Abstract
Neuropeptides constitute a vast and functionally diverse family of neurochemical signaling molecules and are widely involved in the regulation of various physiological processes. The nematode Caenorhabditis elegans is well-suited for the study of neuropeptide biochemistry and function, as neuropeptide biosynthesis enzymes are not essential for C. elegans viability. This permits the study of neuropeptide biosynthesis in mutants lacking certain neuropeptide-processing enzymes. Mass spectrometry has been used to study the effects of proprotein convertase and carboxypeptidase mutations on proteolytic processing of neuropeptide precursors and on the peptidome in C. elegans. However, the enzymes required for the last step in the production of many bioactive peptides, the carboxyl-terminal amidation reaction, have not been characterized in this manner. Here, we describe three genes that encode homologs of neuropeptide amidation enzymes in C. elegans and used tandem LC-MS to compare neuropeptides in WT animals with those in newly generated mutants for these putative amidation enzymes. We report that mutants lacking both a functional peptidylglycine α-hydroxylating monooxygenase and a peptidylglycine α-amidating monooxygenase had a severely altered neuropeptide profile and also a decreased number of offspring. Interestingly, single mutants of the amidation enzymes still expressed some fully processed amidated neuropeptides, indicating the existence of a redundant amidation mechanism in C. elegans. All MS data are available via ProteomeXchange with the identifier PXD008942. In summary, the key steps in neuropeptide processing in C. elegans seem to be executed by redundant enzymes, and loss of these enzymes severely affects brood size, supporting the need of amidated peptides for C. elegans reproduction.
Keywords: neuropeptide, Caenorhabditis elegans (C. elegans), mass spectrometry (MS), copper monooxygenase, peptides, amidation, PAL, PAM, peptidomics, PHM
Introduction
The nervous system of a Caenorhabditis elegans hermaphrodite comprises 302 neurons. These neurons generate behaviors such as locomotion, egg-laying, and foraging and also display functional plasticity such as adaptation and associative learning. Neuropeptides play a central role in all of these aspects of nervous system function, making C. elegans an excellent organism for the study of neuropeptide biology. Despite its anatomical simplicity, C. elegans expresses a large and diverse set of neuropeptides. From studies using both genomic and biochemical techniques, 31 FMRF-amide–like6 peptide genes (flp-1 through flp-28, flp-32, flp-33, flp-34), 60 neuropeptide-like peptide genes (nlp-1 through nlp-57, pdf-1, snet-1, and ntc-1), and 40 insulin-like peptide genes (ins-1 through ins-39 and daf-28) have been annotated in the C. elegans genome (1–3).
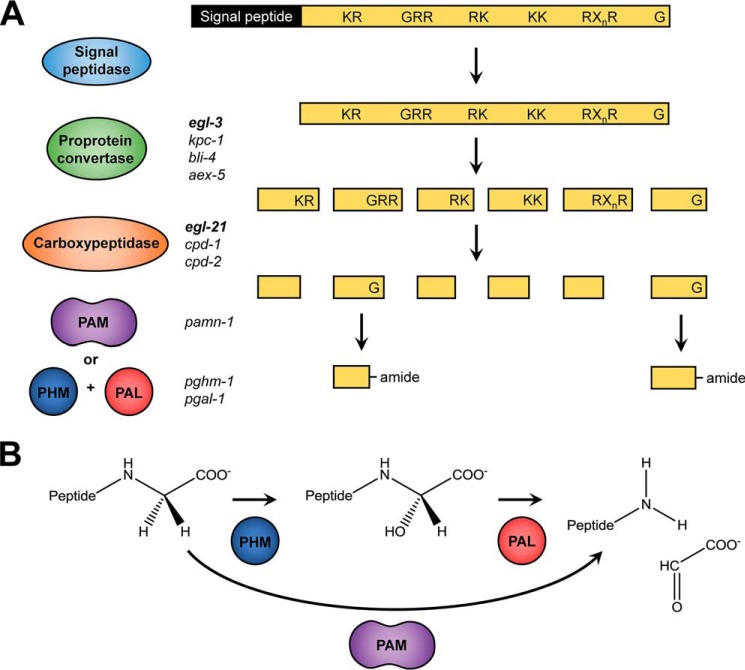
The biochemical pathway responsible for the generation of mature neuropeptides is highly conserved within the animal kingdom. Neuropeptides are synthesized as larger inactive prepropeptide precursors in the endoplasmic reticulum (Fig. 1A). Upon entry of the prepropeptide into the secretory pathway, the signal peptide is removed, and the remaining propeptide is further transferred to the trans-Golgi network. Subsequent processing involves proprotein convertases (PC),7 cleaving the propeptide after dibasic sites containing a combination of arginines and/or lysines. The C. elegans genome encodes four PC genes that can execute this first step: egl-3, kpc-1, bli-4, and aex-5. However, peptidomics experiments showed that only egl-3 mutants display a striking absence of mature neuropeptides, implying that this is the major PC needed for neuropeptide processing (4). Following proteolysis by PCs, the dibasic residues at the C terminus of the neuropeptide precursor are removed by carboxypeptidases, which in C. elegans are encoded by the genes egl-21, cpd-1, and cpd-2. The egl-21 gene has been most extensively studied, and carboxyl-terminally extended peptides were shown to occur in egl-21 mutants (5, 6). Both cpd-1 and cpd-2 were discovered based on their homologies to carboxypeptidase E and D. However, little is known about their exact in vivo function in neuropeptide synthesis (5, 7). For some peptides, the cleavage by the carboxypeptidase is the final biosynthetic step, as their biological activity is acquired upon removal of the basic amino acids. However, many neuropeptides require additional post-translational modifications (PTMs) to gain full biological activity (reviewed in De Haes et al. (8)). One such PTM is the conversion of a carboxyl-terminal glycine into an α-amide. Neuropeptide amidation occurs frequently in all Metazoa, as the neuropeptides that require amidation include the FMRF-amide–like peptides, substance P, oxytocin, α-melanocyte–stimulating hormone, vasopressin, and nematocin, to name just a few (9–12).
Figure 1.
Neuropeptide-processing pathway. Neuropeptides are synthesized as large preproproteins that require post-translational processing, and as exemplified in A, the signal peptide is cleaved upon entry into the secretory pathway by a signal peptidase. Subsequently, a proprotein convertase (mainly egl-3, but also kpc-1, bli-4, and aex-5 in C. elegans) cleaves the remaining part of the precursor protein at specific motifs containing basic amino acids (KR, RR, RK, KK, or RXnR with n = 2, 4, 6, or 8). These residues are then removed by a carboxypeptidase (EGL-21, CPD-1 and CPD-2 in C. elegans) to yield the cleaved peptide. Finally, the carboxyl-terminal glycine residue, if present, is transformed into an amide. B, carboxyl-terminal amidation involves two steps: hydroxylation of the glycine α-carbon by a PHM, followed by a cleavage reaction performed by a PAL. This will generate a glyoxylate molecule and the α-amidated peptide. In vertebrates, these two enzymatic activities are contained in one bifunctional enzyme, PAM.
The oxidative cleavage of the carboxyl-terminal glycine to generate the α-amide moiety is mediated by a multistep reaction (Fig. 1). This process involves hydroxylation of the glycine α-carbon by a peptidylglycine α-hydroxylating monooxygenase (PHM), followed by a cleavage reaction performed by peptidyl–α-hydroxyglycine α-amidating lyase (PAL) to generate a glyoxylate molecule and the α-amidated peptide (Fig. 1B). For these reactions, both PHM and PAL require metal ions bound in their catalytic centers: PHM has two copper-binding elements, whereas PAL binds one zinc ion and one calcium ion. PHM also requires molecular oxygen and ascorbate. In vertebrates, both enzymatic domains responsible for carboxyl-terminal amidation are encoded within the same gene, resulting in a single bifunctional enzyme, peptidylglycine α-amidating monooxygenase (PAM) (13–15). By contrast, many invertebrates express two distinct enzymes from different genes. For example Drosophila melanogaster expresses one PHM gene and two distinct PAL genes, one of which is believed to be secreted and the other integral to cellular membranes (16, 17). Interestingly, Cnidaria, the oldest animal group with an organized nerve net, and the human parasite flatworm Schistosoma mansoni were long believed to encode separate PHM and PAL enzymes only (18–20). However, more recent research provides evidence for the presence of a bifunctional PAM gene in both species (17, 21). The snail Lymnaea stagnalis has a multifunctional PAM gene that encodes one PAL domain and four PHM domains (22). Another mollusc, Aplysia californica, also expresses a PAM zymogen containing both PHM and PAL domains that are separated by a proteolytic cleavage site (23).
C. elegans expresses a large number of amidated peptides, and potential orthologs of the bifunctional PAM as well as the monofunctional PHM and PAL have been observed in the genome (17, 24, 25). However, these C. elegans enzymes have never been characterized. To assess their function and possible redundancies in neuropeptide biosynthesis, we used LC coupled to MS (LC-MS) and compared neuropeptide profiles of mutants deficient in the presumptive C. elegans-amidating enzymes. In addition, functional tests evaluating total brood size of these mutants suggest that at least one of the neurochemical signals that (indirectly) promotes egg-laying is an amidated peptide.
Results
Putative C. elegans PHM, PAL, and PAM enzymes are conserved among species
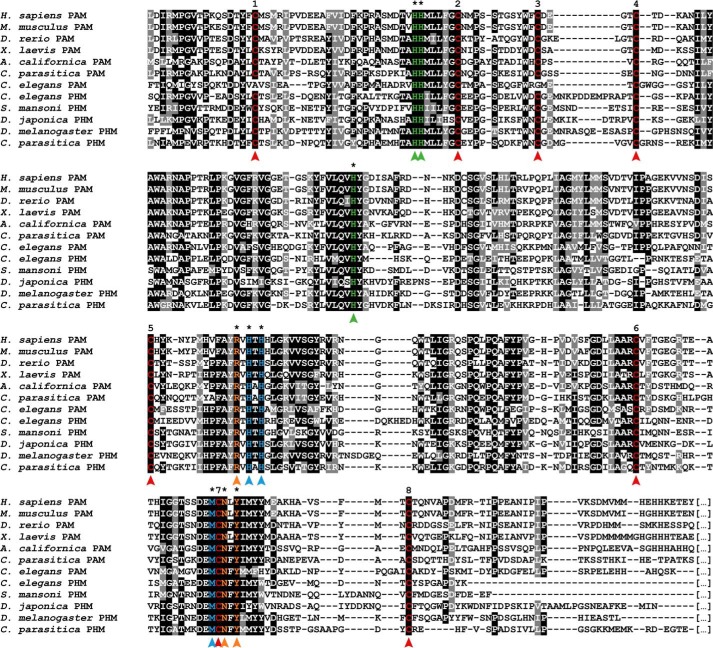
BLAST analyses using human PAM and Drosophila PHM as queries revealed the presence of three different C. elegans genes likely to encode PHM, PAL, and PAM enzymes: Y71G12B.4 (pghm-1), F21F3.1 (pgal-1), and T19B4.1 (pamn-1), respectively. Furthermore, protein sequence alignment of these putative C. elegans-amidating enzymes to those from other species immediately highlights the high degree of conservation of key functional motifs. Notably, the typical copper-binding type II ascorbate-dependent monooxygenase signatures HHMX2FXC and HXFX4HTHX2G, which are highly conserved throughout metazoan evolution (26), are present in both C. elegans PAM as well as PHM proteins (Fig. 2). Additionally, eight cysteines believed to form disulfide bridges (14) are fully present in C. elegans PHM, whereas two of them are absent in PAM. Three residues that mediate interactions with the neuropeptide substrates (14) are also highly conserved between the C. elegans enzymes and PAM and PHM in other species (marked in orange on Fig. 2).
Figure 2.
Inter-phyla sequence comparison of bifunctional PAM and monofunctional PHM enzymes. Multiple sequence alignments of the PHM domain of PAM enzymes are from H. sapiens (NP_000910.2), the mouse M. musculus (P97467.2), the zebrafish D. rerio (XP_699436.4), the African clawed frog X. laevis (NP_001079520.2), the marine snail A. californica (AAF67216.1), the sea anemone C. parasitica (Q9GQN2), and C. elegans (P83388.2) with PHM enzymes from C. elegans (Q95XM2.1), the parasitic platyhelminth S. mansoni (AAO18222.1), the planarian D. japonica (BAD98846.1), the fruit fly D. melanogaster (AAF47127.1), and C. parasitica (AAG24505.1). Whereas eight canonical cysteine residues (numbered 1–8, residues marked in red) that form four putative disulfide bridges are present in PHM enzymes and in the PHM domains of the bifunctional PAM enzymes, cysteines 1 and 3 are missing in C. elegans PAM. All residues marked with an asterisk are involved in the catalytic function of PHM and are highly conserved throughout the species. One copper ion is bound by three histidine residues, marked in green. The second catalytic copper ion is held in place by two histidines and one methionine (marked in blue). The arginine, asparagine, and tyrosine residues marked in orange are part of the substrate-binding site and interact with the peptide to be amidated (14).
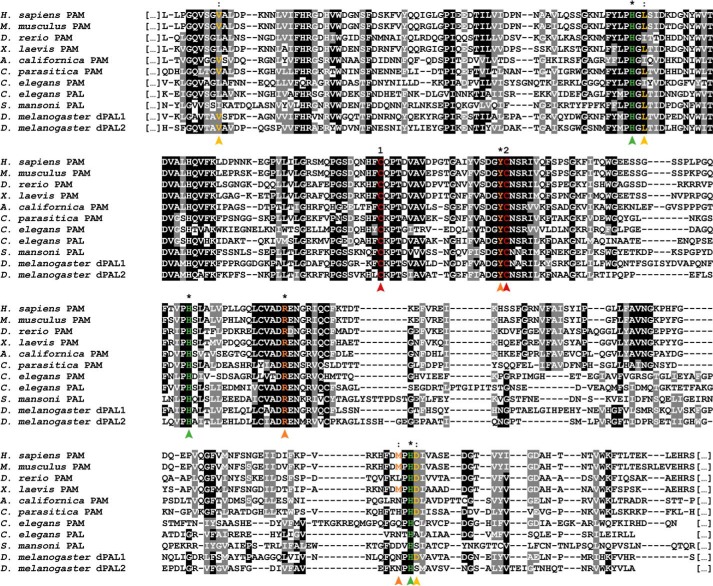
Alignments of the PAL domain from PAM enzymes with monofunctional PAL enzymes from other species revealed two conserved cysteines known to form a structurally crucial disulfide bridge (13), as well as three histidine residues that lock the catalytic Zn2+ in place (Fig. 3). However, the Ca2+-binding domain present in enzymes from other species seems to be incomplete in C. elegans PAL and even completely absent from C. elegans PAM. This finding is consistent with a previous report that demonstrated this calcium-binding site to confer structural stability, as opposed to mediating a critical catalytic function (13). Furthermore, a methionine residue responsible for positioning the substrate in the catalytic site shows very poor conservation and is completely absent in both C. elegans PAM and PAL. The poor conservation of the Ca2+-binding site and the substrate-positioning methionine was also noted by Atkinson et al. (18) in an analysis of S. mansoni PAL sequences. The catalytic dyad itself, responsible for the interaction with the substrate, does show high conservation throughout all species (marked in orange on Fig. 3) (13).
Figure 3.
Inter-phyla sequence comparison of bifunctional PAM and monofunctional PAL enzymes. Multiple sequence alignments of the PAL domain of PAM enzymes are from H. sapiens (NP_000910.2), the mouse M. musculus (P97467.2), the zebrafish D. rerio (XP_699436.4), the African clawed frog X. laevis (NP_001079520.2), the marine snail A. californica (AAF67216.1), the sea anemone C. parasitica (Q9GQN2), and C. elegans (P83388.2) with PAL enzymes from C. elegans (CCD69816.1), the parasitic platyhelminth S. mansoni (ACN42951.1), and the fruit fly D. melanogaster (ACJ13179.1 and AAF47043.2). Two conserved cysteines that form a disulfide bridge are numbered and marked in red. Critical residues for catalytic activity are marked with an asterisk or colon, denoting perfectly conserved or partially conserved residues, respectively. The residues involved in the binding of a catalytic zinc ion are marked in green and are highly conserved throughout species. Residues marked in yellow are responsible for coordination of a calcium ion. These seem to lack a high degree of conservation in other species. Although one Ca2+-binding leucine residue is still present in C. elegans PAL, they appear to be completely absent in C. elegans PAM. Key active site residues are marked in orange. Both the catalytic arginine and tyrosine are highly conserved, and the methionine residue that binds and positions the substrate shows a much lower conservation. The low conservation of this methionine and the Ca2+-binding residues in different species were also observed by Atkinson et al. (18).
pghm-1 and pghm-1;pamn-1 mutants have severe defects in peptide amidation
To determine whether C. elegans PHM, PAL, and PAM homologs function in the post-translational processing of neuropeptide precursors, we devised a differential peptidomics strategy that compared the endogenous peptide content of pghm-1, pgal-1, and pamn-1 single and pghm-1;pamn-1 double mutants. Using quadrupole-Orbitrap LC-MS and subsequent data analysis, we were able to identify a total of 152 C. elegans peptides by mass matching (mass difference ≤5 ppm). From this result, a first selection was made based on peptides that potentially undergo α-amidation. This reduced the list of peptides to 121 sequences. Second, we selected for peptides that across mutants are present in at least one of three modification states relevant to the amidation reaction (carboxyl-terminal glycine-extended, hydroxyglycine intermediate, or amidated state), which allows for comparing peptides between different mutants. This yielded a final list of 52 (out of the initial 152) peptides of interest, with 45 of them MS/MS confirmed. All 52 peptides were subjected to further analysis (Tables S1–S3). The mass readouts for two neuropeptides in WT and pghm-1;pamn-1 strains illustrate the effect of loss of these enzymes on peptide amidation (Figs. S7 and S8).
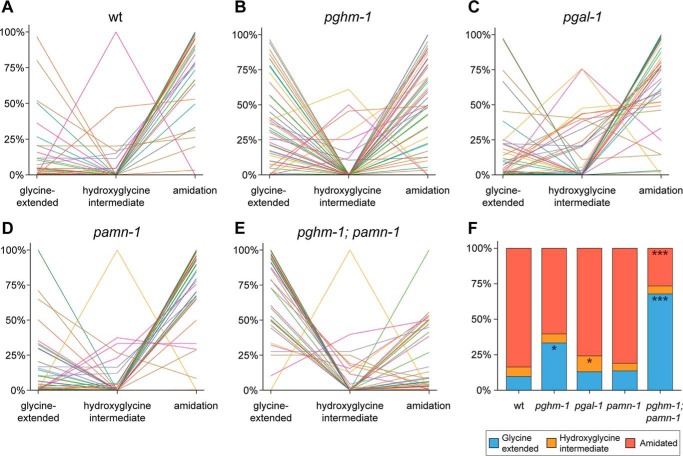
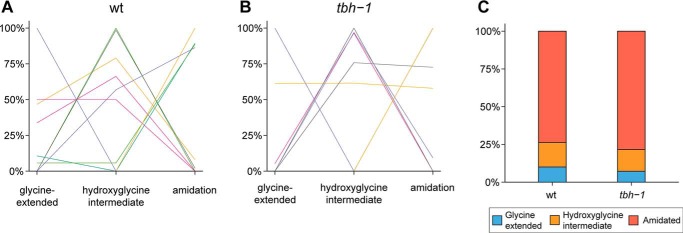
In line with our expectations, we found that most neuropeptides in the WT are fully amidated, with only a minute fraction as glycine-extended or hydroxyglycine intermediates (Fig. 4, A and F). Higher amounts of unprocessed glycine-extended peptides were found in the pghm-1 mutant (Fig. 4, B and F), indicating that loss of this PHM homolog is sufficient to disrupt peptide amidation. Mutation of pgal-1 and pamn-1 separately only had minor effects on neuropeptide amidation. pgal-1 mutants showed an increase in hydroxyglycine intermediates (Fig. 4, C and F), whereas pamn-1 mutants exhibited an almost WT amidation profile (Fig. 4, D and F). A higher amount of hydroxyglycine intermediates in pgal-1 was expected, because the inactive PAL is unable to convert these intermediates into mature amidated peptides. A drastic change in peptide profile was observed in pghm-1;pamn-1 double mutants, in which the first step of the amidation reaction should be completely blocked. Here, the large increase in the amount of unprocessed peptides was accompanied by a corresponding decrease of amidated peptides (Fig. 4, E and F). When combined, the peptide amidation profiles of the pghm-1 and pamn-1 single mutations did not reflect the severe phenotype seen in the pghm-1;pamn-1 double mutant, suggesting functional overlap between the encoded enzymes. Notably, pghm-1;pamn-1 double mutants still expressed some fully amidated neuropeptides, despite completely lacking enzymes homologous to known PAMs.
Figure 4.
Modification states of separate identified neuropeptides (A–E) and distribution of neuropeptide modification states (F) in WT and mutant animals. Each individual line corresponds to an identified neuropeptide (52 in total). The y axis represents the mean values of normalized relative peptide abundances. The majority of neuropeptides in wildtype (wt) (A) are detected as carboxyl-terminal amidated peptides. Mutating pghm-1 seems to disrupt normal amidation, because more neuropeptides are detected with the carboxyl-terminal glycine still present (B). Knockout of pgal-1 seems to have only a small effect on the amount of amidated neuropeptides (C); however, an increase in hydroxyglycine intermediates can be seen. Inactivation of pamn-1 has no severe effects on amidation (D). Finally, mutating both pghm-1 and pamn-1 displays the most severe amidation effects (E). The majority of neuropeptides are found with their carboxyl-terminal glycine still present. When looking at the modification state distribution for all 52 peptides (F), WT animals indeed show a high occurrence of amidation (84.4%) and only low amounts of glycine-extended peptides. Although knockout of pghm-1 results in a rise of glycine-extended peptides (*, p < 0.05), other single mutants seem to be less affected. Although pgal-1 shows a significant increase in hydroxyglycine intermediates (*, p < 0.05), it still contains a high amount of amidated peptides. pamn-1 knockout does not seem to have any major effects on peptide amidation, because it closely resembles WT. A clear effect is seen in the pghm-1;pamn-1 double mutant, where the occurrence of amidated peptides collapses (***, p < 0.001) and is concomitant with a rise in glycine-extended peptides (***, p < 0.001). Because the effect in the double mutant is more severe than the additive effects of each single mutant, this may suggest that pghm-1 and pamn-1 are interchangeable to a certain degree. Data are derived from 52 identified peptides, present in all mutants. For each mutant, three (WT, pgal-1, and pamn-1) or two (pghm-1 and pghm-1;pamn-1) replicates were used. For more detail, including data of individual replicates and annotation of all individual neuropeptides, see Figs. S2 and S3.
To verify that the selection of 52 peptides (see “Experimental procedures”) had no impact on the assessment of the overall modification state of endogenous neuropeptides, we additionally examined the total amount of identified peptides per mutant (Fig. S4). It can be concluded that the ratios for the different modifications are similar for a given mutant, regardless of whether the 52 selected peptides (Fig. 4F) or their overall total amounts (Fig. S4) were considered. We therefore conclude that our method of selecting 52 peptides occurring in all mutant animals did not bias our analysis.
pghm-1;pamn-1 double mutant has defective egg-laying behavior
Because C. elegans carrying nonfunctional alleles of proprotein convertase (egl-3) or carboxypeptidase (egl-21) have defective reproductive behavior (6, 27, 28), and because neuropeptides (including FMRF-amides) have been shown to modulate egg-laying (29–32), we assessed the amidation mutants for defects in egg-laying. No single amidating enzyme mutant displayed severe differences in egg-laying behavior or brood size as compared with the WT (Fig. 5). The slight reduction of amidated peptides that we observed in pghm-1 worms (Fig. 4, B and F) does not seem to greatly affect egg-laying or brood size. The most striking observation was made in the pghm-1;pamn-1 double mutant, for which total brood size is decreased to 57% of the normal amount.
Figure 5.
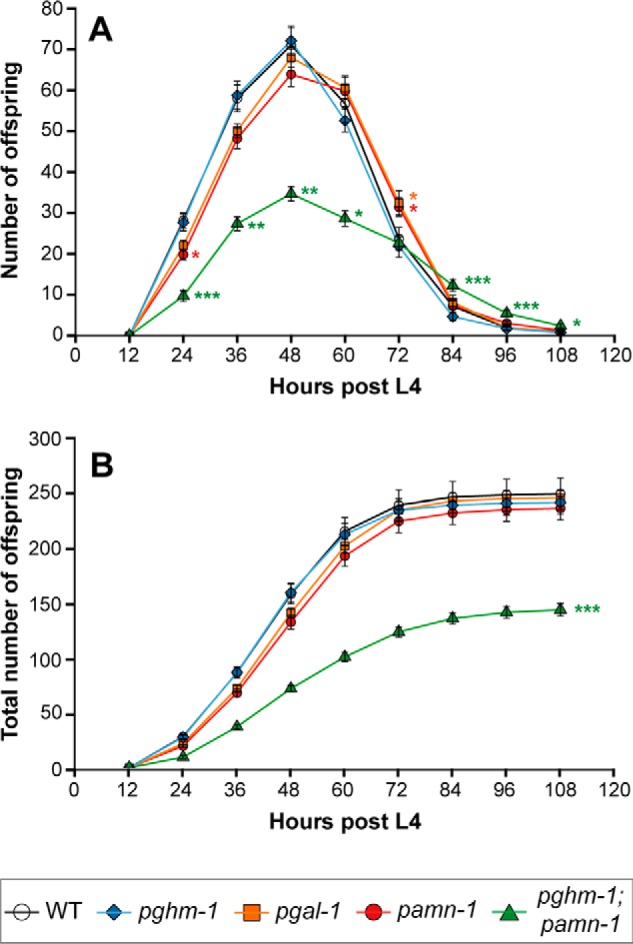
Egg-laying profile (A) and total brood size (B) of WT (n = 27), pamn-1 (n = 35), pgal-1 (n = 32), pghm-1 (n = 22), and pghm-1;pamn-1 (n = 21) mutant worms. The single mutants do not show any severe defects in egg-laying behavior nor a difference in total brood size. Inactivation of both pghm-1 and pamn-1, however, results in a severely diminished brood size that is reduced by 42% compared with WT (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
pghm-1, pgal-1, and pamn-1 are expressed in the nervous system
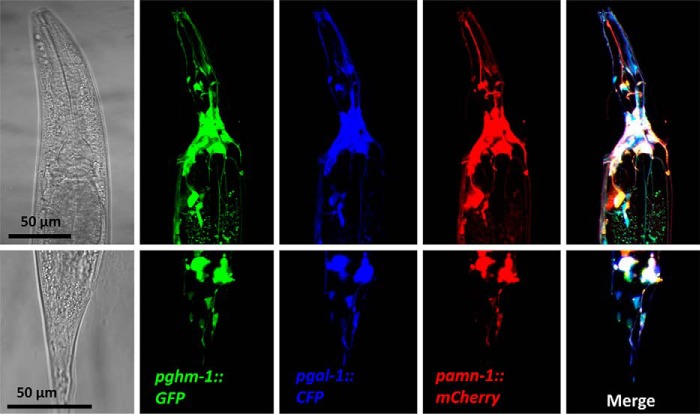
Our peptidomics data indicate a functional overlap of the PHM, PAL, and PAM enzymes in neuropeptide amidation. We therefore assessed the overall expression pattern of these enzymes. Regulatory regions from the pghm-1, pgal-1, and pamn-1 loci were used to drive expression of GFP, CFP, and mCherry, respectively. Reporter constructs were co-injected, and the resulting transgenic strains showed co-expression of all three fluorescent reporters throughout the nervous system (Fig. 7). These data indicate that neuropeptide PHM, PAL, and PAM enzymes are expressed throughout the nematode nervous system, where they likely function redundantly to promote neuropeptide synthesis.
Figure 7.
Co-localization of pghm-1, pgal-1, and pamn-1. Expression of pghm-1::GFP, pgal-1::CFP, and pamn-1::mCherry fusion constructs was monitored in young adults by confocal imaging in multitrack mode. Co-localization of the three reporter constructs could be observed in most of the cell bodies of the nerve ring in the head (upper panels) and the tail ganglia (lower panels).
Copper-binding ascorbate-dependent monooxygenase family member tbh-1 has no major role in peptide amidation
Based on our peptidomics data, pghm-1;pamn-1 double mutant animals still exhibit low levels of mature amidated peptides. Because the putative major amidation enzymes are incapacitated in this double mutant, it is possible that an unidentified enzyme may be the cause of this residual neuropeptide amidation. We further explored the C. elegans genome for similar enzymes. A BLAST search using human PAM revealed tyramine β-hydroxylase (tbh-1), an enzyme involved in the biosynthesis of the monoamine neurotransmitter octopamine in C. elegans, as the only other, be it remotely, related protein. The closest mammalian homolog of tbh-1 is dopamine β-hydroxylase (DBH), which is involved in the biosynthesis of norepinephrine and a known member of the copper-binding ascorbate-dependent monooxygenases (24). Protein sequence alignment of C. elegans TBH-1 with PGHM-1 and PAMN-1 reveals several significant amino acid substitutions within the catalytic site (Fig. S5). The described peptide substitutions between C. elegans PHM/PAM and TBH-1 (Fig. S5) fully correspond with the observations made by Prigge et al. (24) when aligning rat PHM and DBH. These substitutions result in both the destruction of a salt bridge essential for securing the peptidylglycine substrate into place and induce a change in polarity (hydrophilic to hydrophobic) at the peptide backbone recognition site. These severe deviations in the tbh-1 catalytic site suggest that also in C. elegans this enzyme cannot amidate (neuro)peptide substrates. This is further strengthened by the limited expression of tbh-1, being only present in the RIC interneurons and the gonadal sheet cells (33).
Nevertheless, to confirm our hypothesis that tbh-1 is not involved in neuropeptide amidation, we analyzed a tbh-1 mutant strain and compared its amidation profile with that of WT animals. In total, 74 neuropeptides were identified by mass matching. After applying filtering criteria (see “Experimental procedures”), 36 neuropeptides were used for further analysis (Tables S1–3). When comparing the modification states for individual peptides, no apparent differences between WT or tbh-1 mutant animals can be observed (Fig. 6, A and B, and Fig. S6). This is also confirmed by the overall distribution of amidation states, in which no significant differences were measured (Fig. 6C).
Figure 6.
Modification states of separate identified neuropeptides (A and B) and distribution of neuropeptide modification states (C) in WT and tbh-1 animals. Each individual line corresponds to an identified neuropeptide (36 in total). The y axis represents the mean values of normalized relative peptide abundances. The majority of all neuropeptides are detected as carboxyl-terminally amidated peptides in both WT (A) as well as in tbh-1 mutants (B). This is also apparent in the overall modification state distribution of all 36 peptides (C), where no significant differences were observed between WT and tbh-1 mutants. For more detail, including data of individual replicates and annotation of all individual neuropeptides, see Fig. S6.
To fully study the effect of tbh-1 on amidation, we attempted the creation of a pghm-1;pamn-1;tbh-1 triple mutant. Despite our numerous efforts, the creation of such a mutant failed. Because both parental strains (tbh-1 and pghm-1;pamn-1) already display a sick phenotype, we suggest that crippling a major part of neuropeptide signaling together with octopamine signaling may have detrimental effects on worm viability, which therefore results in lethality. However, as mutation of tbh-1 has no effect on the neuropeptide amidation profile, we conclude that it is highly unlikely to be involved in neuropeptide amidation in C. elegans.
Discussion
Neuropeptides are widely involved in the regulation of various physiological processes and in particular in the neuromodulatory control of behavior (34, 35). Hence, knowledge about biologically active peptides and their processing enzymes is crucial for a deeper functional insight into these fundamental processes. Using LC-MS, we previously compared peptide profiles of C. elegans strains having mutations in the presumed neuropeptide precursor cleavage enzymes (4, 5, 36). However, little work has been done on the enzymes responsible for the carboxyl-terminal amidation reaction in nematodes.
Based on its homology to the catalytic domains of vertebrate bifunctional amidation enzyme PAM (Figs. 2 and 3), we now report the C. elegans genome to contain three genes that function in neuropeptide amidation. Two genes (pghm-1 and pgal-1) encode separate PHM and PAL enzymes, and one (pamn-1) encodes a bifunctional PAM (17, 24, 25). Next, by applying MS-based peptidomics approach, we extracted peptides irrespective of their source tissue, enabling us to assess the overall peptide profile of the entire organism.
Using differential peptidomics, we compared the amidation profiles of mutants lacking the putative C. elegans amidation enzymes. We observed that the absence of pghm-1 alone or both pghm-1 and pamn-1 simultaneously resulted in an increase of unprocessed glycine-extended peptides, an effect most pronounced in the double mutant. Because pgal-1 and pamn-1 single mutants both display an amidation profile similar to that of the WT, we propose that a certain degree of functional redundancy exists among the three amidation enzymes. Indeed, their overlapping functions fit into a model in which dysfunction of one gene can be compensated by a combination of the remaining other two (Fig. 1B). Furthermore, our localization data suggest that pghm-1, pgal-1, and pamn-1 promoters show widespread co-expression throughout the nervous system (Fig. 7), supporting a general role of these enzymes in neuronal function. The pghm-1 mutant seems to be the only exception to this proposed system of redundancy. The remaining PGAL-1 and PAMN-1 appear to be inadequate to compensate for loss of PGHM-1; as opposed to the other single mutants, pghm-1 mutants lose their WT-like amidation profile. This observation could imply that some peptides specifically rely on PGHM-1 for processing. Indeed, pghm-1 mutants have a large subpopulation (33 out of a total of 52) of nonamidated peptides that remain amidated in the pamn-1 mutant (Fig. S2). In the context of this model, the ability to shunt amidation intermediates to another redundant enzyme should be lost in the pghm-1;pamn-1 double mutant, in which all alternative routes for the first step of amidation are blocked. As expected, the double mutant exhibits the most severe increase in glycine-extended peptides, as compared with the respective single mutants. Such mechanisms of redundancy are not uncommon and seem to exist at multiple neuropeptide-processing levels. The proprotein convertases (Fig. 1A) EGL-3 and AEX-5, for instance, are likely to act in parallel as egl-3 mutants still produce mature neuropeptides (4, 28). A similar redundant role was found in mice lacking PC1/3 or PC2 (37, 38). Another backup system was hypothesized to exist for the next step in neuropeptide processing, where the presence of fully processed peptides in carboxypeptidase E (CPE) mutant mice has been attributed to the presence of carboxypeptidase D, both being similar in function (39, 40). Redundancy of carboxypeptidases is also hypothesized for C. elegans, because egl-21 mutant worms revealed both extended intermediates as well as mature peptides (5). Other putative carboxypeptidase enzymes have been proposed, but they have not been thoroughly tested yet (3, 7). Here, we suggest a similar redundant system for the neuropeptide amidation reaction in nematodes by identifying C. elegans PHM, PAL, and PAM genes that broadly co-localize in the nervous system. All three enzymes are also closely linked on chromosome I in C. elegans, underscoring the possibility of gene duplication during evolution, leading to families of structurally related proteins (21).
Our analysis of viable offspring production is consistent with the hypothesis that there is redundancy in mechanisms that mediate neuropeptide amidation. Each single mutant displays WT egg-laying behavior in terms of timing and total brood size. The double pghm-1;pamn-1 mutant, however, shows a severe decline in total brood size. Correct neuropeptide processing has already been shown to be essential for normal C. elegans egg-laying, as mutations of egl-3 (PC2) and egl-21 (CPE) resulted in severe defects in this process (27). Additionally, amidated flp-1, flp-10, and flp-17 peptides have already been shown to regulate egg-laying behavior (30, 31). Here, we provide further evidence for the general importance of neuropeptide amidation for this process. The fact that deletion of all amidation enzymes in mice leads to embryonic lethality (41) indicates the essential nature of amidated peptides, and it also explains the importance of functional redundancy of a vital system. Curiously, abolishing nearly all of the peptide amidation in C. elegans does not seem to induce lethality, which could indicate that amidated neuropeptides are less involved in embryonic development in C. elegans as compared with in mice. Importantly, however, there are still a few amidated peptides present in the pghm-1;pamn-1 double mutant (Fig. 4, E and F, and Fig. S2), suggesting the existence of at least one other amidating enzyme. A BLAST search against the C. elegans genome using PGHM-1 and PGAL-1 as queries only revealed the remotely related tyramine β-hydroxylase (tbh-1) to be similar. This enzyme catalyzes the formation of the monoamine neurotransmitter octopamine from tyramine in C. elegans. Although a known member of the copper-binding ascorbate-dependent monooxygenases, it is highly unlikely to be involved in the amidation process. First, sequence alignment of tbh-1 with C. elegans PAMN-1 and TBH-1 revealed several substitutions in the catalytic site (Fig. S5), making it unsuitable for the amidation of peptide substrates. These substitutions were found to be identical to those in rat PHM and DAB (24). Currently, there is no evidence that mammalian DBH possesses any neuropeptide-amidating properties. Second, peptidomics analysis of tbh-1 mutants shows no aberrant amidation profile. Finally, the expression of tbh-1 is limited to one pair of interneurons (RIC) and the gonadal sheath cells, with only the RIC interneurons producing low amounts of neuropeptides. All of this points to the fact that tbh-1 does not play any role in the amidation of neuropeptides in C. elegans.
Additionally, it is worth mentioning that multiple attempts in generating a homozygous pghm-1;pamn-1;tbh-1 mutant remained unsuccessful. We are under the impression that knocking out octopamine synthesis together with the majority of the neuropeptide amidation system may lead to a lethal phenotype. Because both octopamine and FMRF-amide signaling act in concert for several essential biological processes (e.g. egg-laying, locomotion, and feeding behavior), it is not inconceivable that incapacitating both signaling pathways simultaneously can lead to lethality (42, 43).
To summarize, the C. elegans key steps in neuropeptide-processing seem to be executed by redundant enzymes, resulting in minimal physiological and phenotypic effects upon functional loss of a single enzyme. Our work provides biochemical evidence that this redundant mechanism exists for the C. elegans neuropeptide-amidating enzymes and, when nearly completely eliminated, results in a marked decrease of amidated peptides. Although this decrease does not result in lethality, as opposed to what is seen for mice and potentially implying that not all amidation enzymes have been discovered in C. elegans yet, we have shown that the animals' brood size is severely affected. Our mass spectrometric data support the notion that pamn-1, pghm-1, and pgal-1 are bona fide neuropeptide-amidating enzymes in C. elegans.
Experimental procedures
C. elegans strains
WT N2 Bristol and mutant strains VC2129 pamn-1(ok2681), RB1721 pghm-1(ok2189), and MT9455 tbh-1 (n3247) were obtained from the Caenorhabditis Genetics Stock Center (University of Minnesota). All strains were backcrossed with WT N2 to remove potentially interfering background mutations generated by the random mutagenesis procedure. Backcrossing was done six times for all strains, except pgal-1(n5050), which was backcrossed four times. The backcrossed strains are renamed LSC1384, LSC1382, and LSC1383, respectively. LSC1382 and LSC1384 were used to create a pghm-1(ok2189);pamn-1(ok2681) double mutant, named LSC1169. Strains were cultivated on standard nematode growth medium seeded with Escherichia coli OP50. All experiments were performed at 20 °C.
Inter-phyla sequence alignment of PHM, PAL, and PAM
PHM, PAL, and PAM protein sequences from A. californica, C. elegans, Calliactis parasitica, Danio rerio, D. melanogaster, Dugesia japonica, Homo sapiens, Mus musculus, S. mansoni, and Xenopus laevis were acquired from NCBI Protein. Sequences were aligned using the PSI-Coffee algorithm, which is optimized for the alignment of distantly related proteins using homology extension (44). Shading was added using version 3.21 of BOXSHADE (Hofmann and Baron, http://www.ch.embnet.org/software/BOX_form.html).8
Extraction of endogenous peptides
WT, pamn-1, pghm-1, pgal-1, pghm-1;pamn-1, and tbh-1 mixed stage worms from 24 fully grown Petri dishes (inner diameter, 90 mm) were collected, and peptides were extracted using an acidified methanol extraction solvent as described previously (2, 4, 5, 45, 46). This extraction solvent has the advantage of precipitating larger proteins, whereas smaller peptides remain in solution. A size-exclusion column (Sephadex PD MiniTrap G-10, GE Healthcare) and a 10-kDa cutoff filter (Amicon Ultra-4, Merck Millipore) were used to enrich the sample for peptide content by isolating the 700–10,000-Da mass fraction. This enriches for neuropeptides and removes many biomolecules that are not of interest out of the complex samples. Two or three replicates were subjected to this same protocol. Additionally, a pooled sample was created by combining 10% of the volume of all samples. All samples were briefly stored at 4 °C prior to MS analysis.
Quadrupole-Orbitrap LC-MS/MS
Quadrupole-Orbitrap LC-MS/MS experiments were conducted using a Dionex UltiMate 3000 UHPLC coupled on line to a Thermo Fisher Scientific Q Exactive mass spectrometer. The UHPLC is equipped with a guard pre-column (Acclaim PepMap100, C18, 75 μm × 20 mm, 3 μm, 100 Å; Thermo Fisher Scientific) and an analytical column integrated in the nano-electrospray ion source (EASY-Spray, PepMap RSLC, C18, 75 μm ×500 mm, 2 μm, 100 Å; Thermo Fisher Scientific). The sample was separated at a flow rate of 300 nl/min, using a 210-min linear gradient from 3 to 55% acetonitrile containing 0.1% formic acid. MS data were acquired using a data-dependent (dynamic exclusion settings at 15 s) Top10 method choosing the most abundant precursor ions from a full MS survey scan for higher-energy collisional dissociation (HCD) fragmentation. Full MS scans were acquired at a resolution of 70,000 at m/z 200, with a maximum injection time of 256 ms. The resolution for MS/MS scans after higher energy-collisional dissociation fragmentation was set at 17,500 at m/z 200, with a maximum injection time of 64 ms.
Mass spectrometry data analysis
LC-MS data were used to determine the abundance of each neuropeptide in the different samples. In this case, neuropeptide identification is based on matching a measured mass to an in-house library of theoretical neuropeptide masses. Subsequently, LC-MS/MS fragmentation data were used to confirm these identified peptides. Both procedures are described here in more detail.
To correct for inter-run variation causing retention time shifts, all data files were aligned using Progenesis LC-MS software (Nonlinear Dynamics). We reasoned that defects in amidating enzymes potentially result in aberrant peptide profiles between the different mutants and the WT, thereby making a correct run alignment with Progenesis more challenging. To overcome this issue, we manually selected the pooled sample as the reference to which all other runs were aligned. Peak picking was done in automatic mode, using the default sensitivity settings. These results were then filtered on charge state, retaining all features with charges ranging from 2 to 7. All selected features were exported to a .csv file containing the m/z, charge, deconvoluted mass, abundance, and retention times. For peptide annotation, we developed a custom R script (47) that compares all the detected masses in the Progenesis .csv file to an in-house peptide library containing the masses of 352 currently predicted C. elegans neuropeptides and their post-translational modifications (1, 3, 7, 42, 48). Deconvoluted masses that match within an error margin of 5 ppm are interpreted as a positive hit. Only peptides that are possible substrates for amidation, i.e. sequences containing a carboxyl-terminal glycine, as reviewed by Eipper et al. (9), were selected for data analysis, and their abundances were normalized per run using the median abundance of the entire run, including all peptides, as a normalization factor. The total abundance was calculated by summation of all different m/z values of the same peptide. Peptides were grouped per modification (i.e. glycine-extended, hydroxyglycine intermediate, amidation; Fig. 1; henceforward referred to as modification states) for each mutant. Because we want to compare the modification states of each peptide between mutants, only peptides that had been detected at least once (irrespective of modification state) in every mutant were retained (Tables S1 and S2), i.e. peptides that are present in one mutant, but completely undetected in another, are omitted from the analysis. At this point, every identified peptide per run has three abundance values corresponding to every possible modification state. To ensure an equal weight for each peptide and to eliminate otherwise unwanted over-representation of abundant peptides in the results, individual peptide values were normalized by calculating the ratio (in percentages) of the abundance in one certain state to the total sum of all modification states. These relative percentages were computed for every peptide in each sample. The data were statistically analyzed by an analysis of variance followed by a post hoc Dunnett's test. Because peptides were extracted from mixed-stage C. elegans cultures, possible stage-specific effects on peptide levels might occur. However, because no abnormal variation between replicates was observed, differences in contribution of developmental stages do not severely affect peptide levels in our sample set (Fig. S3).
MS/MS fragmentation data were analyzed using PEAKS software (Bioinformatics Solutions) with a custom-made library containing 190 C. elegans peptide precursor proteins. Parent mass error was set at 10 ppm, and fragment mass error was at 0.02 Da. The following variable modifications were taken into account: oxidation (+15.99 Da); glycine loss + amidation (−58.01 Da); pyroglutamation from glutamic acid (−18.01 Da); pyroglutamation from glutamine (−17.03 Da); phosphorylation of serine, threonine, or tyrosine (+79.97 Da); and carboxyl-terminal hydroxylated glycine (+15.99 Da). Data of all peptides confirmed by MS/MS can be found in Table S3. All MS data have been integrally deposited to the ProteomeXchange Consortium (49) via the PRIDE partner repository with the dataset identifier PXD008942 and DOI 10.6019/PXD008942.
Egg-laying assay and determination of brood size
To estimate the progress of egg-laying and total brood size for each strain, synchronized single L4 animals were isolated on nematode growth medium plates (inner diameter, 35 mm) seeded with 10 μl of an E. coli OP50 culture (grown overnight). Every 12 h, each worm was transferred to a new plate until the end of the 4-day period, thereby encompassing the time frame in which most of the eggs are laid. Hatched L3–L4 worms on each plate were counted to determine the viable egg-laying profile and total brood size of each mutant. To meet the criteria of the linear mixed-effects model, data were log-transformed prior to statistical analysis. For the total brood size, the raw data were first subjected to a Box-Cox power transformation to comply with the assumptions of the subsequently fitted linear mixed-effects model.
Molecular biology and transgenic strains
A pghm-1 (Y71G12B.4) promoter fragment of 1115 bp was amplified by PCR using primers CACAAGCTTTGGCGACTTGTAATTTTCAGCA and GGATCCCGGAGCCGAGCCTCACAA and cloned in the multiple cloning site of the empty GFP expression vector pPD.95.75. A 1689-bp upstream fragment of pgal-1 (F21F3.1) was amplified using TACTGCAGAGTTGTGAACTGCAATGGC and ATCCATGGTCCTAGAATGAGTCGGCTTCA and cloned in front of CFP, replacing the Pmyo-3::NpHR of the Pmyo-3:: NpHR::CFP plasmid (A. Gottschalk lab). Similarly, a 2937-bp promoter fragment of pamn-1 (T19B4.1) was amplified using CACAAGCTTGGTCATAGGGAAAGGCCAAG and ACGGATCCACCTGAAAATATAATTTATAAATTGGA and cloned in front of mCherry, replacing the F49H12.4 promoter from the F49H12.4::mCherry plasmid (D. Miller III lab). Specifications of all used promoter regions can be found in Fig. S1. The resulting three plasmids were co-injected at 80 ng/μl by conventional microinjection protocols to generate six independent lines, all showing similar GFP, CFP, and mCherry expression overall.
Confocal imaging
Fluorescence images were recorded using a Zeiss LSM 510 laser-scanning microscope equipped with a Plan-Apochromat ×63/1.4 oil differential interference contrast objective in multitrack mode. CFP, GFP, or mCherry were excited at 405, 488, or 543 nm, and fluorescence was sequentially monitored using BP475-525, LP530, or LP560 filters, respectively. Z-stacks were acquired and subsequently projected using ImageJ software (National Institutes of Health).
Author contributions
S. V. B., L. S., S. H., and L. T. conceptualization; S. V. B., J. W., G. M., N. R., H. R. H., L. S., S. H., and L. T. resources; S. V. B., K. B., W. D. H., and G. M. data curation; S. V. B., K. B., W. D. H., and G. M. software; S. V. B., J. W., W. D. H., G. M., S. H., and L. T. formal analysis; S. V. B. and J. W. validation; S. V. B., J. W., K. B., W. D. H., G. M., N. R., H. R. H., L. S., S. H., and L. T. investigation; S. V. B., J. W., W. D. H., and S. H. visualization; S. V. B., J. W., K. B., and N. R. methodology; S. V. B., J. W., K. B., W. D. H., S. H., and L. T. writing-original draft; S. V. B. project administration; S. V. B., J. W., K. B., W. D. H., N. R., H. R. H., L. S., S. H., and L. T. writing-review and editing; K. B., L. S., S. H., and L. T. supervision; L. S. and L. T. funding acquisition.
Supplementary Material
Acknowledgments
Several strains used in this work were provided by the Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health Office of Research Infrastructure Programs Grant P40 OD010440. We are grateful to Shari Wouters and Matthias Boris Van Hiel for their assistance, and the SyBioMa facility (KU Leuven) for access to their MS equipment. We also thank our lab technician Francisco Naranjo for assistance.
This work was supported in part by KU Leuven Grant C14/15/049, FWO Grants 1511512N, G069713N, and G052217N, and Belspo Grant IAP-P7/44. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The mass spectrometric raw data and spectral libraries associated with this manuscript are available from ProteomeXchange with the accession number PXD008942.
This article contains Figs. S1–S8 and Tables S1–S3.
C. elegans FMRF-amide–like peptides are named after the presence of a carboxyl-terminal RF-amide motif, similar to the mollusk FMRF-amide peptide (Phe-Met-Arg-Phe-NH2).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PC
- proprotein convertase
- PHM
- peptidylglycine α-hydroxylating monooxygenase
- PAM
- peptidylglycine α-amidating monooxygenase
- PTM
- post-translational modification
- DBH
- dopamine β-hydroxylase
- GFP
- green fluorescent protein
- CFP
- cyan fluorescent protein
- CPE
- carboxypeptidase E
- PAL
- peptidyl–α-hydroxyglycine α-amidating lyase.
References
- 1. Li C., and Kim K. (2010) Neuropeptide gene families in Caenorhabditis elegans. Adv. Exp. Med. Biol. 692, 98–137 10.1007/978-1-4419-6902-6_6 [DOI] [PubMed] [Google Scholar]
- 2. Husson S. J., Reumer A., Temmerman L., De Haes W., Schoofs L., Mertens I., and Baggerman G. (2014) Worm peptidomics. EuPA Open Proteomics 3, 280–290 10.1016/j.euprot.2014.04.005 [DOI] [Google Scholar]
- 3. Hobert O. (2013) The neuronal genome of Caenorhabditis elegans. WormBook 2013 10.1895/wormbook.1.161.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Husson S. J., Clynen E., Baggerman G., Janssen T., and Schoofs L. (2006) Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J. Neurochem. 98, 1999–2012 10.1111/j.1471-4159.2006.04014.x [DOI] [PubMed] [Google Scholar]
- 5. Husson S. J., Janssen T., Baggerman G., Bogert B., Kahn-Kirby A. H., Ashrafi K., and Schoofs L. (2007) Impaired processing of FLP and NLP peptides in carboxypeptidase E (EGL-21)-deficient Caenorhabditis elegans as analyzed by mass spectrometry. J. Neurochem. 102, 246–260 10.1111/j.1471-4159.2007.04474.x [DOI] [PubMed] [Google Scholar]
- 6. Jacob T. C., and Kaplan J. M. (2003) The EGL-21 carboxypeptidase E facilitates acetylcholine release at Caenorhabditis elegans neuromuscular junctions. J. Neurosci. 23, 2122–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li C., and Kim K. (2014) Family of FLP peptides in Caenorhabditis elegans and related nematodes. Front. Endocrinol. 5, 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Haes W., Van Sinay E., Detienne G., Temmerman L., Schoofs L., and Boonen K. (2015) Functional neuropeptidomics in invertebrates. Biochim. Biophys. Acta 1854, 812–826 10.1016/j.bbapap.2014.12.011 [DOI] [PubMed] [Google Scholar]
- 9. Eipper B. A., Stoffers D. A., and Mains R. E. (1992) The biosynthesis of neuropeptides: peptide α-amidation. Annu. Rev. Neurosci. 15, 57–85 10.1146/annurev.ne.15.030192.000421 [DOI] [PubMed] [Google Scholar]
- 10. Merkler D. J. (1994) C-terminal amidated peptides: production by the in vitro enzymatic amidation of glycine-extended peptides and the importance of the amide to bioactivity. Enzyme Microb. Technol. 16, 450–456 10.1016/0141-0229(94)90014-0 [DOI] [PubMed] [Google Scholar]
- 11. Beets I., Janssen T., Meelkop E., Temmerman L., Suetens N., Rademakers S., Jansen G., and Schoofs L. (2012) Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science 338, 543–545 10.1126/science.1226860 [DOI] [PubMed] [Google Scholar]
- 12. Kumar D., Mains R. E., and Eipper B. A. (2016) 60 years of POMC: from POMC and α-MSH to PAM, molecular oxygen, copper, and vitamin C. J. Mol. Endocrinol. 56, T63–T76 10.1530/JME-15-0266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chufán E. E., De M., Eipper B. A., Mains R. E., and Amzel L. M. (2009) Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme. Structure 17, 965–973 10.1016/j.str.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prigge S. T., Kolhekar A. S., Eipper B. A., Mains R. E., and Amzel L. M. (1997) Amidation of bioactive peptides: the structure of peptidylglycine α-hydroxylating monooxygenase. Science 278, 1300–1305 10.1126/science.278.5341.1300 [DOI] [PubMed] [Google Scholar]
- 15. Eipper B. A., Milgram S. L., Husten E. J., Yun H. Y., and Mains R. E. (1993) Peptidylglycine α-amidating monooxygenase: a multifunctional protein with catalytic, processing, and routing domains. Protein Sci. 2, 489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kolhekar A. S., Roberts M. S., Jiang N., Johnson R. C., Mains R. E., Eipper B. A., and Taghert P. H. (1997) Neuropeptide amidation in Drosophila: separate genes encode the two enzymes catalyzing amidation. J. Neurosci. 17, 1363–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han M., Park D., Vanderzalm P. J., Mains R. E., Eipper B. A., and Taghert P. H. (2004) Drosophila uses two distinct neuropeptide amidating enzymes, dPAL1 and dPAL2. J. Neurochem. 90, 129–141 10.1111/j.1471-4159.2004.02464.x [DOI] [PubMed] [Google Scholar]
- 18. Atkinson L. E., McVeigh P., Kimber M. J., Marks N. J., Eipper B. A., Mains R. E., Day T. A., and Maule A. G. (2010) A PAL for Schistosoma mansoni PHM. Mol. Biochem. Parasitol. 173, 97–106 10.1016/j.molbiopara.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mair G. R., Niciu M. J., Stewart M. T., Brennan G., Omar H., Halton D. W., Mains R., Eipper B. A., Maule A. G., and Day T. A. (2004) A functionally atypical amidating enzyme from the human parasite Schistosoma mansoni. FASEB J. 18, 114–121 10.1096/fj.03-0429com [DOI] [PubMed] [Google Scholar]
- 20. Hauser F., Williamson M., and Grimmelikhuijzen C. J. (1997) Molecular cloning of a peptidylglycine α-hydroxylating monooxygenase from sea anemones. Biochem. Biophys. Res. Commun. 241, 509–512 10.1006/bbrc.1997.7854 [DOI] [PubMed] [Google Scholar]
- 21. Attenborough R. M., Hayward D. C., Kitahara M. V., Miller D. J., and Ball E. E. (2012) A “neural” enzyme in nonbilaterian animals and algae: preneural origins for peptidylglycine α-amidating monooxygenase. Mol. Biol. Evol. 29, 3095–3109 10.1093/molbev/mss114 [DOI] [PubMed] [Google Scholar]
- 22. Spijker S., Smit A. B., Eipper B. A., Malik A., Mains R. E., and Geraerts W. P. (1999) A molluscan peptide α-amidating enzyme precursor that generates five distinct enzymes. FASEB J. 13, 735–748 10.1096/fasebj.13.6.735 [DOI] [PubMed] [Google Scholar]
- 23. Fan X., Spijker S., Akalal D. B., and Nagle G. T. (2000) Neuropeptide amidation: cloning of a bifunctional α-amidating enzyme from Aplysia. Brain Res. Mol. Brain Res. 82, 25–34 10.1016/S0169-328X(00)00173-X [DOI] [PubMed] [Google Scholar]
- 24. Prigge S. T., Mains R. E., Eipper B. A., and Amzel L. M. (2000) New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell. Mol. Life Sci. 57, 1236–1259 10.1007/PL00000763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Asada A., Orii H., Watanabe K., and Tsubaki M. (2005) Planarian peptidylglycine-hydroxylating monooxygenase, a neuropeptide processing enzyme, colocalizes with cytochrome b561 along the central nervous system. FEBS J. 272, 942–955 10.1111/j.1742-4658.2004.04528.x [DOI] [PubMed] [Google Scholar]
- 26. Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Marchler G. H., Song J. S., Thanki N., et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trent C., Tsuing N., and Horvitz H. R. (1983) Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kass J., Jacob T. C., Kim P., and Kaplan J. M. (2001) The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J. Neurosci. 21, 9265–9272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindemans M., Liu F., Janssen T., Husson S. J., Mertens I., Gäde G., and Schoofs L. (2009) Adipokinetic hormone signaling through the gonadotropin-releasing hormone receptor modulates egg-laying in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 106, 1642–1647 10.1073/pnas.0809881106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ringstad N., and Horvitz H. R. (2008) FMRF-amide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat. Neurosci. 11, 1168–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waggoner L. E., Hardaker L. A., Golik S., and Schafer W. R. (2000) Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics 154, 1181–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang Y.-J., Burton T., Ha L., Huang Z., Olajubelo A., and Li C. (2015) Modulation of locomotion and reproduction by FLP neuropeptides in the nematode Caenorhabditis elegans. PLoS ONE 10, e0135164 10.1371/journal.pone.0135164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alkema M. J., Hunter-Ensor M., Ringstad N., and Horvitz H. R. (2005) Tyramine functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron 46, 247–260 10.1016/j.neuron.2005.02.024 [DOI] [PubMed] [Google Scholar]
- 34. Frooninckx L., Van Rompay L., Temmerman L., Van Sinay E., Beets I., Janssen T., Husson S. J., and Schoofs L. (2012) Neuropeptide GPCRs in C. elegans. Front. Endocrinol. 3, 167 10.3389/fendo.2012.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schoofs L., De Loof A., and Van Hiel M. B. (2017) Neuropeptides as regulators of behavior in insects. Annu. Rev. Entomol. 62, 35–52 10.1146/annurev-ento-031616-035500 [DOI] [PubMed] [Google Scholar]
- 36. Husson S. J., and Schoofs L. (2007) Altered neuropeptide profile of Caenorhabditis elegans lacking the chaperone protein 7B2 as analyzed by mass spectrometry. FEBS Lett. 581, 4288–4292 10.1016/j.febslet.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 37. Hardiman A., Friedman T. C., Grunwald W. C. Jr., Furuta M., Zhu Z., Steiner D. F., and Cool D. R. (2005) Endocrinomic profile of neurointermediate lobe pituitary prohormone processing in PC1/3- and PC2-Null mice using SELDI-TOF mass spectrometry. J. Mol. Endocrinol. 34, 739–751 10.1677/jme.1.01812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan H., Che F.-Y., Peng B., Steiner D. F., Pintar J. E., and Fricker L. D. (2006) The role of prohormone convertase-2 in hypothalamic neuropeptide processing: a quantitative neuropeptidomic study. J. Neurochem. 98, 1763–1777 10.1111/j.1471-4159.2006.04067.x [DOI] [PubMed] [Google Scholar]
- 39. Che F. Y., Yan L., Li H., Mzhavia N., Devi L. A., and Fricker L. D. (2001) Identification of peptides from brain and pituitary of Cpefat/Cpefat mice. Proc. Natl. Acad. Sci. U.S.A. 98, 9971–9976 10.1073/pnas.161542198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song L., and Fricker L. D. (1995) Purification and characterization of carboxypeptidase D, a novel carboxypeptidase E-like enzyme, from bovine pituitary. J. Biol. Chem. 270, 25007–25013 10.1074/jbc.270.42.25007 [DOI] [PubMed] [Google Scholar]
- 41. Czyzyk T. A., Ning Y., Hsu M.-S., Peng B., Mains R. E., Eipper B. A., and Pintar J. E. (2005) Deletion of peptide amidation enzymatic activity leads to edema and embryonic lethality in the mouse. Dev. Biol. 287, 301–313 10.1016/j.ydbio.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 42. Peymen K., Watteyne J., Frooninckx L., Schoofs L., and Beets I. (2014) The FMRF-amide-like peptide family in nematodes. Front. Endocrinol. 5, 90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Churgin M. A., McCloskey R. J., Peters E., and Fang-Yen C. (2017) Antagonistic serotonergic and octopaminergic neural circuits mediate food-dependent locomotory behavior in Caenorhabditis elegans. J. Neurosci. 37, 7811–7823 10.1523/JNEUROSCI.2636-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.-M., Taly J.-F., and Notredame C. (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Husson S. J., Clynen E., Baggerman G., De Loof A., and Schoofs L. (2005) Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem. Biophys. Res. Commun. 335, 76–86 10.1016/j.bbrc.2005.07.044 [DOI] [PubMed] [Google Scholar]
- 46. Husson S. J., Clynen E., Boonen K., Janssen T., Lindemans M., Baggerman G., and Schoofs L. (2010) Approaches to identify endogenous peptides in the soil nematode Caenorhabditis elegans. Methods Mol. Biol. 615, 29–47 10.1007/978-1-60761-535-4_3 [DOI] [PubMed] [Google Scholar]
- 47. RCore Team. (2016) R: A Language and Environment for Statistical Computing., R Foundation for Statistical Computing, R Version 3.4.1, Vienna, Austria [Google Scholar]
- 48. Mirabeau O., and Joly J.-S. (2013) Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci. U.S.A. 110, E2028–E2037 10.1073/pnas.1219956110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P.-A., Xenarios I., Eisenacher M., Mayer G., Gatto L., et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 10.1038/nbt.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.