Abstract
Zinc is an essential metal in bacteria. One important bacterial zinc transporter is AdcA, and most bacteria possess AdcA homologs that are single-domain small proteins due to better efficiency of protein biogenesis. However, a double-domain AdcA with two zinc-binding sites is significantly overrepresented in Streptococcus species, many of which are major human pathogens. Using molecular simulation and experimental validations of AdcA from Streptococcus pyogenes, we found here that the two AdcA domains sequentially stabilize the structure upon zinc binding, indicating an organization required for both increased zinc affinity and transfer speed. This structural organization appears to endow Streptococcus species with distinct advantages in zinc-depleted environments, which would not be achieved by each single AdcA domain alone. This enhanced zinc transport mechanism sheds light on the significance of the evolution of the AdcA domain fusion, provides new insights into double-domain transporter proteins with two binding sites for the same ion, and indicates a potential target of antimicrobial drugs against pathogenic Streptococcus species.
Keywords: ABC transporter, kinetics, protein structure, Streptococcus, thermodynamics, double domain, zinc transporter
Introduction
Zinc is an essential trace element for all pathogenic bacteria because many crucial enzymes and transcription factors require zinc to maintain their native structure and biological functionality. It has been shown that zinc homeostasis is essential to invasion and infection by pathogenic bacteria (1, 2). Zinc directly binds to metalloenzymes or zinc finger proteins and functions as a structural or catalytic element to regulate cellular metabolism and gene expression. It also has an immune function as an anti-inflammatory agent (3, 4). To colonize in different organs, pathogenic bacteria must adapt to changing metal concentrations in various host microenvironments (5). In general and at high concentrations, zinc has been shown to be toxic (6). However, the free zinc concentration in host tissues is usually very low, ∼1 μm in lung alveolar lavages for example (7, 8). Therefore, pathogenic bacteria require an efficient zinc import system for survival, especially in hosts.
Most bacteria have evolved several zinc uptake systems to strictly control zinc concentration within cells, such as ZnuABC in Escherichia coli, the Zur family (YcdH/YceA/YcdI) in Bacillus subtilis, and AdcABC in Streptococcus pyogenes and Streptococcus pneumoniae (9–11). The ABC system in bacteria is usually composed of a zinc-binding lipoprotein, a membrane permease, and an ATPase. In Gram-positive bacteria, the lipoprotein (e.g. AdcA from S. pyogenes) attached to the cell surface acquires zinc from the environment and interacts with the membrane permease to deliver the zinc ions into the cell via ATPase, which provides energy (12).
S. pyogenes is one of the most pathogenic bacteria, causing infectious diseases that can be lethal (13, 14). It must adapt to a wide range of metal concentrations in host microenvironments during the colonization process (1, 15). Two membrane-associated lipoproteins, AdcA and Lbp, have been identified as essential zinc uptake systems as the deletion of these two genes resulted in a requirement for zinc and decreased infectivity in a mouse model. Both proteins belong to the metal-binding receptor family as ABC transporters. Most bacterial zinc-uptake proteins contain only one zinc-binding domain. Indeed, the protein Lbp in S. pyogenes and its homologous protein AdcAII in S. pneumoniae are single-domain, zinc-binding proteins whose structures and functions have been intensively investigated (16–18).
In contrast, AdcA in S. pyogenes is yet not crystallized and characterized. AdcA proteins in S. pneumoniae and S. pyogenes share 61% homology, and both are predicted to have two zinc-binding domains. In vitro studies indicated higher zinc affinity for the N-terminal domain. The S. pyogenes AdcA double-domain organization is rare among other bacterial genera. Due to the distinct translation and mRNA degradation mechanisms, bacteria synthesize large proteins in a much less efficient manner than do eukaryotes (19). Therefore, bacteria tend to maintain proteins as small as possible. Thus, there should be a distinct functional and beneficial role for the double-domain AdcA for zinc uptake in Streptococcus species.
In this study, we applied molecular modeling and molecular dynamics simulation to assess the dynamic structure and functional features of the two domains of Streptococcus AdcA. Based on the computational results, we propose a sequential structural stabilization model for the two fused domains that can conduct an interdomain conformational change when bound to zinc, an exceptional feature that enhances zinc uptake efficiency in zinc-deficient environments as compared with single-domains proteins. We further experimentally validated the predicted features and the double-domain organization that endows Streptococcus with unique survival advantages in zinc-depleted environments.
Results
Double-domain AdcA homologs are conserved and overrepresented in Streptococcus
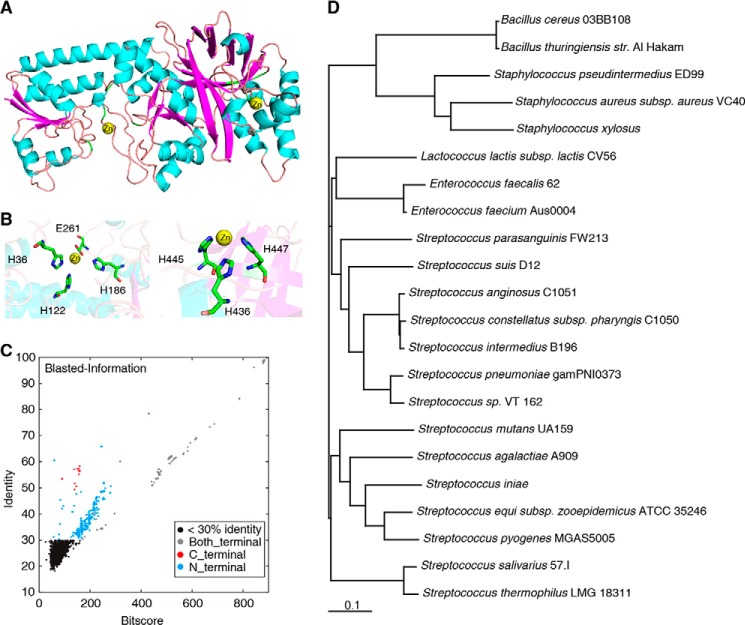
We first constructed the S. pyogenes AdcA structure using homology modeling, taking as templates the most homologous crystal structures of B. subtilis Bsu-YcdH (43.9% sequence identity to the N-terminal domain; Protein Data Bank code 2O1E) and E. coli San-YodA (47.3% sequence identity to the C-terminal domain, Protein Data Bank code 1TXL) (Fig. S1B). The constructed AdcA structure contains the amino acid residues 31–515 (Fig. 1A), which is almost full-length except for the predicted signal peptide (amino acids 1–30; Fig. S1A). The modeled structure gained a discrete optimized potential energy score of −49,727.9, indicating that this structure was approximately at the optimal low-energy state (20). A Ramachandran plot indicated that the modeled three-dimensional structure was reasonable (Fig. S1C). Virtual docking analysis suggested a zinc-binding center in the N-terminal domain (His-36, His-122, His-186, and Glu-261) and a zinc-binding center in the C-terminal domain (His-436, His-445, and His-447) (Fig. 1B). The two predicted zinc-binding sites, located in the highly conserved area, were in conformity with the principle of ConSurf evolutionary conservation patterns (21–23).
Figure 1.
Structure and phylogeny of AdcA. A, the three-dimensional structure of S. pyogenes AdcA, constructed using homology modeling. Two zinc ions are indicated as yellow balls. B, the zinc-binding residues in the N- and C-terminal domains of AdcA. C, homology search with AdcA of 3,449 bacterial genomes using Blastx. Each dot represents the best match within each bacterial genome. Black dots, <30% homology; red dots, single-domain proteins homologous to the C-terminal domain of AdcA; blue dots, single-domain proteins homologous to the N-terminal domain of AdcA; gray dots, double-domain proteins homologous to the entire AdcA. D, phylogenetic tree of the bacterial species with double-domain AdcA homologs.
To determine the universality of the double-domain organization of AdcA homologs in bacteria, we compared S. pyogenes AdcA against 3,449 completely sequenced bacterial genome sequences in the NCBI database using the Blastx tool. We found 649 proteins with more than 30% homology to AdcA. Among these, 31 proteins were single-domain zinc-binding proteins homologous to the C-terminal domain, 408 proteins contained a single zinc-binding domain homologous to the N-terminal domain, and 210 were double-domain proteins (Fig. 1C). These 210 double-domain proteins belonged to 22 species, 14 of which belonged to Streptococcus, showing a significant overrepresentation of this genus (p = 4.3608 × 10−159 by Fisher exact test). A phylogenic tree constructed based on these species showed that the 14 Streptococcus species evolved in two branches and that the evolutionary distances were maintained below 0.22, indicating that these double-domain zinc-binding proteins were highly conserved in Streptococcus during evolution from the common ancestor as compared with the other species (Fig. 1D). These structural results implied that Streptococcus AdcA possesses a unique ability for zinc uptake.
The N-terminal domain stabilizes the protein structure more strongly than does the C-terminal domain upon zinc binding
To predict the properties of S. pyogenes AdcA, we performed molecular dynamics (MD)5 simulations on the homology model. Independent replications of the MD simulation using random initial atomic velocities resulted in similar trajectories. All trajectories reached equilibrated conformation within 5 ns. These timescales are 1–2 orders of magnitude shorter than the timescale for forming any secondary structure. To exclude the formation of new secondary structures, we further performed 300-ns MD simulations for apo-AdcA and Zn2-AdcA, a timescale that is sufficient to represent the conformational change of complex proteins (24, 25). Compared with the initial state, there was no obvious change in conformation or hydrogen bonds in the equilibrium state (Fig. S5, A–C). Moreover, the trend of change in secondary structures, caused by binding of the two zinc ions, in MD simulations was consistent with CD spectrum experimental results (Fig. 2C and Table S2). The merged results from the initial structure and the average structure of the equilibrium state are shown in Fig. S5D. These show that the simulated structure is close to its native state in physiological solutions.
Figure 2.
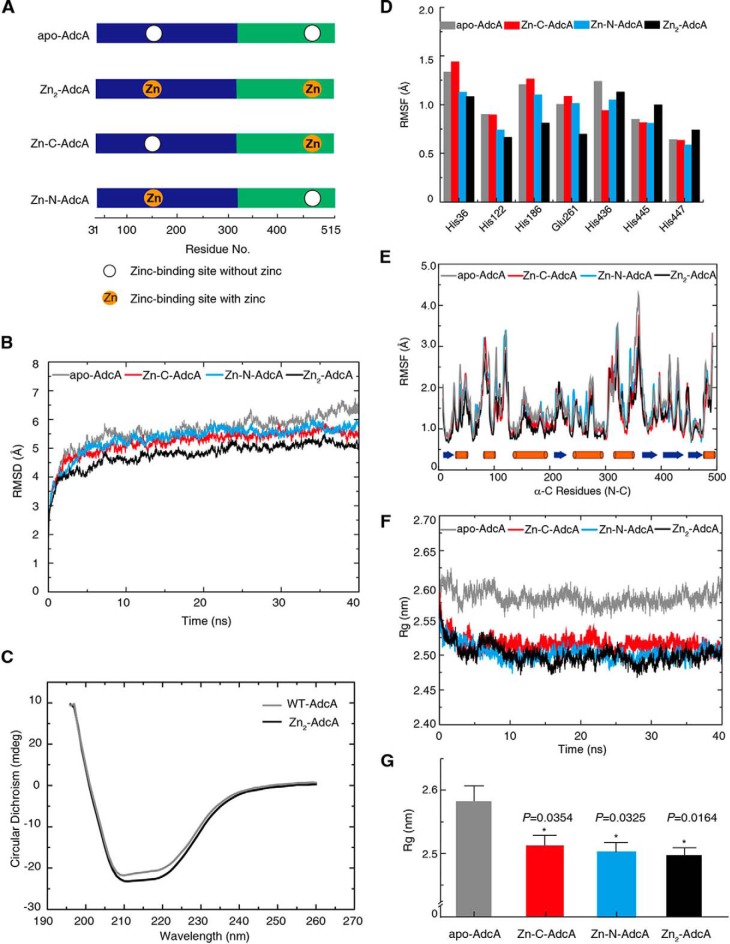
Protein flexibility. A, the four protein model constructs used in the molecular dynamic simulations. B, the time evolution of the average RMSD of the C-α atoms in the apo-AdcA (gray), Zn-C-AdcA (red), Zn-N-AdcA (bright blue), and Zn2-AdcA (black) simulations. Each sample was repetitively analyzed three times (40-ns duration). C, secondary structures of AdcAs with or without zinc ions determined by CD spectrometry. D, the RMSF of the C-α profile of residues in the two active binding sites of apo- and zinc-bound AdcA MD trajectories. E, the RMSF of C-α for all residues calculated over the 40-ns trajectory in the absence and presence of zinc. Orange and blue bars denote α-helices and β-strands, respectively. F, time evolution of the C-α average of the Rg values. G, corresponding p value statistics of Rg shown in F, and error bars represent S.D.
Next, we simulated AdcA structures without zinc atoms (apo-AdcA), with zinc in the N-terminal binding center (Zn-N-AdcA) alone, with zinc in the C-terminal binding center (Zn-C-AdcA) alone, and with zinc in both binding centers (Zn2-AdcA) (Fig. 2A). Trajectories of 40-ns MD simulations revealed the highest root mean square displacement (RMSD) for apo-AdcA, indicating that zinc binding stabilizes the protein structure (Fig. 2B). Indeed, Zn2-AdcA had the lowest RMSD, indicating that it has the highest rigidity among all four forms. This was confirmed by CD spectra (Fig. 2C): the unordered fraction of AdcA was reduced from 19.1 ± 0.14 (apo-AdcA) to 11.2 ± 0.56% (Zn2-AdcA) when saturated by zinc, and the helix fraction slightly increased from 53.2 ± 0.62 to 58.8 ± 0.14% (Table S2).
However, the structural rigidity of the two zinc-binding centers deviated remarkably. We calculated the root mean square fluctuation (RMSF) of the zinc-binding residues; a lower RMSF indicates higher rigidity. The four residues of the N-terminal domain (His-36, His-122, His-186, and Glu-261) showed reduced RMSF values after binding zinc, whereas two of the three residues in the C-terminal domain (His-436, His-445, and His-447) showed increased RMSF values (Fig. 2D) (24). These results indicated that zinc stabilizes only the N-terminal domain and that flexibility of the C-terminal domain is not affected by zinc. This was echoed by the RMSF of all C-α atoms of the two domains (Fig. 2E): 132 of 291 residues (45.5%) in the N-terminal domain showed reduced RMSF after binding zinc, whereas 45 of 194 residues (23.9%) in the C-terminal domain showed reduced RMSF after binding zinc.
Moreover, the radius of gyration (Rg) within the period of equilibrium also suggested a more flexible structure of Zn-C-AdcA, Zn-N-AdcA, and Zn2-AdcA compared with apo-AdcA (Fig. 2, F and G). To further analyze the Rg values between these three curves, we used symbolic aggregate approximation to evaluate the statistical differences in detail (Fig. S6). The major goal of the symbolic aggregate approximation algorithm is to convert time-series data to a symbolic representation, e.g. a < b < c < d. Then, the mean value of each section is calculated (26). This method can be used to accurately distinguish differences in the data (27, 28). The strings, converted from the Rg values of Zn-C-AdcA, Zn-N-AdcA, and Zn2-AdcA, were shown as dcbbcbcb, cbacbabc, and cbaabaab, respectively (Fig. S6). Thus, the Rg of Zn-C-AdcA was considerably greater than that of Zn-N-AdcA, and Zn-N-AdcA had a similar Rg compared with Zn2-AdcA (Fig. 2, F and G, and Fig. S6). Therefore, we posit that the N-terminal domain stabilizes the protein structure more than does the C-terminal domain and possesses a higher affinity for zinc. Interestingly, zinc binding in one domain influenced rigidity in the other domain. Zinc binding to the N-terminal domain (Zn-N-AdcA) reduced the RMSF of 18.7% of the residues in the C-terminal domain, and Zn-C-AdcA reduced the RMSF of 12.4% of the residues in the N-terminal domain (Fig. 2E). This suggested that the two domains, each with distinct zinc-binding properties, may synergize upon zinc binding, creating new conformational features that do not exist in the single domains.
Affinity and speed: New features emerge by synergy of the two domains
To validate the above mentioned postulations, we mutated key binding residues to alanine to abolish the N-terminal (H36A/H122A/H186A/E261A) or C-terminal (H436A/H445A/H447A) zinc-binding sites while maintaining the full lengths of N-AdcA and C-AdcA. To completely remove the interactions between the two domains, we created an N-lobe (residues 31–321) and a C-lobe (residues 322–515) to mimic the single-domain AdcA that exists in most bacterial species (Fig. 3A). These mutants and the WT AdcA were expressed and purified (Fig. S3, A and B). The inductively coupled plasma MS measurement showed that all the purified proteins did not contain any metal ions.
Figure 3.
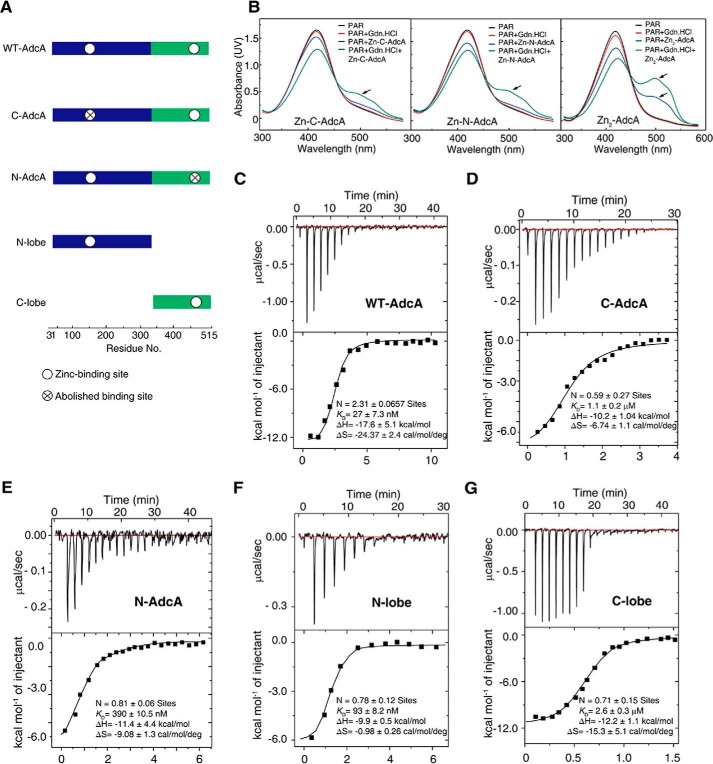
Zinc-binding affinity of AdcAs and PAR competition for zinc ions. A, the five protein constructs used to determine the biophysical properties. The constructs were overexpressed in E. coli BL21 (DE3) strain and purified. B, UV-visible spectra of PAR, which competes for zinc, for Zn-C-AdcA, Zn-N-AdcA, and Zn2-AdcA in the presence or absence of guanidine hydrochloride (Gdn.HCl). C–G, isothermal titration calorimetry binding curves of WT AdcA (C), C-AdcA (D), N-AdcA (E), the N-lobe (F), and the C-lobe (G) at 25 °C. The parameters N, KD, ΔH, and ΔS are shown in the diagrams.
To detect the strength differences of the two binding domains of AdcA, a 4-(2-pyridylazo)resorcinol (PAR) competition test with Zn2-AdcA was performed. As shown in Fig. 3B, PAR could only capture one zinc from Zn2-AdcA under normal conditions, whereas it could capture other zinc atoms only under harsh denaturation conditions, e.g. 6 m guanidine hydrochloride. These results of the competition test showed that the differences in zinc binding strength of the N- and C-terminal domains are significant.
We further experimentally determined the zinc-binding affinities of WT and mutant AdcA using isothermal titration calorimetry (ITC), and the calculated values of N, KD, ΔH, and ΔS are shown in Fig. 3, C–G. The KD of the WT AdcA for Zn2+ was measured as 27 ± 7.3 nm, much lower than that of any mutant (Fig. 3C). The independently expressed N-lobe and C-lobe had KD values of 93 ± 8.2 nm and 2.6 ± 0.3 μm, respectively (Fig. 3, F and G), confirming that the N-lobe has higher zinc-binding affinity than does the C-lobe. Moreover, the full-length expressed mutants N-AdcA and C-AdcA had dissociation constants of 390 ± 10.5 nm and 1.1 ± 0.2 μm (Fig. 3, D and E). Notably, the independently expressed N-lobe had a stronger binding affinity for Zn2+ than did full-length N-AdcA, implying that the two domains interact with each other, influence each other's conformation, and thus regulate zinc affinity. Thermodynamic data on enthalpy changes (ΔH) and entropy changes (TΔS) of AdcAs were detected by ITC. For zinc binding, WT AdcA, C-AdcA, N-AdcA, the N-lobe, and the C-lobe were, respectively, found to have ΔH values of −17.6 ± 5.1, −10.2 ± 1.04, −11.4 ± 4.4, −9.9 ± 0.5, and −12.2 ± 1.1 kcal/mol and TΔS values of −7.3 ± 0.7, −2.0 ± 0.3, −2.7 ± 0.4, −0.3 ± 0.07, and −4.5 ± 1.5 kcal/mol. The values of ΔH and TΔS indicated that zinc binding to the five models of AdcA is both enthalpically and entropically favorable with ΔH being the main driving factor.
Next, we measured the zinc binding kinetics of AdcA. As expected, N-AdcA, with higher affinity-bound zinc, had very fast kinetics and a two-component reaction (k1 = 45.45 ± 0.07 s−1 and k2 = 0.93 ± 0.02 s−1, A1 = 0.099 ± 0.000 and A2 = 0.161 ± 0.002). In contrast, C-AdcA, with a less rigid structure, bound zinc at a much lower rate and had a second-order reaction. The fast reaction was a minor reaction with an amplitude of A1 = 0.154 ± 0.026 and k1 = 5.02 ± 0.03 s−1, whereas the major reaction had the kinetic parameters A2 = 0.271 ± 0.036 and k2 = 0.15 ± 0.01 s−1 (Fig. 4A). WT AdcA, with two binding centers, revealed a similarly fast process (k1 = 41.05 ± 0.01 s−1) as compared with N-AdcA and a much-accelerated slow process (k2 = 0.75 ± 0.02 s−1) as compared with C-AdcA (Fig. 4A). This indicated that, in the presence of a low concentration of zinc, the N-terminal domain rapidly bound a zinc atom and stabilized the entire protein structure. The C-terminal domain was then stabilized to obtain a faster binding speed. Both the zinc binding velocity and affinity constant of the C-terminal domain are 1 order of magnitude slower and weaker, respectively, than that of the N-terminal domain.
Figure 4.
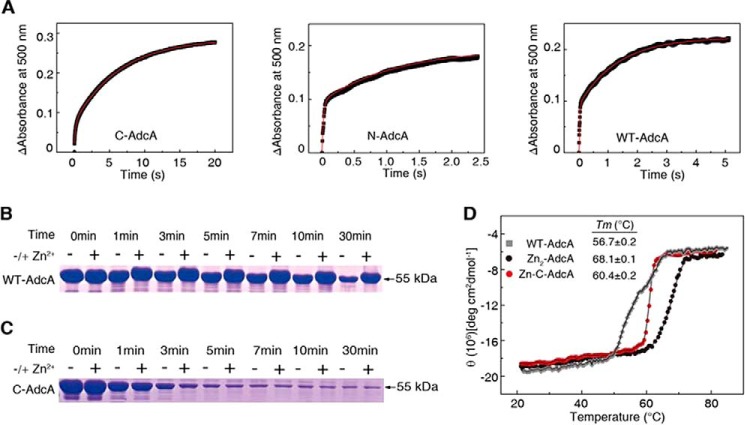
Biochemical, thermodynamic, and kinetic characterization to verify protein stability. A, kinetics of AdcA binding to zinc. Time-dependent reactions were created using 10 μm apo-form proteins mixed with 40 μm Zn(PAR)2. B, proteinase K sensitivity of WT AdcA. Proteins (15 μg) were subjected to proteinase K (30 μg/liter) for 0, 1, 3, 5, 7, 10, and 30 min. C, proteinase K sensitivity of C-AdcA. D, thermal stabilities of WT AdcA, Zn2-AdcA, and Zn-C-AdcA. Thermal unfolding transitions were monitored using far-UV CD spectra at 223 nm. deg, degrees.
To validate the structural stability of the zinc-bound N-terminal domain, we performed proteinase K digestion assays. Zinc-saturated AdcA was almost intact after 30 min of proteinase K digestion, whereas the apo form of AdcA was mostly digested (Fig. 4B). In contrast, C-AdcA was vulnerable to proteinase K attack in both the apo and zinc-bound forms (Fig. 4C) due to the lack of stabilization by the zinc-bound N-terminal domain. This was echoed by the melting temperature of these two proteins. The melting temperature of Zn-C-AdcA was similar to that of WT AdcA and much lower than that of Zn2-AdcA (Fig. 4D). These results suggested that the zinc in the N-terminal domain was crucial for enhancing the zinc binding rate in the C-terminal domain. As the C-terminal domain binds zinc with less affinity, it can pass zinc downstream to other zinc-binding proteins that finally transport zinc into the cell.
Structural alteration upon zinc binding
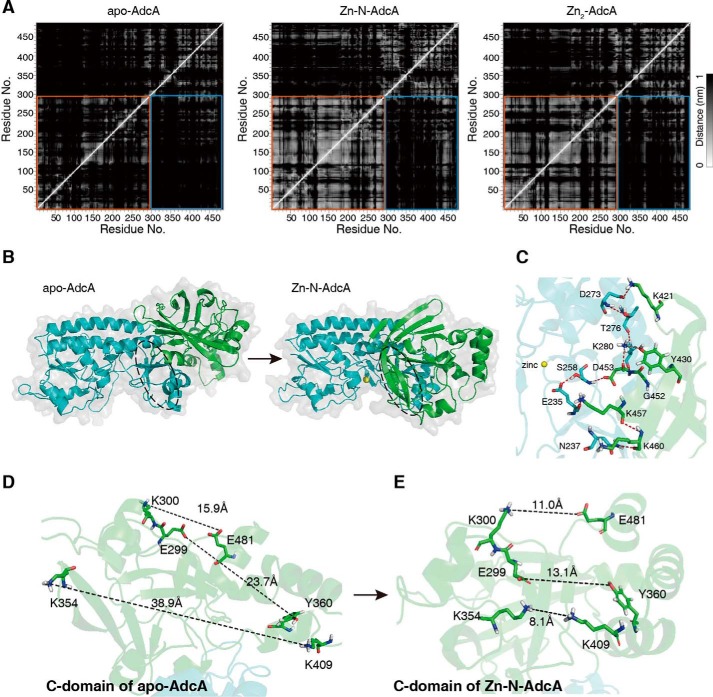
To investigate in detail the structural alterations after zinc binding, we compared the MD simulation trajectories of apo-AdcA, Zn-N-AdcA, and Zn2-AdcA. The mean distance matrix showed remarkably decreased interdomain distances when the N-terminal domain bound a zinc atom (29) (Fig. 5A), which can be visualized in the steady-state three-dimensional conformations (Fig. 5B and Fig. S4). This can be explained by N-terminal domain stabilization of the C-terminal domain via interatomic interactions of the peptide chains. The mean distance matrix of Zn-N-AdcA showed adjacency of the residue pairs Asn-237 and Lys-460, Glu-235 and Lys-457, Ser-258 and Asp-453, Lys-280 and Asp-453, Lys-280 and Tyr-430, and Asp-273 and Lys-421. All these interactions are interdomain interactions (30, 31) spanning the entire interaction surface between the two domains (Fig. 5C).
Figure 5.
Zinc binding in the N-terminal domain of AdcA stabilizes the C-terminal domain conformation for C-terminal zinc binding. A, mean distance matrices for the different states of the proteins are shown for apo-AdcA, Zn-N-AdcA, and Zn2-AdcA. Lighter colors represent closer distances between two amino acid residues. An orange box denotes the N-terminal domain, and the bright blue boxes denote the interdomain distances. B, representative structures of apo-AdcA and Zn-N-AdcA. C, hydrogen bonds (red dashes) on the N- and C-terminal interaction surfaces. D and E, the differences in the C-terminal domain conformations were compared between apo-AdcA and Zn-N-AdcA, and the distances between the selected residues were measured.
Moreover, the interdomain surface is linked by at least 10 hydrogen bonds (Fig. 5C, red dashed lines). This multianchored stabilization mechanism explains the stability of the induced conformational change. The induced conformation dramatically decreased the distance among the residues around the zinc-binding pocket of the C-terminal domain (32), specifically Lys-300 and Glu-481, Glu-299 and Tyr-360, and Lys-354 and Lys-409 (Fig. 5, D and E), facilitating the binding to zinc. This explained the increased affinity of the C-terminal domain when the N-terminal domain was zinc-bound. In summary, these analyses demonstrated the structural basis of the synergy between the two domains for both metal-binding affinity and rate.
Verification of the interaction surface of the N-terminal and C-terminal domains using MD simulations
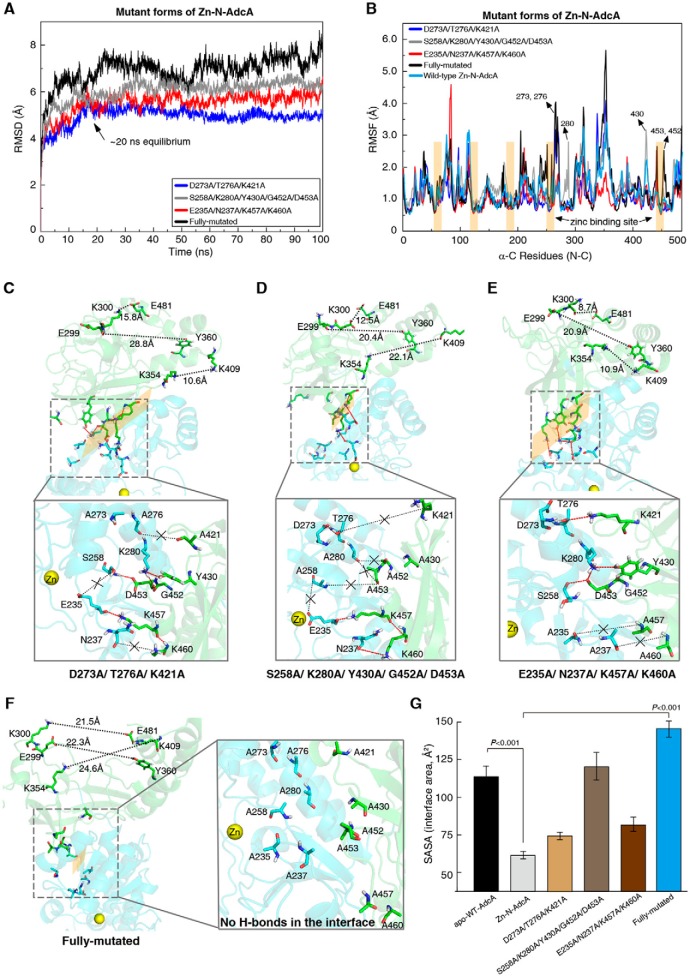
We showed that the interface interactions rely on 10 hydrogen bonds between the N-terminal and C-terminal domains, involving the 12 amino acid residues depicted in Fig. 5C. To evaluate the contributions of the 12 residues to the interface interaction, we successively established 12 single-site mutants of Zn-N-AdcA. However, we found that none of the single-site mutants caused any reduction of the surface formation (Fig. S7). Subsequently, we established four Zn-N-AdcA mutants, mutating groups of the abovementioned residues according to their vicinity: D273A, T276A, and K421A; K280A, S258A, Y430A, D453A, and G452A; E235A, N237A, K460A, and K457A; and a variant mutated for all 12 residues. In the fully mutated protein, only one zinc ion was located in the N-terminal domain of AdcA.
The trajectory analyses of the 100-ns MD simulations showed that these mutant proteins took a much longer time (∼20 ns; Fig. 6A) to reach the equilibrium state than did the WT Zn-N-AdcA (∼5 ns; Fig. 2B). Especially the fully mutated protein did not even reach the equilibrium state in 100 ns (Fig. 6A). The result revealed that the 12 residues of the interaction surface are crucial to stabilizing the skeleton of the whole protein. Among the 12 residues, the six residues at positions 273, 276, 280, 430, 452, and 453 showed increased RMSF values after mutation to alanine (Fig. 6B).
Figure 6.
Identification of the N- and C-terminal interaction surfaces of Fig. 5C. A, the time evolution of RMSD values in the four mutant Zn-N-AdcAs, D273A/T276A/K421A (blue), S258A/K280A/Y430A/G452A/D453A (gray), E235A/N237A/K457A/K460A (red), and the fully mutated variant (black). B, the RMSF of the C-α for all residues of the four mutant proteins calculated over the 100-ns trajectory. The RMSF values of the mutated amino acids became greater. Hydrogen bonds (red dashes), missing hydrogen bonds (black dashes with “×”), interaction surfaces (dotted line frame), and conformation changes of the C-terminal domain are shown in the four mutated forms of Zn-N-AdcA, D273A/T276A/K421A (C), S258A/K280A/Y430A/G452A/D453A (D), E235A/N237A/K457A/K460A (E), and the fully mutated variant (F). G, the calculated SASA values of the interface area, respectively, and error bars represent S.D.
We next assessed whether the hydrogen bonds stabilized the interaction surface, measuring the solvent-accessible surface area (SASA) where a smaller SASA value represents tighter structures. The mutant structures showed that abolishing any group of hydrogen bonds reduced the interaction surface (Fig. 6, C–E). Abolishing all hydrogen bonds completely destroyed the interaction surface. These results were also reflected in the SASA values (Fig. 6G).
To further investigate the role of these hydrogen bonds in the allosteric cross-talk between the two domains, we calculated the average separation distance between representative residue pairs around the zinc-binding pocket of the C-terminal domain (Fig. 6, C–F), specifically Lys-300 and Glu-481, Glu-299 and Tyr-360, and Lys-354 and Lys-409. The three corresponding average separation distances for each of the above mentioned mutants were 15.8, 28.8, and 10.6 Å (Fig. 6C, D273A, T276A, and K421A); 12.5, 20.4, and 22.1 Å (Fig. 6D, K280A, S258A, Y430A, D453A, and G452A); 8.7, 20.9, and 10.9 Å (Fig. 6E, E235A, N237A, K460A, and K457A); 21.5 Å, 22.3 Å, and 24.6 Å (Fig. 6F, fully mutated variant). Significantly, conformational changes in the C-terminal domain induced by the N-terminal domain of these four mutant proteins were reduced to varying extents when compared with WT Zn-N-AdcA (11.0, 13.1, and 8.1 Å; Fig. 5E). Among these, the fully mutated variant had the greatest impact on the conformation of the C-terminal domain. Based on these results, we propose a possible mechanism: the N-terminal domain firmly and rapidly binds a zinc ion and draws the C-terminal domain closer, relying on hydrogen bonds. Subsequently, this stabilizes the C-terminal domain and tightens the zinc-binding pocket, which facilitates zinc binding, especially in zinc-depleted environments.
A synergistic double-domain AdcA endows growth advantages at low zinc concentrations
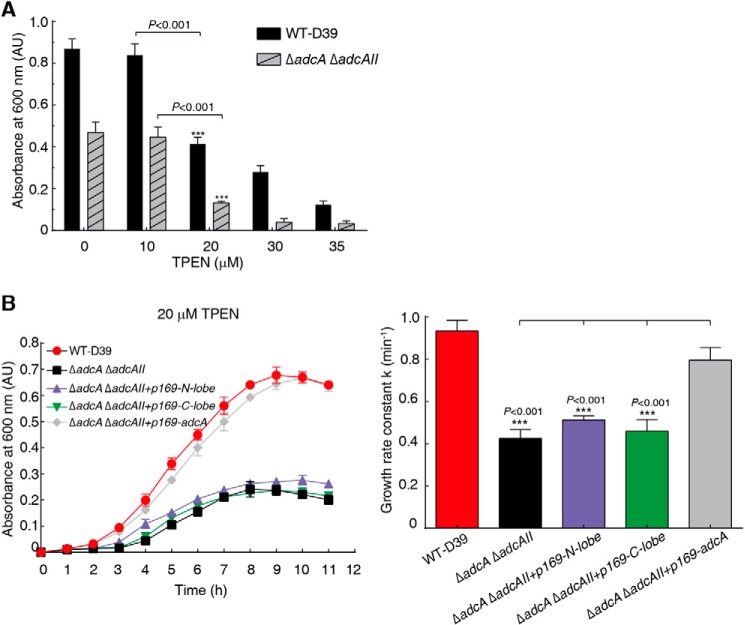
The double-domain AdcA increased both affinity and rate of zinc binding. Therefore, we postulate that bacteria use the synergistic organization of the double-domain AdcA to more efficiently take up zinc, even under conditions of extremely low environmental zinc concentrations, to maintain growth. Due to the lack of genetic manipulation tools in S. pyogenes, we performed experiments in S. pneumoniae. We used N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) to chelate zinc ions in the media and to create a zinc-deficient environment. With increasing TPEN concentrations, significant growth hindrance was observed from 20 μm TPEN (Fig. 7A) where the free zinc concentration was estimated to be ∼150–300 nm. Then we used this concentration to test zinc uptake ability. The deletion strain ΔadcA ΔadcAII of S. pneumoniae, with a deleted zinc transporter AdcA system, grew significantly slower than the WT strain and stopped growing at an A600 of 0.22. Expression of either N-AdcA or C-AdcA in the deletion strain did not rescue growth, whereas expression of the double-domain AdcA almost completely rescued the growth to a rate similar to that of the WT strain (Fig. 7B). This result validated that the double-domain organization facilitated bacterial growth in a zinc-deficient environment.
Figure 7.
The growth of bacteria in a zinc-deficient environment. A, the absorbance at 600 nm of WT S. pneumoniae (D39) and ΔadcA ΔadcAII mutant strains cultured in C+Y medium containing TPEN at different final concentrations for 8 h. B, the growth curves (left) of D39 strains grown in C+Y medium containing 20 μm TPEN and corresponding growth rate constants (right) calculated for WT, ΔadcA ΔadcAII, and ΔadcA ΔadcAII with the N-lobe, C-lobe, and full-length adcA, respectively, and error bars represent S.D. AU, absorbance units.
Discussion
Generally, bacterial genomes tend to encode smaller proteins (267 amino acids long on average) than do eukaryotes (33). Major reasons include that mRNA is degraded by endonucleases in bacteria, making it difficult to translate full-length long proteins (19), and multidomain large proteins need proper translational pausing sites for correct folding, which may be disturbed by environmental and molecular factors (34–36). Indeed, in most bacterial species, the homologs of Streptococcus AdcA are single-domain small proteins. Therefore, the evolution of the double-domain AdcA in Streptococcus (515 amino acids) should be unlikely unless an emergent feature benefits the adaptive survival of Streptococcus in host environments, such as lung tissue (1–2 μm zinc) (7). A previous study suggested that N-terminal domains in two-domain proteins are biased to be shorter and are predicted to fold faster than their C-terminal counterparts (37). However, AdcA is a special case of an overall trend in two-domain proteins as its N-terminal domain is longer than its C-terminal domain.
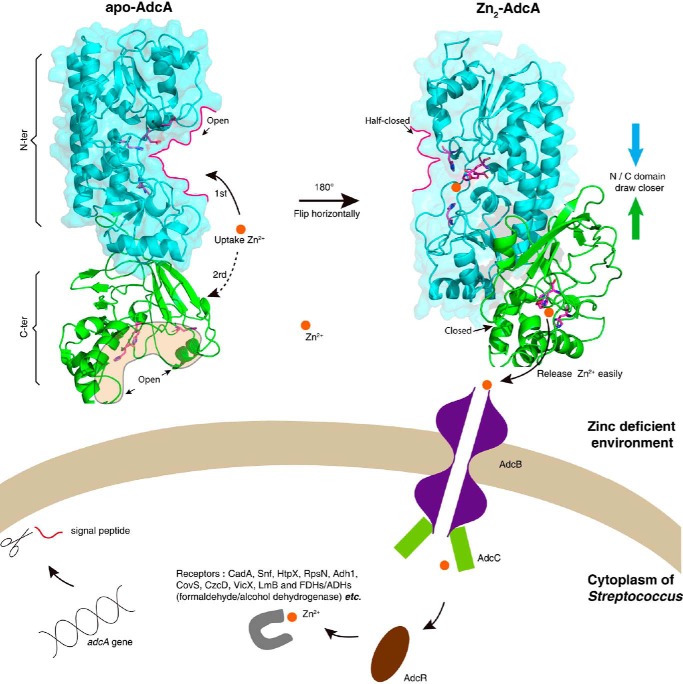
In this study, we found that double-domain AdcA with two zinc-binding sites is significantly overrepresented in Streptococcus species by homology comparison between AdcA and the library of known genomes of bacteria. We have shown evidence that fusion of the two domains creates a new, emergent, structure-based functional advantage that is greater than the sum of the advantages provided by two independent domains. In the presence of zinc, the N-terminal domain firmly and quickly binds a zinc ion and changes its conformation. This conformation change in the N-terminal domain subsequently stabilizes the C-terminal domain and tightens its zinc-binding pocket, facilitating zinc binding, especially in zinc-depleted environments. The relatively less stable conformation and weak binding of the C-terminal domain ensure a rapid transfer of zinc to the downstream transporter, the transmembrane protein AdcB of Streptococcus species (Fig. 8). Otherwise, overly stable and strong binding would limit the cyclic utilization of zinc transporters on the cell surface and the flux of zinc into the cell as shown in Fig. 7B. This interdomain synergy via interatomic interaction of the peptide chains endows Streptococcus with exceptional zinc uptake efficiency in zinc-depleted media, thus benefiting its survival in harsh conditions, e.g. in lung alveolar lavages with low zinc concentration (7). Therefore, the emergent features of the fused domains could be a simple evolutionary response to specific metal-deficient conditions (Fig. 8).
Figure 8.
The model of zinc uptake in Streptococcus via AdcA from a zinc-deficient environment. The N-terminal (N-ter) domain and the C-terminal (C-ter) domain of AdcA are colored cyan and green, respectively. The two binding sites are represented as an open state in apo-AdcA but a half-closed or closed state in Zn2-AdcA. Compared with the C-terminal domain, the N-terminal domain has a faster binding rate and stronger affinity for zinc ions. The release of a zinc ion from the C-terminal domain of AdcA to AdcB is easier than that from the N-terminal domain. The receptor proteins for zinc ions in the Streptococcus cytoplasm are represented schematically by the gray loop.
As organisms need multiple trace elements for physiology, other metal ion transporters are also found to exist in double-domain formats with two binding sites for the same ion. Examples are not restricted to bacteria and include the human transferrin receptor that binds two samarium ions (Protein Data Bank code 1CX8), human copper-lactoferrin that binds two copper ions (Protein Data Bank code 1LFI), Rapana thomasiana hemocyanin that binds two copper ions (Protein Data Bank code 1LNL), lactoferrin that binds two Fe3+ ions (Protein Data Bank code 1B7Z), and others. Our model for emergent interdomain synergy may provide new insights to better understand such cases.
Our finding also emphasizes the interdomain conformation change after metal binding, which may implicate a novel target for antimicrobial drugs against pathogenic Streptococcus species. No AdcA homolog was found in the human genome using the HMMER tool (Fig. S8), suggesting that such a conformation may not exist in the human proteome. Therefore, a rationally designed blocker molecule that binds the interdomain surfaces may abolish the synergy of the two domains, sufficiently reduce the influx of zinc, and inhibit the growth of the pathogen. The action mechanism of the designed blocker molecule would be totally different from that of existing antibiotics. To be noted, the interdomain interaction is stabilized by as many as 10 hydrogen bonds (38, 39), suggesting that mutation of a few amino acids will not abolish the interaction, thus minimizing the probability of the bacteria to evolve resistance by simple point mutations. This finding may help during the treatment of bacterial infections caused by Streptococcus.
In conclusion, we suggest a special zinc transportation mechanism mediated by AdcA in Streptococcus species. An N-terminal binding site preferentially binds a zinc ion and induces stabilization of the overall conformation of the protein via interdomain interaction. This allows the C-terminal binding domain to acquire a zinc ion while also allowing zinc bound by the C-terminal domain to be more likely and preferentially released (Fig. 8). This model elucidates the significance of the evolution of the domain fusion, provides new insights on double-domain transporter proteins with two binding sites for the same ion, and implicates a novel target for antimicrobial drugs against pathogenic Streptococcus species.
Experimental procedures
MD simulation
The preprocessed structures of apo-AdcA, Zn-N-AdcA (mutation of the three zinc-binding ligands in the C-terminal domain), Zn-C-AdcA (mutation of the four binding ligands in the N-terminal domain), and Zn2-AdcA, obtained from homology modeling, were used as starting conformations. These conformations were solvated in cubic periodic boxes containing 0.15 m Na+ and Cl− ions to neutralize the system (40, 41). No zinc ions were added to the simulated solution, representing a zinc-deficient environment. Energy minimization in each AdcA system was performed for the first relaxed energy through 400 steps of the steepest descent energy method and then continued with 25,000 steps of the conjugate gradient method. All MD simulations were simulated at a temperature of 310.15 K (42) and pressure of 1 atm (43) by the Gromacs 4.6.6 package with the simple point charge (SPC) model for liquid water as described previously (44). The zinc ion force field of the Gromacs package has been developed in various aspects (45), including bonds, angles, impropers, metal center, dihedrals, and normal van der Waals, and it is suitable for the protein-zinc simulation (46–48). Therefore, we used the default parameters of Gromos43a1 force field for the simulation of AdcAs with zinc. We set the value of the emtol convergence criterion at 1,000 kJ/mol/nm, and the temperature in the box was determined by the v-rescale temperature coupling method. We used the particle mesh Ewald method to calculate electrostatic interactions within the system. The position restraint simulations for the systems were executed for 50 ps, and then we actualized the 40-ns unrestrained simulation.
We simulated four structures of the AdcA system as mentioned above, and each system was repetitively carried out three times with random initial velocities (49). To avoid equilibration artifacts, we calculated the structural features using trajectory data ranging from 10 to 40 ns, which represents the structures at their equilibrated states. To validate the equilibrium of each structure, we also performed 300-ns MD simulations for apo-AdcA and Zn2-AdcA. We analyzed RMSD, RMSF, secondary structure, and Rg using Gromacs tools g_rms, g_rmsf, do_dssp, and g_gyrate, respectively. The mean distances between residues were calculated by the Gromacs tool g_dist. Protein structures were visualized using PyMOL 1.7. The matrix of residue distance fluctuations was represented by methods described previously (50, 51), and it can be used to describe the plasticity and elasticity of residues in structural fluctuations. To reduce the deviation, all MD trajectories data were derived from three replicates (Fig. S2).
Homology modeling
The NCBI BLAST search tool was used to find several proteins that have the highest homology to AdcA. Then a multiple sequence alignment and cluster analysis were performed using proteins with high scores through the software package ClustalX 2.0. The tertiary structure of AdcA was modeled using MODELLER in Accelrys Discovery Studio Client 4.1 (52, 53). Proteins San-YodA (Protein Data Bank code 1TXL) and Bsu-YcdH (Protein Data Bank code 2O1E), with a high level of amino acid sequence homology to AdcA, were selected as the templates to model the initial stage of the three-dimensional structure of AdcA. Subsequently, flexible molecule docking between AdcA and zinc was processed via the LigandFit module of the software to find the lowest energy conformation combining the ligand and the receptor in the active site (Fig. S1B). The reliability of the model was evaluated by discrete optimized potential energy and Ramachandran plots (54).
Construction, expression, and purification of WT AdcA
Genomic DNA extracted from S. pyogenes MGAS5005 (ATCC BAA-947TM) was used as a template to amplify the adcA gene (1,488 bp without the N-terminal signal). PCR primers were designed to introduce the restriction enzyme sites of BamHI and SalI for the adcA gene (Table S1). Detailed methods were described previously (55). Purified AdcA protein was confirmed using 12% SDS-PAGE and identified using MS (ABI 4800 MALDI-TOF/TOF) according to a method described previously (56) (Fig. S3D).
Proteinase K resistance experiments
To test the proteinase K (Roche Diagnostics) sensitivity of AdcA, the same amounts of apo-, zinc-saturated WT, and mutant AdcA (15 μg) were incubated with 30 μg/liter proteinase K in 20 mm Tris-HCl buffer (pH 8.0) containing CaCl2 (10 mm) at 25 °C for 0, 1, 3, 5, 7, 10, and 30 min. Proteolysis was stopped by addition of 5 μl of 100 mm phenylmethanesulfonylfluoride and boiling for 10 min. Digestion fragments were analyzed by 12% SDS-PAGE. Gels were stained with Coomassie Blue R-250 and scanned with Image Scanner II (GE Healthcare).
Circular dichroism spectroscopy
Far-UV CD studies were performed with a CD spectrometer (Chirascan, Applied Photophysics Ltd., Leatherhead, UK) using a quartz cuvette with a 0.1-cm optical path length at a wavelength range of 260–190 nm at room temperature. CD data were collected for 5 μm apo-, zinc-saturated WT, and mutant AdcA in 20 mm Tris-HCl (pH 7.4) in a data pitch of 1 nm at a scanning rate of 100 nm/min. Each CD spectrum was repeated three times, and a blank containing the same buffer was subtracted as a reference. Analysis of the experimental data was performed using the software CDPro. CD spectra were also used to study the thermal stability of proteins in the absence or presence of zinc ions. Thermal unfolding curves were monitored from 20 to 90 °C using an increase rate of 2 °C/min by detecting the loss of secondary structures at 222 nm. Each data set was obtained three times using steps of 0.5 °C, and Tm values were calculated by the included Glob3 software.
Growth media and growth curve assays
Casein-based semisynthetic liquid culture medium (C+Y medium) was used to culture WT and mutant D39 strains (57). To establish Zn(II) starvation conditions, TPEN (Sigma-Aldrich) was added into C+Y medium at final concentrations of 10, 20, 30, and 35 μm (8, 58, 59). We determined the growth curves for WT and ΔadcA ΔadcAII double-mutant strains cultured in zinc starvation medium at 37 °C with 5% CO2 for 12 h by measuring A600 values at different time points. For the double-mutant strain, 20 μm TPEN significantly inhibited bacterial growth at an A600 of ∼0.15 after 12 h of culture. We selected 20 μm TPEN to add to the medium to create zinc deficiency. Different treated S. pneumoniae strains were inoculated into C+Y medium, and growth curves were determined three times.
Construction, expression, and purification of mutant AdcAs
Based on the predicted structure of AdcA with molecular docking, the N-terminal binding site is composed of His-36, His-122, His-186, and Glu-261, and the C-terminal binding site is composed of His-436, His-445, and His-447. The four amino acids in the N-terminal domain and the three amino acids in the C- terminal domain were simultaneously mutated to alanine to generate the four-residue mutant H36A/H122A/H186A/E261A (C-AdcA) and the three-residue mutant H436A/H445A/H447A (N-AdcA), respectively, using a QuikChange mutagenesis kit (Stratagene) with the original pGEX-4T-adcA plasmid as template. The primers used for constructing the mutants are listed in Table S1. All the constructed plasmids were transformed to E. coli XL1-blue, which were then screened on LB agar plates containing 100 μg/ml ampicillin followed by DNA sequencing (Invitrogen). The plasmids with correct sequences were transformed to E. coli BL21 (DE3) for expression. Expression and purification of mutant AdcAs were conducted as done for WT AdcA.
Construction of the ΔadcA/ΔadcAII double-mutant strain
The primer sequences used to construct mutant strains are listed in Table S1. The double mutant strain was constructed as described previously (60, 61). Long flanking homology PCR products contained an antibiotic resistance cassette (erythromycin or spectinomycin) flanked by 600-bp-long fragments homologous to the end of each target gene, adcA or adcAII. Then the long flanking homology PCR fragments were transformed into S. pneumoniae D39 competent cells. Transformants were selected with antibiotic-containing Columbia sheep blood agar plates after overnight culture at 37 °C with 5% CO2 and confirmed by DNA sequence analysis and PCR (Fig. S3C). The mutant strain was stable after six sequential passages in Todd-Hewitt broth with 0.5% yeast extract (THY) medium in the absence of antibiotics.
Construction of the three types of overexpression strains
To construct overexpression strains of S. pneumoniae D39 for recovery of expression of different types of AdcA, the plasmid pIB169 (p169) was used in this study (62). We constructed three recombinant plasmids, p169-N-lobe (expressing the N-lobe alone), p169-C-lobe (expressing the C-lobe alone), and p169-adcA (expressing full-length adcA) (Fig. S3C). The constructed plasmids were transformed into the ΔadcA ΔadcAII double mutant strain, and the positive clones were screened using Columbia blood plates with 4 μg/ml chloramphenicol. The transformants with the recovered genes were verified by PCR. All primers are listed in Table S1.
Comparison of the zinc binding strength of the two domains of AdcA
To compare the binding strength of the N-terminal and C-terminal domains of AdcA, a special zinc metallochromic indicator, PAR, was used. A final concentration of 100 μm PAR was added to 5 μm Zn-N-AdcA, Zn-C-AdcA, and Zn2-AdcA in 20 mm Tris-HCl (pH 7.4) with or without 6 m guanidine hydrochloride. UV/visible absorbance spectra were obtained from 300 to 600 nm after a 5-min equilibration at room temperature.
Zinc-binding affinity determination
To determine the binding affinities of WT and mutant AdcAs with zinc, ITC experiments were performed at 25 °C using a MicroCalorimeter Auto-ITC 200 (Malvern, UK). Prior to the experiments, the instrument was washed with deionized water, and the ZnCl2 and AdcAs were dissolved in the same solution buffer (20 mm Tris-HCl, 100 mm NaCl (pH 7.4)) (55). Typically, an experiment consisted of loading the syringe with zinc ions at a concentration at least 10-fold higher than the AdcAs samples, which were placed in the cell. The titration parameters were set as follows: 2 μl of ZnCl2 solution were injected into the 300-μl protein sample cell during each titration with 15–20 injections. The delay time between injections was set at 200 s to ensure thermal equilibrium before the next injection. The background heat effect was subtracted by addition of zinc alone to the buffer. All integrated heat data were analyzed using Origin 7.0 software for fitting calculations.
Stopped-flow absorbance kinetics
Stopped-flow spectroscopy was performed on a stopped-flow reaction analyzer (Chirascan SF.3, Applied Photophysics Ltd.) using the absorbance mode to monitor absorbance changes at 495 nm over time. The Zn(PAR)2 complex (200 μm PAR in 20 mm Tris-HCl buffer combined with 40 μm Zn2+) was loaded into the A drive syringe, the B drive syringe was filled with 10 μm apo-AdcAs, and transient mixing of the reaction was driven by bottled nitrogen. Experimental parameters were set as follows: 1-nm bandwidth, 10-mm optical path, 495 nm scanned wavelength with a 475-nm filter, and a 25 °C water bath temperature. Buffer incubated with Zn(PAR)2 was used as the reference. Tests were repeated until consistent results were obtained. The collected data were analyzed using exponential equations.
Author contributions
X. S., G. Z., and Q.-Y. H. conceived and designed the project. K. C., C. W., N. L., J. H., and B. Z. performed the experiments and data analysis. X. C. analyzed data. K. C., N. L., X. S., and G. Z. wrote the manuscript.
Supplementary Material
Acknowledgment
We are grateful for the high-performance platform of Jinan University.
This work was supported by National Natural Science Foundation of China Grants 21571082 (to X. S.) and 21271086 (to Q.-Y. H.), National High-Tech Research and Development Program (863) of China Grant 2014AA020504 (to G. Z.), Guangdong Natural Science Research Grants 2015A030313334 (to X. S.) and 32213027/32215077 (to Q.-Y. H.), and Guangzhou Science and Technology Grant 201607010228 (to X. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8, Tables S1 and S2, and structures constructed using homology modeling.
- MD
- molecular dynamics
- PAR
- 4-(2-pyridylazo)resorcinol
- TPEN
- N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine
- RMSF
- root mean square fluctuation
- RMSD
- root mean square displacement
- Rg
- radius of gyration
- ITC
- isothermal titration calorimetry
- SASA
- solvent-accessible surface area.
References
- 1. Zalewski P. D., Truong-Tran A. Q., Grosser D., Jayaram L., Murgia C., and Ruffin R. E. (2005) Zinc metabolism in airway epithelium and airway inflammation: basic mechanisms and clinical targets. A review. Pharmacol. Ther. 105, 127–149 10.1016/j.pharmthera.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 2. Murakami M., and Hirano T. (2008) Intracellular zinc homeostasis and zinc signaling. Cancer Sci. 99, 1515–1522 10.1111/j.1349-7006.2008.00854.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou X., Cooper K. L., Sun X., Liu K. J., and Hudson L. G. (2015) Selective sensitization of zinc finger protein oxidation by reactive oxygen species through arsenic binding. J. Biol. Chem. 290, 18361–18369 10.1074/jbc.M115.663906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prasad A. S. (2014) Zinc: an antioxidant and anti-inflammatory agent: role of zinc in degenerative disorders of aging. J. Trace Elem. Med. Biol. 28, 364–371 10.1016/j.jtemb.2014.07.019 [DOI] [PubMed] [Google Scholar]
- 5. Ma L., Terwilliger A., and Maresso A. W. (2015) Iron and zinc exploitation during bacterial pathogenesis. Metallomics 7, 1541–1554 10.1039/C5MT00170F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Djoko K. Y., Ong C. L., Walker M. J., and McEwan A. G. (2015) The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J. Biol. Chem. 290, 18954–18961 10.1074/jbc.R115.647099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shafeeq S., Kuipers O. P., and Kloosterman T. G. (2013) The role of zinc in the interplay between pathogenic streptococci and their hosts. Mol. Microbiol. 88, 1047–1057 10.1111/mmi.12256 [DOI] [PubMed] [Google Scholar]
- 8. Tedde V., Rosini R., and Galeotti C. L. (2016) Zn2+ uptake in Streptococcus pyogenes: Characterization of adcA and lmb null mutants. PLoS One 11, e0152835 10.1371/journal.pone.0152835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaballa A., and Helmann J. D. (1998) Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180, 5815–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patzer S. I., and Hantke K. (2000) The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275, 24321–24332 10.1074/jbc.M001775200 [DOI] [PubMed] [Google Scholar]
- 11. Capdevila D. A., Wang J., and Giedroc D. P. (2016) Bacterial strategies to maintain zinc metallostasis at the host-pathogen interface. J. Biol. Chem. 291, 20858–20868 10.1074/jbc.R116.742023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blindauer C. A. (2015) Advances in the molecular understanding of biological zinc transport. Chem. Commun. 51, 4544–4563 10.1039/c4cc10174j [DOI] [PubMed] [Google Scholar]
- 13. Bessen D. E. (2009) Population biology of the human restricted pathogen, Streptococcus pyogenes. Infect. Genet. Evol. 9, 581–593 10.1016/j.meegid.2009.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cunningham M. W. (2000) Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511 10.1128/CMR.13.3.470-511.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Magneson G. R., Puvathingal J. M., and Ray W. J. Jr. (1987) The concentrations of free Mg2+ and free Zn2+ in equine blood plasma. J. Biol. Chem. 262, 11140–11148 [PubMed] [Google Scholar]
- 16. Lawrence M. C., Pilling P. A., Epa V. C., Berry A. M., Ogunniyi A. D., and Paton J. C. (1998) The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure 6, 1553–1561 10.1016/S0969-2126(98)00153-1 [DOI] [PubMed] [Google Scholar]
- 17. McDevitt C. A., Ogunniyi A. D., Valkov E., Lawrence M. C., Kobe B., McEwan A. G., and Paton J. C. (2011) A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog. 7, e1002357 10.1371/journal.ppat.1002357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loisel E., Jacquamet L., Serre L., Bauvois C., Ferrer J. L., Vernet T., Di Guilmi A. M., and Durmort C. (2008) AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: biochemical and structural analysis. J. Mol. Biol. 381, 594–606 10.1016/j.jmb.2008.05.068 [DOI] [PubMed] [Google Scholar]
- 19. Valleriani A., Zhang G., Nagar A., Ignatova Z., and Lipowsky R. (2011) Length-dependent translation of messenger RNA by ribosomes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 83, 042903 10.1103/PhysRevE.83.042903 [DOI] [PubMed] [Google Scholar]
- 20. Unissa A. N., Sudha S., Selvakumar N., and Hassan S. (2011) Binding of activated isoniazid with acetyl-CoA carboxylase from Mycobacterium tuberculosis. Bioinformation 7, 107–111 10.6026/97320630007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Armon A., Graur D., and Ben-Tal N. (2001) ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 307, 447–463 10.1006/jmbi.2000.4474 [DOI] [PubMed] [Google Scholar]
- 22. Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., and Ben-Tal N. (2003) ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164 10.1093/bioinformatics/19.1.163 [DOI] [PubMed] [Google Scholar]
- 23. Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., and Ben-Tal N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Skjaerven L., Grant B., Muga A., Teigen K., McCammon J. A., Reuter N., and Martinez A. (2011) Conformational sampling and nucleotide-dependent transitions of the GroEL subunit probed by unbiased molecular dynamics simulations. PLoS Comput. Biol. 7, e1002004 10.1371/journal.pcbi.1002004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miettinen M. S., Knecht V., Monticelli L., and Ignatova Z. (2012) Assessing polyglutamine conformation in the nucleating event by molecular dynamics simulations. J. Phys. Chem. B 116, 10259–10265 10.1021/jp305065c [DOI] [PubMed] [Google Scholar]
- 26. Keogh E., and Lin J. (2005) Clustering of time-series subsequences is meaningless: implications for previous and future research. Knowl. Inf. Syst. 8, 154–177 10.1007/s10115-004-0172-7 [DOI] [Google Scholar]
- 27. Junejo I. N., and Aghbari Z. A. (2012) Using SAX representation for human action recognition. J. Vis. Commun. Image Represent. 23, 853–861 10.1016/j.jvcir.2012.05.001 [DOI] [Google Scholar]
- 28. Georgoulas G., Karvelis P., Loutas T., and Styliosa D. C. (2015) Rolling element bearings diagnostics using the Symbolic Aggregate approXimation. Mech. Syst. Signal Process. 60–61, 220–242 10.1016/j.ymssp.2015.01.033 [DOI] [Google Scholar]
- 29. He Y., Maisuradze G. G., Yin Y., Kachlishvili K., Rackovsky S., and Scheraga H. A. (2017) Sequence-, structure-, and dynamics-based comparisons of structurally homologous CheY-like proteins. Proc. Natl. Acad. Sci. U.S.A. 114, 1578–1583 10.1073/pnas.1621344114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rose A. S., Elgeti M., Zachariae U., Grubmüller H., Hofmann K. P., Scheerer P., and Hildebrand P. W. (2014) Position of transmembrane helix 6 determines receptor G protein coupling specificity. J. Am. Chem. Soc. 136, 11244–11247 10.1021/ja5055109 [DOI] [PubMed] [Google Scholar]
- 31. Czub J., Wieczór M., Prokopowicz B., and Grubmüller H. (2017) Mechanochemical energy transduction during the main rotary step in the synthesis cycle of F1-ATPase. J. Am. Chem. Soc. 139, 4025–4034 10.1021/jacs.6b11708 [DOI] [PubMed] [Google Scholar]
- 32. Timmins A., Saint-André M., and de Visser S. P. (2017) Understanding how prolyl-4-hydroxylase structure steers a ferryl oxidant toward scission of a strong C-H bond. J. Am. Chem. Soc. 139, 9855–9866 10.1021/jacs.7b02839 [DOI] [PubMed] [Google Scholar]
- 33. Brocchieri L., and Karlin S. (2005) Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 33, 3390–3400 10.1093/nar/gki615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang G., Hubalewska M., and Ignatova Z. (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 16, 274–280 10.1038/nsmb.1554 [DOI] [PubMed] [Google Scholar]
- 35. Zhang G., and Ignatova Z. (2009) Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLoS One 4, e5036 10.1371/journal.pone.0005036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo J., Lian X., Zhong J., Wang T., and Zhang G. (2015) Length-dependent translation initiation benefits the functional proteome of human cells. Mol. Biosyst. 11, 370–378 10.1039/C4MB00462K [DOI] [PubMed] [Google Scholar]
- 37. Jacob E., Unger R., and Horovitz A. (2013) N-terminal domains in two-domain proteins are biased to be shorter and predicted to fold faster than their C-terminal counterparts. Cell Rep. 3, 1051–1056 10.1016/j.celrep.2013.03.032 [DOI] [PubMed] [Google Scholar]
- 38. Monecke T., Haselbach D., Voß B., Russek A., Neumann P., Thomson E., Hurt E., Zachariae U., Stark H., Grubmüller H., Dickmanns A., and Ficner R. (2013) Structural basis for cooperativity of CRM1 export complex formation. Proc. Natl. Acad. Sci. U.S.A. 110, 960–965 10.1073/pnas.1215214110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Graen T., Inhester L., Clemens M., Grubmüller H., and Groenhof G. (2016) The low barrier hydrogen bond in the photoactive yellow protein: a vacuum artifact absent in the crystal and solution. J. Am. Chem. Soc. 138, 16620–16631 10.1021/jacs.6b05609 [DOI] [PubMed] [Google Scholar]
- 40. Sun X., Ågren H., and Tu Y. (2014) Microsecond molecular dynamics simulations Provide Insight into the Allosteric Mechanism of the Gs Protein Uncoupling from the beta2 adrenergic receptor. J. Phys. Chem. B 118, 14737–14744 10.1021/jp506579a [DOI] [PubMed] [Google Scholar]
- 41. Chen W., Lou J., Hsin J., Schulten K., Harvey S. C., and Zhu C. (2011) Molecular dynamics simulations of forced unbending of integrin αvβ3. PLoS Comput. Biol. 7, e1001086 10.1371/journal.pcbi.1001086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pereira A. R., Hsin J., Król E., Tavares A. C., Flores P., Hoiczyk E., Ng N., Dajkovic A., Brun Y. V., VanNieuwenhze M. S., Roemer T., Carballido-Lopez R., Scheffers D. J., Huang K. C., and Pinho M. G. (2016) FtsZ-dependent elongation of a coccoid bacterium. MBio 7, e00908–e00916 10.1128/mBio.00908-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heymann G., Dai J., Li M., Silberberg S. D., Zhou H. X., and Swartz K. J. (2013) Inter- and intrasubunit interactions between transmembrane helices in the open state of P2X receptor channels. Proc. Natl. Acad. Sci. U.S.A. 110, E4045–E4054 10.1073/pnas.1311071110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kumar A., and Purohit R. (2014) Use of long term molecular dynamics simulation in predicting cancer associated SNPs. PLoS Comput. Biol. 10, e1003318 10.1371/journal.pcbi.1003318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van Gunsteren F., Eising B. S. A., Hünenberger P., Krüger P., Mark A., Scott W., and Tironi I. (1996) Biomolecular Simulation: the GROMOS96 Manual and User Guide, pp. 397–434, Verlag der Fachvereine Hochschulverlag AG an der ETH Zurich, Zurich, Switzerland [Google Scholar]
- 46. van Gunsteren W. F., Daura X., and Mark A. E. (2002) GROMOS force field, in Encyclopedia of Computational Chemistry, pp. 1211–1216, John Wiley and Sons, New York [Google Scholar]
- 47. Manzetti S., McCulloch D. R., Herington A. C., and van der Spoel D. (2003) Modeling of enzyme-substrate complexes for the metalloproteases MMP-3, ADAM-9 and ADAM-10. J. Comput. Aided Mol. Des. 17, 551–565 10.1023/B:JCAM.0000005765.13637.38 [DOI] [PubMed] [Google Scholar]
- 48. Park P. S., Sapra K. T., Koliński M., Filipek S., Palczewski K., and Muller D. J. (2007) Stabilizing effect of Zn2+ in native bovine rhodopsin. J. Biol. Chem. 282, 11377–11385 10.1074/jbc.M610341200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li J., Rossetti G., Dreyer J., Raugei S., Ippoliti E., Lüscher B., and Carloni P. (2014) Molecular simulation-based structural prediction of protein complexes in mass spectrometry: the human insulin dimer. PLoS Comput. Biol. 10, e1003838 10.1371/journal.pcbi.1003838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morra G., Potestio R., Micheletti C., and Colombo G. (2012) Corresponding functional dynamics across the Hsp90 Chaperone family: insights from a multiscale analysis of MD simulations. PLoS Comput. Biol. 8, e1002433 10.1371/journal.pcbi.1002433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Best R. B., Hummer G., and Eaton W. A. (2013) Native contacts determine protein folding mechanisms in atomistic simulations. Proc. Natl. Acad. Sci. U.S.A. 110, 17874–17879 10.1073/pnas.1311599110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li B. J., Wang H., Gong T., Chen J. J., Chen T. J., Yang J. L., and Zhu P. (2017) Improving 10-deacetylbaccatin III-10-β-O-acetyltransferase catalytic fitness for Taxol production. Nat. Commun. 8, 15544 10.1038/ncomms15544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yan C., Liu D., Li L., Wempe M. F., Guin S., Khanna M., Meier J., Hoffman B., Owens C., Wysoczynski C. L., Nitz M. D., Knabe W. E., Ahmed M., Brautigan D. L., Paschal B. M., et al. (2014) Discovery and characterization of small molecules that target the GTPase Ral. Nature 515, 443–447 10.1038/nature13713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Anil B., Song B., Tang Y., and Raleigh D. P. (2004) Exploiting the right side of the Ramachandran plot: substitution of glycines by D-alanine can significantly increase protein stability. J. Am. Chem. Soc. 126, 13194–13195 10.1021/ja047119i [DOI] [PubMed] [Google Scholar]
- 55. Zhang L., Li N., Cao K., Yang X. Y., Zeng G., Sun X., and He Q. Y. (2017) Crucial residue Trp158 of lipoprotein PiaA stabilizes the ferrichrome-PiaA complex in Streptococcus pneumoniae. J. Inorg. Biochem. 167, 150–156 10.1016/j.jinorgbio.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 56. Li H., Li N., Xu Q., Xiao C., Wang H., Guo Z., Zhang J., Sun X., and He Q. Y. (2013) Lipoprotein FtsB in Streptococcus pyogenes binds ferrichrome in two steps with residues Tyr137 and Trp204 as critical ligands. PLoS One 8, e65682 10.1371/journal.pone.0065682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lacks S., and Hotchkiss R. D. (1960) A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39, 508–518 10.1016/0006-3002(60)90205-5 [DOI] [PubMed] [Google Scholar]
- 58. Ding B., and Zhong Q. (2017) Zinc deficiency: an unexpected trigger for autophagy. J. Biol. Chem. 292, 8531–8532 10.1074/jbc.H116.762948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mayer L. S., Uciechowski P., Meyer S., Schwerdtle T., Rink L., and Haase H. (2014) Differential impact of zinc deficiency on phagocytosis, oxidative burst, and production of pro-inflammatory cytokines by human monocytes. Metallomics 6, 1288–1295 10.1039/c4mt00051j [DOI] [PubMed] [Google Scholar]
- 60. Bayle L., Chimalapati S., Schoehn G., Brown J., Vernet T., and Durmort C. (2011) Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol. Microbiol. 82, 904–916 10.1111/j.1365-2958.2011.07862.x [DOI] [PubMed] [Google Scholar]
- 61. Wach A. (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12, 259–265 [DOI] [PubMed] [Google Scholar]
- 62. Biswas I., Jha J. K., and Fromm N. (2008) Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154, 2275–2282 10.1099/mic.0.2008/019265-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.