Summary
The gut microbiota play important roles in lipid metabolism and absorption. However, the contribution of the small bowel microbiota of mammals to these diet-microbe interactions remains unclear. We determine that germ-free (GF) mice are resistant to diet-induced obesity and malabsorb fat with specifically impaired lipid digestion and absorption within the small intestine. Small bowel microbes are essential for host adaptation to dietary lipid changes by regulating gut epithelial processes involved in their digestion and absorption. Additionally, GF mice conventionalized with high-fat diet-induced jejunal microbiota, exhibit increased lipid absorption even when fed a low-fat diet. Conditioned media from specific bacterial strains directly upregulates lipid absorption genes in murine proximal small intestinal epithelial organoids. These findings indicate that proximal gut microbiota play key roles in host adaptability to dietary lipid variations through mechanisms involving both the digestive and absorptive phases and that these functions may contribute to conditions of over- and undernutrition.
eTOC Blurb
Martinez-Guryn et al. demonstrate that small intestinal microbiota are critical transducers of dietary signals that allow the host to adapt to variations in lipid digestion and absorption. High fat diet-induced jejunal microbiota directly promote mucosal lipid absorption, and are potentially a factor contributing to conditions of over- and undernutrition.
Introduction
Recent work highlights the role of the trillions of intestinal microbes in health and disease. Our microbial organ is highly sensitive to environmental factors, in particular, dietary stress. Several studies have revealed that high fat (HF) diets profoundly, rapidly, and sustainably alter microbial communities in as little as 24–48 hours in mice and humans (Carmody et al., 2015; David et al., 2014; Howe et al., 2016), resulting in the development of metabolic disturbances. Not only are changes in community membership evident, i.e. abundance of Firmicutes and Proteobacteria as well as decreasing diversity, but also microbial metabolic function is altered, i.e. decreasing short chain fatty acid (SCFA) production (David et al., 2014; Leone et al., 2015). Many of these studies have relied on examining cecal or fecal microbiota. A critical, and unanswered, question is whether diet-mediated changes in the small intestinal microbiome elicit a significant impact on the host in the proximal small intestine, a crucial site for macronutrient absorption and energy acquisition.
The proximal small intestine serves as the major site of macronutrient digestion and absorption. Complex interactions between dietary cues, the gut endocrine response, bile release, exocrine function of the pancreas, and absorptive enterocyte function are needed for efficient absorption of lipids and other dietary nutrients. The small intestine harbors a complex microbial community, albeit with less diversity and abundance (≈103 – 107 microbial cells/gram) than the colonic microbiota (≈1012 cells/gram; (El Aidy et al., 2013; Donaldson et al., 2015). This could be because microbial populations in the small intestine are routinely subjected to adverse host factors affecting assemblage, such as low pH, faster transit time, bile acids, and antimicrobial peptide exposure. Studies suggest that Firmicutes and Proteobacteria, which dominate the small intestine, are more tolerant of these factors (Angelakis et al., 2015; Donaldson et al., 2015). Bacteroidetes are also bile tolerant but the abundance of this phylum is reduced in the small intestine and is increased in abundance in the large intestine (Angelakis et al., 2015; Donaldson et al., 2015; Turnbaugh et al., 2009).
Numerous studies have shown that changes in cecal and stool microbiota are associated with dietary shifts, but it remains unclear whether they faithfully represent alterations of regional microbiota of the small intestine that are more likely responsive to dietary perturbations and elicit a direct impact on host functions related to metabolism and fat absorption. To date, limited studies have evaluated small intestine microbiota structure under obese vs lean states or with dietary manipulation and whether these changes contribute to metabolic disease. Yasuda et al. characterized regional microbiota distal of the jejunum in rhesus macaques. It was concluded that stool microbiota structure was strongly correlated with colonic communities but only moderately with those in the small intestine; however characterization of the duodenal microbiota was not performed (Yasuda et al., 2015). Obese and lean pigs were found to have increased abundance of clostridium and SMB53 in jejunum and ileum, however no clear differences between obese and lean states was evident (Yang et al., 2016). Turnbaugh et al. demonstrated that western diets shifted gut microbiota structure along the length of the gut including the small intestine, however, this region was not further investigated, nor was a comparison made between the different regions (Turnbaugh et al., 2009). Studies have examined microbiota changes or host responses in the duodenum (Angelakis et al., 2015), jejunum, and ileum (Donaldson et al., 2015; Ge et al., 2008; Yang et al., 2016), yet a comparative analysis between small intestinal regions in response to low fat (LF) vs high fat (HF) diets has not been performed.
Some evidence suggests that microbes are important for lipid absorption. For example, germ free (GF) mice consuming a HF diet were shown to have elevated fecal lipid levels compared to conventional mice (Rabot et al., 2010). In addition, conventionalized GF zebrafish displayed increased long and short-chain fatty acid uptake in the intestinal epithelium under fasted and fed conditions (Semova et al., 2012). More recently, antibiotic-treated rats displayed reduced lymphatic lipids following fat challenge (Sato et al., 2016). Despite these findings, diet-microbe interactions in the small intestine and mechanisms driving lipid digestion and absorption have not been thoroughly examined. To address this, we postulated that proximal small intestinal microbiota play an important role in regulating lipid digestion and absorption and are vulnerable to HF diet-induced alterations. Lastly, we hypothesized that these alterations promote lipid absorption, contributing to development of obesity. Thus, the goals of this study were to 1) determine if microbes are required for proper digestion and absorption of dietary lipid using GF animals, 2) examine the impact of HF diets on small intestinal microbiota membership using 16s rRNA gene profiles, 3) determine the functional impact of HF diet-induced jejunal microbiota on fat absorption, and 4) test proof-of-concept that microbially-derived metabolites or products from specific microbial strains directly affect lipid absorption. Here, we demonstrate that gut microbes act as indispensable signal transducers of dietary lipid, allowing the host to adapt fat absorption. Consumption of a HF diet profoundly alters the microbiota in the small intestine, and this community increases fat absorption in conventionalized animals. Lastly, we show that these effects may be promoted by specific bacterial strains through microbe-derived components or small molecules.
Results
GF mice are protected from high fat (HF) diet-induced obesity, in part by decreased dietary lipid digestion and absorption
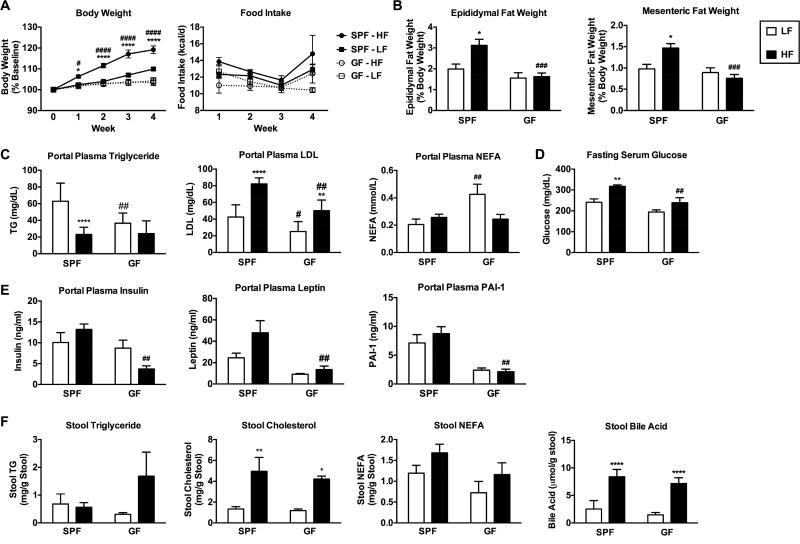
To establish whether GF mice display signs of fat malabsorption as previously reported, SPF and GF mice were fed a purified LF or HF diet (Table S1) for four weeks. Metabolic indices and stool lipid content were measured. Consistent with previous reports, GF mice fed HF diet were resistant to weight gain (Figure 1A; Leone et al. 2015) and exhibited reduced epididymal and mesenteric fat pad mass (Figure 1B). Food consumption was not significantly different between groups on purified diets (Figure 1A). Markers of insulin resistance, gut peptide hormones that regulate insulin signaling (i.e., GLP-1), and circulating lipids were measured in portal and peripheral plasma. GF mice exhibited profoundly reduced levels of portal plasma triglyceride (TG) compared to SPF mice on a LF diet and reduced low density lipoprotein (LDL) levels compared to SPF mice on both LF and HF diet (Figure 1C). Non-esterified fatty acids (NEFA) were elevated in GF mice on LF, but not HF diet. (Figure 1C). Similar trends in lipid profiles were found in peripheral plasma (data not shown). In addition, GF mice fed HF diet displayed reduced levels of fasting blood glucose (Figure 1D), insulin, leptin, and plasminogen activator inhibitor-1 (PAI-1) compared to SPF mice (Figure 1E). Gut peptide hormones ghrelin and GLP-1 were higher in GF vs SPF mice under LF conditions, which are indicative of a state of energy deficit and improved insulin signaling (Pannacciulli et al., 2006; Pinkney, 2014), respectively (Figure S1A). Concurrently, stool triglyceride levels tended to be elevated in GF compared to SPF mice fed a HF diet (Figure 1F). Levels of non-esterified fatty acids (NEFA) tended to be lower in GF mice fed a HF diet compared to SPF mice. Stool bile acids and total cholesterol were significantly increased following HF diet in both SPF and GF mice (Figure 1F). We noted that GF mice fed HF diet excreted significantly more stool (grams of dry weight) over 24 hours compared to HF-fed SPF mice (Figure S1B–C). Based on these observations, we speculated that GF mice fed HF diet have impaired fat absorption and digestive capacity, thereby conferring protection from HF diet-induced adiposity, elevated plasma lipids and markers of insulin resistance. Therefore, we sought to further examine if GF mice display an active impairment in lipid absorption and to identify the mechanisms involved.
Figure 1. GF mice are resistant to diet-induced obesity.
SPF and GF mice were fed a low fat (LF) or high fat (HF) diet for 4 weeks (Table S1). A) Body weight was measured weekly and expressed as a percentage relative to baseline. B) Epididymal and mesenteric fat pad weights were collected at the end of the study and expressed as a percentage of total body weight. C) Triglycerides (TG), low density lipoprotein (LDL), and non-esterified fatty acid (NEFA) levels were measured in portal plasma. D) Fasting serum glucose levels. E) Markers of insulin resistance were measured in portal plasma. See also Figure S1. F) Stool TG, NEFA, bile, and total cholesterol levels were measured. Data were pooled across 2–3 independent experiments (each with 2–6 animals per group) and are shown as means +/− SEM (n=10–13 A; n=6–9 B; n=5–12 C; n=4–9 D; n=5–6 E; n=5–9 F). * p ≤ 0.05 (LF vs HF), # p ≤ 0.05 (SPF vs GF).
GF mice have severely impaired lipid absorption
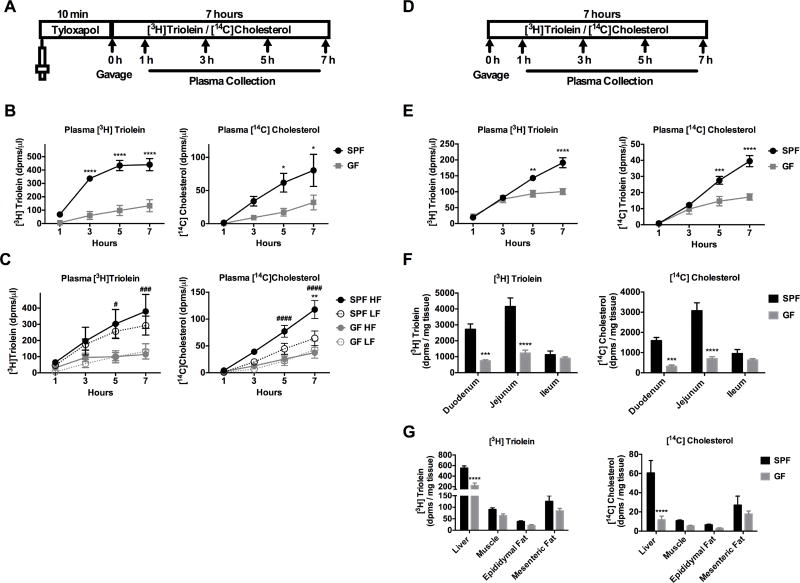
Transport of dietary lipids from the gut lumen into the systemic circulation requires both digestive and absorptive functions of the gastrointestinal (GI) tract (Hui and Howles, 2005; Iqbal and Hussain, 2009). To determine if GF mice exhibit impaired lipid digestion and absorption, GF and SPF mice maintained on a standard chow diet (Table S1) were gavaged with [3H]triolein (triglyceride consisting of oleic acid) and [14C]cholesterol, and the amount of radiolabeled lipid accumulating in the plasma was measured over seven hours. Prior to gavage, mice were injected retro-orbitally with tyloxapol, a peripheral lipoprotein lipase (LPL) inhibitor which blocks peripheral uptake of lipid thereby allowing for the detection of accumulating radiolabeled lipid in the blood over time (Figure 2A, Sontag et al. 2013). GF mice displayed a dramatically decreased rate of [3H]triolein and [14C]cholesterol absorption compared to SPF mice (Figure 2B). To examine if this was due to slower small intestinal transit time, SPF and GF mice were gavaged with corn oil plus activated charcoal for 2 hours to track the distance traveled. There was not a significant difference in small intestinal transit time between SPF and GF animals (Figure S2A–B).
Figure 2. GF mice have impaired lipid absorption and transport compared to SPF mice.
A) Schematic of experimental procedure is shown. Mice were treated for 10 minutes with (A–C) or without (E–G) tyloxapol followed by gavage with [3H]triolein and [14C]cholesterol. B) Radiolabeled lipid absorption was measured in SPF and GF mice that were fed a standard chow diet. C) Radiolabeled lipid absorption was measured in mice that were fed a low fat (LF) or high fat (HF) diet for 4 weeks. D) Schematic of experimental procedure without tyloxapol is shown. E) Radiolabeled lipid absorption was measured in SPF and GF mice fed a standard chow diet. F–G) Radiolabeled lipid was measured in intestinal epithelium or metabolic tissues. See also Figure S2. Data were pooled across 1–3 independent experiments and are shown as means +/− SEM (n= 7–9 B; n=5–11 C; n=5–6 E–G). B, E–G) * p ≤ 0.05 (SPF vs GF). C) * p ≤ 0.05 (LF vs HF), # p ≤ 0.05 (SPF vs GF).
To determine if lipid absorption could be restored by HF diet feeding, radiolabeled lipid absorption was examined in SPF and GF mice fed a purified LF or HF diet (Table S1). HF-fed SPF mice had significantly higher plasma levels of [3H]triolein and [14C]cholesterol compared to GF mice fed HF diet at 5 and 7 hours (Figure 2C). SPF mice fed HF diet displayed higher [14C]cholesterol levels compared to SPF mice fed a LF diet at 7 hours (Figure 2C). There was no increase in [3H]triolein and [14C]cholesterol absorption in GF animals fed HF vs. LF diet (Figure 2C). Thus, HF diet alone was not sufficient to restore lipid absorption in GF mice, indicating that the presence of microbes is necessary for adequate intestinal lipid absorption.
To examine the level of peripheral [3H]triolein and [14C]cholesterol uptake, an identical experiment was conducted in chow-fed SPF and GF mice but without tyloxapol pretreatment (Figure 2D). Consistent with previous results, GF mice had significantly lower plasma levels of [3H]triolein and [14C]cholesterol compared to SPF mice at 5 and 7 hours (Figure 2E). Notably, SPF mice had lower [3H]triolein and [14C]cholesterol levels without tyloxapol compared to when tyloxapol was used (Figure S2C), but levels in GF animals are relatively similar between the two conditions (Figure S2C), suggesting that there is more rapid peripheral uptake in SPF mice compared to GF mice. In addition, we observed that GF mice have significantly lower levels of radiolabeled lipid in the duodenal and jejunal epithelium (Figure 2F) and in the liver (Figure 2G) as compared to SPF mice.
It should be noted that in these experiments, the SPF and GF mice were not maintained in identical bedding conditions, as SPF mice were maintained in corn cob bedding and GF mice were housed in pine shavings. To address this issue, we repeated the radiolabeled lipid absorption assay using tyloxapol in SPF vs GF mice raised in pine shavings. Consistent with our previous results, we found that GF mice had significantly reduced [3H]triolein and [14H]cholesterol absorption compared to SPF mice (Figure S2D).
GF mice exhibit both impaired lipid digestion and absorption within the small intestine
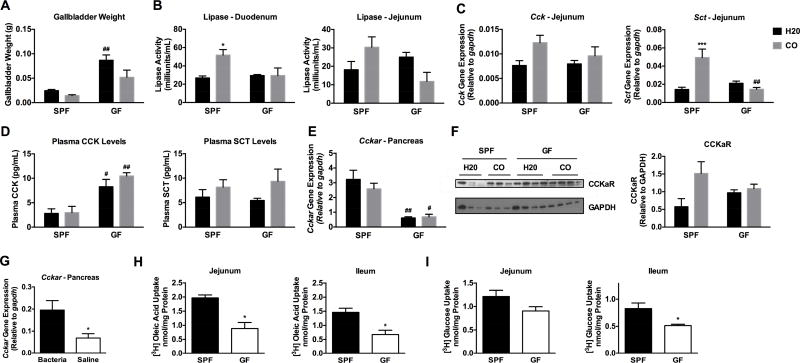
To better understand the mechanisms behind decreased fat absorption in GF mice, fat digestion and transport was assessed in GF vs SPF mice. Fat digestion is largely facilitated by the enteroendocrine hormones cholescystokinin (CCK) and secretin (SCT) (Hui and Howles, 2005). These peptide hormones are produced in I and S endocrine cells, respectively, of the epithelium lining the proximal small intestine and are released upon feeding. CCK stimulates the release of bile from the gallbladder for emulsification of fat as well as pancreatic secretion of lipase. Similarly, SCT stimulates pancreatic secretion of digestive enzymes and bicarbonate for neutralization of chyme and proper enzymatic function (Iqbal and Hussain, 2009). Notably, enteroendocrine cell number is not altered in the duodenum or jejunum of GF compared to SPF animals (Duca et al., 2013), but we expected that a breakdown in enteroendocrine signals may explain reduced lipid absorption observed in GF mice. To evaluate the involvement of impaired enteroendocrine signaling in GF mice, GF and SPF mice were fasted for four hours and challenged with a lipid bolus of corn oil (CO) or H2O control for 2 hours (Drover et al., 2005). Gallbladder weights were significantly higher in GF vs. SPF mice under control conditions but not following CO gavage (Figure 3A). Gene expression of the receptor for CCK (Cckar) was not different in SPF vs GF mice (data not shown). Interestingly, CO significantly increased lipase activity in the duodenum of SPF, but not GF mice, implicating an impairment in CCK signaling to the pancreas in GF mice (Figure 3B). A similar trend was seen in the jejunum. GF mice exhibited reduced gene expression of Cck and Sct in the jejunum compared to SPF mice following CO gavage (Figure 3C). Unexpectedly, reduced lipase activity in GF mice could not be explained by circulating CCK levels as plasma CCK levels were elevated in GF mice (Figure 3D). This compelled us to investigate whether pancreatic expression of the Ccka receptor (Cckar) was reduced in GF mice. Indeed, GF mice had significantly lower mRNA of Cckar in the pancreas compared to SPF mice (Figure 3E). Western blot analysis revealed that SPF mice gavaged with CO exhibited elevated CCKaR protein levels compared to H2O control whereas GF mice did not display an elevated CCKaR response to CO challenge relative to H2O control (Figure 3F). However, this effect was not significant based on densitometry analysis. Circulating SCT levels were not significantly different in GF vs SPF mice (Figure 3D) nor were expression levels of secretin receptor (Sctr) in the pancreas (data not shown). Next, GF mice were gavaged with or without a combination of heat-killed gram-negative (Bacteroides thetaiotamicron) and gram-positive (Lactobacillus rhamnosus gg) bacteria and pancreatic Cckar expression was measured. Exposure to both gram-negative and gram-positive heat-killed bacteria increased expression of Cckar in the pancreas compared to GF controls (Figure 3G). Taken together, these results suggest that reduced lipid absorption in GF mice involves a breakdown in CCKa receptor-mediated stimulation of the pancreas.
Figure 3. GF mice exhibit impaired lipid digestive and absorptive function.
A–E) SPF and GF mice were fasted for 4 hours and gavaged with corn oil (CO) or water (H2O) for 2 hours. A) Gallbladder weights. B) Duodenum and jejunum lipase activity. C) Jejunal gene expression of cholecystokinin (Cck) and secretin (Sct). D) Plasma CCK and SCT levels were measured via commercially available EIA assays. E) Pancreatic gene expression of ccka receptor (Cckar), F) western blot of CCKaR, and densitometry of CCKaR. G) Pancreatic Cckar gene expression was determined in GF mice following gavage with vehicle (PBS) or heat-killed Bacteroidetes thetaiotamicron plus Lactobacillus rhamnosus gg. (H) Levels of [3H]oleic acid and of (I) [3H]glucose uptake in brush border membrane vesicles from jejunum and ileum of SPF vs GF mice. Data presented in A–F represent two independent experiments, with 3–4 animals each and are shown as means +/− SEM (n=4 A-D; n=3–4 E-G; n=4–5 H; n=3 I). A–F) * p ≤ 0.05 (LF vs HF), # p ≤ 0.05 (SPF vs GF). G–I) * p ≤ 0.05 (SPF vs GF).
To examine if reduced lipid transport was occurring at the site of absorptive enterocytes (e.g., apart from the enteroendocrine signaling required for digestion), brush border membrane vesicles were isolated from the jejunum and ileum of GF and SPF mice, followed by incubation with [3H]oleic acid. Brush border membrane vesicles from GF mice exhibited decreased [3H]oleic acid (Figure 3H) as well as [3H]glucose uptake compared to those from SPF counterparts (Figure 3I), implicating a role for microbes in regulating processes related to membrane transport. Gene expression of fatty acid transporters and esterification enzymes (Figure S1D) was measured in the duodenum and jejunum of SPF and GF mice under LF and HF conditions. The fatty acid translocase Cd36 was upregulated under HF vs LF diet conditions and in GF vs SPF mice in the jejunum. Dgat1 was upregulated in GF vs SPF mice under LF conditions in the duodenum and jejunum (Figure S1D). Genes related to lipogenesis and fat oxidation were also measured in the duodenum and jejunum (Figure S1E). The transcriptional regulator Pparα, involved in fat oxidation, was upregulated in GF vs SPF mice under LF and HF conditions in the jejunum (Figure S1E). Intriguingly, in the duodenum, Dgat2 was induced by HF diet in SPF mice, but not GF mice, suggesting that the regulation of Dgat2 may involve diet-microbe interactions. To better elucidate mechanisms behind potential host-microbe interactions, we next sought to identify the role that HF dietinduced gut microbes play in lipid absorption.
Gut microbes regulate lipid absorption
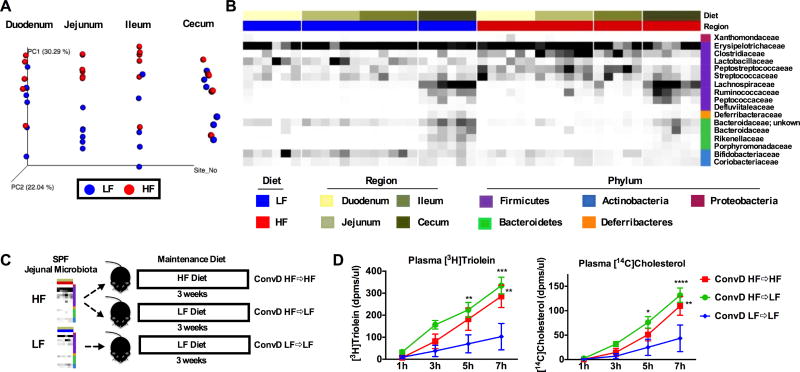
Understanding small intestinal microbiota composition is integral to identifying key host-microbe interactions that drive fat absorption. Thus, we characterized the microbial community structure of the small intestine in SPF mice under LF and HF conditions. Sequencing of 16S rRNA gene amplicons was performed in mucosal scrapings collected from the duodenum, jejunum, ileum, as well as cecal contents of mice fed either a LF or HF diet for four weeks. Differences between LF and HF microbiota were apparent in the jejunum and ileum based on Bray Curtis beta diversity analyses as indicated in the PCoA plot expressed on a forced axis for regional site (Figure 4A) and in PCoA plots for individual intestinal regions (Figure S3A). Adonis and Anosim tests were performed for Bray Curtis and Canberra diversity metrics to examine differences between LF and HF diet groups in each intestinal region (Table 1). HF diet increased the relative abundance of the family Clostridiaceae compared to LF diet in all regions, especially in the jejunum and ileum (Figure 4B). Similar results were found in a pilot study conducted early in the development of this project (data not shown). HF diet also decreased abundance of the families Bifidobacteriaceae and Bacteroidaceae in all intestinal regions (Figure 4B). Analysis between MED oligotypes from LF or HF fed mice revealed significant differences in the jejunum (Table 2) and duodenum in which the abundance of the family Peptostreptococcaceae was significantly increased (Bonferroni p value = 0.0472, data not shown). Significant differences in MED oligotypes were not found between LF and HF diets in the ileum and cecum. MED oligotypes found to be significantly increased by HF diet in the jejunum belonged to the Clostridium and Turicibacter genera and Peptostreptococcaceae family, whereas oligotypes that were significantly decreased by HF diet include those mapping to Bifidobacterium, Allobaculum, and Bacteroidales (Table 2). Abundance of the 16S rRNA gene increased along the length of the proximal intestine, but did not significantly differ in HF vs. LF conditions (Figure S2B). Altogether, microbiota structure differs along the length of the small intestine and HF diet has a dramatic impact on microbial structure and function particularly in the small intestine.
Figure 4. HF diet-induced jejunal microbiota promotes lipid absorption.
SPF mice were fed LF or HF diet for four weeks. Small intestinal mucosal scrapings and cecal contents were collected for 16S rRNA amplicon sequencing. A) PCoA plot showing Bray Curtis distances of microbial communities between LF and HF diet on a forced axis for intestinal region. B) Heat map displaying relative abundance of taxa in small intestine and cecal contents between LF and HF diets. C) Experimental design for conventionalization of jejunal microbiota D) Absorption of [3H]Triolein and [14C]Cholesterol is shown over time and expressed as dpms/µl plasma. See also Figure S3. Data are shown as means +/− SEM (n= 6 A–B; n=3–5 D). * p ≤ 0.05 (LF vs HF), # p ≤ 0.05 (SPF vs GF).
Table 1.
R statistics and P values from Adonis and Anosim tests from Bray Curtis and Canberra metrics between diets within each region and comparing across region within each diet.
| BRAY CURTIS LF VS HF | CANBERRA LF VS HF | |||||||
| Adonis | Anosim | Adonis | Anosim | |||||
| R2 | P value | R | P value | R2 | P value | R | P value | |
| DUODENUM | 0.108 | 0.248 | 0.004 | 0.372 | 0.096 | 0.323 | 0.035 | 0.268 |
| JEJUNUM | 0.544 | 0.004 | 0.826 | 0.004 | 0.263 | 0.006 | 0.754 | 0.003 |
| ILEUM | 0.321 | 0.018 | 0.403 | 0.02 | 0.192 | 0.007 | 0.472 | 0.002 |
| CECUM | 0.241 | 0.035 | 0.369 | 0.032 | 0.198 | 0.003 | 0.532 | 0.002 |
| BRAY CURTIS REGIONAL COMPARISONS | CANBERRA REGIONAL COMPARISONS | |||||||
| Adonis | Anosim | Adonis | Anosim | |||||
| R2 | P value | R | P value | R2 | P value | R | P value | |
| LF | 0.308 | 0.001 | 0.236 | 0.003 | 0.314 | 0.001 | 0.458 | 0.001 |
| HF | 0.406 | 0.001 | 0.384 | 0.001 | 0.323 | 0.001 | 0.570 | 0.001 |
Table 2.
Oligotypes generated via Minimum Entropy Decomposition (MED) that are significantly different between low fat (LF) and high fat (HF) diets in the jejunum are shown. Taxonomic level is indicated as “g” = genus; “f” = family; “o” = order; “_1” arbitrary value assigned to different oligotypes at the same taxonomic level.
| MED Oligotypes | FDR Corrected |
Bonferroni | LF Mean | HF Mean |
|---|---|---|---|---|
| gClostridium_sensu_stricto_1 | 0.002 | 0.002 | 9051 | 269457 |
| gTuricibacter_1 | 0.015 | 0.042 | 19969 | 100701 |
| gClostridium_sensu_stricto _6 | 0.015 | 0.059 | 106 | 3175 |
| gBifidobacterium; sAGR2158_1 | 0.015 | 0.060 | 105593 | 30976 |
| fPeptostreptococcaceae _4 | 0.017 | 0.095 | 308 | 1767 |
| gBifidobacterium; sAGR2158_2 | 0.017 | 0.118 | 244 | 0 |
| oBacteroidales_7 | 0.017 | 0.121 | 1365 | 0 |
| gAllobaculum_12 | 0.022 | 0.174 | 681 | 24 |
| fPeptostreptococcaceae_2 | 0.032 | 0.289 | 8545 | 52757 |
| fPeptostreptococcaceae_1 | 0.045 | 0.468 | 16617 | 88104 |
| gAllobaculum_12 | 0.045 | 0.500 | 845 | 208 |
| gClostridium_ sensu_stricto _3 | 0.045 | 0.538 | 139 | 2215 |
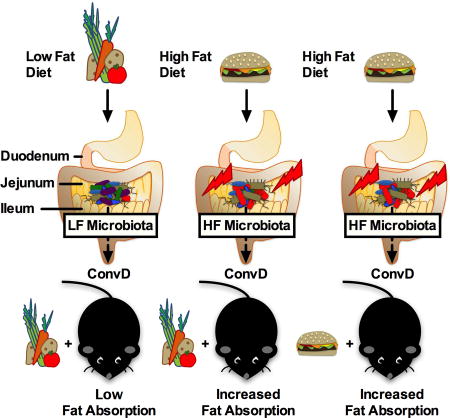
We next determined if conventionalization of GF animals with HF mucosa-associated jejunal microbiota increased lipid absorption compared to conventionalization with LF jejunal microbiota and if these changes required the selective pressure of the donor diet (Figure 4C). GF mice were conventionalized with HF diet jejunal microbiota and maintained on a HF diet (ConvD HF⇒HF), conventionalized with HF jejunal microbiota and maintained on a LF diet (ConvD HF⇒LF), or conventionalized with LF jejunal microbiota and maintained on a LF diet (ConvD LF⇒LF) for three weeks (Figure 4C). Prior to conventionalization, GF mice were acclimated to their respective diets for one week. HF jejunal microbes increased lipid absorption to the same degree, regardless of whether they were maintained of LF or HF diet. Both groups that received HF microbes exhibited increased lipid absorption as compared to mice receiving LF microbes (ConvD LF⇒LF) (Figure 4D). Notably, 16S rRNA gene abundance did not differ between groups at any time point following conventionalization from day 3 post conventionalization through day 21 or at the completion of the study (Figure S2D). We calculated transplantation efficiency by determining the percentage of donor oligotypes represented in recipient stool at day 3 (d3) and day 21 (d21). At d3, ~79% donor oligotypes were represented in the recipients and at d21, 50–60% donor oligotypes were represented in the recipients (Figure S3E). Despite this, microbial community structure remained similar between groups receiving HF microbes (i.e., ConvD HF⇒HF and ConvD HF⇒LF groups) until day 21, where community structure became more similar between ConvD HF⇒LF and ConvD LF⇒LF based on Bray Curtis beta diversity analyses as indicated in the PCoA plot shown in Figure S2C. Thus, even though HF⇒LF community structure was shifted toward a LF⇒LF community by day 21, the HF microbes influenced the level of lipid absorption in these mice, perhaps through differentially reprograming of the GF mouse small intestine. These results suggest that small bowel microbes from HF diet conditions increase lipid absorption compared to LF-derived microbes independent of post-conventionalization dietary pressure.
To directly compare the impact of jejunal vs cecal microbiota on lipid absorption, GF mice were colonized with microbiota from these regions under chow-fed conditions in both the donor and recipient mice. Conventionalization with chow-fed microbiota did not restore lipid absorption compared to GF mice from either region (Figure S2E). Interestingly, this lack of restoration was similar to findings with purified LF jejunal microbiota shown in Figure 4D. Thus, HF diet-induced microbiota may be necessary to elicit this host response. Future studies are needed to further interrogate this theory using HF jejunal microbiota vs HF cecal microbiota for conventionalization of GF mice.
A member of Clostridiaceae increases Dgat2 expression in vitro and in vivo
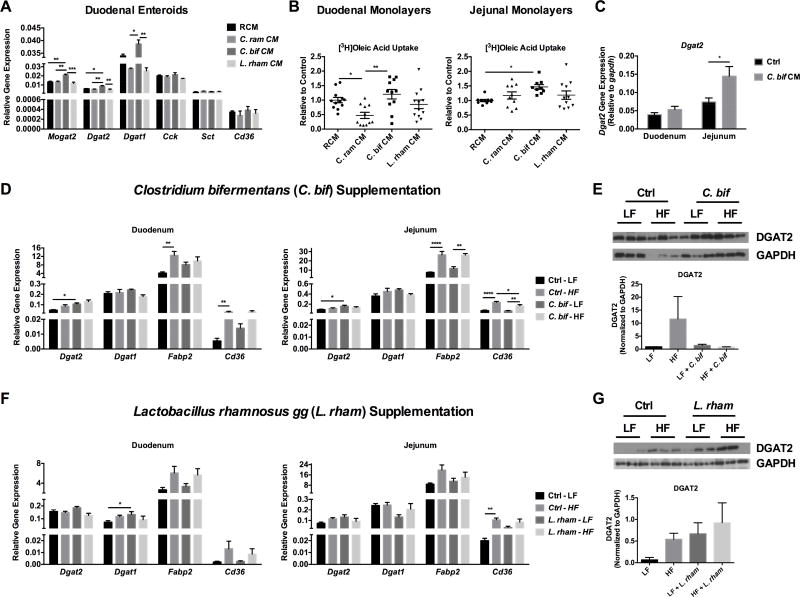
To establish proof-of-concept that specific strains of bacteria effect lipid absorption pathways, a reference strain belonging to Clostridiaceae, Clostridium bifermentans, was tested in an in vitro model of small intestinal organoid cultures. This microbe was selected based on its fermentative activity and identification in human sewage and feces. C. bifermentans is a spore-forming gram positive anaerobe that has been shown to produce a wide range of metabolites including acetate, lactate, carbon dioxide, and hydrogen that have potential in eliciting host responses (Wong et al., 2014). However, a role of C. bifermentans in regulating host metabolism or small intestinal lipid absorption has not been previously described. Therefore, this bacterium was grown in anaerobic conditions after which conditioned media (CM), presumably containing bioactive metabolites, was collected and used to treat duodenal and jejunal cultures of enteroids derived from mice (Sato et al., 2009). We also selected Clostridium ramosum due to previous findings that this bacteria was found to be associated with human obesity (Le Chatelier et al., 2013) and promoted diet-induced obesity in gnotobiotic mice (Woting et al., 2014). Lactobacillus rhamnosus gg CM was also selected as a comparison strain based on the finding from our previous experiment that this strain could impact Cckar expression in the pancreas indicating an effect related to digestion but not absorption (Figure 3G). After 24 hours of treatment, C. bifermentans CM selectively induced the expression of critical esterification enzymes (i.e, monacylglycerol O-acyltransferase, Mogat2, and diacylglycerol O-acyltransferases, Dgat1, Dgat2) involved in lipid transport but not Cck, Sct, or Cd36 (Figure 5A). In addition, C. bifermentans CM increased oleic acid uptake as compared to C. ramosum CM in duodenal monolayers or reinforced clostridial media (RCM) control in jejunal monolayers (Figure 5B). Consistent with these findings, we noted that CM from C. bifermentans increased Dgat2 mRNA levels in the jejunum of LF-fed mice in vivo (Figure 5C). Based on these results, we further examined C. bifermentans in a separate in vivo study. SPF mice were treated with an antibiotic cocktail for fourteen days followed by weekly gavage with C. bifermentans (1×109 CFUs) under either LF or HF conditions for four weeks. Mice fed a HF diet supplemented with C. bifermentans gained significantly more weight compared to mice fed a LF diet with C. bifermentans (Figure S4A), however supplementation of HF diet-fed mice with C. bifermentans does not further increase body weight significantly compared to HF diet alone (Figure S4A). C. bifermentans did not significantly increase body fat or plasma lipids compared to control groups (i.e., HF+C. bif vs HF or LF+C. bif vs LF) (Figure S4A–C). Supplementation with C. bifermentans significantly increased mRNA levels of Dgat2, but not Dgat1, in the duodenum and jejunum (Figure 5D) under LF conditions with a trend toward increased levels under HF conditions. No appreciable effects of C. bifermentans on DGAT2 protein levels were observed, however, this could be due to intra-group variation (Figure 5E). Gene expression of Fabp2 and Cd36 were significantly elevated by HF diet regardless of supplementation (Figure 5D). C. bifermentans did not significantly impact processes related to lipid digestion including no change in gallbladder weight, plasma CCK, or SCT levels (Figure S4E–G). No differences were detected across groups in 16S rRNA gene abundance in cecal contents at the end of the study (Figure S4D). However, taxonomic changes were apparent with C. bifermentans supplementation compared to controls, including an increase in the abundance of Clostridiaceae under HF conditions (Figure S4E). While it remains unclear if C. bifermentans elicits a direct increase in adiposity under HF conditions, both our in vitro and in vivo results suggest that this Clostridiaceae member increases oleic acid uptake and the expression of Dgat2 involved in TAG synthesis, possibly through C. bifermentans-derived bioactive components or molecules.
Figure 5. Clostridium bifermentans and Lactobacillus rhamnosus gg induces Dgat2 expression.
A) Gene expression of esterification enzymes (monoacylglycerol O-acyltransferase (Mogat2), and diacylglcerol O-acyltransferase (Dgat1 and Dgat2), cholescystokinin (Cck), secretin (Sct), and fatty acid translocase (Cd36) following conditioned media (CM) treatment from Clostridium bifermentans (C. bif; green), Clostridium ramosum (C. ram; red), Lactobacillus rhamnosus gg (L. rham; purple) or reinforced clostridial media (RCM; blue) control in duodenal organoids. B) Uptake of [3H]oleic acid in CM-treated cultures of duodenal and jejunal organoids. C–G) Antibiotic-treated SPF mice were maintained on either a LF or HF diet and gavaged weekly for four weeks with vehicle control or 1×109 CFUs Clostridium bifermentans (D–E), C. bifermentans CM (C), or L. rhamnosus gg (F–G). Expression of esterification enzymes (Dgat2 and Dgat1) and fat transport genes (fatty acid binding protein, Fabp2 and Cd36) were measured in the duodenum and jejunum via qRT-PCR in the C. bifermentans study (C) and the L. rhamnosus gg study (E). DGAT2 protein levels were measured in the C. bifermentans study (D) and the L. rhamnosus gg study (F). See also Figure S4 and S5. Data are shown as means +/− SEM (n=3 A; n=9–12 B; n=5 C–G). Data shown in panels A and B are representative of two independent experiments; * p ≤ 0.05.
To determine if other microbes had similar capabilities or worked through alternate mechanisms in vivo, we evaluated the effect of Lactobacillus rhamnosus gg supplementation using the same experimental protocol and examined changes in weight gain, adiposity, and markers of lipid absorption. Mice fed a HF diet supplemented with L. rhamnosus gg gained significantly more weight compared to mice fed a LF diet with L. rhamnosus gg (Figure S5A). Supplementation of HF diet with L. rhamnosus gg does not further increase body weight significantly compared to HF diet alone (Figure S5A). L. rhamnosus gg significantly increased epididymal and mesenteric fat under HF diet conditions compared to LF fed mice supplemented with L. rhamnosus gg (Figure S5B). Mice fed HF diet and supplemented with L. rhamnosus gg displayed increased LDL and cholesterol levels compared to mice fed LF diet and supplemented with L. rhamnosus gg (Figure S5C). L. rhamnosus gg induced Dgat1 under LF conditions compared to control in the duodenum and Dgat2 gene expression and protein levels in the jejunum, although not significantly (Figure 5F–G). Similar to C. bifermentans, L. rhamnosus gg did not have a significant impact on processes related to lipid digestion including gallbladder weight, plasma CCK, or SCT levels in this study (Figure S5E–G). No differences were detected across groups in 16S rRNA gene abundance in cecal contents at the end of the study (Figure S5D). Although a significant increase in L. rhamnosus gg itself was not detected, taxonomic changes were apparent with L. rhamnosus gg supplementation compared to controls (Figure S5E). Collectively, our findings show that specific microbial strains effect expression of genes involved in re-esterification of TG, such as Dgat1 and Dgat2 but the exact mechanisms are yet to be explored.
Discussion
Most studies examining the relationship between gut microbiota and host metabolism have primarily relied on fecal or colonic luminal samples. In contrast, few studies have considered the roles of small bowel microbiota, particularly for nutrient digestion and absorption, which largely occur in the small intestine. This study finds that small bowel microbiota associated with the intestinal mucosa are highly sensitive to dietary cues and play an important role in nutrient assimilation, particularly in the physiological and possibly pathophysiological regulation of host lipid digestive and absorptive pathways. Even though the small bowel microbiota are fewer in number and less diverse than their colonic microbiota counterpart, they are most proximate in timing and exposure to nutrient-rich dietary cues which may dramatically affect their composition and function relative to downstream microbiota and nearest to tissues most important for digestion and absorption (most affected by and affective to diet and metabolism). Indeed we show that HF diet feeding dramatically impacts the small bowel microbiota. Due to the highly unstable environment of the duodenum, with an influx of bile and pancreatic secretions, microbes could elicit a less influential role on host responses in this region. Downstream of this in the jejunum, host absorption is still highly active, yet the environment may allow for important host-microbe interactions. Therefore, we propose that along with dietary residuals, host-driven cues in this region may have a great impact on shaping the microbiota.
The influence of altered community membership in the small intestine on host outcomes has not been previously examined using microbiota transplant specific to the jejunum under HF vs LF conditions. We show that conventionalization of GF mice with HF diet-induced, mucosa-associated, jejunal microbiota increased radiolabeled lipid absorption compared to LF diet-induced, mucosa-associated, jejunal microbiota, independent of the diet that mice were consuming (Figure 4D). In other words, maintaining the GF mice on a LF diet was not protective against the lipid absorption promoting-effects of the HF microbiota. We speculate that microbial communities derived from HF conditions differentially program the GF small intestine, promoting increased lipid absorption upon lipid challenge, as compared to LF microbes. Given the decreased lipid absorption found in GF mice maintained on a high fat diet (Figure 2C), we conclude that microbes can play an essential role in regulating host lipid absorption. Therefore, changes in the small bowel microbiota could have significant consequences for the functional role of the small intestine in regulating host macronutrient digestion and absorption. This has important implications in malnutrition and obesity, as a specific restructuring of the small bowel microbiota may be required to either increase or decrease lipid transport, respectively.
Our findings are similar to previous reports demonstrating elevated TG in the stool of GF compared to SPF mice (Rabot et al., 2010) and reduced lymphatic transport of lipid in antibiotic-treated rats (Sato et al., 2016). We further demonstrate an active impairment of lipid absorption in GF mice using radiolabeled lipid absorption assays (Figure 2). We showed that impaired CCK signaling may be partly responsible for these effects. While Duca et al. (Duca et al., 2013) demonstrated decreased CCK protein levels in the proximal intestine, we found that GF mice have elevated circulating CCK levels. However, we found that the defect in CCK signaling may be due to reduced expression of the Ccka receptor (Cckar) in the pancreas as opposed to changes in the amount of CCK protein produced, a finding that has not been previously reported (Figure 3E). These differences might be explained by an overall improper development of the GF mouse. However, it was previously demonstrated that enteroendocrine cell number was not altered in the duodenum or jejunum of GF animals (Duca et al., 2013) and our results show that heat-killed bacteria directly regulate the pancreatic expression of Cckar (Figure 3F). In addition to altered digestive function, we demonstrate that GF animals have reduced brush border membrane transport of free oleic acid (Figure 3G). Taken together, these findings indicate that gut microbes regulate fat uptake locally in the gut and impact extra-intestinal organs such as the pancreas.
An alternative explanation for the resistance of GF mice to HF diet-induced obesity is increased fat oxidation in the gut and peripheral tissues such as liver and muscle. El Aidy et al. (El Aidy et al., 2013) demonstrated that fecal transplantation in GF mice resulted in significant changes in lipid metabolic pathways of peripheral tissues occurring as early as 1 day post transplantation. Genes involved in fatty acid oxidation were downregulated (i.e., Pparα) in the jejunum while genes involved in glycolysis were upregulated suggesting that conventionalization shifted energy utilization (El Aidy et al., 2013). We also show that GF mice display an upregulation of genes involved in fat oxidation, such as Cd36 and Pparα compared to SPF mice (Figure S1). Thus, increased oxidation of any absorbed fats in the intestine would prevent their incorporation into chylomicrons and delivery to the periphery. Earlier work by Bäckhed et al. (Bäckhed et al., 2007) showed that GF mice exhibit increased phosphorylated AMPK levels in the muscle and liver, as well as carnitine-palmitoyltransferase involved in fatty acid oxidation. Our results expand on this finding by showing that intestinal absorption as well as peripheral uptake of lipid in the liver and adipose tissue is reduced in GF animals (Figure S2C). Altogether, GF mice may be resistant to diet-induced obesity due to a combination of impaired fat digestion and absorption, disrupted circadian rhythms, as well as increased fat oxidation in the gut and other metabolically active tissues.
To examine a direct interaction between microbial cues and lipid absorption, we examined the capacity of the reference strains, C. bifermentans and L. rhamnosus gg, to impact lipid absorption in vitro and in vivo. Our in vitro studies suggest that different strains have specific actions and targets in duodenal organoid cultures. For example, soluble mediators or components from C. bifermentans have a selective impact on Dgat2 expression (Figure 5A). Notably, DGAT2 is an enzyme critical for lipid TAG synthesis and storage (Yen et al., 2008). Overexpression of DGAT2 increases the number and total area of cytoplasmic lipid droplets compared to wild type (WT) mice in the jejunum (Hung et al., 2017). DGAT2 deficiency leads to severely reduced plasma and liver TG levels (Stone, 2004; Yen et al., 2008) and DGAT2−/− mice only survive a few hours after birth, where carcasses have ~90% less TG than WT counterparts (Stone, 2004; Yen et al., 2008), demonstrating the importance of this enzyme for lipid homeostasis and life. Thus, control of Dgat2 expression by microbe-derived components or molecules may have significant implications for host physiology. The specific microbe-derived components or molecules driving host fat absorption also remain elusive. Future studies will focus on interrogating these bioactive bacterial components or molecules using classical digestion, filtration, and biochemical fractionation approaches. At this time, our study establishes proof-of-concept that microbes play a role in in dietary lipid processing facilitated, in part, through regulation of DGAT2. It should be noted, however, that this is only one of many collective mechanisms necessary for fully functional lipid digestion and absorption. The use of these candidate strains does not recapitulate the entire host response to the microbiota, where a complete restoration of lipid absorption likely involves the complex integration of diverse microbial taxa and microbial signals. Thus, the entire community of microbes and their corresponding gene function and metabolite production is likely required to influence other processes underlying the complexities of lipid digestion and transport. In addition, the interactive effects of diet, particularly those high in fat, or the direct effect of diet likely influences the degree of fat absorption. Notably, high fat diets directly impact host responses. For instance, long-chain fatty acids can mediate activation of nuclear hormone receptors such as PPARγ (Ferré, 2004), that lead to the upregulation of fat transport genes like Cd36. Further study is warranted to understand how different microbial taxa, alone or in concert, versus dietary cues differentially or additively impact host lipid digestion and absorption pathways.
Limitations to this study include bedding conditions between SPF and GF mice. In most cases SPF mice were maintained in corn cob bedding and GF mice were maintained in pine shavings. We addressed this caveat by demonstrating that GF mice displayed a significant reduction in lipid absorption compared to SPF mice when maintained in identical bedding conditions. Another limitation to the study was the use of antibiotics in the experiments conducted to address the impact of specific microbial strains on lipid absorption (Figure 5), as antibiotics may elicit direct effects on host pathways and may prove difficult to replicate in future studies. Notably, all mice in these experiments were treated with the same antibiotic regimen prior to placement on LF or HF diet and supplementation of the microbial strains, and thus the differences observed should reflect that of the dietary effects (LF vs HF) and the presence/absence of the bacterial strains used.
Overall, our study shows that 1) microbes are essential for proper lipid digestion and absorption in the small intestine mediated, in part, through systemic control of enteroendocrine signaling as well as a local impact on fatty acid transport in enterocytes, and 2) small bowel microbes are particularly sensitive to dietary pressures that promote increased macronutrient absorption (Summarized in Figure S6). Together, this work has important implications in developing approaches to combat obesity. Our findings would suggest that it is likely more important to develop interventions specifically targeting the small bowel microbiota, either through decreasing the abundance or activity of certain microbes that may otherwise promote fat absorption or through increasing the abundance of microbes that may inhibit fat uptake. Conversely, the same approaches could be used to promote more efficient nutrient digestion and absorption in conditions of intestinal failure (i.e., small bowel resection or scarring in Crohn’s disease) or environmental enteropathy. Continued efforts in understanding host-microbe interactions in the proximal small intestine is warranted for improved treatment options in either case of nutritional status.
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eugene Chang (echang@medicine.bsd.uchicago.edu).
Experimental Model and Subject Details
Animals
All murine experimental procedures were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC). C57BL/6 mice were bred and maintained under standard 12:12 h light/dark conditions at the University of Chicago. Age and litter-matched specific pathogen free (SPF) or germ free (GF) male C57Bl/6 mice between 8–12 weeks old were randomized into experimental groups. Mice were either fed a standard chow diet (Envigo 2018S; Leone et al. 2015) for the duration of the experiment (Figures 2B, 3), a purified low fat diet (Envigo TD.00102 customized diet; Table S1; Howe et al. 2016), or purified high saturated milk fat diet (Envigo TD.97222 customized diet; Table S1; Howe et al. 2016) for four weeks ad libitum. Diets were formulated by Envigo (Figures 1, 2C, 4, 5) and compositions shown in Table S1 and published previously (Howe et al., 2016; Leone et al., 2015). Gnotobiotic diets were gamma irradiated at a dose of 20–50kGy. and tested for sterility prior to use. SPF mice shown in Figure 4 and 5 were individually housed on standard cage racks in corn cob bedding. GF mice were housed in gnotobioitc isolators in standard conditions with pine shavings. All mice shown in Figure S2D–E including SPF, GF, and conventionalized mice were housed on pine shavings. Weekly body and food consumption weights were recorded. Mice were anesthetized using 10 mg/ml ketamine/xylazine (Figures 1, 4A–B), sevoflurane (Figures 3, 5), or isoflurane (Figures 2, 4D) followed by exsanguination and cervical dislocation. Plasma and collected tissues were snap-frozen and stored at −80°C. Beginning at the base of the stomach, the proximal 4 cm of the small intestine was collected as the duodenum. The next 2 cm were disposed, and the following 6 cm were collected as the jejunum. Ileum was harvested beginning at 8 cm proximal to the cecum where the most distal 2 cm were disposed. For the study shown in Figure 3A, GF and SPF mice were fasted for 4 hours, gavaged with corn oil (CO) or H2O control, and sacrificed 2 hours later. CO was used based on its diverse fatty acid profile consisting of ~25% saturated and monounsaturated fat and the remainder being comprised of polyunsaturated fat (Kostik et al., 2013). For the study in Figure 4C–D, a 3-week conventionalization period was chosen due to findings that the transcriptional program in the jejunum of colonized germ-free mice occurs after only 1 day (El Aidy et al., 2013) and based on our unpublished data showing that microbial communities begin to shift based on the recipient diet after 2 to 3 weeks post-conventionalization.
Bacteria
For the heat-killed bacterial gavage study shown in Figure 3F, Bacteroides thetaiotaomicron (ATCC®29148™) and Lactobacillus rhamnosus GG (ATCC®53103™) were grown in BHIS media (#14315, Sigma) and MRS media (Difco, Detroit, MI), respectively, at 37°C for 48 hours. Bacterial cultures at 50 mL were then centrifuged at 10,000 × g at 4°C for 10 min an d resuspended in 5 mL sterile PBS prior and heat-killed at 100°C for 15 minutes and k illing confirmed by streaking out a sample on Lactobacillus agar plates. Sterilized culture solutions were then mixed 1:1 for a total of 10 mL and germ-free mice were gavaged with 200 µL 3×/week for two weeks. Conventionalization (Figure 4D) was performed using jejunal intestinal mucosal scrapings that were pooled across mice from Figure 4A–B within diet group in an anaerobic chamber and were resuspended in 10 mL sterile PBS (20mg/mL). Mice were gavaged with 200 µl gavage solution per mouse and maintained in flexible film gnotobiotic isolators during the 3-week conventionalization period. Transplantation efficiency was calculated as the percentage of donor oligotypes present in the total taxa detected in recipients and has been reported in Figure S3. The transplantation efficiency was 78–79% for each group when comparing taxa present in recipients at d3 following conventionalization and ~50–60% when comparing taxa present in recipients at d21 following conventionalization. Similar to the methods described for Figure 4D, mice conventionalized in Figure S2E were gavaged with 200 µl jejunal or cecal slurry (20 mg/mL) prepared in sterile PBS. For the Clostridium bifermentans (ATCC®638™; Figure 5C–E) and Lactobacillus rhamnosus gg (ATCC®53103™; Figure 5F–G) supplementation studies, mice were acclimated to a purified LF diet for four days and mice were gavaged with 200 µl antibiotics containing metronidazole 1mg/ml, cefaperazone 1 mg/ml, neomycin 0.5 mg/ml, vancomycin 0.5 mg/ml per day for 14 days. Upon the last antibiotic treatment, cage bedding was changed, mice were placed on experimental LF or HF diets (Howe et al. 2016; Table S1), and two days later bedding was changed again, and the mice were gavaged with or without 1×109 CFUs C. bifermentans or L. rhamnosus gg weekly for four weeks.
For generation of conditioned media used for experiments shown in Figure 5, Clostridium bifermentans (ATCC®638™), Clostridium ramosum (ATCC®25582™), and Lactobacillus rhamnosus gg (ATCC®53103™) were purchased from ATCC and were streaked out and cultured for 2 days on RCM agar plates in an anaerobic chamber. A single colony was picked from each plate and used to inoculate 10 mL Reinforced Clostridium Media (RCM; Fisher Scientific) for 24 hours. One tube without bacteria was prepared and served as control. Bacteria were spun at 10,000 × g for 10 min and supernatant was filter sterilized using a 0.2 micron filter, aliquoted, and frozen at −80°C. Filtered RCM media was also collected for vehicle control treatments.
Primary Duodenal and Jejunal Organoid Cultures
Cultures of duodenal and jejunal organoids were prepared from 10–12 week old SPF C57Bl/6 male mice and cultured following the procedure by Sato et al. 2009 (Sato et al., 2009). Organoids were plated in 50µl matrigel beads in 24 well plates. Complete culture media is composed of advanced DMEM/F12 (Life Technologies), 1× Glutamax (Life Technologies), 1× HEPES buffer (Life Technologies), 1× Pen/Strep (Life Technologies), 1× N2 supplement (Life Technologies), 1× B-27 Supplement Minus Vitamin A (Life Technologies), murine epidermal growth factor (50 ng/ml; Peprotech), Noggin (100 ng/ml; Peprotech), R-spondin-1 (500 ng/ml; Peprotech), Jagged-1 (1 µM; Anaspec), Y-27632 (10 µM; Selleck Chemicals via Fisher) and media was refreshed every two days. Cultures were treated with 10% bacterial conditioned media (preparation described below) for 24 hours. Upon harvest, organoids were released from matrigel via gentle pipetting, added to a microcentrifuge tube, and spun at 150 × g at 4°C. Supernatant was removed and Trizol added for RNA extraction, cDNA synthesis, and qPCR analyses.
Method Details
Plasma Insulin Resistance Markers and Gut Peptide Hormones
Mice were injected I.P. with 10 mg/ml ketamine/xylazine. Blood was collected from the portal vein using an insulin syringe and collected into ETDA-coated microfuge tubes. Blood was centrifuged at 5000 rpm for 10 minutes at 4°C and plasma supernatant was stored at −80°C. Insulin resistance markers and gut peptide hormones were measured using Bio-Rad’s Bio-Plex Pro™ Mouse Diabetes Panel (Hercules, CA) following manufacturer’s instructions.
Portal Plasma Lipids
Plasma triglyceride, low density lipoprotein, and non-esterified free fatty acids were measured using commercially available colorimetric kits from Wako Chemicals (Richmond, Va) following manufacturer’s microplate protocols.
Fasting Plasma Glucose
Mice were fasted for four hours and blood collected via cheek bleed. Blood was centrifuged at 10000 × g for 10 minutes. Serum glucose was measured using an Accu-Chek®glucometer (Roche, Indianapolis, IN).
Stool Lipids and Total Bile Acids
Lipids from 50–100 mg stool were extracted using the Folch method of lipid extraction (Folch et al. 1957). Triglyceride and cholesterol levels were measured from lipid extract using Wako Chemicals (Richmond, VA) colorimetric assays following manufacturer’s microplate instructions. Total bile acids were extracted from 10 mg dried stool using a 1:1 ratio of tert:butanol to water and measured using Diazyme Total Bile Acids Assay from Diazyme Laboratories (Poway, CA).
In Vivo Radiolabeled Lipid Absorption Assay
SPF and GF mice were injected retro-orbitally with or without 10% tyloxapol (Sigma Aldrich) prepared in PBS for 10 minute and subsequently gavaged with a single dose of 200 µl corn oil containing 2 µCi [3H]triolein (Perkin Elmer) and 2 µCi [3H]cholesterol (Perkin Elmer) for a total of seven hours as previously described (Sontag et al. 2013). Mice were anesthetized with isoflurane and blood was collected retro-orbitally after 1, 3, 5, and 7 hours. The amount of radioisotope in plasma was measured via scintillation counting and expressed as dpms/µl.
Gene Expression Analyses
Gene expression analyses were performed as previously described (Leone et al., 2015). Briefly, RNA was extracted from 25 mg tissue and homogenized in Trizol. Reverse transcription was performed using Roche Transcriptor Reverse Transcriptase Reagents (Indianapolis, IN). Real time quantitative PCR was performed using iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA) and a Roche Lightcycler 480 system (Roche Applied Science, Indianapolis, IN). Forward and reverse primer sequences are available in Table S2. Gene expression levels were determined using the comparative CT method normalizing target mRNA to GAPDH endogenous control. All samples were checked for quality and had a A260/A280 ratio of 1.8–2.0. All primer sequences were blasted to confirm matches to intended gene targets. PCR efficiencies were calculated and shown in Table S3.
Protein Expression
Western blots were performed following previously described methods (Laemmli et al. 1970). Briefly, protein concentration of samples was determined using a BCA assay with BSA as a standard. Protein samples were separated on 10% acrylamide gels with 30 µg protein per lane and blotted onto nitrocellulose membranes. Membranes were blocked in a blocking buffer consisting of 5% milk and 1% BSA in TBST. Membranes were probed with the following primary and secondary antibodies in blocking buffer: anti-CCK-AR (N-20) (Santa Cruz, SC-16172, goat polyclonal IgG) at 1:200 and with swine anti-goat secondary antibody at 1:15,000 (Southern Biotech, 6300-05), anti-GAPDH MAb, (Ambion, AM4300) at 1:100,000 followed by anti-mouse secondary antibody at 1:50,000 (Cell Signaling Technology, 7076S), and lastly anti-DGAT2 (4C1) (Santa Cruz, SC-293211, mouse monoclonal IgG) at 1:500 followed by anti-mouse secondary antibody probed at 1:2,500 (Cell Signaling Technology, 7076S). Densitometry was performed using Image Studio Lite by LI-COR Biosciences (https://www.licor.com/bio/products/software/image_studio_lite/).
Measurement of Lipase Activity
Mucosal scrapings containing luminal contents were collected from the duodenum and jejunum. The region of the duodenum collected spanned from the base of the stomach to 4 cm distal of the stomach. The jejunum spanned a region 6 cm distal from the stomach and 12 cm distal from the stomach (total 6 cm collected). Lipase activity was measured using a commercially available kit from Sigma-Aldrich (St. Louis, MO) based on a coupled enzyme reaction using glycerol as a standard (Cat# MAK046) following manufacturer’s instructions. Endogenous glycerol in the sample is accounted for by running a sample blank without lipase substrate and subtracting that value from the total. The colorimetric product is proportional to the amount of lipase activity expressed as milliunits/mL, where one unit generates one µM glycerol.
Cholecystokinin and Secretin Levels
Circulating CCK and SCT concentrations were measured using EIA kits from RayBiotech, Inc (Cat# EIAM-CCK-1; Norcross, GA) and Phoenix Pharmaceuticals, Inc (Cat# EK-067-04; Burlingame, CA), respectively, following manufacturer’s instructions.
Oleic Acid and Glucose Uptake in Brush Border Membranes
Brush border membrane (BBM) vesicles were prepared from SPF and GF jejunal and ileal tissue as previously described (Prieto et al., 1996). First, fifteen centimeters of jejunum or ileum were taken, washed with ice cold saline and mucosal scrapings removed by glass slides. Scrapings were placed in 20 mls homogenization buffer (10 mM Tris pH 7.4, 2 mM EDTA, 1 mM PMSF with Roche Complete Protease Inhibitor) and homogenized at medium speed using a Polytron. Nuclei were removed by centrifugation at 500 g for 5 min at 4°C, mitochondria removed by centrifuging the supernatant at 10,000 × g for 10 min at 4°C and supernatant remove d. MgSO4 was added to 10 mM and allowed to sit on ice for 30 min, mixing every 10 min. Basolateral, Golgi, and ER membranes were removed by centrifugation at 10,000 × g for 10 min at 4°C and the supernatant was centrifuged at 45,000 × g for 30 min at 4°C to obtain brush border membrane vesicles. These were resuspended in 300 mM mannitol, 0.1 mM MgSO4, 20 mM HEPES-Tris pH 7.4 and homogenized first in a Teflon pestle homogenizer followed by passing through a 25- gauge needle.
Uptakes were performed by adding vesicles (from 40–90 µg protein in 20µl volume for oleic acid uptake or 10 µg in 20 µl for glucose uptake) to specific uptake buffer (100 mM NaSCN, 100 mM mannitol, 0.1 mM MgSO4, 20mM HEPES-Tris pH 7.4) with added substrate. For oleic acid uptake, BBM were incubated with 1.5 nmoles [3H]oleic acid (Perkin Elmer, Billerica, MA) for 10 minutes (specific activity 6902.67 cpms/nmol) delivered in 150 µM/L non-radioactive oleate complexed to 400 µM bovine serum albumin (BSA). Samples were run in triplicate. For glucose uptake, BBM were incubated with 1.5 nM or 5µCi/ml [3H]glucose for 10 seconds (specific activity 1666.67 cpms/nmol) delivered in 150 µM glucose solution in sodium-containing buffer. Samples were run in duplicate with or without 0.2 mM phorizin (to determine uptake specific to sodium-dependent glucose transporter, SGLT1, activity), washed, and collected for scintillation counting. Incubation times for radiolabeled compounds were determined by conducting preliminary time courses to identify the linear range of uptake. Uptakes were terminated by dilution to 1 ml with ice-cold saline with 0.1% albumin, filtration on HAWP 0.45 μ filters on a fritted glass support (both Millipore, Medford, MA) and washed with ice-cold saline containing 0.1% albumin three times rapidly with 3 ml each. Filters were removed, placed in vials, and counted by liquid scintillation spectroscopy. Data are expressed as nmol/mg protein.
[3H]Oleic Acid Uptake in Enteroid Monolayers
Duodenal and jejunal monolayers were grown as previously described (In et al., 2016). Transwells (24-well inserts, 0.33 cm2 surface area, 0.4 µM pore polyester membrane, Sigma Aldrich) were coated with human collagen type IV solution (final concentration of 10µg/cm2) and incubated at 37°C for a minimum of 2 hours. C oated transwells were washed 3 times with base media (DMEM/F12, Hepes, P/S, glutamine). Enteroids were collected and washed as explained previously, and mechanically disrupted by pipetting at least 30 times. Next, 50 µL of the disrupted enteroid mixture was seeded onto the transwell and incubated for 1 hour at 37°C. Base me dia (DMEM/F12, Hepes, P/S, glutamine) including growth factors was added onto and beneath the transwell. Media was replaced every two days. Enteroid monolayers were cultured until 90–100% confluency was reached and were treated for 24 hours with 10% CM of RCM, C. bifermentans, C. ramosum and L. rhamnosus gg. Time course experiments with [3H]oleic acid were performed, after which a 10 minute treatment time was chosen for further experiments. Monolayers were treated with 0.05 µCi/µL [3H]oleic acid (Perkin Elmer). Hot media was removed and washed 3 times with ice-cold termination buffer. Termination buffer was removed completely after the third wash and 0.5 mL lysis buffer (0.1% SDS in PBS) was added. Each well was scraped with a pipette tip and the solution was transferred into scintillation vials. Additionally, 250 µL of lysis buffer was added to each well to ensure no remaining cells were on the transwell. Radioactivity was quantified by liquid scintillation counting.
DNA Extraction and 16S rRNA Sequencing
Mucosal scrapings from the small intestine and cecal contents were collected at time of sacrifice and immediately snap frozen in liquid nitrogen and stored at −80 C. DNA was extracted using previously published protocols (Wang et al., 2009). To assess bacterial community structure, primers specific for 16S rRNA V4–V5 region (Forward: 338F: 5’-GTGCCAGCMGCCGCGGTAA-3’ and Reverse: 806R: 5’-GGACTACHVGGGTWTCTAAT-3’) that contained Illumina 3' adapter sequences as well as a 12-bp barcode were used. Sequences were generated by an Illumina MiSeq DNA platform at Argonne National Laboratory. The raw sequencing data were de-multiplexed, and partially overlapping paired-end reads were merged using the Illumina Utilities library (available at https://github.com/merenlab/illumina-utils; Eren et al., 2013a; Eren et al. 2015). Mismatches at the overlapping regions of pairs were resolved using the base with the higher Q-score, and the merged sequences were kept for downstream analyses only if they contained three or less mismatches at the overlapping region, and if 66% of the bases in the first half of each read had an average Q-score of 30. All merged reads with >0 mismatches were filtered. The quality filtered reads were partitioned into ecologically relevant units using Minimum Entropy Decomposition (MED) (Eren et al., 2015a) with default parameters. MED resolves a given amplicon dataset iteratively into high-resolution oligotypes using Shannon entropy (Eren et al., 2013b). GAST was used to assign taxonomy to oligotypes (Huse et al., 2008). Oligotype community data were normalized and taxonomic relative abundances in LF and HF groups were compared within each region. Differences across intestinal regions were compared within each diet. QIIME software was utilized to determine significant changes in relative oligotype abundance were assessed using parametric t-tests (FDR correction p ≤ 0.05). ADONIS and ANOSIM tests were performed to detect significance between LF and HF based on Bray Curtis and Canberra metrics (Caporaso et al., 2010). Data were imported into QIIME to generate PCoA plots on a forced axis for intestinal region. Heat maps were generated using Anvi’o (Eren et al., 2015b).
16S rRNA Gene Quantification
16S rRNA gene copy number was determined from small intestinal mucosal scrapings, luminal cecal contents, or stool as previously described (Louis et al., 2010; Vital et al., 2013). Genes were quantified by determining a standard curve for gene copy number by cloning primer sequences into pCR4-TOPO plasmids. Forward (F) and reverse (R) primer sequences for 16S rRNA: F-TCCTACGGGAGGCAGCAGT; RGGACTACCAGGGTATCTAATCCTGTT.
Quantification and Statistical Analyses
Statistical analyses were performed using GraphPad Prism v6.0 using two-way ANOVA with Tukey’s multiple comparisons test (Figure 1, Figure S1, Figure 2C, Figure S2C–E, Figure S3B,D–E, Figure 3A–E, Figure S3, Figure 4D, Figure S4, Figure S5), two-way ANOVA with Sidak’s multiple comparisons test (Figure 2B,D–G, Figure 5C–F), unpaired t test (Figure 3F–H, Figure S2A), one-way ANOVA with Tukey’s multiple comparisons test (Figure 5A–B). Data are presented as means +/− SEM. Differences between group means were considered significant at p < 0.05. Sample sizes can be found in the figure legends, where n represents the number of animals or number of cell culture wells used in the experiment. Outliers were removed based on ROUT method of outlier detection.
Data and Software Availability
The 16S rRNA sequencing files have been deposited into the NCBI database at (https://www.ncbi.nlm.nih.gov/sra/SRP128454).
Supplementary Material
Highlights.
Small bowel microbiota regulate host dietary fat digestion and absorption.
Gut microbes and their mediators drive lipid absorption through multiple mechanisms.
Specific bacterial strains influence processes underlying intestinal lipid absorption.
High fat diet-induced jejunal microbiota directly increase gut lipid absorption.
Acknowledgments
We thank Cierra Howard, Uzoamaka Gloria Aniagba, Kali Deans, and Dafna Meyers for their technical assistance, and we would like to thank Mrinalini Rao for her critical review of the manuscript. We extend our gratitude to the UChicago Gnotobiotic Research Animal Facility animal care staff for germ-free work, Argonne National Laboratory for sequencing services, and to our administrator Fran Vukovich for her efforts in coordinating various logistical aspects of this work. This work was supported by the NIH NIDDK under grants DK42086 for the Digestive Diseases Research Center Core (DDRCC), DK097268, and T32DK007074.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, K.M., C.R., M.M., V.L., N.H., E.B.C.; Methodology, K.M., N.H., T.S.; Data Curation, K.M., N.H., Investigation K.M., K.F., J.P., C.C., S.U., P.O., J.M., T.S., V.L.; Supervision, M.M., V.L., E.B.C.; Writing - Original Draft, K.M., Writing - Review and Editing, K.M., N.H., J.P. M.M, V.L., E.B.C. Funding Acquisition, E.B.C.; Resources, E.B.C.
Declaration of Interests
The authors declare no competing interests.
We have no conflicts of interest to disclose.
References
- El Aidy S, Merrifield CA, Derrien M, van Baarlen P, Hooiveld G, Levenez F, Doré J, Dekker J, Holmes E, Claus SP, et al. The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut. 2013;62:1306–1314. doi: 10.1136/gutjnl-2011-301955. [DOI] [PubMed] [Google Scholar]
- Angelakis E, Armougom F, Carriere F, Bachar D, Laugier R, Lagier JC, Robert C, Michelle C, Henrissat B, Raoult D. A metagenomic investigation of the duodenal microbiota reveals links with obesity. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0137784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drover VA, Ajmal M, Nassir F, Davidson NO, Nauli AM, Sahoo D, Tso P, Abumrad NA. CD36 deficiency impairs intestinal lipid secretion and clearance of chylomicrons from the blood. J. Clin. Invest. 2005;115:1290–1297. doi: 10.1172/JCI21514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca FA, Sakar Y, Covasa M. The modulatory role of high fat feeding on gastrointestinal signals in obesity. J. Nutr. Biochem. 2013;24:1663–1677. doi: 10.1016/j.jnutbio.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Eren AM, Vineis JH, Morrison HG, Sogin ML. A Filtering Method to Generate High Quality Short Reads Using Illumina Paired-End Technology. PLoS One. 2013a;8:6–11. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 2013b;4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. Isme J. 2015a;9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015b;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré P. The Biology of Peroxisome Proliferator-Activated Receptors. Diabetes. 2004;53:43–50. doi: 10.2337/diabetes.53.2007.s43. [DOI] [PubMed] [Google Scholar]
- Ge H, Li X, Weiszmann J, Wang P, Baribault H, Chen J-L, Tian H, Li Y. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008;149:4519–4526. doi: 10.1210/en.2008-0059. [DOI] [PubMed] [Google Scholar]
- Howe A, Ringus DL, Williams RJ, Choo Z-N, Greenwald SM, Owens SM, Coleman ML, Meyer F, Chang EB. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J. 2016;10:1217–1227. doi: 10.1038/ismej.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, Chang EB. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. JPEN. J. Parenter. Enteral Nutr. 2013;37:746–754. doi: 10.1177/0148607113486931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DY, Howles PN. Molecular mechanisms of cholesterol absorption and transport in the intestine. Semin. Cell Dev. Biol. 2005;16:183–192. doi: 10.1016/j.semcdb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Hung Y-H, Carreiro AL, Buhman KK. Dgat1 and Dgat2 regulate enterocyte triacylglycerol distribution and alter proteins associated with cytoplasmic lipid droplets in response to dietary fat. 2017 doi: 10.1016/j.bbalip.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring Microbial Diversity and Taxonomy Using SSU rRNA Hypervariable Tag Sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In J, Foulke-Abel J, Zachos NC, Hansen AM, Kaper JB, Bernstein HD, Halushka M, Blutt S, Estes MK, Donowitz M, et al. Enterohemorrhagic Escherichia coli Reduces Mucus and Intermicrovillar Bridges in Human Stem Cell-Derived Colonoids. C. Cell. Mol. Gastroenterol. Hepatol. 2016;2:48–62.e3. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Hussain MM. Intestinal lipid absorption. Am. J. Physiol. Metab. 2009;296:E1183–E1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostik V, Memeti S, Bauer B. Fatty acid composition of edible oils and fats. J. Hyg. Eng. Des. 2013:112–116. [Google Scholar]
- LAEMMLI UK. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoAtransferase gene. Environ. Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- Pannacciulli N, Bunt JC, Koska J, Bogardus C, Krakoff J. Higher fasting plasma concentrations of glucagon-like peptide 1 are associated with higher resting energy expenditure and fat oxidation rates in humans. Am. J. Clin. Nutr. 2006;84:556– 560. doi: 10.1093/ajcn/84.3.556. [DOI] [PubMed] [Google Scholar]
- Pinkney J. The role of ghrelin in metabolic regulation. Curr. Opin. Clin. Nutr. Metab. Care. 2014;17:497–502. doi: 10.1097/MCO.0000000000000101. [DOI] [PubMed] [Google Scholar]
- Prieto RM, Stremmel W, Sales C, Tur JA. Oleic acid uptake by jejunal and ileal rat brush border membrane vesicles. Eur. J. Med. Res. 1996;1:199–203. [PubMed] [Google Scholar]
- Rabot S, Membrez M, Bruneau A, Gerard P, Harach T, Moser M, Raymond F, Mansourian R, Chou CJ. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. Faseb J. 2010;24:4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- Sato H, Zhang LS, Martinez K, Chang EB, Yang Q, Wang F, Howles PN, Hokari R, Miura S, Tso P. Antibiotics Suppress Activation of Intestinal Mucosal Mast Cells and Reduce Dietary Lipid Absorption in Sprague-Dawley Rats. Gastroenterology. 2016;151:923–932. doi: 10.1053/j.gastro.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Semova I, Carten JD, Stombaugh J, MacKey LC, Knight R, Farber SA, Rawls JF. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag TJ, Chellan B, Getz GS, Reardon Ca. Differing rates of cholesterol absorption among inbred mouse strains yield differing levels of HDL-cholesterol. J. Lipid Res. 2013;54:2515–2524. doi: 10.1194/jlr.M040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SJ. Lipopenia and Skin Barrier Abnormalities in DGAT2-deficient Mice. J. Biol. Chem. 2004;279:11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Penton CR, Wang Q, Young VB, Antonopoulos Da, Sogin ML, Morrison HG, Raffals L, Chang EB, Huffnagle GB, et al. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome. 2013;1:8. doi: 10.1186/2049-2618-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YM, Juan JC, Gan HM, Austin CM. Draft Genome Sequence of Clostridium perfringens Strain JJC, a Highly Efficient Hydrogen Producer Isolated from Landfill Leachate Sludge. Genome Announc. 2014;2:1–2. doi: 10.1128/genomeA.00064-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woting A, Pfeiffer N, Loh G, Klaus S, Blaut M. Clostridium ramosum promotes High-Fat diet-induced obesity in Gnotobiotic Mouse Models. MBio. 2014;5:1–10. doi: 10.1128/mBio.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Huang X, Fang S, Xin W, Huang L, Chen C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Nat. Publ. Gr. 2016:1–11. doi: 10.1038/srep27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C-LE, Stone SJ, Koliwad S, Harris C, Farese RV. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C-LE, Nelson DW, Yen M-I. Intestinal triacylglycerol synthesis in fat absorption and systemic energy metabolism. J, Lipid Res. 2015;56:489–501. doi: 10.1194/jlr.R052902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.