the only organism for which all four bioinformatic tools used predict a mitochondrial targeting sequence is Plasmodium knowlesi (Table S2). However, examination of the N terminus of PfFH shows the presence of proximal and distal basic residues interspersed with hydroxylated amino acids, a feature usually present in mitochondrial targeting presequences (26, 28), and this may serve as the targeting signal sequence.
Figure 1.
Generation of PfFH-GFP strain encoding FH-GFP and localization of PfFH. a, scheme showing the integration locus with the regulatable fluorescent affinity (RFA) (GFP + EcDHFR degradation domain + HA) tag in tandem with the fh gene in the strain PfFH-GFP. Oligonucleotides P1 and P2 and P3 and P4 were used for checking the 5′ and 3′ integration, respectively (Table S1). P1 and P4 are beyond the sites of integration in the genome. b, left panel, genotyping by PCR for validating 5′ integration. The templates used in different lanes are as follows: L1, genomic DNA from PfFH-GFP; L3, P. falciparum PM1KO genomic DNA. A band size of 2483 bp validates 5′ integration. Right panel, genotyping by PCR for validating 3′ integration. The templates used in different lanes are as follows: L2, genomic DNA from PfFH-GFP; L3, P. falciparum PM1KO genomic DNA. A band size of 2467 bp validates 3′ integration. Molecular weight markers are in lanes L2 and L1 in left and right panels, respectively. c, upper panel shows a trophozoite, and the lower panel shows a schizont. As evident from the merge, PfFH localizes to the mitochondrion. The Pearson correlation coefficients for the images are 0.8972 (upper panel) and 0.8954 (lower panel).
PfFH complements fumarase deficiency in E. coli
To recombinantly express organellar proteins in E. coli, it is preferable to use the DNA sequence corresponding to only the mature protein with the signal peptide deleted. Because none of the bioinformatic prediction tools was able to identify an unambiguous signal sequence in P. falciparum FH, we resorted to multiple sequence alignment with bacterial single-subunit type and archaeal two-subunit type FH for generating N-terminal deletion constructs. Examination of the multiple sequence alignment shows a 120-amino acid insertion at the N terminus in Plasmodium FHs that is absent in bacterial and archaeal FH sequences (Fig. S1). Of the N-terminal 120 amino acid residues in plasmodial FHs, the first 40 residues are diverse, whereas residues 40–120 show a high degree of conservation (Fig. S2) within the genus. Hence, for functional complementation in the E. coli fh null mutant, three different expression constructs of PfFH protein in pQE30 were generated: those expressing the full-length (PfFHFL); N-terminal 40 residues deleted (PfFHΔ40); and N-terminal 120 residues deleted (PfFHΔ120) enzymes.
E. coli has three genes that encode fumarate hydratase; fumA and fumB of the class I type and fumC of the class II type. fumA and fumC genes are in tandem and are driven by a common promoter (7, 29). Starting with JW4083-1, a ΔfumB strain of E. coli, a triple knockout ΔfumACB strain, in which all three major fum genes (fumA, fumC, and fumB) are deleted, was generated and validated by PCR (Fig. S3). As expected, although the strain was able to grow normally in malate-containing minimal medium (Fig. 2a), it was unable to grow on minimal medium containing fumarate as the sole carbon source (Fig. 2b). As expected, all transformants (containing pQE-PfFHFL, pQE-PfFHΔ40, pQE-PfFHΔ120, and pQE30) of ΔfumACB strain of E. coli grew well on malate-containing minimal medium plates (Fig. 2c). In fumarate-containing M9 plates, the cells expressing PfFHΔ40 and PfFHFL grew faster, whereas the growth rate of cells expressing PfFHΔ120 was slower, and no growth of cells carrying just pQE30 was observed (Fig. 2d). This shows that the PfFH can functionally complement the deficiency of fumarate hydratase activity in the ΔfumACB strain and validates that the P. falciparum enzyme is indeed fumarate hydratase. The slow growth of PfFHΔ120-expressing ΔfumACB E. coli strain indicates that residues 40–120 play a role in the structure and/or function of PfFH despite these residues being conserved only in plasmodial fumarate hydratase sequences and not in others (Fig. S1).
Figure 2.
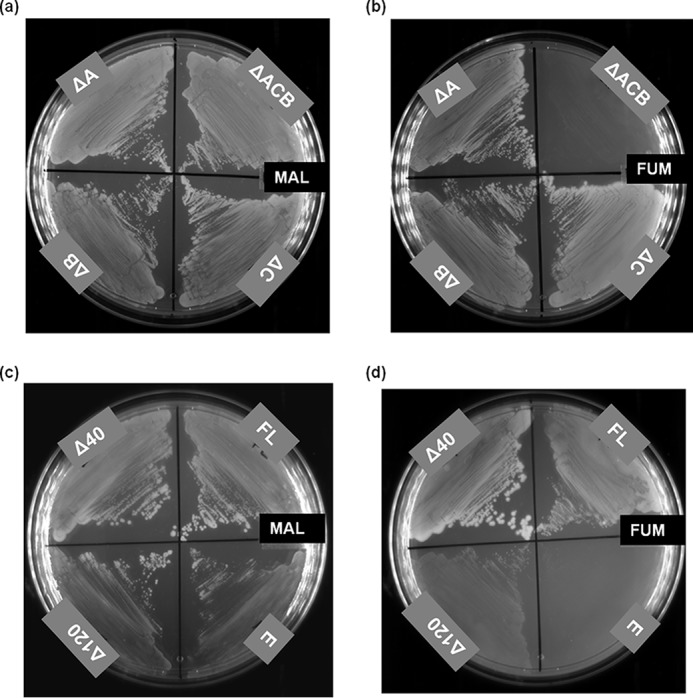
Phenotyping of the E. coli strain ΔfumACB and functional complementation by P. falciparum FH. Growth phenotype of the E. coli strains with at least one copy of fumarate hydratase gene deleted (ΔA, ΔB, and ΔC) and with all three genes deleted (ΔACB) on malate- (MAL) (a) and fumarate (FUM) (b)-containing minimal medium. As evident from the phenotype, ΔfumACB strain (ΔACB) is not able to grow on fumarate-containing minimal medium. c and d, the growth of ΔfumACB strains expressing either PfFHFL (FL) or PfFHΔ40 (Δ40) or PfFHΔ120 (Δ120) of P. falciparum fumarate hydratase on malate- (c) and fumarate (d)-containing minimal medium. The plates were scored after 48 h of incubation at 37 °C. ΔfumACB strain containing just pQE30 (E) was used as a control. The experiment was repeated three times, and the images shown correspond to one of the replicates.
Activity of PfFHΔ40
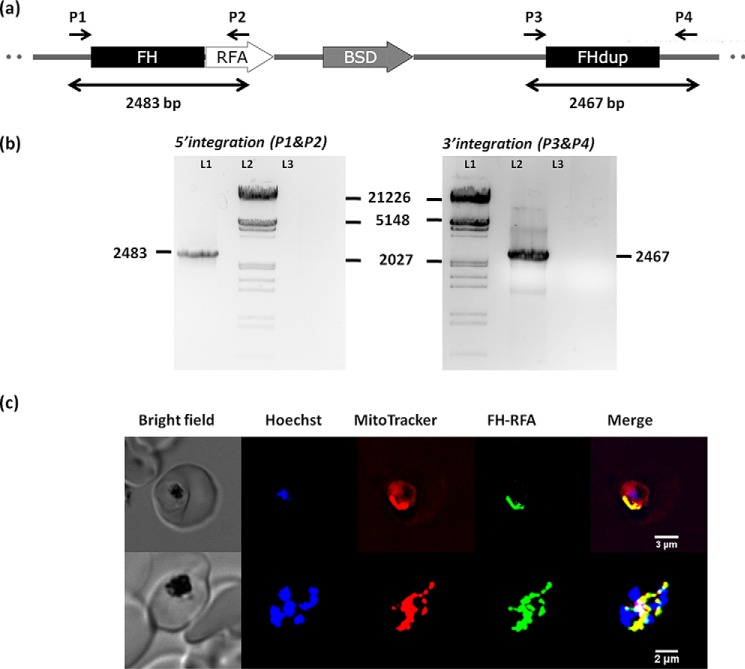
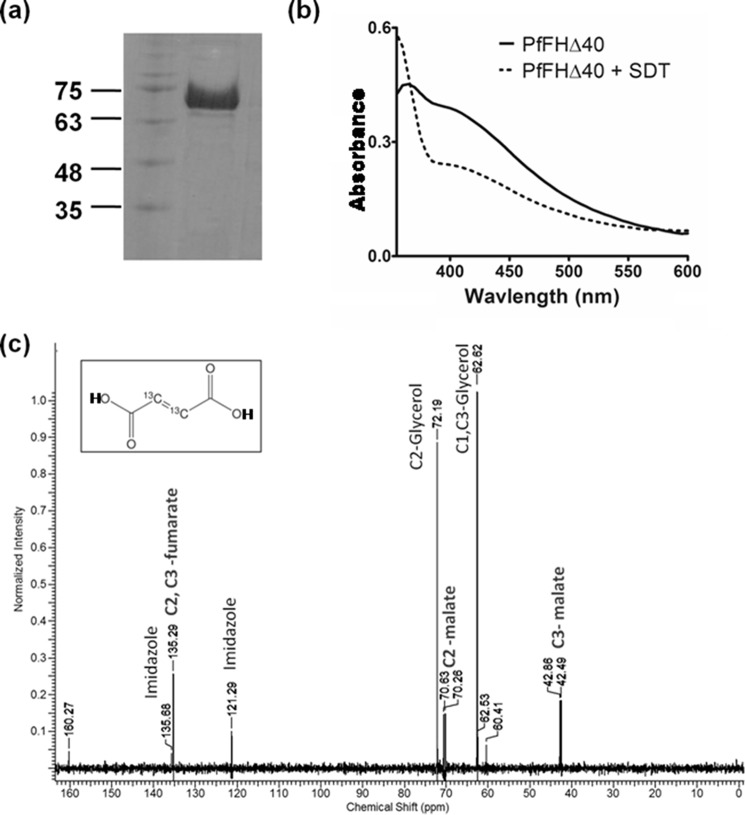
PfFHΔ40 was expressed with an N-terminal His6 tag in Codon plus BL21 (DE3) RIL, purified using Ni-NTA-affinity chromatography (Fig. 3a) and reconstituted in vitro with the Fe–S cluster. The UV-visible spectrum with absorption maxima at 360 and 405 nm indicates the presence of the 4Fe-4S cluster in the enzyme and 78% reconstitution efficiency using an ϵ value of 16,000 m−1 cm−1 at 410 nm (30). The addition of sodium dithionite lowered the absorption intensity at 405 nm indicating a reduction of the cluster (Fig. 3b) (31–34). The enzyme lacking the reconstituted cluster was devoid of any activity. The activity of PfFHΔ40, when examined at 240 nm, showed a time-dependent decrease in absorbance with fumarate as the substrate, and with malate an increase was observed. To confirm the chemical identity of the product formed, an NMR spectrum was recorded with 2,3-[13C]fumarate as the substrate. The appearance of two doublets with chemical shift values 70.63, 70.26 ppm and 42.86, 42.49 ppm corresponding to C2 and C3 carbons, respectively, of malate confirmed that PfFHΔ40 has in vitro fumarase activity (Fig. 3c). The specific activity value calculated from progress curves monitored by UV absorption spectroscopy using 32 mm fumarate as substrate was 138.5 ± 5.8 μmol min−1 mg−1 and was consistent across multiple batches of purified protein. Substrate saturation plots for PfFHΔ40 for both fumarate and malate were hyperbolic in nature indicating the absence of cooperativity. Fit to Michaelis-Menten equation yielded Km and Vmax values that are summarized in Table 2. The Km values for PfFHΔ40 for fumarate and malate in the low millimolar range are similar to that of class I FH from L. major (21) and T. cruzi (22), although for those from bacteria and archaea, the values are in the micromolar range. The catalytic efficiency (kcat/Km) of PfFHΔ40 for fumarate is similar to L. major FH but 10–100-fold lower than that reported for other class I FHs (Table 2).
Figure 3.
Purification and activity of PfFHΔ40. a, 1st lane, protein molecular mass markers (numbers indicated are in kDa); 2nd lane, Ni-NTA-purified PfFHΔ40. b, UV-visible absorption spectrum of purified and reconstituted PfFH shows characteristic peaks at 360 and at 405 nm that indicate the presence of a 4Fe-4S cluster. The spectrum of the protein with oxidized iron–sulfur cluster is shown as a solid line and that with reduced iron–sulfur cluster upon addition of 1 mm sodium dithionite (SDT) is shown as a dashed line. c, validation of malic acid formation by 13C NMR. The NMR spectrum of assay mixture consisting of 50 μm 2,3-[13C]fumaric acid in 100 mm potassium phosphate, pH 7.4, incubated with 100 μg of purified PfFHΔ40 enzyme, shows the presence of peaks corresponding to [13C]malic acid. Unreacted [13C]fumaric acid is also present. The inset shows the chemical structure of [13C]fumaric acid. The spectrum is an average of 3000 scans acquired using Bruker 400-MHz NMR spectrometer. The peaks corresponding to imidazole and glycerol are from the protein solution.
Table 2.
Kinetic parameters of PfFHΔ40 and other class I FH
The units for Km, kcat, and kcat/Km are mm, s−1, and s−1 m−1, respectively.
| Organism/enzyme name | Substrate | Km | kcat | kcat/Km | Refs. |
|---|---|---|---|---|---|
| P. falciparum | |||||
| PfFHΔ40 | Fumarate | 2.6 ± 0.3 | 182 ± 8 | 7.0 × 104 | This study |
| Malate | 1.2 ± 0.1 | 159 ± 9 | 1.3 × 105 | ||
| Mesaconate | 3.2 ± 0.3 | 60 ± 2 | 1.9 × 104 | ||
| T. cruzi | |||||
| TcFHca | Fumarate | 0.8 ± 0.2b | 400 ± 100 | 8.2 × 106c | 22 |
| Malate | 2.5 ± 0.6b | 290 ± 40 | 1.3 × 106c | ||
| TcFHmd | Fumarate | 1.5 ± 0.4 | 2300 ± 500 | 1.5 × 106 | |
| Malate | 2.8 ± 0.2 | 1050 ± 40 | 0.4 × 106 | ||
| E. colie | |||||
| fumA | Fumarate | 0.09 ± 0.02 | 617 | 6.6 × 106 | 14 |
| Malate | 0.40 ± 0.05 | 352 | 8.7 × 105 | ||
| Mesaconate | 0.22 ± 0.02 | 56 | 2.5 × 105 | ||
| fumB | Fumarate | 0.21 ± 0.03 | 655 | 3.1 × 106 | |
| Malate | 0.78 ± 0.13 | 290 | 3.7 × 105 | ||
| Mesaconate | 0.10 ± 0.01 | 58 | 5.8 × 105 | ||
| L. major | |||||
| LMFH-1f | Fumarate | 2.5 ± 0.4 | 28.3 ± 4.7 | 1.1 × 104 | 21 |
| Malate | 2.3 ± 0.3 | 12.9 ± 1.3 | 5.6 × 103 | ||
| LMFH-2g | Fumarate | 5.7 ± 1.4 | 204.2 ± 52.1 | 3.6 × 104 | |
| Malate | 12.6 ± 2.7 | 151.4 ± 20.5 | 1.2 × 104 | ||
| P. furiosuse | |||||
| FH | Fumarate | 0.34 | 1101 | 3.2 × 106 | 69 |
| Malate | 0.41 | 1514 | 3.7 × 106 | ||
| P. thermop.h | |||||
| MmcBCi | Fumarate | 0.43 | 219 | 5.1 × 105 | 8 |
| Malate | 0.59 | 25.2 | 4.3 × 104 | ||
| B. xenovoranse | |||||
| Bxe_A3136j | Fumarate | 0.10 ± 0.01 | 296 | 2.8 × 106 | 10 |
| Malate | 0.28 ± 0.02 | 118 | 3.98 ×105 | ||
| Mesaconate | 0.03 ± 0.01 | 117 | 3.6 × 106 | ||
a This is cytosolic T. cruzi FH.
b TcFHc exhibits cooperativity and hence k0.5 values are reported. The value of Hill coefficient is 1.4 for both substrates.
c The values reported are kcat/kh0.5 with units being s−1 m−1.4.
d This is mitochondrial T. cruzi FH.
e The kcat values provided in the table for FH from E. coli, P. furiosus, and B. xenovorans are calculated from the Vmax values reported in the reference provided.
f LMFH-1 is mitochondrial L. major FH.
g LMFH-2 is cytosolic L. major FH.
h P. thermopropionicum is abbreviated as P. thermop.
i Putative FH in P. thermopropionicum has been annotated as MmcBC (8).
j UniProt id is for FH from B. xenovorans (10).
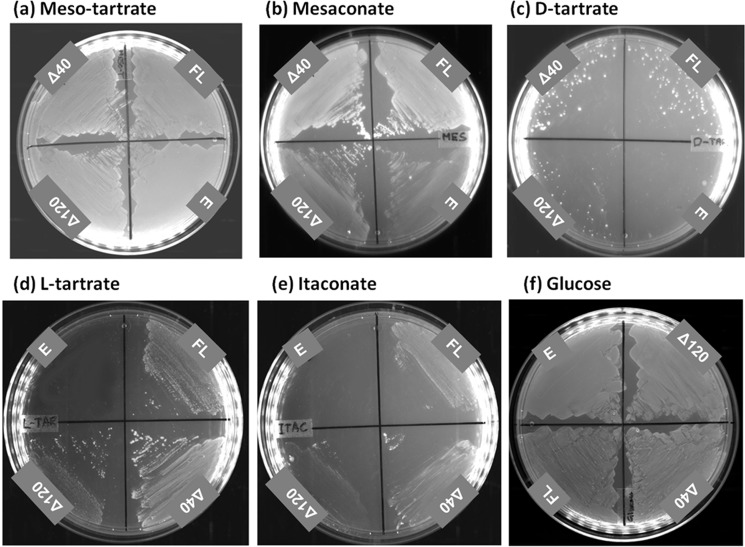
The substrate promiscuity of PfFHFL, PfFHΔ40, and PfFHΔ120 for other dicarboxylic acids was examined using growth complementation in the E. coli strain, ΔfumACB (Fig. 4). Growth on l-tartrate, d-tartrate, and itaconate was conditional to the presence of PfFH, whereas growth on meso-tartrate was independent. All three PfFH constructs greatly enhanced the growth of the ΔfumACB strain of E. coli on mesaconate over the control. In vitro activity measurements showed that the parasite enzyme utilizes mesaconate as a substrate converting it to S-citramalate with Km and kcat/Km values of 3.2 ± 0.3 mm and 1.9 × 104 m−1 s−1, respectively, with the latter value 3.7- and 6.8-fold lower than that for fumarate and malate, respectively (Table 2). The in vitro activity on d-tartrate was measured by a coupled enzyme assay using PfMDH. This activity at 2 mm d-tartrate was 7.8 μmol min−1 mg−1, which is 9.4-fold lower than that on malate at a similar concentration. The poor growth of ΔfumACB strain expressing PfFH constructs on this substrate correlates with the weak in vitro activity. Inhibition of PfMDH (the coupling enzyme) at higher concentrations of d-tartrate precluded estimation of kcat and Km values for this substrate. PfFHΔ40 failed to show in vitro activity on itaconate (a succinate analog), R-malate and R,R-tartrate (l-tartrate) even at a concentration of 10 mm, indicating that the enzyme is highly stereospecific in recognition of substrates. The growth phenotype of ΔfumACB on R,R-tartrate and itaconate could arise from PfFH playing a secondary but critical role required for cell growth. These results show that the substrate promiscuity profile of PfFH is similar to class I enzymes from other organisms (7, 9, 14, 35) with the order of preference being fumarate followed by mesaconate and the least preferred being d-tartrate. However, the significance of the extended substrate specificity of PfFH for mesaconate and d-tartrate with regard to Plasmodium cellular biochemistry is unclear at this stage.
Figure 4.
Growth of E. coli strain ΔfumACB expressing PfFH on different carbon sources. a–f, the growth of ΔfumACB strains expressing either PfFHFL (FL) or PfFHΔ40 (Δ40) or PfFHΔ120 (Δ120) were tested on meso-tartrate (a), mesaconate (b), d-tartrate (c), l-tartrate (d), itaconate (e), and glucose-containing minimal medium (f). The plates were scored after 48 h of incubation at 37 °C. ΔfumACB strain containing just pQE30 (E) was used as a control. The experiment was repeated three times, and the images correspond to one of the replicates. The glucose-containing plate served as a control for the number of cells plated across the different constructs.
Mercaptosuccinic acid is class I FH-specific inhibitor
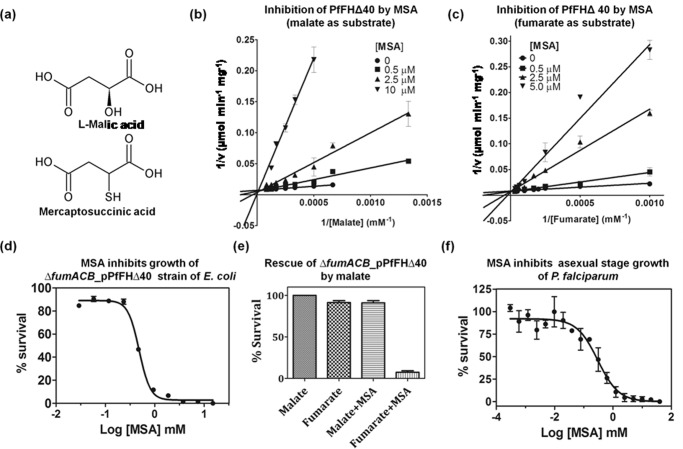
Analogs of fumarate, malate, and intermediates of the TCA cycle (including their analogs) were tested for their effect on PfFHΔ40 (Text S1). Of these, the only molecules that inhibited PfFH activity were dl-mercaptosuccinic acid (dl-MSA, Fig. 5a) and meso-tartrate. Double-reciprocal plots of initial velocity as a function of varied substrate (fumarate and malate) concentrations at different fixed dl-MSA concentrations yielded lines that intersected on the 1/v axis indicating the competitive nature of inhibition (Fig. 5, b and c). The Ki values for dl-MSA for PfFHΔ40 with malate and fumarate as substrates are 321 ± 26 and 548 ± 46 nm, respectively. MSA used in our study is an enantiomeric mixture of dl-isomers, and as d-malate does not inhibit PfFH, the Ki value is expected to be half of that determined. The absence of inhibition of EcFumC even at a 10 mm concentration of dl-MSA suggests the exclusive specificity of this compound for class I FH. dl-MSA inhibition of EcFumA with a Ki of 2.9 ± 0.22 μm (fumarate as substrate) indicates that the molecule is a general class I FH inhibitor. Interestingly, dl-MSA is not a substrate for PfFH as seen by spectrophotometric assays at 240 nm with 10 mm dl-MSA and 1 μm enzyme that failed to show either formation of the enediolate intermediate or the product fumarate through the liberation of H2S.
Figure 5.
Specificity of dl-MSA for class I FH. a, structures of l-malic acid and mercaptosuccinic acid. b, Lineweaver-Burk plot of initial velocity at varied malate and different fixed MSA concentrations. c, Lineweaver-Burk plot of the initial velocity at varied fumarate and different fixed MSA concentrations. d, inhibition of the growth of the ΔfumACB_pPfFHΔ40 strain of E. coli by MSA. e, rescue of MSA-mediated growth inhibition of ΔfumACB_pPfFHΔ40 upon addition of malate. f, inhibition of the in vitro growth of intra-erythrocytic asexual stages of P. falciparum by MSA.
Albeit slightly less effective as an inhibitor, meso-tartrate competitively inhibited PfFHΔ40 with a Ki value of 114 ± 17 μm. This compound inhibited both EcFumA and EcFumC with similar Ki values of 625 and 652 μm, respectively. Meso-tartrate has two chiral carbons with S-configuration at C2 and R-configuration at C3. It should be noted that in the case of both EcFumC and EcFumA, S,S-tartrate (d-tartrate) is a substrate (14). The inhibition by meso-tartrate of both class I and II FH indicates relaxed stereospecificity of these enzymes at the C3 carbon of the substrates. Pyromellitic acid, a known potent inhibitor of class II FH, had no effect on the activity of the two class I enzymes tested (PfFHΔ40 and EcFumA), while completely abolishing the activity of the class II enzyme (EcFumC).
Growth inhibition by dl-MSA
Because the growth of E. coli ΔfumACB strain on minimal medium containing fumarate as the sole carbon source is conditional to the presence of functional FH, the effect of dl-MSA on the growth of ΔfumACB_pPfFHΔ40 was examined. dl-MSA inhibited the growth of ΔfumACB_pPfFHΔ40 with an IC50 of 482 ± 4 μm (Fig. 5d), and the addition of malate completely rescued the inhibition (Fig. 5e). This shows that the toxicity of dl-MSA is indeed due to specific inhibition of the metabolic conversion of fumarate to malate. The ΔfumACB E. coli strain can serve as a facile primary screening system for small molecules acting as inhibitors of PfFH as it circumvents in vitro assays with the oxygen-sensitive labile enzyme. With dl-MSA as an inhibitor of PfFH under in vitro and in vivo conditions, the molecule was checked for its toxicity on asexual intra-erythrocytic stages of P. falciparum in in vitro culture. dl-MSA was found to kill parasites in culture with an IC50 value of 281 ± 68 μm (Fig. 5f). Although dl-MSA is a potent inhibitor of PfFHΔ40 with a Ki value of 547 ± 47 nm (with fumarate as substrate), the IC50 values for the inhibition of both ΔfumACB_pPfFHΔ40 and P. falciparum are significantly higher.
Essentiality of fumarate hydratase for P. berghei is host strain-dependent
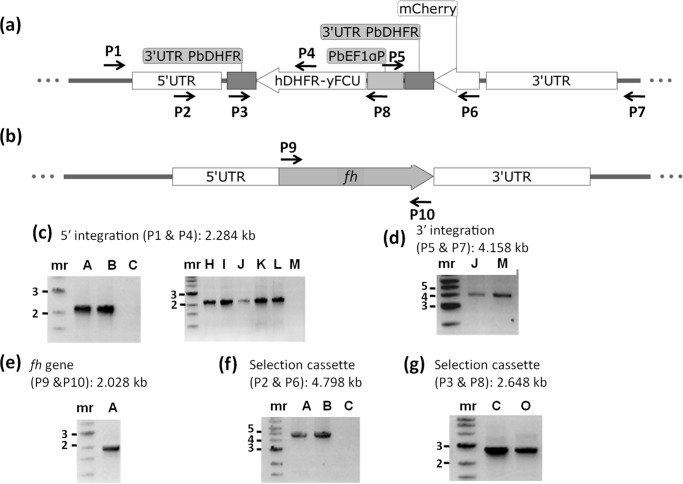
An earlier attempt at knockout of fumarate hydratase gene in P. falciparum was not successful (23). Our attempts at knockdown of PfFH levels by removal of trimethoprim from cultures of the FH-regulatable fluorescent affinity tag-expressing strain, where FH is fused to EcDHFR degradation domain, did not result in lowering of protein levels. It has been shown that proteins targeted to organelles and possessing a signal sequence lack accessibility to proteasomal degradation machinery, and hence the conditional degradation approach may be unsuitable for achieving knockdown of these protein levels (36). Therefore, the essentiality of FH was examined in P. berghei with BALB/c mice as host. For this, the fh gene knockout construct generated through the recombineering-based strategy was used (Fig. S4). Transfected parasites were injected into mice, selected on pyrimethamine, and drug-resistant parasites that appeared 10 days after infection were subjected to limiting dilution cloning. All the 17 P. berghei clones (A to Q) obtained by limited dilution cloning of the drug-resistant parasites were examined by PCR to confirm the presence of the integration cassette and the absence of the fh gene. Oligonucleotides used for genotyping of the clones are listed in Table S1. The expected genomic locus upon integration of the selectable marker cassette by double-crossover recombination and the wildtype (with fh gene) are shown schematically in Fig. 6, a and b. Genotyping by PCR was performed to confirm integration of the selectable marker cassette at the expected locus, the presence/absence of the fh gene, and the presence of the selection cassette (Fig. 6 and Fig. S5). The results of the PCRs showed that though all the parasite clones carried the selectable marker, hDHFR-yFCU cassette in their genomic DNA (Fig. 6, f and g and Fig. S5, f and g), they also retained the fh gene (Fig. 6e and Fig. S5e). In two of the clones (C and O), the integration of the cassette was at a random site as they failed to answer for both 5′ and 3′ integration PCRs. The 12 clones that yielded the expected PCR-amplified fragment for 5′ integration (Fig. S5c) did not yield a band of the expected size for 3′ integration PCR. One clone (M) yielded expected PCR-amplified fragment for only 3′ integration (Fig. 6d) and not for 5′ integration (Fig. 6c and Fig. S5c). Integration of the selection cassette through single crossover recombination using either 5′ or 3′ homology arm with the intact fh gene present downstream or upstream, respectively, would yield this PCR result. As the DNA used for transfection was linear, circularization of the fragment must have enabled this single crossover recombination. Only two clones (J and Q) were positive for both 5′ and 3′ integration PCRs while continuing to harbor fh (Fig. S5, c–e). These two clones must have arisen from a double crossover recombination event in a population of parasites harboring a duplicated copy of fh. Although parasites with gene duplication are thought to be unstable, the existence of duplication has been noted earlier in the case of rio2 (37) and dhodh (38). The variation in the genotype across the 17 clones that we have obtained shows that the parasites have not multiplied from a single wrong event of homologous recombination. On the contrary, the clonal lines with different genotypes, continuing to harbor fh, suggest a strong selection pressure for the retention of this gene. The PCRs with primers P9 and P10 (Fig. S5e) encompassing the full-length gene yielded the expected size band with genomic DNA from all 17 clones, indicating that all clones contain the full-length fh gene.
Figure 6.
Genotyping of P. berghei clones of knockout of fumarate hydratase. a, schematic representation of the selectable marker cassette inserted into the fh gene locus of P. berghei genome. Primers (P1–P8) used for diagnostic PCRs are indicated. b, schematic representation of the fh gene (PBANKA_0828100) flanked by 5′ UTR and 3′ UTR showing the location of primers P9 and P10. Shown is agarose gel electrophoresis of PCRs with genomic DNA from clones A–C (c, left panel) and clones H–M (c, right panel) for detection of 5′ integration; d, clones J and M for detection of 3′ integration (other clones did not answer for this PCR); e, clone A for the detection of fh gene; f, clones A–C for the presence of selectable marker cassette; g, clones C and O using primers P3 and P8. Shown in this figure is the genotyping of representative P. berghei clones, and the data on all 17 clones that were characterized are provided in Fig. S5. Clones C and M and O did not answer for 5′ integration, and only clones J, M, and Q answered for 3′ integration (c and d here and Fig. S5, c and d). All clones answered for the presence of the fh gene (e here and Fig. S5e). All clones except C and O answered by PCR with primers P2 and P6 indicating the integration of the entire selectable marker cassette into the genome (f here and Fig. S5f). Clones C and O answered for a shorter fragment of the selectable marker cassette covered by primers P3 and P8 (g here and Fig. S5g). hDHFR-yFCU, human DHFR-yeast cytosine and uridyl phosphoribosyltransferase; mr, molecular weight marker; numbers to the left of c–g are the sizes of the marker DNA fragments in kbp.
A study that appeared recently (39) reports on the knockout of the fh gene in P. berghei, with the knockout parasites exhibiting a slow growth phenotype. Although the authors show the absence of fh gene expression in the knockout strain, the genotyping for confirmation of knockout that was carried out with oligonucleotide primers corresponding to the homology arm used for recombination cannot confirm the site of integration. Apart from the length of the homology arm used for recombination, the key difference is with regard to the strains of mice used. Although our study has used BALB/c, Niikura et al. (39) have used the C57BL/6 strain of mice. P. berghei is known to exhibit differences in growth and infectivity across different strains of mice (40–42). This prompted us to examine the essentiality of fh gene for P. berghei when grown in the two different mouse strains, C57BL/6 and BALB/c. For this, a single transfection mixture was split into two halves and injected into C57BL/6 and BALB/c mice, and the whole experiment was performed twice. In the two experiments, intravenous injection of the transfected P. berghei cells yielded parasites in both strains of mice, C57BL/6 and BALB/c. However, in one of the attempts, upon pyrimethamine selection, drug-resistant parasites appeared only in the C57BL/6 mouse and not in the BALB/c strain even after 20 days of observation. In the second attempt, pyrimethamine-resistant parasites were obtained in both C57BL/6 and BALB/c mice. Genotyping by PCR of drug-selected parasites using diagnostic oligonucleotides was performed. Drug-selected parasites obtained from C57BL/6 mice of both attempts of transfection showed the right integration of the marker cassette along with the absence of the fh gene (results of genotyping performed from the second attempt of transfection is shown in Fig. 7). On the contrary, genotyping by PCR of drug-selected parasites obtained from BALB/c mice, revealed the presence of the marker cassette (Fig. 7b) along with the fh gene (Fig. 7a). Results from the transfection experiments, taken together, indicate that the fh gene in P. berghei can be knocked out when the parasites are grown in C57BL/6 strain of mice and not when BALB/c mouse is the host. It should be noted that a slow-growing FHKO line in BALB/c could be outgrown by parasites carrying the random or single crossover integration events, strongly suggesting that FH is required for robust growth. Similar mouse strain-specific essentiality of a P. berghei gene is seen in the case of purine nucleoside phosphorylase (PNP). Although P. berghei PNP has been shown to be refractory to knockout in transfectants grown in BALB/c mice as deposited in the PhenoPlasm database by Rayner and co-workers (43, 44), Niikura et al. (45) have successfully deleted the gene in P. berghei when transfected and grown in C57BL/6 mice. It should be noted that the exact nature of the plasmid constructs used for knockout are different across the two studies. To our knowledge, the study reported here is the first where simultaneously the same knockout construct has been used for deletion of fh gene using two different strains of mice as hosts. The variation that we observed across the two hosts used suggests the role of mouse strain in determining the essentiality of a parasite gene.
Figure 7.
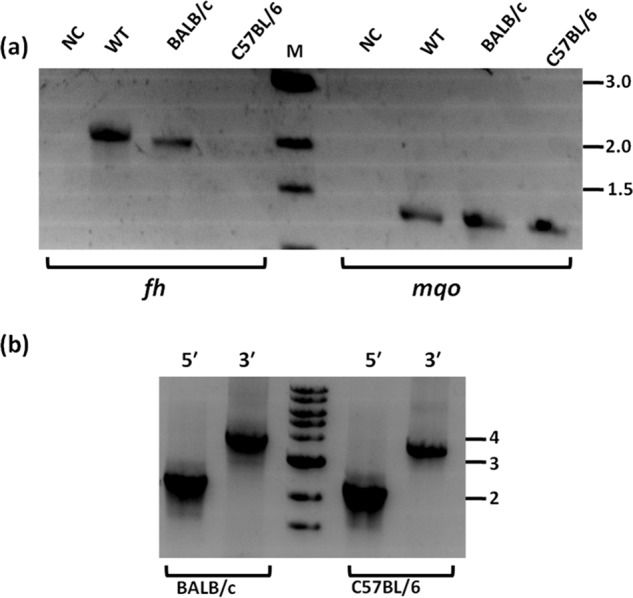
Genotyping of transfectants grown and drug-selected in different mouse strains. a, presence/absence of fumarate hydratase gene was validated by PCR using oligonucleotides P9 and P10 and genomic DNA isolated from respective parasites as template (lanes bracketed as fh). As a positive control for the presence of genomic DNA, PCR was performed with oligonucleotides corresponding to a segment of mqo gene loci (lanes bracketed as mqo). b, validation of 5′ and 3′ integration using primer pairs P1 and P4 and P5 and P7, respectively. Primer locations are as in Fig. 6. NC, negative control lacking template DNA; WT, P. berghei ANKA wildtype genomic DNA; BALB/c, genomic DNA from transfectants grown in BALB/c mouse; C57BL/6, genomic DNA from transfectants grown in C57BL/6 mouse; M, marker. Numbers to the right of figure are the sizes of the marker DNA fragments in kbp. The sequences of the oligonucleotides used are provided in Table S1.
Discussion
This study is the first report on the biochemical characterization of P. falciparum fumarate hydratase, an essential protein for the growth of the asexual stages of the parasite. Unlike higher eukaryotes, including humans that have only class II fumarate hydratase, most parasitic protozoa have only the class I enzyme. The purified Fe-S cluster reconstituted P. falciparum fumarate hydratase catalyzes the reversible conversion of fumarate and malate and also exhibits substrate specificity for d-tartrate and mesaconate. In vivo, the parasite enzyme complements fumarase deficiency in E. coli forming a basis for screening of inhibitors acting through this enzyme. The screening of small molecules as potential inhibitors of PfFH led to the identification of MSA as a class I FH-specific inhibitor. The specificity exhibited by dl-MSA for class I FH and pyromellitic acid and S-2,3-dicarboxyaziridine (46) for class II FH supports the presence of different active-site environments in the two classes of enzymes. This provides a framework for developing class I PfFH (and in general for class I FH)-specific inhibitors that will have no effect on the human enzyme. A recent study has reported the inhibition of TcFH by dl-MSA with a Ki value of 4.2 ± 0.5 μm, although no effect was observed on the class II human FH (22). The reason for dl-MSA's high specificity for class I FH must stem from the presence of 4Fe-4S cluster that interacts with the C2-hydroxyl group of malate (11). Replacement of the hydroxyl group with a thiol probably leads to tight binding through the Fe–S interaction. The 4Fe-4S cluster containing quinolinate synthase (NadA) is strongly inhibited by dithiohydroxyphthalic acid (DTHPA), the thio-analog of the transition-state intermediate of the reaction catalyzed, in a manner similar to MSA inhibition of class I FH. Interactions of the thiol groups of DTHPA with the iron atom of the cluster leads to a strong binding affinity for the enzyme (47). In this context, we expect thiomesaconate and S,S-dithiotartrate, analogs of the substrates mesaconate and S,S- tartrate (d-tartrate) to be also strong inhibitors of class I FH.
Despite the low Ki values for dl-MSA for PfFHΔ40, the IC50 value for parasite killing was higher, which may be due to partial functional substitution by host fumarate hydratase present in the infected erythrocyte compartment. Earlier studies have shown that fumarate in P. falciparum is metabolized to malate, oxaloacetate, aspartate, pyruvate, and lactate with the involvement of enzymes in parasite mitochondrial and cytosolic compartments (3). Under conditions of PfFH inhibition, the flux through these metabolic reactions would require transport of intermediates across three compartments (erythrocyte, parasite, and the mitochondrion), evidence for which is absent. Therefore, human fumarase may not completely substitute the PfFH function. Interestingly, the IC50 value for PfFH-dependent E. coli cell death by dl-MSA is also higher than the Ki value for the enzyme. This common observation of higher IC50 values observed for both cell types could be due to the low intracellular availability of the drug (because of the hydrophilic nature of the molecule and hence poor transport) and/or due to metabolism leading to degradation of MSA. MSA dioxygenase, an enzyme that converts mercaptosuccinic acid to succinate, is present in the bacterium Variovorax paradoxus (48, 49). However, we could not find homologs of the enzyme in E. coli or P. falciparum.
The non-viability of FH-deficient intra-erythrocytic asexual parasites suggests metabolic perturbations leading to lethality. The major source of intracellular fumarate in Plasmodium is from the synthesis of AMP. From the context of metabolism, the absence of fumarate hydratase would result in the accumulation of fumarate. The possible metabolic consequences of this are schematically shown in Fig. 8. The last reaction in AMP synthesis catalyzed by ASL is a reversible process with similar catalytic efficiencies for the forward and reverse reactions. Accumulation of fumarate could lead to an increased flux through the reverse reaction catalyzed by ASL resulting in accumulation of succinyl-AMP and lowered levels of AMP, eventually resulting in compromised cell growth. Subversion of ASL activity through 5-aminoimidazole-4-carboxamide ribonucleotide has been shown to result in parasite death (50). Apart from perturbing AMP synthesis, high levels of fumarate can result in succination of cysteinyl residues in proteins and glutathione (51), thereby compromising cellular homeostasis (52). Succinated proteome in human cell lines (53–55) and Mycobacterium tuberculosis (56) has been examined, and these studies highlight the toxic effects of high levels of fumarate. Fumarate is recycled to aspartate through the action of enzymes FH, malate-quinone oxidoreductase, and aspartate aminotransferase. In the absence of FH, the levels of the malate and oxaloacetate intermediates in this pathway would be perturbed leading to lower levels of recycling. Furthermore, with lowered levels of OAA due to the absence of FH, the generation of NAD+ through MDH would also be impaired. All these biochemical requirements could make FH in Plasmodium essential.
Figure 8.
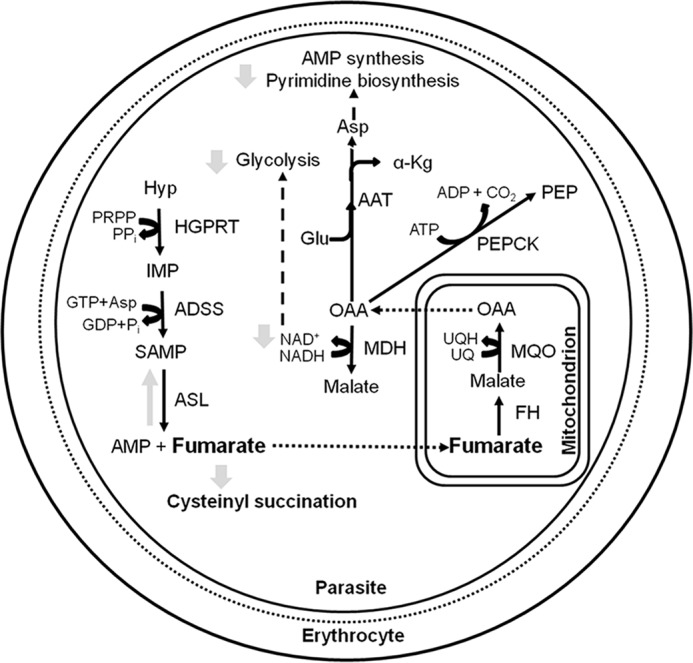
Metabolic consequences of fumarate hydratase gene deletion in Plasmodium. Dashed arrows indicate the flow of metabolites into a pathway, and the dotted arrows indicate transport across compartments. Gray arrows show possible metabolic consequences of fh gene deletion (see text for explanation). AAT, aspartate aminotransferase; HGPRT, hypoxanthine-guanine phosphoribosyltransferase; MDH, malate dehydrogenase, MQO, malate-quinone oxidoreductase; PEPCK, phosphoenolpyruvate carboxykinase; α-Kg, α-ketoglutarate; Hyp, hypoxanthine; PEP, phosphoenolpyruvate; PRPP, phosphoribosyl 5′-pyrophosphate, SAMP, succinyl-AMP, UQ, ubiquinone, UQH, ubiquinol.
Experimental procedures
Materials
RPMI 1640 medium, components of cytomix, and all chemical reagents used were obtained from Sigma. MitoTracker Red CM-H2XRos, Hoechst 33342, AlbuMAX I, Ni-NTA-conjugated agarose, and Phusion high-fidelity DNA polymerase were procured from Thermo Fisher Scientific, Inc. Restriction enzymes and T4 DNA ligase were from New England Biolabs. Primers were custom-synthesized from Sigma, Bangalore. Media components were from Himedia Laboratories, Mumbai, India. 2,3-[13C]Fumarate was procured from Isotec, and dl-mercaptosuccinic acid (dl-MSA) was obtained from Sigma.
Sequence analysis
E. coli FumA (EcFumA) protein sequence (UniProt ID: P0AC33) was used as a query in BLASTP (57) to retrieve all eukaryotic class I FH sequences by restricting the search to eukaryotes. Each of these eukaryotic organisms with class I FH was individually searched for the presence of class II FH using BLASTP and E. coli FumC (EcFumC) (UniProt ID: P05042) as the query sequence. Hits with an e-value lower than 10−10 were considered significant.
Generation of plasmid constructs
Sequences of all oligonucleotide primers used for cloning and for genotyping of E. coli and Plasmodium mutants are given in Table S1. For recombinant expression of PfFH, the DNA fragment corresponding to a protein segment lacking the N-terminal 40 amino acids (Δ40) was amplified by PCR using parasite genomic DNA as template, appropriate oligonucleotides, and Phusion DNA polymerase. The fragment was cloned into modified pET21b (Novagen, Merck) using restriction enzyme sites BamHI and SalI to obtain the construct pET-PfFHΔ40 that encodes the protein with an N-terminal His6 tag. For functional complementation in the fh null strain of E. coli, the pQE30 plasmid (Qiagen, Germany) containing full-length (PfFHFL) and two different N terminus-deleted constructs (PfFHΔ40, PfFHΔ120) of the P. falciparum fh gene were used. The generation of different expression constructs involved amplification by PCR of appropriate fragments followed by cloning into the pQE30 plasmid using restriction sites BamHI and SalI. The plasmids thus obtained are pQE-PfFHFL, pQE-PfFHΔ40, and pQE-PfFHΔ120. For 3′-tagging of the endogenous fh gene with GFP in the P. falciparum strain PM1KO (27), the nucleotide fragment corresponding to the full-length fh gene without the terminator codon was amplified from P. falciparum 3D7 genomic DNA using appropriate oligonucleotides (PfFHpGDB-Xho1-FP and PfFHpGDB-AvrII-RP; Table S1) and cloned into the plasmid, pGDB (27), using the restriction sites XhoI and AvrII to yield the plasmid pGDB-PfFH. For recombinant expression of E. coli FumC and FumA enzymes, the nucleotide sequence corresponding to the full-length genes were PCR-amplified using E. coli genomic DNA as template and cloned in pQE30 and pET-DUET (Novagen, Merck), respectively, using the restriction sites BamHI and SalI. The resulting plasmids are pQE-EcFumC and pET-EcFumA. All the clones were confirmed by DNA sequencing.
Protein expression, purification, and reconstitution of iron–sulfur cluster
For recombinant expression of PfFHΔ40 and EcFumA, the E. coli strain BL21(DE3)-RIL was transformed with pET-PfFHΔ40/pET-EcFumA and selected on Luria-Bertani agar (LB agar) plate containing ampicillin (100 μg ml−1) and chloramphenicol (34 μg ml−1). Multiple colonies were picked and inoculated into 10 ml of LB broth. The culture was grown for 6 h at 37 °C, and the cells were pelleted, washed with antibiotic-free LB broth, and then used for inoculating 800 ml of Terrific broth (TB). The cells were grown at 30 °C until A600 reached 0.5, thereafter induced with IPTG (0.05 mm for PfFHΔ40 and 0.3 mm for EcFumA), and grown for further 16 h at 16 °C for PfFHΔ40 and 4 h at 30 °C for EcFumA. Cells were harvested by centrifugation and resuspended in lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 5 mm β-mercaptoethanol, and 10% glycerol. Cell lysis was achieved by four cycles of French press at 1000 p.s.i., and the lysate was cleared by centrifugation at 30,000 × g for 30 min. The supernatant was mixed with 1 ml of Ni-NTA–agarose slurry pre-equilibrated with lysis buffer and incubated in an anaerobic chamber for 30 min at room temperature. The tube was sealed air-tight within the chamber and transferred to 4 °C. Binding of the His6-tagged PfFHΔ40/EcFumA to Ni-NTA–agarose was continued with gentle shaking for 3 h. The tube was transferred back to the chamber, and the beads were washed with 50 ml of lysis buffer followed by washes with 10 and 20 mm imidazole containing lysis buffer (10 ml each), and the protein was eluted directly with 500 mm imidazole in lysis buffer. An equal volume of 100% glycerol was added to the eluate such that the final concentration of glycerol is 50%. EcFumC was purified under aerobic conditions using Ni-NTA–affinity chromatography.
Reconstitution of the cluster in PfFHΔ40 and EcFumA was performed under anaerobic conditions. For PfFHΔ40, the procedure was initiated by incubation of the protein solution with 5 mm DTT for 30 min. Following this, 0.5 mm each of sodium sulfide and ferrous ammonium sulfate was added. The reconstitution was allowed to proceed overnight following which the protein was used for activity measurements. For EcFumA, reconstitution was achieved by the addition of 5 mm DTT for 30 min followed by the addition of 0.5 mm ferrous ammonium sulfate.
Activity measurements
For recording NMR spectra, the purified recombinant PfFHΔ40 was incubated with 2,3-[13C]fumarate for 30 min at 37 °C in 20 mm sodium phosphate, pH 7.4. The protein was precipitated with TCA, and the supernatant, neutralized with 5 n KOH, was used for recording the 13C NMR spectrum in a 400 MHz Bruker NMR machine. D2O was added to a final concentration of 10% to the sample before acquiring the spectrum.
All initial velocity measurements were performed at 37 °C using a spectrophotometric method and initiated with the addition of the enzyme. The activity of EcFumC was measured using a reported method (11) in a solution containing 100 mm MOPS, pH 6.9, 5 mm MgCl2, and 5 mm DTT. For EcFumA, the assays were performed in 50 mm potassium phosphate, pH 7.4 containing 2 mm DTT. The activity of PfFHΔ40 was found to be maximal at pH 8.5, and all assays were carried out at this pH in 50 mm Tris-HCl. The conversion of fumarate to malate was monitored spectrophotometrically as a drop in absorbance caused by the depletion of fumarate. Depending upon the initial concentration of fumarate, the enzymatic conversion was monitored at different wavelengths as follows: 240 nm (ϵ240 = 2440 m−1 cm−1) (9) for fumarate concentrations of up to 500 μm; 270 nm (ϵ270 = 463 m−1 cm−1) for concentrations ranging from 0.5 to 1.2 mm; 280 nm (ϵ280 = 257 m−1 cm−1) for concentrations from 1.2 to 2.6 mm; 290 nm (ϵ290 = 110 m−1 cm−1) for concentrations from 2.6 to 6 mm; 300 nm (ϵ300 = 33 m−1 cm−1) for concentrations from 6 to 20 mm; and 305 nm (ϵ305 = 18 m−1 cm−1) for concentrations from 20 to 40 mm in a quartz cuvette of 1-cm path length. The conversion of mesaconate to citramalate was monitored as a drop in absorbance at wavelengths 240 nm (ϵ240 = 3791 m−1 cm−1) for concentrations up to 250 μm, 280 nm (ϵ280 = 142 m−1 cm−1) for concentrations from 250 to 4000 μm, and at 290 nm (ϵ290 = 40 m−1 cm−1) for concentrations from 4 to 16 mm. The use of different wavelengths ensured that the sensitivity of the detection of conversion of fumarate to malate was maximal. The conversion of malate to fumarate was monitored spectrophotometrically as an increase in absorbance at 240 nm due to the synthesis of fumarate. Activity on tartrate was monitored by a coupled enzyme assay using P. falciparum malate dehydrogenase (PfMDH) purified in-house from an E. coli expression clone (3). The assay was carried out at 37 °C in 50 mm Tris-HCl, pH 8.5, containing 100 μm NADH, 4 μg of PfMDH, and 2 mm d-tartrate. The reaction was initiated with 3.4 μg of PfFH.
For testing the effect of small molecules on the activity of PfFHΔ40, fumarate was used as the substrate at a concentration of 3 mm. The molecules were tested at a concentration of 0.5 mm. For estimating Ki values for dl-MSA and meso-tartrate, the initial velocity was measured at varying concentrations of malate (46 μm to 12 mm)/fumarate (24 μm to 25 mm) with dl-MSA/meso-tartrate fixed at different concentrations. The mode of inhibition was inferred from the type of intersection pattern of lines in the Lineweaver-Burk plot. The Ki value for dl-MSA was obtained from a global fit of the data by non-linear regression analysis to a competitive model for enzyme inhibition using GraphPad Prism5. All assays were performed at least three times. Data points in the plots and the values derived are mean ± S.E.
Generation and phenotyping of ΔfumACB strain of E. coli
To generate a fumarate hydratase-null strain of E. coli, a fumB-null strain (JW4083-1, fumB748 (del)::kan), derived from the E. coli strain BW25113, was obtained from Coli Genetic Stock Centre (CGSC), Yale University, New Haven, CT (58). To remove the kanamycin cassette flanked by FRT sites at the fumB gene locus and to subsequently knock out fumA and fumC, standard protocols were followed (59), and this is described in the supporting Methods S1 and Fig. S3). Knockout of the genes was validated by PCR using genomic DNA of the mutant strain as template and appropriate oligonucleotides (Table S1). M9 minimal medium agar plates containing either fumarate or malate (0.4%) as the sole carbon source and supplemented with trace elements were used to check the phenotype of the strains ΔfumACB, ΔfumA, ΔfumB, and ΔfumC. An equal number of cells of each of these strains was spread on both malate- and fumarate-containing M9 agar plates, and the growth phenotype was scored at the end of 48 h of incubation at 37 °C under aerobic conditions.
Complementation of FH deficiency in ΔfumACB strain with PfFH and growth inhibition with MSA
The ΔfumACB strain of E. coli was transformed with the plasmids pQE-PfFHFL, pQE-PfFHΔ40, pQE-PfFHΔ120, and pQE30 and selected on LB plate containing 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin. A single colony from the plate was inoculated into 10 ml of LB broth and allowed to grow overnight. An aliquot of each of the cultures was washed three times with sterile M9 medium to remove traces of LB broth. The cells were resuspended in M9 medium, and an A600-normalized aliquot of the suspensions was spread on an M9 agar plate containing the appropriate carbon source and antibiotics. It should be noted that the parent strain BW25113 has a single copy of the lacI+ allele and not the lacIq (60), and hence, for induction of protein expression in this strain using pQE30-based constructs, the addition of IPTG is optional.
To check the effect of MSA on the E. coli strain ΔfumACB with the plasmid pQE-PfFHΔ40, the culture was grown overnight in 10 ml of LB medium, and 1 ml of the culture was washed twice with M9 minimal medium and resuspended in 1 ml of M9 minimal medium. 150 μl of this suspension was added to tubes containing 5 ml of M9 minimal medium with 10 mm fumarate as the sole carbon source and appropriate antibiotics (50 μg ml−1 kanamycin and 100 μg ml−1 ampicillin). Varied concentrations of dl-MSA ranging from 1 μm to 15 mm were added to the tubes. The growth of the cultures was monitored by measuring A600 at the end of 10 h. The experiment was performed three times. Data points in the plot and the value derived are mean ± S.E.
P. falciparum culture, transfection, and growth inhibition with MSA
Intra-erythrocytic stages of P. falciparum 3D7 strain (procured from MR4) were grown by the method established by Trager and Jensen (61). The parasites were grown in medium containing RPMI 1640 buffered with 25 mm HEPES and supplemented with 20 mm sodium bicarbonate, 0.5% AlbuMAX I, 0.5% glucose, and 100 μm hypoxanthine. O-positive erythrocytes from healthy volunteers were added to the culture to a final hematocrit of 2% for regular maintenance. For examining the localization of PfFH, PM1KO strain (27) was transfected with the plasmid pGDB-PfFH. For this, preloading of erythrocytes (62) with plasmid DNA was carried out by electroporation using a square wave pulse (eight pulses of 365 V each lasting 1 ms with a gap of 100 ms) program in XL electroporator (Bio-Rad). Briefly, 100 μg of plasmid DNA dissolved in cytomix (62) was used for transfection of uninfected erythrocytes resuspended in cytomix. After electroporation, the cells were washed with incomplete media to remove cell debris, and 1 ml of infected erythrocytes (2% hematocrit and 6–8% parasitemia) containing late schizont stage parasites was added. The parasites were allowed to reinvade, and when the parasitemia reached 6–8%, drug selection was started by the addition of trimethoprim (10 μm) and blasticidin S (2.5 μg ml−1). This strain of P. falciparum is referred to as PfFH-GFP. The strain was subjected to three rounds of drug cycling, which included growing the cultures on and off blasticidin (10 days each) in the continuous presence of trimethoprim following which the genotyping of the strain was performed by PCR using genomic DNA as template and primers P1–P4 (Table S1).
The IC50 value for dl-MSA for inhibition of parasite growth was determined by a serial 2-fold dilution. Briefly, the effect of dl-MSA on the viability of the 3D7 strain of P. falciparum was determined by counting the number of parasites in at least 1000 erythrocytes in Giemsa-stained smears of cultures grown in the presence of increasing concentrations (3 μm to 40 mm) of the drug. The experiment was performed three times. Data points in the plot and the value derived are mean ± S.E.
Mitochondrial staining and microscopy
For mitochondrial staining of PfFH-GFP parasites, the culture containing mixed stages of parasites was washed twice with incomplete medium to remove any traces of AlbuMAX I, and the cells were resuspended with incomplete medium containing 100 nm MitoTracker CM-H2XRos and incubated at 37 °C in a candle jar for 30 min. For nuclear staining, Hoechst 33342 was added to the culture to a final concentration of 5 μg ml−1 and incubated for an additional 5 min at 37 °C. For imaging, 500 μl of this culture was washed once with incomplete medium, and the cells were resuspended in 50% glycerol/PBS solution. 5 μl of the suspension was placed under a coverslip and imaged using DeltaVision Elite wide-field microscope (GE Healthcare) at room temperature. The images were processed using ImageJ (63, 64). The Pearson's correlation coefficient was obtained using the Coloc2 plugin from Fiji (65).
P. berghei culturing and genetic manipulation
Intra-erythrocytic asexual stages of P. berghei ANKA (procured from MR4) were maintained in BALB/c mice. For the generation of the knockout construct and for the transfection of parasites, established procedures were followed (66, 67). All transfection experiments were performed twice. Starting from fh genomic library clone (Clone ID: PbG01-2466a09) obtained from PlasmoGEM repository (Wellcome Trust Sanger Institute, UK), the fh gene knockout construct was generated by using the recombineering strategy described by Pfander et al. (66) (Fig. S4). Flanking the resistance marker, this construct has 1395- and 2049-bp DNA segments corresponding to regions upstream and downstream, respectively, of the fh gene to enable gene knockout by double-crossover recombination. For transfection of P. berghei, the parasites were harvested from infected mice at a parasitemia of around 1–3%. Around 0.8–1.0 ml of blood was obtained from each mouse, and the parasites were synchronized at schizont stage by in vitro growth at 36.5 °C with constant shaking at an optimal speed of 120–150 rpm in medium containing RPMI 1640 medium with glutamine, 25 mm HEPES, 10 mm NaHCO3, and 20% fetal bovine serum under a gassed environment (5% oxygen, 5% carbon dioxide, and 90% nitrogen). The schizonts were purified by density gradient centrifugation on Nycodenz and transfected with NotI-digested linear DNA of the fh gene knockout construct using a 2D-nucleofector (Lonza, Switzerland). Pyrimethamine selection was started 1 day after transfection (67). Limiting dilution cloning of the drug-resistant parasites was performed using 16 mice in two batches (32 mice in total). Genomic DNA was isolated from 17 individual parasite lines and subjected to a series of diagnostic PCRs to check for integration and loss of fh gene. Mouse-strain–dependent essentiality of fh for P. berghei was examined by transfection of wildtype parasites harvested from infected BALB/c mice with fh gene knockout construct. An equal volume of parasite suspension from this single transfection reaction was injected intravenously into BALB/c and C57BL/6 mice. Genotyping of drug-selected parasites obtained from both mice was performed by PCR using gene- and integration-specific oligonucleotides. The sequences of the oligonucleotides used are provided in Table S1.
Ethics statement
All animal experiments involving BALB/c and C57BL/6 mice adhered to the standard operating procedures prescribed by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), a statutory body under the Prevention of Cruelty to Animals Act of 1960 and Breeding and Experimentation Rules of 1998, Constitution of India. The study was a part of the project numbered HB004/201/CPCSEA and is approved by the Institutional animal ethics committee (IAEC) that comes under the purview of CPCSEA. Whole blood for P. falciparum culturing was collected from healthy volunteers with written informed consent.
Author contributions
V. J., A. S., H. B. conceived and designed the experiments. V. J., A. S., P. K., J. K. performed the experiments. V. J., A. S., H. B. analyzed the data. V. J., A. S., H. B. wrote the paper.
Supplementary Material
This work was supported in part by Department of Biotechnology, Ministry of Science and Technology, Government of India, Grants BT/PR11294/BRB/10/1291/2014 and BT/PR13760/COE/34/42/2015; Science and Engineering Research Board, Department of Science and Technology, Government of India, Grant EMR/2014/001276, and Institutional funding from Jawaharlal Nehru Centre of Advanced Scientific Research, Department of Science and Technology, India. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5, Tables S1–S2, Methods S1, Text S1, and Refs. 1–5.
- TCA
- tricarboxylic acid
- OAA
- oxaloacetate
- PfFH
- P. falciparum fumarate hydratase
- Ec
- E. coli
- ADSS
- adenylosuccinate synthetase
- ASL
- adenylosuccinate lyase
- MDH
- malate dehydrogenase
- DHFR
- dihydrofolate reductase
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- dl-MSA
- dl-mercaptosuccinic acid
- DTHPA
- dithiohydroxyphthalic acid
- CPCSEA
- Committee for the Purpose of Control and Supervision of Experiments on Animals
- PNP
- purine nucleoside phosphorylase.
References
- 1. Roth E. (1990) Plasmodium falciparum carbohydrate metabolism: a connection between host cell and parasite. Blood Cells 16, 453–460 [PubMed] [Google Scholar]
- 2. MacRae J. I., Dixon M. W., Dearnley M. K., Chua H. H., Chambers J. M., Kenny S., Bottova I., Tilley L., and McConville M. J. (2013) Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol. 11, 67 10.1186/1741-7007-11-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bulusu V., Jayaraman V., and Balaram H. (2011) Metabolic fate of fumarate, a side product of the purine salvage pathway in the intra-erythrocytic stages of Plasmodium falciparum. J. Biol. Chem. 286, 9236–9245 10.1074/jbc.M110.173328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Teipel J. W., Hass G. M., and Hill R. L. (1968) The substrate specificity of fumarase. J. Biol. Chem. 243, 5684–5694 [PubMed] [Google Scholar]
- 5. Chen B.-S., Otten L. G., and Hanefeld U. (2015) Stereochemistry of enzymatic water addition to C=C bonds. Biotechnol. Adv. 33, 526–546 10.1016/j.biotechadv.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 6. Resch V., and Hanefeld U. (2015) The selective addition of water. Catal. Sci. Technol. 5, 1385–1399 10.1039/C4CY00692E [DOI] [Google Scholar]
- 7. Woods S. A., Schwartzbach S. D., and Guest J. R. (1988) Two biochemically distinct classes of fumarase in Escherichia coli. Biochim. Biophys. Acta 954, 14–26 10.1016/0167-4838(88)90050-7 [DOI] [PubMed] [Google Scholar]
- 8. Shimoyama T., Rajashekhara E., Ohmori D., Kosaka T., and Watanabe K. (2007) MmcBC in Pelotomaculum thermopropionicum represents a novel group of prokaryotic fumarases. FEMS Microbiol. Lett. 270, 207–213 10.1111/j.1574-6968.2007.00665.x [DOI] [PubMed] [Google Scholar]
- 9. Flint D. H. (1994) Initial kinetic and mechanistic characterization of Escherichia coli fumarase A. Arch. Biochem. Biophys. 311, 509–516 10.1006/abbi.1994.1269 [DOI] [PubMed] [Google Scholar]
- 10. Kronen M., Sasikaran J., and Berg I. A. (2015) Mesaconase activity of class I fumarase contributes to mesaconate utilization by Burkholderia xenovorans. Appl. Environ. Microbiol. 81, 5632–5638 10.1128/AEM.00822-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feliciano P. R., Drennan C. L., and Nonato M. C. (2016) Crystal structure of an Fe-S cluster–containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold. Proc. Natl. Acad. Sci. U.S.A. 113, 9804–9809 10.1073/pnas.1605031113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beinert H., Kennedy M. C., and Stout C. D. (1996) Aconitase as iron–sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96, 2335–2374 10.1021/cr950040z [DOI] [PubMed] [Google Scholar]
- 13. Lloyd S. J., Lauble H., Prasad G. S., and Stout C. D. (1999) The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex. Protein Sci. 8, 2655–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kronen M., and Berg I. A. (2015) Mesaconase/fumarase FumD in Escherichia coli O157:H7 and promiscuity of Escherichia coli class I fumarases FumA and FumB. PLoS ONE 10, e0145098 10.1371/journal.pone.0145098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sacchettini J. C., Meininger T., Roderick S., and Banaszak L. J. (1986) Purification, crystallization, and preliminary X-ray data for porcine fumarase. J. Biol. Chem. 261, 15183–15185 [PubMed] [Google Scholar]
- 16. Weaver T. M., Levitt D. G., and Banaszak L. J. (1993) Purification and crystallization of fumarase C from Escherichia coli. J. Mol. Biol. 231, 141–144 10.1006/jmbi.1993.1264 [DOI] [PubMed] [Google Scholar]
- 17. Weaver T., Lees M., Zaitsev V., Zaitseva I., Duke E., Lindley P., McSweeny S., Svensson A., Keruchenko J., Keruchenko I., Gladilin K., and Banaszak L. (1998) Crystal structures of native and recombinant yeast fumarase. J. Mol. Biol. 280, 431–442 10.1006/jmbi.1998.1862 [DOI] [PubMed] [Google Scholar]
- 18. Weaver T. (2005) Structure of free fumarase C from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 61, 1395–1401 10.1107/S0907444905024194 [DOI] [PubMed] [Google Scholar]
- 19. Pereira de Pádua R. A., and Nonato M. C. (2014) Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of recombinant human fumarase. Acta Crystallogr. F Struct. Biol. Commun. 70, 120–122 10.1107/S2053230X13033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacot D., Waller R. F., Soldati-Favre D., MacPherson D. A., and MacRae J. I. (2016) Apicomplexan energy metabolism: carbon source promiscuity and the quiescence hyperbole. Trends Parasitol. 32, 56–70 10.1016/j.pt.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 21. Feliciano P. R., Gupta S., Dyszy F., Dias-Baruffi M., Costa-Filho A. J., Michels P. A., and Nonato M. C. (2012) Fumarate hydratase isoforms of Leishmania major: subcellular localization, structural and kinetic properties. Int. J. Biol. Macromol. 51, 25–31 10.1016/j.ijbiomac.2012.04.025 [DOI] [PubMed] [Google Scholar]
- 22. de Pádua R. A. P., Kia A. M., Costa-Filho A. J., Wilkinson S. R., and Nonato M. C. (2017) Characterisation of the fumarate hydratase repertoire in Trypanosoma cruzi. Int. J. Biol. Macromol. 102, 42–51 10.1016/j.ijbiomac.2017.03.099 [DOI] [PubMed] [Google Scholar]
- 23. Ke H., Lewis I. A., Morrisey J. M., McLean K. J., Ganesan S. M., Painter H. J., Mather M. W., Jacobs-Lorena M., Llinás M., and Vaidya A. B. (2015) Genetic investigation of tricarboxylic acid metabolism during the Plasmodium falciparum life cycle. Cell Rep. 11, 164–174 10.1016/j.celrep.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carey M. A., Papin J. A., and Guler J. L. (2017) Novel Plasmodium falciparum metabolic network reconstruction identifies shifts associated with clinical antimalarial resistance. BMC Genomics 18, 543 10.1186/s12864-017-3905-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oborník M., Modrý D., Lukeš M., Cernotíková-Sříbrná E., Cihlář J., Tesařová M., Kotabová E., Vancová M., Prášil O., and Lukeš J. (2012) Morphology, ultrastructure and life cycle of Vitrella brassicaformis n. sp., n. gen., a novel chromerid from the Great Barrier Reef. Protist. 163, 306–323 10.1016/j.protis.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 26. Fukasawa Y., Tsuji J., Fu S.-C., Tomii K., Horton P., and Imai K. (2015) MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics 14, 1113–1126 10.1074/mcp.M114.043083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muralidharan V., Oksman A., Iwamoto M., Wandless T. J., and Goldberg D. E. (2011) Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc. Natl. Acad. Sci. U.S.A. 108, 4411–4416 10.1073/pnas.1018449108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bender A., van Dooren G. G., Ralph S. A., McFadden G. I., and Schneider G. (2003) Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum. Mol. Biochem. Parasitol. 132, 59–66 10.1016/j.molbiopara.2003.07.001 [DOI] [PubMed] [Google Scholar]
- 29. Bell P. J., Andrews S. C., Sivak M. N., and Guest J. R. (1989) Nucleotide sequence of the FNR-regulated fumarase gene (fumB) of Escherichia coli K-12. J. Bacteriol. 171, 3494–3503 10.1128/jb.171.6.3494-3503.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Antonkine M. L., Koay M. S., Epel B., Breitenstein C., Gopta O., Gärtner W., Bill E. L., and Lubitz W. (2009) Synthesis and characterization of de novo designed peptides modelling the binding sites of [4Fe–4S] clusters in photosystem I. Biochim. Biophys. Acta 1787, 995–1008 10.1016/j.bbabio.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 31. Jervis A. J., Crack J. C., White G., Artymiuk P. J., Cheesman M. R., Thomson A. J., Le Brun N. E., and Green J. (2009) The O2 sensitivity of the transcription factor FNR is controlled by Ser24 modulating the kinetics of [4Fe-4S] to [2Fe-2S] conversion. Proc. Natl. Acad. Sci. U.S.A. 106, 4659–4664 10.1073/pnas.0804943106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crack J. C., Smith L. J., Stapleton M. R., Peck J., Watmough N. J., Buttner M. J., Buxton R. S., Green J., Oganesyan V. S., Thomson A. J., and Le Brun N. E. (2011) Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins. J. Am. Chem. Soc. 133, 1112–1121 10.1021/ja109581t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crack J. C., Stapleton M. R., Green J., Thomson A. J., and Le Brun N. E. (2013) Mechanism of [4Fe-4S](Cys)4 cluster nitrosylation is conserved among NO-responsive regulators. J. Biol. Chem. 288, 11492–11502 10.1074/jbc.M112.439901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakamaru-Ogiso E., Yano T., Ohnishi T., and Yagi T. (2002) Characterization of the iron–sulfur cluster coordinated by a cysteine cluster motif (CXXCXXXCX27C) in the Nqo3 subunit in the proton-translocating NADH-quinone oxidoreductase (NDH-1) of Thermus thermophilus HB-8. J. Biol. Chem. 277, 1680–1688 10.1074/jbc.M108796200 [DOI] [PubMed] [Google Scholar]
- 35. van Vugt-Lussenburg B. M., van der Weel L., Hagen W. R., and Hagedoorn P.-L. (2013) Biochemical similarities and differences between the catalytic [4Fe-4S] cluster containing fumarases FumA and FumB from Escherichia coli. PLoS ONE. 8, e55549 10.1371/journal.pone.0055549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hallée S., and Richard D. (2015) Evidence that the malaria parasite Plasmodium falciparum putative rhoptry protein 2 localizes to the Golgi apparatus throughout the erythrocytic cycle. PLoS ONE 10, e0138626 10.1371/journal.pone.0138626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gomes A. R., Bushell E., Schwach F., Girling G., Anar B., Quail M. A., Herd C., Pfander C., Modrzynska K., Rayner J. C., and Billker O. (2015) A genome-scale vector resource enables high-throughput reverse genetic screening in a malaria parasite. Cell Host Microbe 17, 404–413 10.1016/j.chom.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guler J. L., Freeman D. L., Ahyong V., Patrapuvich R., White J., Gujjar R., Phillips M. A., DeRisi J., and Rathod P. K. (2013) Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLoS Pathog. 9, e1003375 10.1371/journal.ppat.1003375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Niikura M., Komatsuya K., Inoue S.-I., Matsuda R., Asahi H., Inaoka D. K., Kita K., and Kobayashi F. (2017) Suppression of experimental cerebral malaria by disruption of malate:quinone oxidoreductase. Malar. J. 16, 247 10.1186/s12936-017-1898-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eling W., van Zon A., and Jerusalem C. (1977) The course of a Plasmodium berghei infection in six different mouse strains. Z. Parasitenkd. 54, 29–45 10.1007/BF00380634 [DOI] [PubMed] [Google Scholar]
- 41. Scheller L. F., Wirtz R. A., and Azad A. F. (1994) Susceptibility of different strains of mice to hepatic infection with Plasmodium berghei. Infect. Immun. 62, 4844–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amani V., Boubou M. I., Pied S., Marussig M., Walliker D., Mazier D., and Rénia L. (1998) Cloned lines of Plasmodium berghei ANKA differ in their abilities to induce experimental cerebral malaria. Infect. Immun. 66, 4093–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bushell E., Gomes A. R., Sanderson T., Anar B., Girling G., Herd C., Metcalf T., Modrzynska K., Schwach F., Martin R. E., Mather M. W., McFadden G. I., Parts L., Rutledge G. G., Vaidya A. B., et al. (2017) Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272 10.1016/j.cell.2017.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanderson T., and Rayner J. C. (2017) PhenoPlasm: a database of disruption phenotypes for malaria parasite genes. Wellcome Open Res. 2, 45 10.12688/wellcomeopenres.11896.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Niikura M., Inoue S.-I., Mineo S., Yamada Y., Kaneko I., Iwanaga S., Yuda M., and Kobayashi F. (2013) Experimental cerebral malaria is suppressed by disruption of nucleoside transporter 1 but not purine nucleoside phosphorylase. Biochem. Biophys. Res. Commun. 432, 504–508 10.1016/j.bbrc.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 46. Ueda Y., Yumoto N., Tokushige M., Fukui K., and Ohya-Nishiguchi H. (1991) Purification and characterization of two types of fumarase from Escherichia coli. J. Biochem. 109, 728–733 10.1093/oxfordjournals.jbchem.a123448 [DOI] [PubMed] [Google Scholar]
- 47. Chan A., Clémancey M., Mouesca J.-M., Amara P., Hamelin O., Latour J.-M., and Ollagnier de Choudens S. (2012) Studies of inhibitor binding to the [4Fe-4S] cluster of quinolinate synthase. Angew. Chemie Int. Ed. Engl. 51, 7711–7714 10.1002/anie.201202261 [DOI] [PubMed] [Google Scholar]
- 48. Brandt U., Waletzko C., Voigt B., Hecker M., and Steinbüchel A. (2014) Mercaptosuccinate metabolism in Variovorax paradoxus strain B4—a proteomic approach. Appl. Microbiol. Biotechnol. 98, 6039–6050 10.1007/s00253-014-5811-7 [DOI] [PubMed] [Google Scholar]
- 49. Brandt U., Schürmann M., and Steinbüchel A. (2014) Mercaptosuccinate dioxygenase, a cysteine dioxygenase homologue, from Variovorax paradoxus strain B4 is the key enzyme of mercaptosuccinate degradation. J. Biol. Chem. 289, 30800–30809 10.1074/jbc.M114.579730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bulusu V., Thakur S. S., Venkatachala R., and Balaram H. (2011) Mechanism of growth inhibition of intra-erythrocytic stages of Plasmodium falciparum by 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR). Mol. Biochem. Parasitol. 177, 1–11 10.1016/j.molbiopara.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 51. Alderson N. L., Wang Y., Blatnik M., Frizzell N., Walla M. D., Lyons T. J., Alt N., Carson J. A., Nagai R., Thorpe S. R., and Baynes J. W. (2006) S-(2-Succinyl)cysteine: a novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch. Biochem. Biophys. 450, 1–8 10.1016/j.abb.2006.03.005 [DOI] [PubMed] [Google Scholar]
- 52. Sullivan L. B., Martinez-Garcia E., Nguyen H., Mullen A. R., Dufour E., Sudarshan S., Licht J. D., Deberardinis R. J., and Chandel N. S. (2013) The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol. Cell 51, 236–248 10.1016/j.molcel.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Frizzell N., Rajesh M., Jepson M. J., Nagai R., Carson J. A., Thorpe S. R., and Baynes J. W. (2009) Succination of thiol groups in adipose tissue proteins in diabetes: succination inhibits polymerization and secretion of adiponectin. J. Biol. Chem. 284, 25772–25781 10.1074/jbc.M109.019257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Merkley E. D., Metz T. O., Smith R. D., Baynes J. W., and Frizzell N. (2014) The succinated proteome. Mass Spectrom. Rev. 33, 98–109 10.1002/mas.21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang M., Ternette N., Su H., Dabiri R., Kessler B. M., Adam J., Teh B. T., and Pollard P. J. (2014) The succinated proteome of FH-mutant tumours. Metabolites 4, 640–654 10.3390/metabo4030640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruecker N., Jansen R., Trujillo C., Puckett S., Jayachandran P., Piroli G. G., Frizzell N., Molina H., Rhee K. Y., and Ehrt S. (2017) Fumarase deficiency causes protein and metabolite succination and intoxicates Mycobacterium tuberculosis. Cell Chem. Biol. 24, 306–315 10.1016/j.chembiol.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 58. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., and Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006 2, 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yamamoto N., Nakahigashi K., Nakamichi T., Yoshino M., Takai Y., Touda Y., Furubayashi A., Kinjyo S., Dose H., Hasegawa M., Datsenko K. A., Nakayashiki T., Tomita M., Wanner B. L., and Mori H. (2009) Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5, 335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Trager W., and Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 10.1126/science.781840 [DOI] [PubMed] [Google Scholar]
- 62. Rug M., and Maier A. G. (2012) Transfection of Plasmodium falciparum. Methods Mol. Biol. 923, 75–98 [DOI] [PubMed] [Google Scholar]
- 63. Abràmoff M. D., Magalhães P. J., and Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 64. Hartig S. M. (2013) Basic image analysis and manipulation in ImageJ. Curr. Protoc. Mol. Biol. Chapter 14, Unit14.15 [DOI] [PubMed] [Google Scholar]
- 65. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pfander C., Anar B., Schwach F., Otto T. D., Brochet M., Volkmann K., Quail M. A., Pain A., Rosen B., Skarnes W., Rayner J. C., and Billker O. (2011) A scalable pipeline for highly effective genetic modification of a malaria parasite. Nat. Methods. 8, 1078–1082 10.1038/nmeth.1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Janse C. J., Ramesar J., and Waters A. P. (2006) High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 1, 346–356 10.1038/nprot.2006.53 [DOI] [PubMed] [Google Scholar]
- 68. Woo Y. H., Ansari H., Otto T. D., Klinger C. M., Kolisko M., Michálek J., Saxena A., Shanmugam D., Tayyrov A., Veluchamy A., Ali S., Bernal A., del Campo J., Cihlář J., Flegontov P., et al. (2015) Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. Elife 4, e06974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van Vugt-Lussenburg B. M., van der Weel L., Hagen W. R., and Hagedoorn P.-L. (2009) Identification of two [4Fe-4S]-cluster–containing hydro-lyases from Pyrococcus furiosus. Microbiology 155, 3015–3020 10.1099/mic.0.030320-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.