Abstract
Translocation of lipid II across cytoplasmic membrane is essential in peptidoglycan biogenesis. Although most steps are understood, identifying the lipid II flippase has yielded conflicting results, and the lipid II binding properties of two candidate flippases, MurJ and FtsW, remain largely unknown. Here we apply native mass spectrometry to both proteins and characterise lipid II binding. We observed lower levels of lipid II binding to FtsW compared to MurJ, consistent with MurJ having a higher affinity. Site directed mutagenesis of MurJ suggests that mutations at A29 and D269 attenuate, and chemical modification of A29 eliminate, lipid II binding to MurJ. The antibiotic ramoplanin dissociates lipid II from MurJ whereas vancomycin binds to form a stable complex with MurJ:lipid II. Furthermore, we reveal cardiolipins associate with MurJ but not FtsW, and exogenous cardiolipins reduce lipid II binding to MurJ. These observations provide insights into determinants of lipid II binding to MurJ and suggest roles for endogenous lipids in regulating substrate binding.
Introduction
The peptidoglycan (PG) sacculus is an essential and specific component of the bacterial cell wall found on the outside of the cytoplasmic membrane. The main function of the PG is to preserve the integrity of the cell by providing structural support and protection from osmotic lysis (1). Lipid II is the precursor building block of the PG and is composed of undecaprenyl-diphosphate linked to N-acetylglucosamine-N-acetylmuramic acid pentapeptide. Following synthesis on the inner face of the cytoplasmic membrane, lipid II is translocated to the outer face of the membrane (2). The disaccharide-peptide monomer is then incorporated into the existing PG by the glycotransferase (GTase) and transpeptidase (TPase) activities of penicillin-binding proteins (PBPs) together with additional factors (3) (Figure 1a).
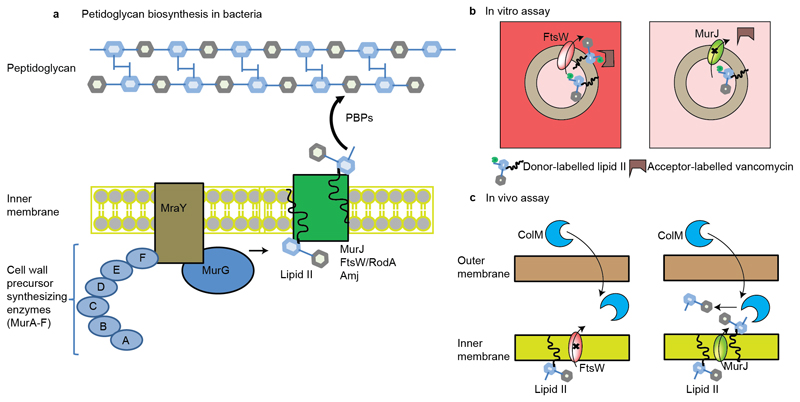
Figure 1. Schematic representation of peptidoglycan biosynthesis and assays used to measure the translocation of lipid II.
a, Synthesis of the peptidoglycan begins with the formation of soluble precursors, produced from a series of cytoplasmic reactions, catalyzed by MurA-MurF. MraY-MurG complete the synthesis of the precursor lipid II. Lipid II is then translocated to the periplasm by different families of flippases (such as FtsW or RodA, MurJ, and Amj), where it is incorporated into the pre-existing cell wall by penicillin binding proteins and additional factors (1–3). b, Assay used to measure the in vitro translocation of lipid II in liposomes (12). In this case, liposomes are loaded with donor-labeled lipid II (lipid II with green bulb) and when lipid II is flipped across the membrane (left panel with FtsW loaded), then an acceptor-labeled vancomycin derivative (U shaped box in brown) binds to it and a fluorescence signal is created. In this assay, fluorescence signal was created for lipisomes loaded with FtsW (left panel) but not when loaded with MurJ (right panel). c, Assay used to measure the in vivo translocation of lipid II in living E. coli (13). In this case, purified ColM toxin was added to actively growing E. coli cells. ColM cleaved lipid II product (disaccharide-pentapeptide) was then quantified in the periplasm. In this assay, flipping was observed only for MurJ (right panel) but not for FtsW (left panel).
The structure of lipid II is highly conserved, which makes it an ideal target for antibiotics. Consequently lipid II has been the focus of considerable attention over the past 50 years (4). Glycopeptides and unmodified peptides, including vancomycin and the defensins, target lipid II (5,6). Furthermore, several recently identified antibiotics also target lipid II, for example nisin (7); and the depsipeptides, including Teixobactin, which is a gram-positive antibiotic with no known resistance mechanism (8).
While the steps involved in the synthesis and incorporation of lipid II into the existing PG that take place in the cytoplasm are well characterized, the mechanism that translocates lipid II across the membrane has long remained a mystery, reviewed in (9–11). Moreover, the identification of potential lipid II flippases has been the subject of controversy. Recent results from elegant studies in Escherichia coli identified two different candidates as lipid II flippases: MurJ and FtsW (and its orthologs RodA, SpoVE) (12–16). The integral membrane protein FtsW belongs to the SEDS protein family (Shape, Elongation, Division and Sporulation) and plays an essential role in cell division. The integral membrane protein MurJ, on the other hand, belongs to the MOP exporter superfamily (Multidrug/Oligosaccharidyl-lipid/Polysaccharide) and is essential for viability and peptidoglycan synthesis in E. coli.
Experimental evidence for FtsW functioning as a lipid II flippase comes from a biochemical assay (Figure 1b), wherein the reconstituted FtsW is capable of flipping a fluorescent analogue of lipid II and phospholipids across the membrane of proteoliposomes (12). The authors propose that these lipids are transported through different mechanisms of facilitated diffusion through a pore-like structure of FtsW. Further mutagenesis studies of FtsW showed that residues R145 and K153 of transmembrane helix 4 are essential for flippase activity (14). Unlike FtsW, MurJ showed no activity in this in vitro assay leading the authors to conclude that FtsW is therefore the essential lipid II flippase (12).
Evidence for MurJ as the lipid II flippase comes from observations that depletion of MurJ causes a decrease in PG synthesis and an accumulation of PG nucleotide and lipid intermediates (15). These observations are supported by a combination of whole cell assays and genetic analysis (13, 16) (Figure 1c). In this assay the authors used the Colicin M toxin (ColM), a lipid II degrading enzyme acting exclusively at the outer face of the cytoplasmic membrane and monitored living E. coli. They identified five functional cysteine-containing MurJ variants which can be inhibited by a sulfhydral-reactive reagent, 2-sulfonatoethyl methanethionsulfonate (MTSES) and showed that upon MurJ inhibition, the newly synthesized lipid II was no longer accessible to ColM. In addition, a recent model of a lipid II bound state generated using the inward facing conformation of the structure of MurJ from Thermosipho africanus (MurJTA) is consistent with its transport function (17). A role of MurJ in lipid II flipping is further supported by complementation assays where MurJTA can complement E. coli in which the native MurJ expression was downregulated point to a role of MurJ in lipid II flipping.
In contrast to the above observations, MurJ is not essential in the Gram-positive bacterium Bacillus subtilis, and studies in this bacterium have identified a new potential lipid II flippase Amj (18). Interestingly, from sequence analyses Amj bears no similarity to either MurJ or FtsW. Furthermore, Amj from B. subtilis was able to support the growth of MurJ-inactivated E. coli cells, suggesting that Amj, having evolved independently, is functionally redundant with MurJ. In addition, recent genetic data and assays with a purified B. subtilis RodA suggested that SEDS proteins, like FtsW, function as GTases and not as lipid II flippases (19, 20). However, further studies on E. coli FtsW did not show GTase activity. Rather, FtsW formed a trimeric complex with the accessory proteins PBP3 and PBP1B, which function to incorporate lipid II into the PG, and control lipid II polymerization of PBP1B (21).
These conflicting results highlight the need for further experiments to clarify the roles and lipid II interaction properties of MurJ and FtsW. Here we apply native mass spectrometry (MS), a technique that has been optimized to study intact membrane proteins and their lipid- and drug-binding properties directly (22–27). Following purification of both MurJ and FtsW, we compared their affinity for lipid II and deduced their relative binding constants. We then analysed the molecular details of binding using site directed mutagenesis, a lipid II precursor and the addition of antibiotics that target lipid II. Combining this information with lipidomics we link the effect of co-purified lipids on lipid II binding and provide new insights into lipid II recognition by MurJ.
Results
MurJ and FtsW are monomeric in solution
Initially we performed expression trials of six different constructs of MurJ and FtsW to maximise the amount of protein produced (Supplementary Table 1). Our initial screening produced good protein yields for constructs of MurJ with a C-terminal 6×His tag and FtsW with a N-terminal 6×His tag. We then used these constructs to express MurJ and FtsW and purified both proteins using similar protocols (see Methods). We extracted both proteins from membranes using n-dodecyl-β-D-maltopyranoside (DDM) and then optimized a delipidation protocol (26) such that it was possible to apply relatively mild mass spectrometry conditions while still obtaining well-resolved spectra (28, 29). Employing a detergent screen (22) to the delipidated proteins, selecting detergents from different classes, yielded resolvable spectra of MurJ and FtsW in DDM, OGNG (octyl glucose neopentyl glycol), and LDAO (n-dodecyl-N,N-dimethylamine-N-oxide). In the case of DDM and OGNG high-energy mass spectrometry conditions were required to release the proteins from the detergent micelles which carried them into the gas phase (28) (Supplementary Figure 1), which are unsuitable to retain non-covalent interactions. For MurJ and FtsW in LDAO, well-resolved charge states were observed with the anticipated masses corresponding to the delipidated proteins (Figure 2a, 2c). The lower charge state series observed using LDAO is consistent with the charge-reducing effects of this detergent (28), and coupled with the lower energy required to record resolved spectra makes using LDAO ideal for carrying out our lipid II binding studies. Moreover, the spectra of MurJ and FtsW indicate that both proteins are monomeric in solution for all the detergents tested in this study, consistent with the recent MurJTA crystal structure (17).
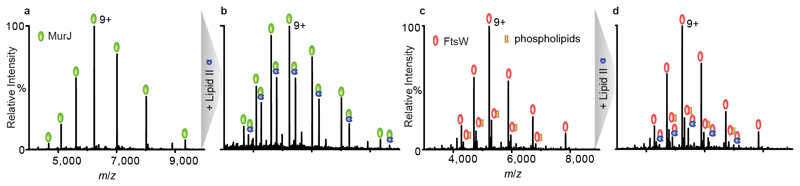
Figure 2. Mass spectra of MurJ and FtsW before and after addtion of lipid II.
a, c, Mass spectra of MurJ and FtsW proteins (5 μM) in 0.05% (w/v) LDAO reveal a charge state series consistent with a single monomeric protein MurJ (green) and with some phospholipid binding (orange) to FtsW (red). Theoretical and observed masses for MurJ and FtsW are 56090 Da and 56089 Da, and 47172 Da and 47039 Da respectively. b,d Addition of lipid II (blue) at a final concentration of 5 μM to MurJ and 10 μM to FtsW, respectively, leads to formation of a complex with both proteins. A much greater degree of complex formation is observed for MurJ than FtsW at lower lipid II concentration. We find that lipid II binding does not vary significantly in LDAO concentrations ranging from 0.05 (2×CMC)–0.1% (4×CMC).
Lipid II binding to MurJ and FtsW
Having optimised delipidation and mass spectrometry protocols we then performed lipid II binding experiments to probe the relative affinity of both MurJ and FtsW to lipid II. To a solution of 5 µM MurJ in 200 mM ammonium acetate (pH 8.0) and 0.05% (w/v) LDAO, we added lipid II (to a final concentration of 5 µM) and recorded mass spectra under our optimised conditions (see Methods and Supplementary Table 2). The mass spectrum shows a charge state series for a second higher mass species at ~60% intensity of the peak corresponding to MurJ. This mass difference (1875 Da) is consistent with the binding of one molecule of lipid II (Figure 2b). The same concentration of lipid II was then added to FtsW, however no binding of lipid II was evident at this lipid II concentration. We then doubled the concentration of lipid II added to FtsW and recorded a mass spectrum. Additional peaks with a low relative intensity (~10%) compared to the unbound form were observed, with a mass difference also corresponding to lipid II binding to FtsW (Figure 2d). Comparing the mass spectra of the two proteins, and the lipid II concentrations necessary to observe binding, we conclude that the binding affinity of lipid II is higher for MurJ.
To strengthen our lipid II binding experiments, we also performed lipid II binding experiments on an architecturally similar protein Pyrococcus furiosus MATE (PfMATE). We purified PfMATE in a similar manner to the other proteins but observed no appreciable lipid II binding at the concentrations used for MurJ and FtsW (Supplementary Figure 3C). This observation suggests that lipid II binding to MurJ, and to a lesser extent FtsW, is representative of MurJ possessing lipid II binding capabilities, beyond that of other proteins, implying an innate lipid II-associated role.
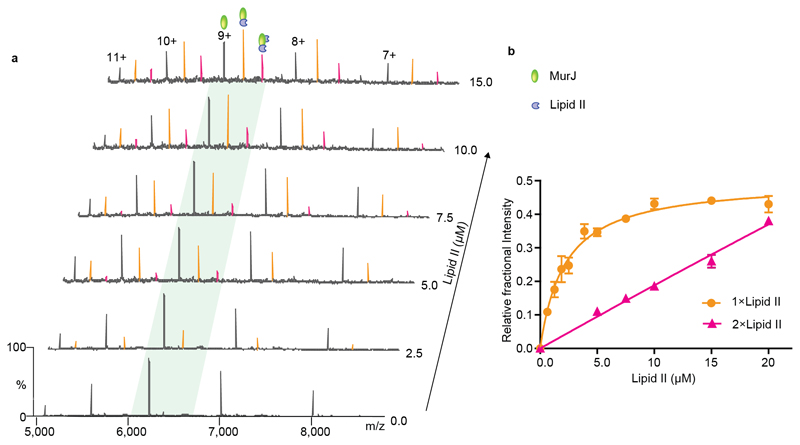
To investigate in further detail the affinity of lipid II for MurJ we incubated the protein with increasing concentrations of lipid II (Figure 3a and Supplementary Table 2). As well as an increase in the intensity of the one lipid II-bound species, at concentrations above 5 µM we observed evolution of an additional charge state series, corresponding to binding of a second molecule of lipid II to MurJ. At 10 µM of lipid II the lipid-bound species predominate over the unbound form of the protein. From this data set we extracted and plotted the relative intensity of lipid II-bound forms as a function of lipid concentration (Figure 3b and Supplementary Methods). Using these data, we calculated dissociation constant (Kd) of 2.9 ± 0.6 µM for the first lipid II binding. We tested further the specificity of lipid II binding by subjecting the protein lipid complex to further purification steps (Supplementary Figure 2) and a wide range of delipidating detergents (Supplementary Figure 3a and 3b). The fact that the complex survives these additional stages confirms that the complex remains associated in solution, even when the pool of available lipid II is depleted,
Figure 3. Determination of dissociation constants for lipid II binding to MurJ.
a Mass spectra recorded for solutions of MurJ with increasing concentrations of lipid II. At 2.5 μM lipid II a charge state series is observed (orange), corresponding to bound lipid II, which increases in intensity as the lipid II concentration is increased to 15 μM. A second lipid II binding peak (pink) emerges at concentrations above 5.0 μM. b A plot of the relative fractional intensity of the lipid binding peaks over the total peak intensity against the lipid II concentration (see Supplementary Methods) yields a curve for the first binding event and linear like fit for the second consistent with non-specific lipid II binding. Each data point and standard deviations are calculated from the average of the five observed charge states in three independent experiments. Error bars represent standard deviations (n=3).
Carrying out the same experiments, but in this case with FtsW, resulted in significantly lower relative intensities of peaks assigned to lipid II binding. With this limited extent of binding, it was not possible to obtain a Kd value for lipid II binding to FtsW since the high concentrations of lipid II required to observe binding are impractical for mass spectrometry. Given the low cellular levels of lipid II (2000 molecules per cell) (30), the relatively similar levels of the two proteins (estimated as MurJ:FtsW:lipid II 1.2:1:45) (see Supplementary Methods) and our anticipated high Kd value for FtsW, under normal conditions FtsW would not compete with MurJ for lipid II in the membrane.
Within the membrane, and among the MOP transporter superfamily, MATE family proteins are thought to couple transport with influx of either Na+ or H+ (31–35). Although not direct evidence of the driving force for lipid II transport by MurJ, we reasoned that if binding were sensitive to pH or sodium ions this could be indicative of a possible energy source. We addressed the sensitivity of lipid II binding to MurJ under a range of different solution conditions, including pH and sodium ion concentration. We then recorded native mass spectra over the pH range 5 to 9 (Supplementary Figure 4a and Supplementary Table 2) and observed low intensity lipid II binding at both low and high pH (5 and 9 respectively). We observed maximal binding of lipid II to MurJ between pH 6 and 8.
We next explored the influence of the sodium ion concentration on lipid II binding. Using Na+ concentrations from 0–750 µM we recorded lipid II binding and plotted the amount of lipid II binding at each concentration (Supplementary Figure 4b and Supplementary Table 2). Although the spectral quality is decreased with Na+ concentrations > 500 µM, the relative intensity of the lipid II bound state did not change significantly across the entire range of Na+ concentrations. These results imply that lipid II binding to MurJ is not sensitive to Na+ concentration but is responsive to changes in H+ concentration with optimal binding at pH 7.0 in line with the pH encountered in the E. coli cytoplasm (pH 7.2 to 7.8) where the majority of lipid II binding events would take place (36).
Antibiotic binding to the MurJ:lipid II complex
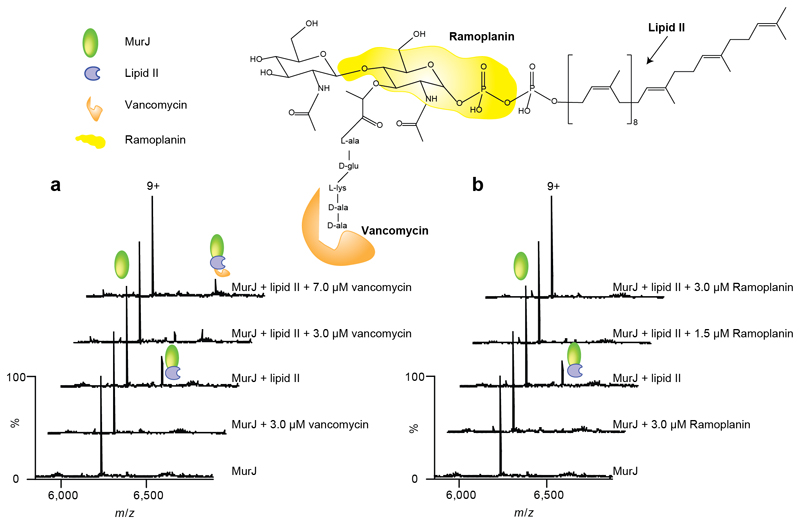
To gain further insight into how lipid II binds to MurJ, we selected two antibiotics that specifically target lipid II. We selected vancomycin, which interacts with the N-terminal L-lys-D-ala-D-ala segment of the pentapeptide, and ramoplanin, which interacts with the MurNAc and pyrophosphate moieties of lipid II (37–39) (Figure 4 and Supplementary Figure 5). Vancomycin and ramoplanin bind to different regions of lipid II, so performing binding assays in their presence will reveal details of the accessibility of the lipid II molecule when bound to MurJ, in particular the binding motifs that are either buried or exposed within the complex formed with MurJ.
Figure 4. Structure of lipid II showing binding sites for antibiotics and mass spectra recorded after addition of antibiotics to MurJ and to the Mur:lipid II complex.
Regions of the lipid II structure are coloured according to the vancomycin and ramoplanin binding sites (36–38) (orange and yellow respectively). a Mass spectra of MurJ with vancomycin shows the 9+ charge state of MurJ and the absence of any appreciable adduct peaks corresponding to bound vancomycin. The mass spectra of MurJ and lipid II (at final concentrations of 5 μM and 3 μM respectively) confirm complex formation and addition of vancomycin (3 μM and 7 μM) leads to formation of the ternary complex. b Analogous spectra were recorded for ramoplanin. Addition of ramoplanin at low concentrations (1.5 μM and 3.0 μM) lead to disappearance of the MurJ:lipid II complex. All experiments were performed three times and spectra shown are representative.
We first added vancomycin to MurJ in the absence of lipid II to detect any possible interactions between the protein and the antibiotic; no binding was detected in the mass spectrum (Figure 4a). The interaction between lipid II and vancomycin without MurJ was also confirmed by the formation of a 1:1 complex (Supplementary Figure 6).
We then attempted to form a ternary complex incubating MurJ and lipid II together (at final concentrations of 5 µM and 3 µM respectively) and then adding vancomycin (at a 3 µM final concentration) to this MurJ:lipid II complex. The resulting mass spectrum revealed an additional charge series of increased mass corresponding to the binding of vancomycin to the MurJ:lipid II complex. At this concentration of vancomycin, approximately equal amounts of the lipid II:MurJ and lipid II:MurJ:vancomycin complexes are formed. Increasing the vancomycin concentration to 7 µM increases the amount of ternary complex formed with no MurJ:lipid II complex remaining. The formation of this ternary complex confirms that the N-terminus of the pentapeptide of bound lipid II is accessible to vancomycin and further implies that this pentapeptide is not involved in lipid II binding to MurJ.
We carried out analogous experiments with ramoplanin, first establishing no direct binding to MurJ (Figure 4b). We then formed the MurJ:lipid II complex (at the same final concentrations as before) and added ramoplanin (1.5 µM final concentration) to this complex. No ternary complex was observed and instead the majority of lipid II dissociated from MurJ. We increased the concentration of ramoplanin to 3 µM and found that no MurJ-lipid II complex remained. Even in the presence of a 5-fold excess of lipid II (15 µM) we were unable to observe the ternary complex and detected a decrease in the amount of the MurJ:lipid II complex (Supplementary Figure 7). There are three probable explanations for our observations (i) the MurNAc and pyrophosphate ramoplanin lipid II binding site is not accessible when MurJ is bound to lipid II or (ii) the ramoplanin:lipid II complex is too large to bind to the MurJ lipid II binding site or (iii) ramoplanin undergoes lipid II induced aggregation. Since we do not observe lipid II bound to MurJ in the presence of ramoplanin the second and third scenarios are most likely. We conclude from the different binding properties of these two antibiotics to the MurJ:lipid II complex that the terminus of the pentapeptide is accessible to vancomycin while MurJ and ramoplanin compete for MurNac pyrophosphate sites on lipid II.
Investigating further the specificity of lipid II binding to MurJ we recruited a second potential lipid substrate, undecaprenyl phosphate (C55-P), which is a precursor in the synthesis of lipid II (5). Under similar MS conditions, we did not observe binding of C55-P even at high concentrations (30 µM) (Supplementary Figure 3d). This result, together the antibiotic binding data above, confirm that while the pentapeptide and undecaprenyl chain are not critical for binding, the pyrophosphate, MurNac and GlcNac moieties of lipid II are important for recognition by MurJ.
Addition of MTSES to the A29C variant abrogates lipid II binding
The in vivo assay for MurJ flippase activity identified a set of 39 cysteine variants found to have different flippase activities (13). It was reported that the A29C mutant (MurJA29C) was functional but that following addition of the cysteine-reactive agent MTSES, flippase activity was inhibited and cell lysis ensued. A total of five positions were identified [A29, N49, S263, D269, and E273 (with only partial inhibition in the case of E273)] from this set of 39 cysteine variants that showed a similar MTSES-dependent behaviour. However, the molecular detail of this inhibition could not be deduced in that study (13). To try to unravel these and identify residues that might be important for lipid II binding we investigated lipid II binding to MurJ variants with and without treatment by MTSES.
We began by substituting A29 to cysteine and purified this variant protein to homogeneity. We then acquired mass spectra both in the presence and absence of lipid II and found that A29C binds to lipid II to a similar extent as wild-type MurJ (Figure 5b). We then added MTSES. In addition to the cysteine residue that we introduced at position 29, there are two native cysteine residues present in the protein (C314 and C419). If all cysteine residues are accessible to the reagent we anticipate a maximum of two MTSES reaction sites in the wild-type protein and three in the A29C variant. The spectra recorded following reaction with the reagent show a maximum of two and three MTSES modifications for wild type and the A29C variant respectively, as anticipated (Figure 5c and 5d). Considering the effect of MTSES addition to these cysteine residues on lipid II binding we found that for the wild-type protein there was little difference in the distribution of lipid II bound states whether or not cysteine residues were modified. In contrast, for the MurJA29C variant, lipid II was unable to bind following labelling by MTSES at the A29C site. We conclude that MTSES-labelled MurJA29C blocks the binding site of lipid II on MurJ.
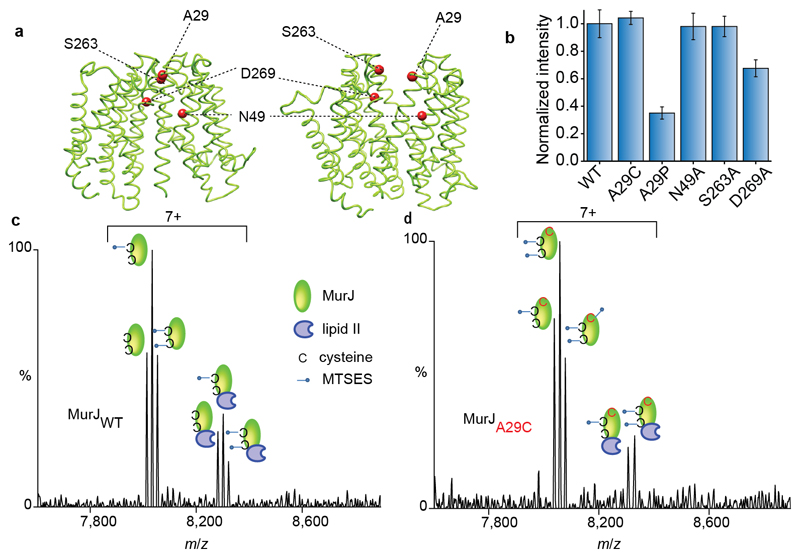
Figure 5. Structures of MurJ with residues investigated through mutation, their effects on lipid II binding and spectra recorded for wild type and A29C after treating with MTSES in the presence of lipid II.
a Structures of MurJ in inward and outward facing forms were generated using I-TASSER and the structure PDB 5T77 and outward parameters of MurJTA (17). Mutated residues are shown as red spheres. b The effect of mutations on lipid II binding normalised to the wild type. N49A and S263A have no effect whereas A29P and D269A both have attenuated lipid II binding. Error bars represent standard deviations (n=3). c Mass spectrum recorded for MTSES treated wild-type MurJ. Both native cysteine residues are modified (second and third peak in the left set) and some unmodified protein remains leading to a triplet. All forms of the protein retain competency to bind lipid II (three peaks in the right hand set). d Mass spectrum for A29C MurJ variant with all three cysteine residues modified by MTSES (first, second and third peaks in the left set). In this case only two of the three modified sites are capable of binding to lipid II (first and second peaks in the right set) implying that the A29C mutant is unable to bind lipid II. All experiments were performed three times and spectra are representative.
To test the importance of A29 for lipid II binding, we mutated A29 to proline to introduce a break in the helical structure of transmembrane 1 (TM1) (Figure 5a). We tested lipid II binding to the A29P variant and observed a significant reduction in binding but not complete disruption (Figure 5b). This result suggests that A29 is not involved directly in the binding site but that the protein structure near A29 is important. This is in agreement with the recent MurJTA crystal structure, wherein A29 is located at an interface where the protein undergoes a conformational change from an inward to outward-facing form and is in the vicinity of the modelled lipid II binding site (17).
Since we observed a clear reduction in lipid II binding for the A29P variant, we carried out analogous experiments by mutating in turn N49, S263, and D269 to alanine and recording their lipid II binding properties individually. We saw no significant loss of binding affinity for N49A or S263A and only a slight attenuation in lipid II binding for D269A (Figure 5b and Supplementary Figure 8). This suggests that N49 and S263 residues are either not directly involved in lipid II binding or are minor contributors. Interestingly, S263 is in close proximity to A29 in the inward facing conformation of the MurJTA crystal structure but both are exposed in models of the outward facing state (17) (Figure 5a). We conclude therefore that changes to A29 have the greatest effect, with mutations to D269 being more disruptive than to either S263 or N49. The close proximity of D269 and A29 implies that they are in the vicinity of a potential lipid II pyrophosphate binding site with both residues becoming accessible in the outward binding state.
Presence of cardiolipin affects lipid II binding to MurJ
Having established the lipid II binding of MurJ we were keen to establish the propensity of MurJ and FtsW to bind to other lipids, particularly endogenous lipids that co-purify. We applied lipidomics protocols to MurJ and FtsW following exposure to mildly delipidating conditions. We found that MurJ co-purifies with a high number of cardiolipins (Supplementary Figure 9b) and other lower molecular mass phospholipids including phosphoethanolamines [PE 16:0/17:1(9Z) and PE 18:1(11E)/17:1(9Z)] (data not shown). The acyl chain length distribution observed for cardiolipins bound to MurJ ranged from 62 to 73 (Supplementary Figure 9c) in line with the normal distribution of cardiolipins in the E. coli membrane (40). In addition, we also quantified the cellular levels of cardiolipins with and without induction of MurJ expression and found twice the concentration of cardiolipins when MurJ was overexpressed (Supplementary Figure 10a). By contrast, lipidomics analysis of FtsW revealed only peaks for phospholipids, no cardiolipins associated with FtsW could be detected (Supplementary Figure 9e).
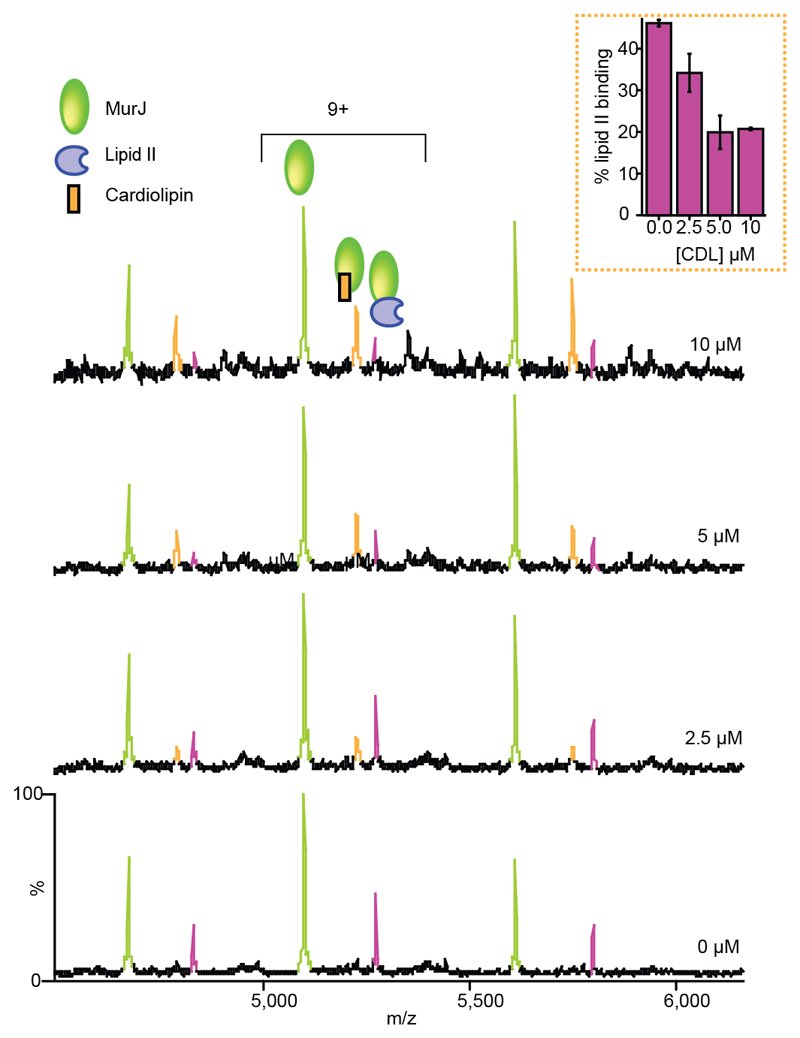
These different lipid binding properties are intriguing and to test the possibility that the presence of co-purified lipids influences lipid II binding properties of MurJ we added cardiolipins at different concentrations (see Supplementary Methods) to solutions containing the MurJ:lipid II complex (Figure 6). At 5 µM cardiolipin (CDL) lipid II binding is diminished and CDL forms the predominant lipid complex with MurJ. At 10 µM CDL only a very small population of lipid II remains bound, and the cardiolipin bound complex further predominates. In similar experiments, when we added the phospholipid PE 16:0/18:1(9z) at 40 µM (see Supplementary Methods) to the solution containing the MurJ:lipid II complex we observed no change in lipid II binding to MurJ (Supplementary Figure 12). This suggests that the reducing effects on lipid II binding are unique to CDL.
Figure 6. Mass spectra reveal the effect of increasing cardiolipin concentration on lipid II binding to MurJ.
In the absence of cardiolipin the MurJ:lipid II (5 μM:3 μM) complex is formed (relative intensity ~45% of the MurJ peak). As the concentration of cardiolipin is increased (from 2.5 μM to 10 μM) the lipid II binding peak decreases and the MurJ:CDL complex predominates. All experiments were performed three times and standard deviations are calculated from the average of the five observed charge states in three independent experiments. Error bars represent standard deviations (n=3).
Having observed the effects of CDL on lipid II binding, we then measured the Kd of CDL binding to MurJ. The Kd value of 13 ± 5 µM (Supplementary Figure 10b) is only moderately higher than the apparent dissociation constant determined here for lipid II (2.9 ± 0.6 µM) and enables us to rationalise the ability of CDL to compete with lipid II for binding, assuming the same binding site for CDL and lipid II. To probe further whether cardiolipin binds at the same binding site as lipid II, or to MurJ in the transmembrane region of the protein we looked for differences in the extent of binding that would be apparent with the removal of a potential binding site. Incubating CDL with MTSES treated A29C (which we hypothesise to sterically block the lipid II binding site) we find that the protein retains the ability to bind CDL, and, importantly, under the same experimental conditions the same amount of CDL binding is observed as binds to the MurJA9C protein without MTSES (Supplementary Figure 11). This suggests that the addition of increasing amounts CDL decreases lipid II binding to MurJ, via a form of allostery that may restrict the conformational landscape available to MurJ (Figure 6). These observations along with the presence of higher amounts of CDL (2×106) when MurJ was overexpressed compared to low levels of lipid II (30), imply that CDL may have a regulatory role in lipid II binding to (or release from) MurJ.
Discussion
The translocation of lipid II across the cytoplasmic membrane is an essential step in PG biogenesis. While the majority of steps in this process are understood, the controversy surrounding the identification of lipid II flippases remains and has hampered progress in understanding the mechanism of lipid II translocation. Specifically, controversy exists over the roles of MurJ and FtsW in E. coli (10–16). To compare the molecular details of lipid II interaction with these two proteins, we used native MS and observed lower levels of lipid II binding to FtsW compared to MurJ, consistent with MurJ having a higher affinity for lipid II than FtsW.
We took advantage of the native MS ability to detect lipid II binding to understand which regions of lipid II are involved in the binding to MurJ. We selected two antibiotics that interact with different regions of lipid II and demonstrated formation of a stable ternary complex in the case of vancomycin and disruption of lipid II binding with ramoplanin. The binding of vancomycin demonstrates that the terminus of the pentapeptide of lipid II remains accessible, even when lipid II is bound to MurJ (Figure 7). Ternary complex formation with vancomycin is only possible when the terminal lysine and alanines of lipid II are exposed, which would not be possible if lipid II is bound to an inward conformation based on the model generated in (17). However, if the protein is in an outward conformation vancomycin could be bound with these residues available for interaction with lipid II, thus enabling formation of the observed ternary complex. Our further binding experiments with C55-P and MurJ suggest no binding between these two. This result together with vancomycin data confirms that the pentapeptide and undecaprenyl chain are not critical for binding.
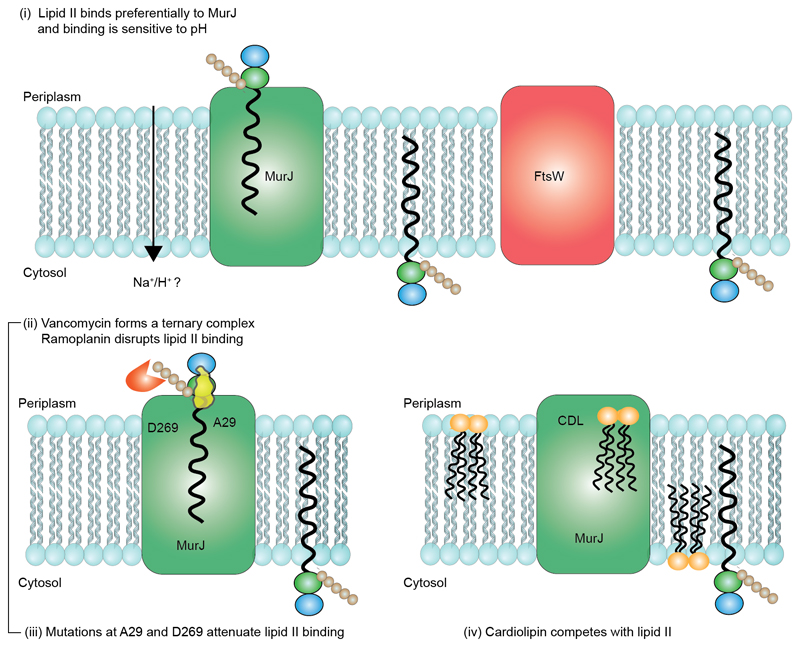
Figure 7. Schematic representation of the competition for lipid II binding between proteins, formation of a ternary complex with the antibiotic vancomycin and interplay between cardiolipin and lipid II binding to MurJ.
(i) Preferential binding to MurJ as opposed to FtsW was demonstrated through lipid II titration experiments. The potential energy source is likely a proton gradient. (ii) Vancomycin binds to the MurJ:lipid II complex whereas ramoplanin induces dissociation of lipid II. Mutations at A29 or D269 potentiate lipid II binding and chemical modification with MTSES abrogates the A29C lipid interaction. (iii) Cardiolipin (CDL) reduces the affinity of MurJ for lipid II but CDL binding, unlike lipid II, is not disrupted by mutation of A29. An allosteric binding mechanism in which the conformational access of MurJ is restricted by bidentate CDL binding is therefore consistent with all experimental data shown here.
Previous chemical genetics studies, wherein a single-cysteine variant of MurJ (A29C) was found to be functional and yet addition of the cysteine-reactive agent MTSES inhibited flippase activity (13), were carried out at the molecular level. Using native MS we found that the A29C variant exhibited no change in lipid II binding compared to the wild type but following labelling of A29C with MTSES, lipid II binding was obliterated. We also mutated this residue to proline (A29P) and observed reduced binding to lipid II. When we mutated residues N49 and S263 to alanine, this did not affect the lipid II binding significantly although some attenuation of binding was observed for the D269A variant. In the recent MurJTA crystal structure (17), A29 is positioned at an interface where the protein undergoes conformational changes, suggesting changes in this region of the protein perturb the lipid II binding capabilities.
Interestingly our lipidomics data and cardiolipin quantification data demonstrate the presence of co-purified cardiolipins and high levels of CDLs when MurJ was expressed. Further addition of exogenous cardiolipin interfered with lipid II binding to MurJ. It is conceivable that CDL binding may contribute to the observation that MurJ was inactive in the in vitro biochemical study (12), and this would correlate with the native gel analysis (21) wherein direct binding of lipid II to MurJ was not observed. Given the bidentate headgroup of cardiolipin and its typical binding motifs within membrane proteins across interfaces or channels it may act as a regulator changing the conformation of MurJ either bridging the open state or preventing access to the lipid II binding site via an alternative mechanism (Figure 7). Whatever the mechanism of regulation the fact that cardiolipin concentrations in the membrane are known to change in response to various external triggers, and to regulate other transporter activities, leads us to speculate that competition between cardiolipin and lipid II may have a regulatory role in flippase activity (41). Further experiments to quantify CDLs present in the inner and outer leaflets (42–44) of the cytoplasmic membrane, may shed light on how the asymmetric distribution of CDL across membrane modulates the lipid II binding to MurJ.
From our data it is not possible to rule out the possibility that both FtsW and MurJ flip lipid II but under different conditions or at discrete stages in PG biogenesis. It may be necessary for FtsW to form a ternary complex with PBP1b and PBP3 first to observe lipid II binding in vitro, as reported recently (21). Our data yield a micromolar affinity for lipid II binding to MurJ alone (Kd ≈3 μM) and a much weaker affinity for FtsW without its accessory proteins. The difference in these values may be related to the different conditions experienced by the proteins, for example by interaction with different accessory proteins, the kinetic requirements at various stages of the biogenesis pathway, and the regulatory roles of lipids, which could act in concert to regulate the activity of different flippases. This orchestration may be required to fine-tune the delivery of this important precursor into the periplasm.
Taken together, our mass spectrometry data suggest a favourable Kd value for MurJ:lipid II binding and provides insights into the key parts of lipid II involved in binding. Our results also expose the competitive binding of endogenous cardiolipins and antibiotics on MurJ and lipid II interactions. The ability of mass spectrometry to uncover endogenous lipid binding, which is variable and not always apparent in different protein preparations, may help to resolve existing controversies in the lipid II flippase field. This mass spectrometry platform could also help reveal the therapeutic potential of other classes of compound that target lipid II, an important consideration given the growing problem of bacterial resistance to many current drugs, including vancomycin.
Methods
Full details of the employed methods are given in Supplementary Information.
Purification of E. coli MurJ and FtsW
Both MurJ and FtsW were purified in the similar manner according to the following procedure at 4 °C. Cells were pelleted by centrifugation at 5,000g and resuspended in a buffer containing 50mM Tris-HCl (pH 8.0), 300mM NaCl and EDTA-free protease inhibitor Cocktail (Roche). The cells were then disrupted by a microfluidizer (Microfluidics). After centrifugation (20,000g for 20 mins), the supernatant was ultra-centrifuged (200,000g), and the membrane fractions were collected. The proteins were solubilized from the membrane fraction with 20 mM Tris (pH 8.0), 150 mM NaCl, 20% glycerol, 2% DDM (Anatrace) for 2 h at 4 °C. The insoluble material was removed by ultracentrifugation. Supernatant was filtered before loading onto a 5 ml His Trap-HP column (GE Healthcare) equilibrated in 20 mM Tris (pH 8.0), 150 mM NaCl, 20 mM imidazole, 10% glycerol, 0.03% DDM. After the clarified supernatant was loaded, the column was then initially washed with 50 ml of 20 mM Tris (pH 8.0), 150 mM NaCl, 20 mM imidazole, 10% glycerol, 0.03% DDM and washed again with 50 ml of 20 mM Tris (pH 8.0), 150 mM NaCl, 80 mM imidazole, 10% glycerol, 0.03% DDM. The bound protein was then eluted with 20 mM Tris (pH 8.0), 150 mM NaCl, 500 mM imidazole, 10% glycerol, 0.03% DDM. Peak fractions were pooled, dialysed and concentrated in a buffer containing 20 mM Tris (pH 8.0) and 150 mM NaCl, 10% glycerol and 0.03 % DDM. Concentrated protein was either used immediately or flash-frozen in liquid nitrogen and stored at -80 °C. Protein concentration was measured using a Biomate UV detector at 280 nm with the calculated coefficient of 1.23 M-1cm-1 and 1.528M-1cm-1 for MurJ and FtsW, respectively.
Native MS
Prior to mass spectrometry analysis, protein solutions were delipidated in detergent using OGNG following a previously established protocol (26). Delipidated samples were then buffer exchanged into 200 mM ammonium acetate pH 8.0, with 2× CMC (critical micelle concentration) of detergent of interest using a Biospin-6 (BioRad) column and introduced directly into the mass spectrometer using gold-coated capillary needles prepared in-house (45). Data were collected on a modified QExactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) optimized for analysis of high mass complexes, using methods previously described for membrane proteins (24). The instrument parameters were as follows: capillary voltage 1.2 kV, S-lens RF 100%, quadrupole selection from 2,000 to 20,000 m/z range, collisional activation in the HCD cell 100-200 V, argon UHV pressure 1.12× 10-9 mbar, temperature 60 °C, resolution of the instrument at 17,500 at m/z = 200 (a transient time of 64 ms) and ion transfer optics (injection flatapole, inter-flatapole lens, bent flatapole, transfer multipole: 8, 7, 6, 4 V respectively). The noise level was set at 3 rather than the default value of 4.64. No in source dissociation was applied. Where required baseline subtraction was performed to get a better quality mass spectrum. All the concentrations used in our MS studies are listed in Table S2.
Lipid analysis
Lipidomic analysis was performed on both MurJ and FtsW samples purified in DDM in order to gain insights on the lipids that co-purify with these proteins. Co-purified lipids have been identified in a similar manner that has been described previously (24, 46). Briefly, protein was digested with trypsin overnight at 37 °C, lyophilized and re-dissolved in 68% solution A (Acetonitrile:H2O 60:40, 10 mM ammonium formate and 0.1% formic acid) and 32% solution B (Isopropanol:Acetonitrile 90:10, 10 mM ammonium formate and 0.1% formic acid) for analysis by reverse phase liquid chromatography tandem MS (RP LC-MS/MS). LC-MS/MS was performed using a Dionex UltiMate 3000 RSLC Nano system coupled to an LTQ Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific). The peptide/lipid mixture was loaded onto a C18 column (Acclaim PepMap 100, C18, 75 µm × 15 cm; Thermo Scientific) at a flow rate of 300 nl min-1. After 10 min solvent B was ramped to 65% over 1 min, then 80% over 6 min, before being held at 80% for 10 min, then ramped to 99% over 6 min and held for 7 min. Typical MS conditions were spray voltage of 1.6 kV and capillary temperature of 275 °C. The LTQ-Orbitrap XL was set up in negative ion mode and in data-dependent acquisition mode to perform five MS/MS scans per MS scan. Survey full-scan MS spectra were acquired on the Orbitrap (m/z 400–2,000) with a resolution of 60,000. Collision-induced dissociation (CID) fragmentation in the linear ion trap was performed for the five most intense ions at an automatic gain control target of 30,000 and a normalized collision energy of 38% at an activation of q = 0.25 and an activation time of 30 ms. Lipid analysis was replicated 3 times.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request
Supplementary Material
Acknowledgements
We acknowledge with thanks funding from a MRC programme grant (MR/N020413/1), an ERC Advanced Grant ENABLE (641317), and a Wellcome Trust Investigator Award (104633/Z/14/Z). We thank Waldemar Vollmer for providing C55-P and Hsin-Yung Yen, Joseph Gault and Mark Agasid for useful discussions.
Footnotes
Author Contributions
J.R.B. and C.V.R. conceived and designed the experiments. J.R.B designed primers and generated the constructs for protein expression. J.R.B and J.S. expressed and purified the proteins. J.R.B. optimized MS conditions and obtained all the MS measurements. J.R.B. performed lipidomics analysis with the help of D.W. S.M. and T.M.A assisted with the data analysis. J.R.B. and C.V.R wrote the paper. All of the authors discussed the results and commented on the manuscript.
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Kruijff B, van Dam V, Breukink E. Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot Essent Fatty Acids. 2008;79:117–121. doi: 10.1016/j.plefa.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 3.van Heijenoort J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol Mol Biol Rev. 2007;71:620–635. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider T, Sahl H-GG. An oldie but a goodie - cell wall biosynthesis as antibiotic target pathway. Int J Med Microbiol. 2010;300:161–169. doi: 10.1016/j.ijmm.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Galley NF, O’Reilly AM, Roper DI. Prospects for novel inhibitors of peptidoglycan transglycosylases. Bioorg Chem. 2014;55:16–26. doi: 10.1016/j.bioorg.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Münch D, Sahl H-GG. Structural variations of the cell wall precursor lipid II in Gram-positive bacteria - Impact on binding and efficacy of antimicrobial peptides. Biochim Biophys Acta. 2015;1848:3062–3071. doi: 10.1016/j.bbamem.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Draper LA, Cotter PD, Hill C, Ross RP. Lantibiotic resistance. Microbiol Mol Biol Rev. 2015;79:171–191. doi: 10.1128/MMBR.00051-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheffers DJ, Tol MB. LipidII: Just Another Brick in the Wall? PLoS Pathog. 2015;11:e1005213. doi: 10.1371/journal.ppat.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz N. Lipid Flippases for Bacterial Peptidoglycan Biosynthesis. Lipid Insights. 2016;8:21–31. doi: 10.4137/LPI.S31783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz N. Filling holes in peptidoglycan biogenesis of Escherichia coli. Curr Opin Microbiol. 2016;34:1–6. doi: 10.1016/j.mib.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohammadi T, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sham L-TT, et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohammadi T, et al. Specificity of the transport of lipid II by FtsW in Escherichia coli. J Biol Chem. 2014;289:14707–14718. doi: 10.1074/jbc.M114.557371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue A, et al. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J Bacteriol. 2008;190:7298–7301. doi: 10.1128/JB.00551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuk AC, Mashalidis EH, Lee SY. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat struct Mol Biol. 2017;24:171–176. doi: 10.1038/nsmb.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meeske AJ, et al. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci USA. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meeske AJ, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emami K, et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol. 2017;2:16253. doi: 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclercq S, et al. Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep. 2017;7:43306. doi: 10.1038/srep43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison TM, et al. Quantifying the stabilizing effects of protein–ligand interactions in the gas phase. Nat Commun. 2015;6:8551. doi: 10.1038/ncomms9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gault J, et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods. 2016;13:333–336. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehmood S, et al. Mass spectrometry captures off-target drug binding and provides mechanistic insights into the human metalloprotease ZMPSTE24. Nat Chem. 2016;8:1152–1158. doi: 10.1038/nchem.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bechara C, et al. A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat Chem. 2015;7:255–262. doi: 10.1038/nchem.2172. [DOI] [PubMed] [Google Scholar]
- 27.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reading E, et al. The role of the detergent micelle in preserving the structure of membrane proteins in the gas phase. Angew Chem Int Ed. 2015;54:4577–4581. doi: 10.1002/anie.201411622. [DOI] [PubMed] [Google Scholar]
- 29.Reading E, et al. The effect of Detergent, Temperature, and Lipid on the Oligomeric State of MscL Constructs: Insights from Mass Spectrometry. Chem Biol. 2015;22:593–603. doi: 10.1016/j.chembiol.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Heijenoort Y, Gómez M, Derrien M, Ayala J, van Heijenoort J. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J Bacteriol. 1992;174:3549–3557. doi: 10.1128/jb.174.11.3549-3557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X, et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467:991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M, Radchenko M, Symersky J, Nie R, Guo Y. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat Struct Mol Biol. 2013;20:1310–1317. doi: 10.1038/nsmb.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu M, et al. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc Natl Acad Sci USA. 2013;110:2099–2104. doi: 10.1073/pnas.1219901110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka Y, et al. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature. 2013;496:247–251. doi: 10.1038/nature12014. [DOI] [PubMed] [Google Scholar]
- 35.Jin Y, Nair A, van Veen HW. Multidrug transport protein NorM from Vibrio cholerae simultaneously couples to sodium- and proton-motive force. J Biol Chem. 2014;289:14624–14632. doi: 10.1074/jbc.M113.546770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zilberstein D, Agmon V, Schuldiner S, Padan E. Escherichia coli intracellular pH, membrane potential, and cell growth. J Bacteriol. 1984;158:246–252. doi: 10.1128/jb.158.1.246-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker S, et al. Chemistry and biology of ramoplanin: a lipoglycodepsipeptide with potent antibiotic activity. Chem Rev. 2005;105:449–476. doi: 10.1021/cr030106n. [DOI] [PubMed] [Google Scholar]
- 38.Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat Rev Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 39.Hamburger JB, et al. A crystal structure of a dimer of the antibiotic ramoplanin illustrates membrane positioning and a potential Lipid II docking interface. Proc Natl Acad Sci USA. 2009;106:13759–13764. doi: 10.1073/pnas.0904686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrett TA, O'Neill AC, Hopson ML. Quantification of cardiolipin molecular species in Escherichia coli lipid extracts using liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2012;26:2267–2274. doi: 10.1002/rcm.6350. [DOI] [PubMed] [Google Scholar]
- 41.Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kagan VE, et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of deploarized mitochondria by mitophagy. Cell Death Diff. 2016;23:1140–1151. doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukopadhyay K, et al. In vitro susceptibility of Staphyococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmtery. Microbiology. 2007;153:1187–1197. doi: 10.1099/mic.0.2006/003111-0. [DOI] [PubMed] [Google Scholar]
- 44.Rosado H, Turner RD, Foster SJ, Taylor PW. Impact of the β-lactam resistance modifier (-)-epicatechin gallate on the non-random distribution of phospholipids across the cytoplasmic membrane of Staphylococcus aureus. Int J Mol Sci. 2015;16:16710–16727. doi: 10.3390/ijms160816710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 46.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal Chem. 2011;83:940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request