Abstract
Site-selective protein modification is a key step in facilitating protein functionalization and manipulation. To accomplish this, genetically engineered proteins were previously required, but the procedure was laborious, complex, and technically challenging. Herein we report the development of aptamer-based recognition-then-reaction to guide site-selective protein/DNA conjugation in a single step with outstanding selectivity and efficiency. As models, several proteins, including human thrombin, PDGF-BB, Avidin, and His-tagged recombinant protein, were studied, and the results showed excellent selectivity under mild reaction conditions. Taking advantage of aptamers as recognition elements with extraordinary selectivity and affinity, this simple preparation method can tag a protein in a complex milieu. Thus, with the aptamer obtained from cell-SELEX, real-time modification of live-cell membrane proteins can be achieved in one step without any pre-treatment.
Keywords: aptamer template, cell-surface modification, protein conjugation, site-selective
The newest trend in protein chemistry is termed site-selective protein-modification chemistry.[1–5] As the name implies, a chemical group, or modification, is labeled at a known site for enhanced biological or therapeutic applications. Bioorthogonal chemistry, defined as a chemical reaction that occurs inside living systems, but with no interference in their biochemistry, has become an important strategy in achieving these objectives.[6–9] The bioorthogonal strategy is highly selective, but slow reactivity and complicated bioengineering limit its utility. Thus, new and simpler methods for selective protein modification with feasible conditions are needed.
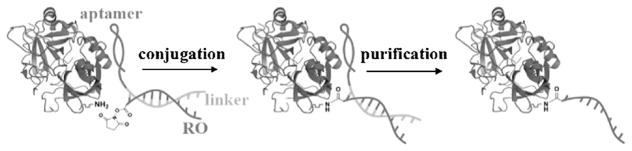
Herein we report an aptamer-templated synthesis (ATS) for site-selective protein/DNA recognition and conjugation. Compared with other DNA-templated technologies, such as DNA template synthesis (DTS) and DNA template protein conjugation (DTPC), the use of an aptamer as the recognition element demonstrates the principle that close proximity of reactants enables the selective reaction.[10–12] Aptamers are single-stranded oligonucleotides selected by SELEX (systematic evolution of ligands by exponential enrichment) to bind specifically to a target molecule,[13–16] and they can be used as templates to introduce a reactive functionality on a protein.[17] In this case, the template is a DNA aptamer lengthened with linker at one end. It can hybridize with a partially complementary reacting oligonucleotide (RO) and bring the RO close to the target protein, thus facilitating conjugation (Figure 1). Site-selectivity is achieved by the high local concentration of amine residues in close range of the aptamer binding site and an activated carboxyl residue at the end of the RO sequence. Once the site is functionalized, cDNA fully complementary to the template is added to remove the aptamer from the protein and generate the protein–RO conjugate. DNA synthesis and modification increase the number of available conjugation reactions and allow the protein to react in its natural state under mild and biocompatible conditions. For any protein, aptamer ligands can be generated by SELEX to serve as templates in ATS. Thus, this method offers a platform for precise site-selective protein conjugation.
Figure 1.
Scheme of ATS for protein/DNA conjugation. The aptamer template partially hybridizes with the reacting oligonucleotide (RO) and brings it close to the target protein, thus facilitating conjugation.
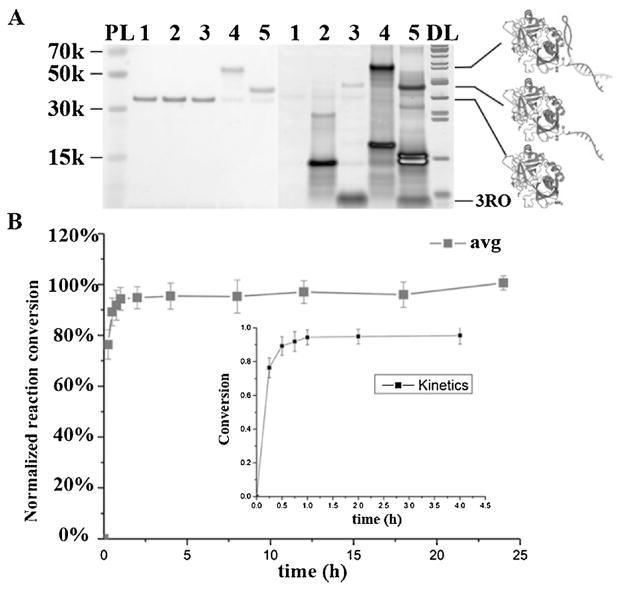
As a demonstration model, human alpha thrombin was used as the target protein, since two aptamers targeting this protein have already been developed: HD22 (29mer DNA aptamer) and TBA (15mer DNA aptamer).[13,18,19] We first tested HD22 for template construction based on its high binding affinity (dissociation constant (Kd) = 0.7 nM).[18] Since the linker was modified at the 3′-terminus of the aptamer, the template is termed aptamer-3′. And the N-hydroxysuccinimide (NHS) ester, which can react with a primary amine group with high efficiency and rapid kinetics in mildly basic aqueous solution was incorporated at the 3′-terminus of the RO strand, the reacting DNA is termed 3RO.[20] The HD22-3′ template, as a heterobifunctional compound, brings together thrombin and 3RO, enabling the reaction, which was monitored with SDS-PAGE (Figure 2A) and non-denaturing PAGE (Figure S1 in the Supporting Information). Without HD22-3′, the nonspecific conjugation between 3RO and thrombin was negligible. However, in the presence of template HD22-3′, specific conjugation was realized with a repeatable high conversion of >85% within 1 hour (Figure 2B, Figure S2), as determined by ImageJ. It is notable that the DNA protein conjugate showed about 32% reduction in band intensity based on Coomassie staining.[11]
Figure 2.
Aptamer-binding enables selective recognition and conjugation on thrombin with HD22-3′ template. A) SDS-PAGE (4–12%) with protein and DNA staining showing aptamer-templated conjugation on thrombin by template HD22-3′. 1. thrombin; 2. thrombin+HD22-3′ (2 equiv.); 3. thrombin+3RO (2 equiv.); 4. thrombin +3RO (2 equiv.) +HD22-3′ (2 equiv.); 5. thrombin+3RO (2 equiv.) +HD22-3′ (2 equiv.), and cDNA replacement (4 equiv. HD22-3′ cDNA) after conjugation. PL: Protein ladder. DL: DNA ladder. (left: protein stained by GelCode Blue; right: DNA stained by Sybr Gold). B) ATS reaction time analysis showing that the reaction was completed within 1 hour. ATS on thrombin with different reaction times was analyzed and collected on 4–12% SDS-PAGE with ImageJ five times (Figure S2). Inset shows the detail in the first 4 hours. The average reaction conversion at 24 hours was set as the maximum (100%) for comparison.
For thrombin, nonspecific conjugation is mainly triggered by electrostatic interaction between negatively charged DNA and the positively charged protein surface. We demonstrated that at 400 mM NaCl and higher, only negligible nonspecific binding of 3RO and thrombin was observed (Figure S3), since the high ion concentration could neutralize the surface charges.[11] Thus, 400 mM NaCl was used in all subsequent experiments, except the cell reaction. To demonstrate sequence dependency of ATS, two mismatched templates were designed, and neither could guide the templated reaction (Figure S4). The purified thrombin conjugate produced by ATS was extracted from non-denaturing PAGE, and the extraction yield was about 56% by comparing to a calibration plot of absorbance versus free RO concentration (Figures S1,S5). As site-selective functionalization of protein is the purpose of ATS, we proceeded to verify its site-selectivity by treating the reaction product with trypsin and using electrophoresis to isolate the digested peptide covalently bound with RO strand (Figure S6). Indeed, mass spectral analysis showed that the conjugate was formed only on Lysine 57 and Lysine 288 (Figure S7), demonstrating the precision of ATS.
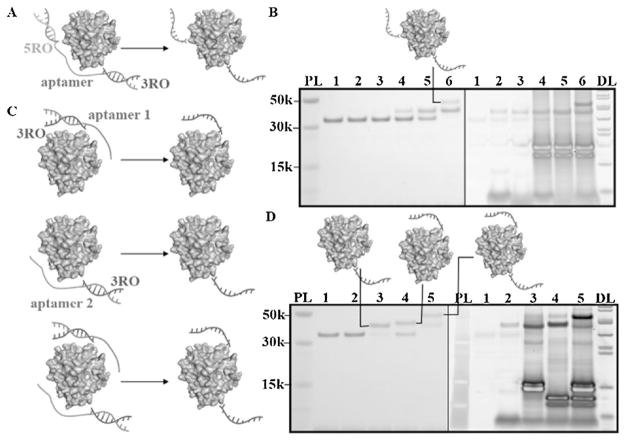
Apart from site-selectivity, another major advantage of ATS is DNA adaptability, such that two templates can be prepared by extending two ends of the same aptamer with different linkers. Thus, it is possible to achieve two conjugations at different positions with one aptamer. To test this, we prepared two templates, aptamer-3′ and aptamer-5′, with two corresponding ROs, 3RO and 5RO. With the assistance of template HD22-3′5′, which represents two linkers on the HD22 aptamer, thrombin with 3RO and 5RO could be synthesized based on the same procedure as that described above (Figure 3A,B). We also conjugated 3RO to thrombin with two templates, HD22-3′ and TBA-3′, respectively, to produce a conjugate with two 3ROs when using two templates at the same time (Figure 3C,D). By using HD22-5′ and TBA-5′, similar results confirmed the concept of multiple site-selective conjugations (Figure S8). When using aptamer TBA to form the template, it was interesting that the conjugation conversions for both 3RO and 5RO exceeded 50% (Figure 3D, Figure S8, ImageJ). However, under the same reaction conditions, less than half of thrombin could bind to the aptamer, since TBA has relatively poor binding ability (Kd = 450 nM).[15] This could be explained by the dynamic equilibrium of protein-aptamer binding. Conjugation using an aptamer with low affinity is driven to the right because the electrostatic interaction is converted into a covalent bond.
Figure 3.
Double conjugation on thrombin using aptamer template. A) Scheme of double conjugation with single aptamer templates. B) SDS-PAGE (4–12%) result. 1. thrombin; 2. thrombin+3RO (2 equiv.); 3. thrombin+5RO (2 equiv.); 4. thrombin+3RO (2 equiv.) +HD22-3′5′ (2 equiv.); 5. thrombin +5RO (2 equiv.) +HD22-3′5′ (2 equiv.) 6. thrombin +3RO (2 equiv.) +5RO (2 equiv.) +HD22-3′5′ (2 equiv.). cDNA replacement (4 equiv.) after each conjugation. PL: Protein ladder. DL: DNA ladder. (left: protein stained by GelCode Blue; right: DNA stained by Sybr Gold) C) Scheme of multiple conjugation with different aptamer templates. D) SDS-PAGE (4–12%) result. 1. thrombin; 2. thrombin and 3RO (2 equiv.); 3. thrombin, 3RO (4 equiv.) and HD22-3′ (2 equiv.), with cDNA of HD22-3′ replacement (4 equiv.) after conjugation; 4. thrombin, 3RO (2 equiv.) and TBA-3′ (2 equiv.), with cDNA of TBA-3′ replacement (4 equiv.) after conjugation; 5. thrombin, 3RO (4 equiv.), HD22-3′ (2 equiv.) and TBA-3′ (2 equiv.), with cDNA of HD22-3′ (4 equiv.) and cDNA of TBA-3′ (4 equiv.) replacement after conjugation. (left: protein stained by GelCode Blue; right: DNA stained by Sybr Gold). The conversions are 80% (5RO, HD22-5′), 55% (3RO, TBA-3′), 40% (5RO, TBA-5′), respectively.
Several other protein/aptamer pairs were also tested with similar reaction conditions. PDGF-BB, a dimeric glycoprotein composed of two B chains, can bind to two aptamers based on its symmetrical structure.[21] By mixing different ratios of DNA (template and RO) to protein, the number of ROs conjugated on PDGF-BB (one or two) could be adjusted (Figure S9). ATS of Streptavidin with template was also achieved with excellent selectivity (Figure S10).[22] Antibody-drug conjugates comprise a significant class of highly potent anticancer therapies, and their synthesis is a challenging topic in bioconjugation.[4,23] However, such conjugation could be easily accomplished with ATS, since an aptamer targeting Human IgG Fc domain resulted in covalent binding of RO to the constant domain on the antibody (Figure S11).[24] Therefore, ATS offers an efficient method of modifying an antibody, but without affecting its recognition ability, and, hence, has high potential in targeted cancer therapy and antibody labeling.
At the same time, ATS does have some limitations, due to the lack of crystal structures for crucial binding site data. Since aptamer binding is a function of molecular recognition, the reaction position cannot be predicted, thus risking potential interference with protein activity. To solve this problem and enlarge the candidate protein repertoire for more inclusive use of ATS, even without known aptamers, we used a His tag aptamer for recombinant protein conjugation.[25] His tag is a peptide tag of polyhistidine present on many recombinant proteins for isolation and purification.[26] Using His tag aptamer, we could enlarge the library of candidate proteins for site-selective conjugation and perform the DNA conjugation on a protein without loss of function, since the His tag is normally far from the active domain. And successful conjugations were realized on N-terminus His-tagged Midkine, a basic heparin-binding growth factor of low molecular weight, as a proof of concept (Figure S12,S13).
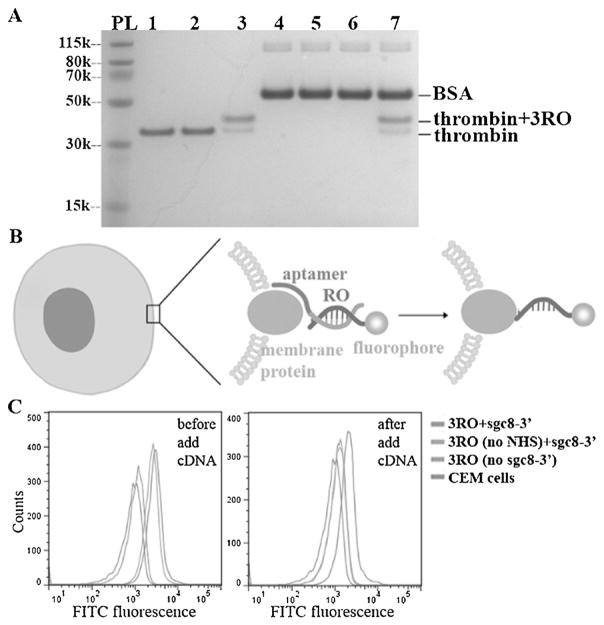
During the development of ATS, we attained similar, or even better, outcomes compared with some reported methods.[9,11] Aptamer selectivity can direct conjugation to a target protein in a complex mixture in vitro, which, compared to bioorthogonal method, does not require any pre-modification of target protein. To demonstrate this, we chose thrombin and BSA as positive and negative proteins and HD22-3′ as the template. The results showed that the conjugation was present only on thrombin in a mixture with equal or even higher concentrations of BSA and thrombin (Figure 4A, Figures S14,S15).
Figure 4.
Aptamer-templated synthesis shows selectivity in protein mixture and on live cells. A) SDS-PAGE (4–12%) result shows selectivity of aptamer-templated conjugation. 1. thrombin; 2. thrombin and 3RO (2 equiv.); 3. thrombin, 3RO (2 equiv.), and HD22-3′ (2 equiv.); 4. BSA; 5. BSA and 3RO (2 equiv.); 6. BSA, 3RO (2 equiv.), and HD22-3′ (2 equiv.); 7. thrombin, BSA, 3RO (2 equiv.), HD22-3′ (2 equiv.). For lanes 3, 6, and 7, cDNA (4 equiv.) were added after conjugation to replace aptamer. Gel is stained for protein by GelCode Blue. B) The design of cellular aptamer-templated conjugation. C) Flow cytometry results show that the aptamer-templated conjugation was achieved on CEM cells with sgc8-3′ template.
Selectivity, biocompatibility and rapid kinetics allow us to implement ATS in a complicated biological system like live cells. It is extremely difficult to modify a target protein in its natural state. However, with the development of cell-SELEX in our laboratory, we have selected aptamers able to bind specifically with target live cells.[27] In particular, Sgc8, a well-studied aptamer, was confirmed as a binder of PTK7 protein on the CCRF-CEM cell membrane. Therefore, we chose Sgc8 for template construction in reactions with CEM cells, while Ramos cells were used as negative control,[28] and FITC-labeled RO (Figure S16) was used for cell conjugation. Based on careful design (Figure 4B, S17), we demonstrated conjugation on target membrane protein (Figure 4C), but no conjugation on control Ramos cells having low levels of PTK7 (Figure S18). Thus, the real-time modification of cellular membrane proteins can indeed be achieved in one-step ATS without the need for pre-modification.
In conclusion, we have developed a method for real-time aptamer-templated synthesis (ATS) that specifically conjugates DNA to proteins. We have demonstrated the feasibility and generality of recognition-then-reaction for bioconjugation as well as the selectivity required for modification on living cells. ATS is distinct compared to other site-selective protein conjugation methods. The ATS template is easy to design and prepare, and it can be easily removed after reaction. Aptamer-templated synthesis: 1) can be performed at a precise position on a protein in its natural state without any genetic engineering; 2) can be performed under biocompatible conditions with rapid kinetics and high conversion yield; 3) is simple and general; and 4) can recognize the target protein in complex biological systems.
Supplementary Material
Acknowledgments
We thank Dr. Kathryn R. Williams for manuscript proof reading and Dr. Kari B. Basso for technical assistance in mass spectrometry. This work was supported by grants awarded by the National Institutes of Health (GM079359 and CA133086) and by the NSFC (Grants 21325520, 21521063).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information (detailed experimental methods and descriptions of protein conjugations, gel electrophoresis, mass spectrum and cell conjugations) and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/anie.201706285.
Contributor Information
Cheng Cui, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA).
Prof. Hui Zhang, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA). Jiangsu Key Laboratory of New Power Batteries, Jiangsu Collaborative Innovation Center of Biomedical Functional Materials, College of Chemistry and Materials Science, Nanjing Normal University Nanjing, 210023 (China)
Dr. Ruowen Wang, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA). Molecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Life Sciences and College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University Changsha, Hunan, 410082 (China)
Dr. Sena Cansiz, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA)
Xiaoshu Pan, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA).
Shuo Wan, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA).
Weijia Hou, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA).
Long Li, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA).
Meiwan Chen, State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau Macau, 999078 (China).
Dr. Yuan Liu, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA). Molecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Life Sciences and College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University Changsha, Hunan, 410082 (China)
Xigao Chen, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA).
Prof. Qiaoling Liu, Molecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Life Sciences and College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University Changsha, Hunan, 410082 (China)
Prof. Weihong Tan, Center for Research at Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics, Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida, Gainesville, FL 32611 (USA). Molecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Life Sciences and College of Chemistry and Chemical Engineering, Aptamer Engineering Center of Hunan Province, Hunan University Changsha, Hunan, 410082 (China)
References
- 1.Krall N, da Cruz FP, Boutureira O, Bernardes GJL. Nat Chem. 2016;8:103–113. doi: 10.1038/nchem.2393. [DOI] [PubMed] [Google Scholar]
- 2.Spicer CD, Davis BG. Nat Commun. 2014;5:4740. doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 3.Zhang C, Welborn M, Zhu T, Yang NJ, Santos MS, Van Voorhis T, Pentelute BL. Nat Chem. 2016;8:120–128. doi: 10.1038/nchem.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chudasama V, Maruani A, Caddick S. Nat Chem. 2016;8:114–119. doi: 10.1038/nchem.2415. [DOI] [PubMed] [Google Scholar]
- 5.Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin JW. Nat Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott TS, Townsley FM, Bianco A, Ernst RJ, Sachdeva A, Elsässer SJ, Davis L, Lang K, Pisa R, Greiss S, Lilley KS, Chin JW. Nat Biotechnol. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Chen PR. Nat Chem Biol. 2014;12:129–137. doi: 10.1038/nchembio.2024. [DOI] [PubMed] [Google Scholar]
- 8.Hang HC, Yu C, Kato DL, Bertozzi CR. Proc Natl Acad Sci USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Sachdeva A, Cox DJ, Wilf NM, Lang K, Wallace S, Mehl RA, Chin JW. Nat Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Liu DR. Angew Chem Int Ed. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2004;116:4956–4979. [Google Scholar]
- 11.Rosen CB, Kodal AL, Nielsen JS, Schaffert DH, Scavenius C, Okholm AH, Voigt NV, Enghild JJ, Kjems J, Tørring T, Gothelf KV. Nat Chem. 2014;6:804–809. doi: 10.1038/nchem.2003. [DOI] [PubMed] [Google Scholar]
- 12.AkÅay G, Belmonte MA, Aquila B, Chuaqui C, Hird AW, Lamb ML, Rawlins PB, Su N, Tentarelli S, Grimster NP, Su Q. Nat Chem Biol. 2016;11:931–936. doi: 10.1038/nchembio.2174. [DOI] [PubMed] [Google Scholar]
- 13.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 14.Ellington AD, Szostak JW. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 15.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 16.Tan W, Donovan MJ, Jiang J. Chem Rev. 2013;113:2842–2862. doi: 10.1021/cr300468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Lu D, Bai H, Jin C, Yan G, Ye M, Qiu L, Chang R, Cui C, Liang H, Tan W. Chem Sci. 2016;7:2157–2161. doi: 10.1039/c5sc02631h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tasset DM, Kubik MF, Steiner WJ. Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Cao Z, Tan W. Proc Natl Acad Sci USA. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattson G, Conklin E, Desai S, Nielander G, Savage MD, Morgensen S. Mol Biol Rep. 1993;17:167–183. doi: 10.1007/BF00986726. [DOI] [PubMed] [Google Scholar]
- 21.Green LS, Jellinek D, Jenison R, Ostman A, Heldin CH, Janjic N. Biochemistry. 1996;35:14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 22.Ruigrok VJ, van Dujin E, Barendregt A, Dyer K, Tainer JA, Stoltenburg R, Strehlitz B, Levisson M, Smidt H, van der Oost J. ChemBioChem. 2012;13:829–836. doi: 10.1002/cbic.201100774. [DOI] [PubMed] [Google Scholar]
- 23.Mullard A. Nat Rev Drug Discovery. 2013;12:329–332. doi: 10.1038/nrd4009. [DOI] [PubMed] [Google Scholar]
- 24.Miyakawa S, Nomura Y, Sakamoto T, Yamaguchi Y, Kato K, Yamazaki S, Nakamura Y. RNA. 2008;14:1154–1163. doi: 10.1261/rna.1005808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan X, Chen W, Lu S, Zhu Z, Chen T, Zhu G, You M, Tan W. Anal Chem. 2012;84:8272–8276. doi: 10.1021/ac301764q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochuli E, Bannwarth W, Dçbeli H, Gentz R, St4ber D. Nat Biotechnol. 1988;6:1321–1325. [Google Scholar]
- 27.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Malikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shangguan D, Cao Z, Meng L, Malikaratchy P, Sefah K, Wang H, Li Y, Tan W. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.