Abstract
Using several HIV-1 tat exon 1 amino acid sequences available from public databases and additional sequences derived from a southern Indian clinical cohort, we compared the profile of the signature amino acid residues (SAR) between two different time periods, 1986–2004 and 2005–2014. The analysis identified eight positions as signature residues in subtype C Tat and demonstrated a changing pattern at four of these positions between the two periods. At three locations (histidine 29, serine 57, and proline 60), there appears to be a nonuniform negative selection against the SAR. The negative selection appears to be severe, especially against histidine 29 (p < .0001) and moderate against proline 60 (p < .0001). The negative selection against serine 57 is statistically insignificant and appears to have begun recently. At position 63, the frequency of signature residue glutamic acid increased over the past decade, although the difference was not significant. Importantly, at the three locations where the negative selection is in progress, the substitute amino acids are the generic residues present in most of the other HIV-1 subtypes. Our data demonstrate that viral evolution can subject specific amino acid residues to subtle and progressive selection pressures without affecting the prevalence of other amino acid residues.
Introduction
Among the various genetic subtypes of HIV-1, subtype C by itself is responsible for half of all global infections.1 HIV-1 subtypes differ from one another in terms of genetic identity, geographical distribution, coreceptor usage, and the potential for pathogenesis, replication, and transmission.2,3 It is increasingly appreciated that the subtype-unique genetic variations have a significant impact on the viral biological properties that in turn may influence the differences in the rates of prevalence of viral subtypes.
Signature sites are particular sites in amino acid or nucleic acid alignments of variable sequences that are distinctly representative of a query set of sequences relative to a background set.4 In other words, signature residues are amino acids or nucleotides that are unique to one or a small number of viral genetic subtypes or present in a majority of the viral isolates of a genetic subtype, in either case, absent from other genetic subtypes or present in a small minority of the other genetic subtypes. The signature amino acid residues (SAR) identified in various proteins of HIV-1, including Tat, are thought to contribute to biological differences among the subtypes.5–9 For the SAR profile analysis, we used HIV-1 Tat as a model viral factor given its importance for viral infectivity and pathogenesis and the moderate magnitude of genetic diversity among the diverse HIV genetic subtypes.10
Based on the structural and functional studies, Tat exon 1 can be divided into five domains.11 In addition to the transactivation of the viral promoter, Tat can regulate several other functions, significantly influencing viral proliferation and replication competence, as well as modulating many cellular functions. Experimental evidence over the past years from many laboratories elucidated in much detail the various biological functions regulated by different functional domains of Tat. The N-terminal domain 1 (aa 1–20) is rich in proline and acidic amino acid residues. The cysteine-rich domain (CRD, 21–40) of Tat plays a critical role in protein dimerization and recruiting P-TEFb, a complex consisting of Cyclin T1 and CDK 9, to the RNA pol complex thereby enhancing transactivation at the level of elongation.12–14 The core domain (CD, 41–48) is rich in hydrophobic residues. The Tat regions, CRD and CD, collectively can interact with tubulin binding proteins leading to apoptosis.15 The first three domains of Tat (aa 1–48) comprise the minimal region for the transactivation potential of the viral protein. The basic domain (BD, 49–59) is necessary for the cellular uptake of Tat, nuclear localization of Tat, and TAR binding.11,16 In addition, the BD is also known to be essential for the recruitment of the p300/CBP transcriptional coactivator.17–19 The C-terminal region (aa 73–101) appears to be mostly dispensable for transactivation. The exon 2 of HIV-1 Tat is majorly dispensable for many known biological functions of Tat. Although a highly conserved RGD (arg-gly-asp) motif is present in exon 2 of many HIV-1 subtypes, the involvement of the motif in binding cellular integrins to influence angiogenesis is controversial.20,21 Of note, in subtype C, the RGD motif is missing in a majority of the viral strains by natural variation. The numerous modulatory effects Tat exerts on the cellular functions and the immune responses have been elegantly reviewed.2,22
In this study, we intend to examine the HIV-1 Tat SAR, where subtype C (query) differs from most of the other subtypes of the HIV-1 M group (background). The Tat protein of HIV-1 is a small polypeptide of 101 amino acid residues encoded by two exons. Five conserved functional domains have been identified in exon 1 of Tat of 72 amino acids.23 The amino acid or RNA sequences of tat can be used to classify the diverse viral subtypes into distinct phylogenetic groups, and such a classification is consistent with a similar analysis performed using pol or env sequences of HIV-1.24 Distinct SAR patterns emerge within Tat subtypes that appear to be broadly nonoverlapping between subtypes. These genetic differences between Tat subtypes can manifest as important differences in biological functions, including Tat's potential for transactivation, cytokine induction, receptor binding, and modulation in the levels of receptor expression.10 To what extent the SAR in a particular Tat sequence can modulate the biological functions of the viral protein is not understood.
We previously demonstrated the presence of seven SAR in Tat of subtype C.25 A detailed examination of one of these residues, serine 31, revealed the loss of an important biological function with a potential impact on clinical manifestation. The C31S variation, seen in more than 90% of subtype C HIV-1 isolates, disrupts the CC dicysteine motif at positions 30 and 31. As a consequence of the variation, subtype C Tat lost its capacity to drive monocyte chemotaxis.25 We hypothesized that the compromised monocyte chemokine function of subtype C perhaps could underlie the reportedly low prevalence of HIV-1-associated dementia (HAD) in India.26,27 Subtype-specific biological differences in Tat have since been reported by many groups, including our own.28–31 The significance of the dicysteine motif for Tat function was subsequently bolstered by a study where a recombinant Tat protein containing the dicysteine motif (CC-Tat), but not the protein lacking the motif (CS-Tat), was cytotoxic to fetal astrocytes and neurons derived from human neuronal progenitor cells.32 A Tat peptide of subtype B containing the dicysteine motif (CysL24–51) has been implicated in direct monocyte chemotaxis in vitro.33 Importantly, Tat of subtype B, but not subtype C, induced apoptosis in neurons by binding to the N-methyl-d-aspartic acid (NMDA) receptor.34 Furthermore, unique biological functions have been ascribed to two other SAR residues in subtype C Tat, leucine 35, and glutamine 39.35 While incompletely studied, the available experimental evidence collectively alludes to the critical role that the SAR play in regulating subtype-specific biological properties of the Tat protein.
Given the high magnitude of genetic diversity, rapid turnover rate, the error-prone nature of HIV-1 reverse transcriptase, and its short generation time, HIV-1 has a great potential for generating a huge genetic variation. Within this backdrop, it is not known if the SAR in Tat are stable over time or whether they undergo a significant magnitude of variation themselves within the context of ongoing genetic sampling by the virus. We undertook this analysis to examine the stability of the signature amino acid profile in subtype C Tat by comparing the global tat exon 1 sequences available in the databases and additionally examining the tat sequences from a southern Indian cohort. We grouped the sequences into two different time frames 1986–2004 and 2005–2014 to match with our previous analysis.25 We demonstrate that specific signature locations can undergo variations, while others remain stable. The negative selection pressure can be quite severe against the presence of certain amino acid residues, such as a histidine at position 29 in subtype C Tat.
Materials and Methods
Clinical samples
The clinical samples for the study were collected from 76 subjects attending the clinic at Y. R. Gaitonde Centre for AIDS Research and Education (YRGCARE), Chennai between September 2011 and November 2013. The study participants consisted of only adult subjects over 18 years of age. The subjects were all drug naive and believed to have acquired the infection primarily through heterosexual transmission. We used the following criteria for the inclusion of the subjects in the study: at the time of enrolment, the subjects should be free of AIDS-related symptoms, their CD4 count should be above 500, and the subjects should be seropositive for at least 5–8 years since diagnosis. As the date of infections of most of the subjects is not known in the Indian clinics, we do not have information regarding the duration of the seropositive status of the subjects. Furthermore, in a cross-sectional study design, we collected a single blood sample from a set of 41 subjects nonoverlapping with the other subjects of this study. The inclusion criteria for this subset of subjects remained the same as described above. Six of the 41 subjects, however, contained a CD4 count between 350 and 500 cells and only one subject below 350 cells (INDO-SA-NLR2061, 338 CD4 cells). The clinical profile of all the study subjects is summarized (Table 1).
Table 1.
The Clinical Profile of the Study Subjects
| S. no. | Subject-ID | Accession no. | Date of visit | Age | Gender | CD4 count (cells/μl of plasma) | Plasma viral load (copies/ml of plasma) |
|---|---|---|---|---|---|---|---|
| 1 | INDO-SA-NLR2001 | KM496583-84 | 26-Sep-11 | 23 | F | 608 | 25,149 |
| 2 | INDO-SA-NLR2002 | KM496585-86 | 26-Sep-11 | 32 | F | 589 | 26,186 |
| 3 | INDO-SA-NLR2004 | KM496587-88 | 26-Sep-11 | 43 | F | 1,012 | 6,070 |
| 4 | INDO-SA-NLR2005 | KM496589-91 | 26-Sep-11 | 35 | F | 599 | 5,713 |
| 5 | INDO-SA-NLR2006 | KM496592 | 26-Sep-11 | 46 | F | 659 | 662 |
| 6 | INDO-SA-NLR2007 | KM496593-95 | 26-Sep-11 | 40 | M | 546 | 11,986 |
| 7 | INDO-SA-NLR2008 | KM496596 | 26-Sep-11 | 46 | M | 1,289 | 3,687 |
| 8 | INDO-SA-NLR2010 | KM496597-99 | 26-Sep-11 | 32 | F | 989 | 251 |
| 9 | INDO-SA-NLR2011 | KM496600 | 26-Sep-11 | 28 | F | 582 | 2,704 |
| 10 | INDO-SA-NLR2012 | KM496602 | 28-Sep-11 | 28 | F | 1,087 | 6,070 |
| 11 | INDO-SA-NLR2014 | KM496603-04 | 28-Sep-11 | 55 | M | 766 | 88,645 |
| 12 | INDO-SA-NLR2015 | KM496605 | 28-Sep-11 | 34 | F | 1,019 | 21,108 |
| 13 | INDO-SA-NLR2016 | KM496607-08 | 28-Sep-11 | 29 | F | 631 | 26,363 |
| 14 | INDO-SA-NLR2017 | KM496609-10 | 28-Sep-11 | 39 | M | 581 | 8,972 |
| 15 | INDO-SA-NLR2018 | KM496611-12 | 28-Sep-11 | 35 | F | 825 | 6,493 |
| 16 | INDO-SA-NLR2019 | KM496613 | 28-Sep-11 | 35 | F | 1,085 | 185 |
| 17 | INDO-SA-NLR2020 | KM496614-16 | 28-Sep-11 | 38 | M | 570 | 70,025 |
| 18 | INDO-SA-NLR2022 | KM496617 | 28-Sep-11 | 38 | M | 1,489 | <150 |
| 19 | INDO-SA-NLR2024 | KM496618 | 14-Oct-11 | 30 | F | 984 | 340 |
| 20 | INDO-SA-NLR2025 | KM496619-20 | 14-Oct-11 | 30 | M | 500 | 3,423 |
| 21 | INDO-SA-NLR2026 | KM496621 | 14-Oct-11 | 18 | M | 1,015 | 8,733 |
| 22 | INDO-SA-NLR2028 | KM496622-23 | 14-Oct-11 | 34 | F | 711 | 24,480 |
| 23 | INDO-SA-NLR2032 | KM496624-25 | 14-Oct-11 | 45 | M | 671 | 9,468 |
| 24 | INDO-SA-NLR2036 | KM496626-28 | 14-Oct-11 | 27 | M | 860 | 3,332 |
| 25 | INDO-SA-NLR2037 | KM496629 | 14-Oct-11 | 29 | M | 732 | 1,505 |
| 26 | INDO-SA-NLR2038 | KM496630 | 31-Oct-11 | 33 | M | 669 | 26,542 |
| 27 | INDO-SA-NLR2039 | KM496631-32 | 31-Oct-11 | 47 | M | 925 | 45,63,295 |
| 28 | INDO-SA-NLR2041 | KM496633 | 31-Oct-11 | 45 | M | 888 | 56,445 |
| 29 | INDO-SA-NLR2043 | KM496634 | 6-Sep-13 | 49 | M | 446 | — |
| 30 | INDO-SA-NLR2044 | KM496635 | 13-Sep-13 | 35 | M | 596 | — |
| 31 | INDO-SA-NLR2049 | KM496636 | 23-Oct-13 | 30 | M | 475 | — |
| 32 | INDO-SA-NLR2050 | KM496637 | 11-Nov-13 | 48 | M | 1,047 | — |
| 33 | INDO-SA-NLR2051 | KM496638 | 11-Nov-13 | 35 | F | 504 | — |
| 34 | INDO-SA-NLR2053 | KM496639 | 11-Nov-13 | 29 | F | 390 | — |
| 35 | INDO-SA-NLR2054 | KM496640 | 11-Nov-13 | 30 | F | 597 | — |
| 36 | INDO-SA-NLR2055 | KM496641 | 11-Nov-13 | 42 | F | 495 | — |
| 37 | INDO-SA-NLR2056 | KM496642 | 11-Nov-13 | 29 | F | 360 | — |
| 38 | INDO-SA-NLR2058 | KM496643 | 11-Nov-13 | 27 | M | 980 | — |
| 39 | INDO-SA-NLR2060 | KM496644 | 11-Nov-13 | 27 | M | 386 | — |
| 40 | INDO-SA-NLR2061 | KM496645 | 11-Nov-13 | 50 | M | 338 | — |
| 41 | INDO-SA-NLR2062 | KM496646 | 11-Nov-13 | 50 | F | 700 | — |
| 42 | INDO-SA-NLR2063 | KM496647 | 11-Nov-13 | 32 | F | 812 | — |
| 43 | INDO-SA-NLR2064 | KM496648 | 11-Nov-13 | 35 | F | 516 | — |
| 44 | INDO-SA-NLR2066 | KM496649 | 11-Nov-13 | 28 | F | — | — |
| 45 | INDO-SA-NLR2067 | KM496650 | 11-Nov-13 | 32 | F | — | — |
| 46 | INDO-SA-NLR2068 | KM496651 | 11-Nov-13 | 34 | M | 1,300 | — |
| 47 | INDO-SA-NLR2072 | KM496652 | 14-Nov-13 | 34 | M | 466 | — |
| 48 | INDO-SA-NLR2073 | KM496653 | 14-Nov-13 | 32 | M | 700 | — |
| 49 | INDO-SA-NLR2076 | KM496654 | 14-Nov-13 | 45 | M | — | — |
| 50 | INDO-SA-NLR2078 | KM496655 | 14-Nov-13 | — | F | — | — |
| 51 | INDO-SA-NLR2080 | KM496656 | 14-Nov-13 | 35 | F | — | — |
| 52 | INDO-SA-NLR2081 | KM496657 | 15-Nov-13 | 30 | F | — | — |
| 53 | INDO-SA-NLR2082 | KM496658 | 14-Nov-13 | — | M | — | — |
Date of visit refers to the enrollment (baseline) time point. The CD4 count and plasma viral load values were determined at the time of visit and not available for some of the patients. The GenBank accession numbers of the Tat sequences are presented.
A single vial of 6–8 ml of peripheral blood was collected after obtaining informed consent from each of the participants. From a few select subjects, additional blood samples were collected at 6 and 12 months. Blood samples collected in EDTA vacutainers (Becton Dickinson) were centrifuged to isolate plasma that was stored in a deep freezer in aliquots. In addition, genomic DNA was extracted from 0.2 ml of whole blood by using a commercial DNA extraction kit (NucleoSpin Blood; Macherey-Nagel GmbH), eluted in a 100 μl volume, and stored at 4°C until use. All the methods were carried out in accordance with Institutional Ethics Committee for Human Research guidelines by Indian Council for Medical Research (ICMR), and the experimental protocols were approved by the Institutional Ethics Committees for Human Studies at Y. R. Gaitonde Care, Chennai and Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore. The clinical profile of the subjects, including the subject ID, sampling date, age, gender, viral load, and the CD4 count, has been provided in Table 1.
RNA isolation and cDNA synthesis
RNA was extracted from 150 μl of plasma using a commercial Viral RNA isolation kit (NucleoSpin® RNA Virus, Ref. No. 740956.50; MACHEREY-NAGEL GmbH & Co. KG). Alternatively, the viral RNA was extracted from 1 ml of plasma using the NucliSENS miniMAG nucleic acid extraction kit (Ref. No. 200 293; BioMerieux). The complementary DNA was synthesized using random hexamers and SuperScript® III Reverse Transcriptase and using a commercial kit (Cat. No. 18080-051; Invitrogen). The samples were incubated at 25°C for 10 min and 50°C for 50 min. The reactions were terminated at 85°C for 5 min followed by RNaseH treatment.
Amplification of Tat exon 1 and DNA sequencing
Tat exon 1 was amplified from the cDNA using the nested PCR format using a long-range kit commercially available (XT-20 PCR system; Merck Genei). A fragment of 911 bp was amplified in the first round of PCR using external primers N1331 (5′-TAGTAGAGGATAGATGGAACAAGSCCCCAG-3′) and N1332 (5′-TCTGTGGGTACACAGGCATGTGTRGCCCA-3′). An alternative external primer pair targeting a fragment of 608 bp was used for a small number of samples when the above primer pair failed to amplify Tat. The primers of the alternative pair consisted of N2242 (5′-GCTTAGGACAATATATCTATGAAACCTATGG-3′) and N2243 (5′-ACACAGGTACCCCATAATAGACTGTGACC-3′). The PCR vials were incubated at 94°C for 2 min before the amplification. The amplification was performed for 35 cycles, each cycle consisting of melting at 94°C for 30 s, annealing at 50°C for 50 s, and extension at 72°C for 1 min, followed by final extension at 72°C for 5 min. Two microliters of the first-round PCR products were transferred to the second-round PCR to amplify a 446-bp fragment containing tat exon 1 using primers N111 (5′-GGGTGYCARCATAGCAGAATAGGCATT-3′) and N1156 (5′-TCATTGCCACTGTCTTCTGCTCT-3′). The cycling conditions were identical to the first round of amplification. Carryover contamination was prevented by adherence to strict procedural and physical safeguards that included reagent preparation and PCR setup, amplification, and post-PCR processing of samples in separate rooms. The PCR products were purified using a commercial DNA purification kit (Cat. No. YDF100; Real Biotech Corporation) and subjected to sequencing. The sequencing was performed on ABI PRISM® 3130xl Genetic Analyzer (Applied Biosystems) using the internal primers N111 and N1156. The sequences are available from GenBank under the accession Nos. KM496583-KM496600, KM496602-KM496605, and KM496607-KM496658.
Phylogenetic analysis
The multiple sequence alignment was performed using ClustalW in the BioEdit software (version 7.0.5.3). The phylogenetic tree was constructed using the neighbor-joining method (Kimura two-parameter model) in 1,000 bootstrapped data sets, using the Molecular Evolutionary Genetics Analysis software version 5.05 (MEGA 5). The sequences were manually edited using Bio-Edit software version 7.0.5.3. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. All the positions containing gaps and missing data were eliminated. There were a total of 201 positions in the final data. In addition, the 74 Tat exon 1 sequences were also analyzed using the RIP 3.0 (Recombinant Identification Program) (www.hiv.lanl.gov) as a part of the quality control, before submission of these sequences to the GenBank, and it confirmed all the sequences to be clustering with subtype C with no recombination events being detected.36
The identification of the SAR and statistical evaluation
The SAR in subtype C and subtype B tat were identified using the Viral Epidemiological Signature Pattern Analysis (VESPA) tool from the HIV sequence database (www.hiv.lanl.gov).4 The database was searched for global HIV-1 tat sequences for subtype A, B, C, D, and CRF01AE. The filters applied while downloading the available HIV-1 exon 1 tat sequences from the LANL sequence database were as follows: exclusion of problematic sequences and one sequence per patient. Importantly, since the information regarding the date of isolation of the clinical sample is not available from the database, we considered the date of sequence submission instead for the analysis. It should be noted that there could be a significant delay between the actual dates of sample collection and sequence submission. In addition, the analyses of the tat sequences of the present cohort and the sequences downloaded from the LANL database were independent of each other without an overlap between the two sets of sequences.
Amino acid sequences downloaded in FASTA format were divided into two groups representing two distinct time periods, 1986–2004 and 2004–2014, for the analysis. The sequence pools were then manually edited using the software MEGA 6.06 to exclude the sequences containing insertions and deletions. The resulting sequences were used as the query and background sequence pools for the signature analysis using the VESPA tool. While the pool of subtype C tat sequences served as the query, the pool of tat sequences derived from subtypes A, B, D and CRF01_AE served as the background for the analysis. The threshold for the analysis was set at 1.0 to identify the signature residues for C-Tat that requires the signature amino acid be included in every sequence in the query set to be considered. The VESPA results were summarized using the contingency tables for each SAR position and analyzed using Fischer's exact test (GraphPad Prism version 5.02). The two groups for the statistical analysis were constituted by the two time periods (1986–2004) and (2005–2014). The categorical variables were the number of sequences containing the signature residue at a given position and the number of sequences having the next best residue that may have the potential to replace the SAR.
Evaluation of the selection pressure
The selection pressure acting on Tat exon 1 was evaluated at the individual codon level by subjecting the 74 Tat sequences of the clinical cohort along with the Indian subtype C reference 93IN101 (accession No. AB023804) to the dn/ds analysis using the SNAP version 2.1.1 (Synonymous Nonsynonymous Analysis Program) tool (www.hiv.lanl.gov).37 The output file of the data per codon generated by the SNAP tool was used to evaluate the dn/ds values at each codon.
Results
Viral evolution being a dynamic process, the primary objective of this work was to examine how stable the profile of SAR is as a function of time. To address this question, we accessed all the available subtype C tat exon 1 amino acid sequences from the extant databases and divided them into two groups: the period covering the previous analysis between the years 1986–200425 and the latter 10 years, 2005–2014. The total number of HIV-1 subtype C Tat sequences available from the extant databases was quite small. There were only 414 and 480 total subtype C Tat sequences available for periods 1986–2004 and 2005–2014, respectively, from the databases. To make the statistics more reliable, we performed the analyses by dividing the sequences into only two categories. Tat exon 1 sequences from other major viral subtypes A, B, D, and CRF01_AE were also acquired for the analysis. Using the software VESPA, the amino acid residues of Tat differentially conserved in subtype C compared with the other subtypes have been identified.4 The data summarized in Figure 1 reinforce our previous analysis regarding the SAR profile in subtype C Tat.25 All the seven amino acid residues (histidine 29, serine 31, leucine 35, glutamine 39, serine 57, proline 60, and glutamic acid 63) that were identified as SAR in our previous analysis have also been confirmed as signature residues in the present analysis for both the periods. In addition to these seven positions, the present analysis also identified an additional residue at position 68 as SAR for subtype C Tat. A leucine residue was present at this position in 215 of 414 (52%) of the viral sequences in subtype C. While only 5 of the 757 (<1%) nonsubtype C Tat sequences have a leucine at this location, a majority of the nonsubtype C Tat sequences instead have a serine residue at this position (437 of 757, 58%).
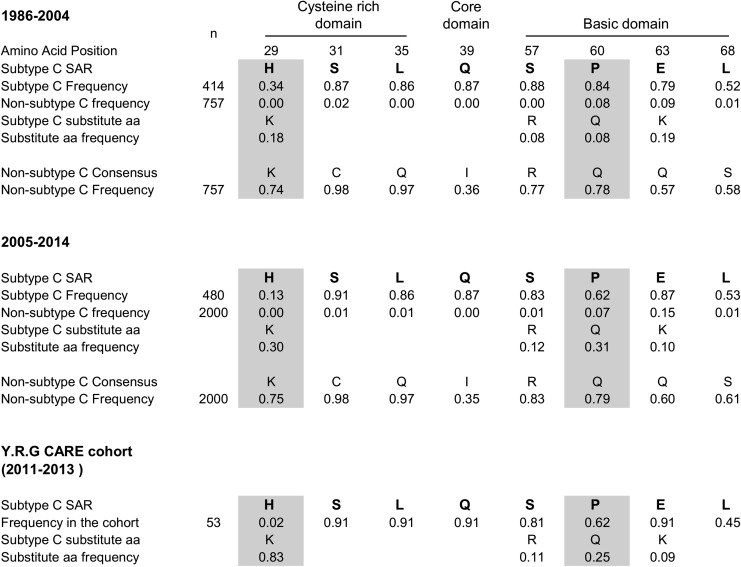
FIG. 1.
HIV-1 subtype C Tat signature amino acid profile. n represents the total number of tat exon 1 amino acid sequences available from the databases. In the global analysis, the change in the prevalence of only histidine 29 and proline 60, but not the other signature amino acid residue (SAR), was found to be statistically significant (shaded, p < .0001).
A similar analysis of the signature residues from the latter period of 2005–2014 and comparison of the signature residue profile between the two periods revealed a changing profile of the signature residues in subtype C Tat. Several key observations can be made. First, at four of eight positions (serine 31, leucine 35, glutamine 39, and leucine 68), the nature of the signature residues and their relative frequencies remained largely unchanged between 1986–2004 and 2005–2014. For instance, at position 31, a serine residue was found during both the periods at a comparable prevalence of 87% and 91% during 1986–2004 and 2005–2014, respectively. Second, at the other four positions (histidine 29, serine 57, proline 60, and glutamic acid 63), the prevalence of the respective signature amino acid appears to be undergoing a change between the two periods of evaluation. While the prevalence of the SAR at three of these locations (29, 57, and 60) is progressively decreasing, at position 63, the frequency of the SAR is on the rise. Third, the rate of replacement of the SAR is not uniform. Histidine 29 is being replaced at a much faster rate than proline 60. In contrast, the substitution of serine 57 appears to be progressing at a very low rate. Alternatively, the variations in the frequency of serine 57 could be transient fluctuations between the time periods. During 1986–2004, 34% (140 of 414) of the subtype C tat sequences contained a histidine at position 29. During the next 10 years covering 2005–2014, the proportion of subtype C Tat sequences containing a histidine at this position dropped significantly to 13% (63 of 480, p < .0001). Last, at all the three positions where the SAR frequency is decreasing, the residues being substituted are the generic amino acids present in all the other viral subtypes. Thus, histidine 29, serine 57, and proline 60 are tending to gradually being replaced with lysine, arginine, and glutamic acid, respectively. Of note, at position 29, as the frequency of histidine diminished, a concomitant rise in the proportion of lysine from 18% (76 of 414) to 30% (142 of 480) was observed between these two time frames. Thus, the signature position 29 in subtype C Tat appears to be progressively obliterated by the substitution of the signature residue histidine by a generic residue lysine, common to all subtypes of HIV-1.
A transformation of a similar nature appears to be in progress at a different signature residue, position 60, in subtype C Tat. At this position, ∼84% (347 of 414) of subtype C tat sequences contained a proline during 1986–2004, whereas only 8% (61 of 757) of non-C tat sequences contained a proline. During the latter period of 2005–2014, the proportion of subtype C Tat containing a proline at this position lessened to 62% (296 of 480, p < .0001). Importantly, as in the case of histidine 29, the signature residue proline 60 is being replaced by the generic amino acid glutamine, which is present at this location in all other subtypes. The prevalence of glutamine increased from 8% (32 of 414) during 1986–2004 to 31% (148/480) during 2005–2014 in subtype C Tat.
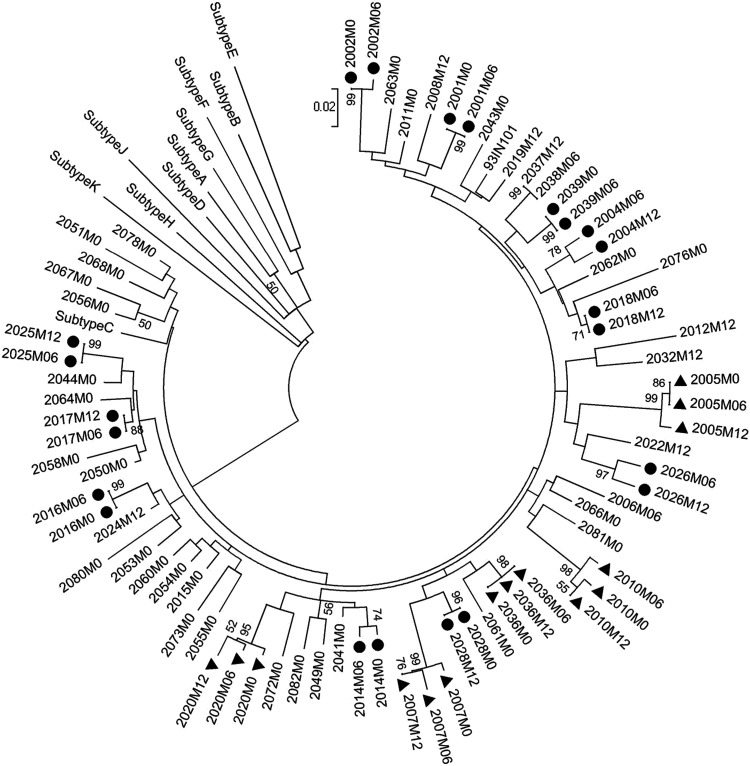
To further corroborate these findings, we examined the genetic variation of Tat exon 1 using a defined clinical cohort derived from southern India. The clinical samples for the cross-sectional analysis were obtained from a cohort of 76 drug-naive subjects between September 2011 and November 2013. The study participants consisted of adult subjects, over 18 years of age, and believed to have acquired the infection primarily through heterosexual transmission. Tat exon 1 could be successfully amplified from the plasma viral RNA in only 53 of 76 subjects (Table 1). The tat sequence was determined directly from the PCR fragment. From 37 of the subjects, the tat sequence was determined at a single time point, and from 11 other subjects, the sequence information was available at two different time points spaced ∼6 months apart. From the remaining five subjects, the sequences were available at three different time points. The phylogenetic analysis, therefore, is based on a total of 74 tat sequences generated from 53 individual subjects. A phylogenetic analysis of the tat sequences revealed a tight clustering of all the tat sequences with subtype C (Fig. 2). In addition, the sequences derived at different time points from the same subject clustered together confirming the absence of intersample contamination. The phylogenetic analysis thus confirmed the subtype C origin of the tat sequences.
FIG. 2.
The phylogenetic analysis of the tat sequences. A total of 74 tat exon 1 sequences derived from 53 subjects are used in the analysis. Filled circles and filled triangles represent subjects from whom samples were available at two (n = 11) and three (n = 5) different time points, respectively. From all of the other subjects, samples at a single time point were collected. The reference sequences of tat from various genetic subtypes—A, B, C, D, E, F, G, H, J, K were downloaded from the Los Alamos HIV database. After gap stripping, there are a total of 201 nucleic acid positions in the final dataset. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site.
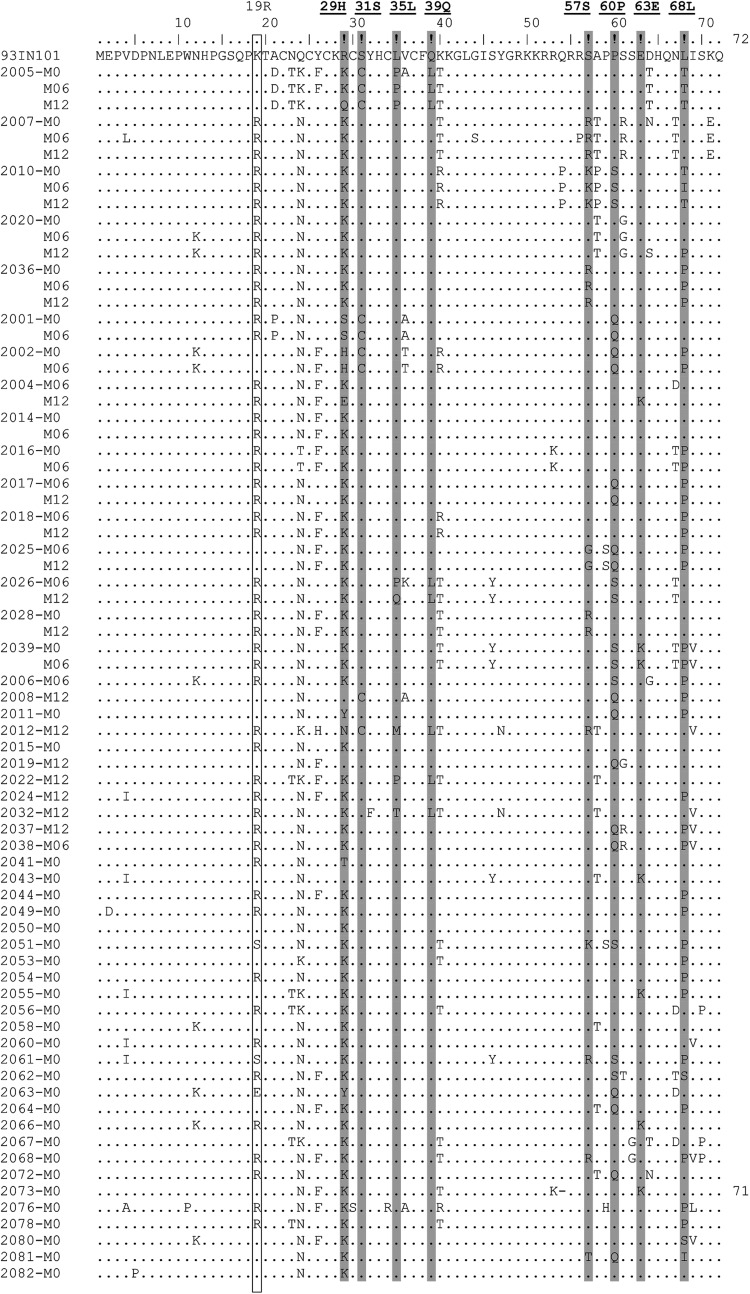
The 74 tat sequences derived from 53 subjects have been compared in multiple-sequence alignment (Fig. 3). In addition, the profile of the SAR was examined by considering only one representative sequence per subject. Thus, the signature residue analysis was based on only 53 of the 74 available tat sequences (Fig. 1). The analysis of tat sequences of the clinical cohort confirmed the changing profile of the subtype C Tat. The SAR at four positions (serine 31, leucine 35, glutamine 39, and leucine 68) remained stable and at proportions comparable to the global analysis. Importantly, the replacement of histidine 29 appears to be taking place at a faster pace compared with proline 60. Only 2 of the 74 Tat sequences, both from the same subject, contained a histidine at position 29. In other words, only 1 of the 53 subjects of the clinical cohort contained the SAR histidine, while most of the others (83%, 44 of 53) contained the generic amino acid lysine. Thus, the prevalence of histidine reduced from 34% during 1986–2004 to 13% during 2005–2014 and to only 2% or less during 2011–2013 in the southern Indian cohort. There appears to be a strong negative selection pressure against the histidine residue at position 29 in subtype C Tat. Negative selection of a similar nature also appears to be operating against proline 60 in subtype C Tat, although not at the same magnitude. The prevalence of proline at position 60 was 62% in our clinical cohort at a magnitude comparable to that of the period 2005–2014 at the global level. In a similar fashion, serine at position 57 has reduced from 88% during 1986–2004 to 83% during 2005–2014 to 81% in the YRG CARE clinical cohort during 2011–2013. The generic amino acid arginine demonstrated a concomitant increase from 8% to 12% during the same period at this residue.
FIG. 3.
Multiple amino acid sequence alignments of tat exon 1. A total of 74 sequences derived from 53 subjects are aligned with the subtype C reference sequence of 93IN101 on top (accession no. AB023804). The label at the left margin identifies the sample and M0, M06, and M12 represent the month the sample was collected for the analysis. The dots represent sequence identity. The eight positions of the protein identified as subtype C signature positions are shaded. Lysine 19 is highlighted using a square box.
To examine the selection pressure acting on Tat at the individual codon level, we subjected the 74 Tat sequences of the clinical cohort along with the Indian subtype C reference 93IN101 (accession No. AB023804) to dn/ds analysis using the SNAP version 2.1.1 tool (www.hiv.lanl.gov).37 The average dn/ds value for Tat exon 1 was estimated to be 0.2503 with the values ranging from 0.0235 to 2.2. The output file of the data per codon generated by the SNAP tool was used to evaluate the dn/ds values at each codon. All the Tat amino acid codons in exon 1 that demonstrated a purifying selection or a positive or negative selection are summarized (Table 2). While a large number of Tat codons showed neutral selection, the codons at 6 different positions 5, 12, 30, 32, 44, and 56 were found to be under the influence of purifying selection with the dn/ds ratio being less than 1. The codons at 17 other positions in Tat were found to be under the influence of positive selection with the dn/ds ratio being greater than 1. Importantly, codons for six of the eight SAR, positions 29, 31, 35, 39, 57, and 60, were found to be under positive selection. The codon 29 demonstrated a positive selection for lysine (K) confirming the replacement of the SAR histidine at this location as identified using the VESPA tool. Likewise, codon 60 showed a positive selection of glutamine (Q) leading to the substitution of the SAR proline at this position, again confirming the VESPA. Three other codons encoding subtype C SAR, S31, L35, and Q39, too demonstrated a positive selection confirming their conservation in the context of the SAR in subtype C Tat. Taken together, the VESPA and the dn/ds analyses were consistent with each other and confirmed the negative selection of the two SAR histidine and proline at positions 29 and 60, respectively.
Table 2.
dn/ds Ratio at the Per Codon Level for Tat Exon 1 Sequences
| SAR | ||||||
|---|---|---|---|---|---|---|
| Amino acid position | Syn | Nonsyn | dn/ds ratio | Evolutionary selection | Amino acid | selection |
| D5 | 0.13 | 0.05 | 0.38 | Purifying | — | |
| N12 | 0.29 | 0.21 | 0.72 | Purifying | — | |
| A21 | 0.03 | 0.13 | 4.33 | Positive | — | |
| N23 | 0.08 | 0.19 | 2.38 | Positive | — | |
| N24 | 0.28 | 0.33 | 1.18 | Positive | — | |
| Y26 | 0.17 | 0.49 | 2.88 | Positive | — | |
| K29 | 0.06 | 0.6 | 10.00 | Positive | H | Negative |
| C30 | 0.31 | 0.03 | 0.10 | Purifying | — | |
| S31 | 0.05 | 0.21 | 4.20 | Positive | S | Positive |
| Y32 | 0.23 | 0.03 | 0.13 | Purifying | ||
| L35 | 0.06 | 0.24 | 4.00 | Positive | L | Positive |
| V36 | 0.04 | 0.29 | 7.25 | Positive | — | |
| Q39 | 0.07 | 0.21 | 3.00 | Positive | Q | Positive |
| K40 | 0.15 | 0.55 | 3.67 | Positive | — | |
| G44 | 0.19 | 0.03 | 0.16 | Purifying | — | |
| Q54 | 0.04 | 0.12 | 3.00 | Positive | — | |
| R56 | 0.05 | 0.03 | 0.60 | Purifying | — | |
| T57 | 0.05 | 0.52 | 10.40 | Positive | S | a |
| Q60 | 0.47 | 0.61 | 1.30 | Positive | P | Negative |
| S61 | 0.1 | 0.25 | 2.50 | Positive | — | |
| S62 | 0.03 | 0.05 | 1.67 | Positive | — | |
| E63 | 0.13 | 0.17 | 1.31 | Positive | E | Positive |
| D64 | 0.15 | 0.32 | 2.13 | Positive | — | |
| I68 | 0 | 0.72 | — | L | a | |
The dn/ds ratio at each codon was evaluated using the SNAP version 2.1.1 (Synonymous Nonsynonymous Analysis Program) tool available at HIV-LANL. The selection pressure deduced by the dn/ds ratio was compared with the selection that was suggestive from the VESPA of the changing profile of the SAR at specific locations.
T57 and I68 are seen only in the case of one subject each, that is, 2067-M0 and 2081-M0, respectively.
SAR, signature amino acid residue; VESPA, Viral Epidemiological Signature Pattern Analysis.
Interestingly, in the first batch of 28 clinical samples collected for this study (NLR2001-2041, Table 1), we found that five different subjects (2001, 2002, 2005, 2008, and 2012) contained a cysteine at the signature position 31 instead of the serine typical in subtype C HIV-1, and especially prevalent in Indian clinical samples. The presence of the cysteine residue was consistent with the repeat samples collected from the subjects at different points. The proportion of clinical samples containing an intact dicysteine motif in (C30C31) Tat is significantly smaller in India compared to the Southern African countries.38 While only 1%–3% of the Indian tat sequences contained the dicysteine motif, 11%–26% of Southern African tat sequences contained the motif. Tat containing the dicysteine motif is not only more neurotoxic31,32 but also is associated with a higher prevalence of HAD.25 Given the pathogenic significance of Tat C30C31, we wondered if a viral subpopulation containing this dicysteine motif was emerging in the geographical region from where the clinical samples have been collected. To explore this possibility, we collected a second set of clinical samples (NLR2043-2082, Table 1), nonoverlapping with the first set, from the same geographical location, and determined the Tat sequences from the blood samples. However, none of the 25 viral isolates from the second set contained a cysteine residue at position 31 (Fig. 3). Thus, it appears that the higher frequency of the dicysteine motif-containing Tat in the first set of the clinical samples was a chance event, and the prevalence of the C30C31 in India continues to remain low as reported previously.25,38 However, the number of sequences analyzed from the clinical cohort remains a limiting factor to draw an assertive inference from this observation. In addition, in the clinical cohort, the position 19 of Tat appears to be unique for the Indian viral isolates, although the significance of this variation is not known (Fig. 3, highlighted using an open box). Most of the cohort clinical samples (33 of 53) contained an arginine at this location instead of the lysine residue typical of subtype C.
Discussion
The long-term objective of our work is to understand the biological significance of the SAR that is unique for different genetic subtypes of HIV-1. The SAR are believed to govern the differential biological properties of viral factors such as Tat that in turn may influence subtype-specific characteristics of viral genetic subtypes. The immediate aim of this work is to examine the genetic stability of the SAR in exon 1 of subtype C Tat of HIV-1 when compared between two different time frames that are separated by 10 years. Our analysis identified that the SAR could fall into two different categories—the variable residues that are subjected to evolutionary changes in time and the stable residues that cannot undergo a variation. The data presented here collectively suggest that the profile of SAR in subtype C tat is stable at certain positions (serine 31, leucine 35, glutamine 39, and leucine 68), but undergoing a change at four other positions at a rate that may be defined as rapid (histidine 29), moderate (proline 60), or slow (serine 57 and glutamic acid 63). Given the limitations of this study, the small sample size, short observation period, and cross-sectional nature of the sequences used in the analysis, it is not possible to draw an assertive conclusion on the speed at which the genetic changes are implemented at specific positions in a viral factor such as Tat. The rate at which a signature residue is replaced could be a reflection of the selection pressure itself. Alternatively, the slow rate of substitution of a SAR, such as serine 57 or glutamic acid 63, could be the consequence of a delayed beginning of the replacement process. This analysis only offers important leads, and future studies must attempt to address this question by a stringent experimental design that includes the collection of longitudinal samples from well-defined clinical cohorts.
In a preliminary analysis, we identified only one SAR, serine 84, in the exon 2 of Tat of subtype C, as opposed to a proline at this position in all other subtypes. The sequences from the databases did not show a significant difference in the frequency of this residue in subtype C. Given that a significant function has not been ascribed to Tat exon 2 and the invariable nature of serine 84 in our preliminary analysis, we primarily focused on exon 1 of Tat.
It is interesting to note that the selection pressures can operate against a specific SAR without affecting the prevalence of other signature residues in the same protein. The impact of the negative selection appears to be particularly severe against the presence of a histidine at position 29. During 1986–2004, 34% of the subtype C tat sequences contained a histidine at position 29, while none of the non-C subtypes contained a histidine at this position. During the same period, nearly 18% of the subtype C Tat sequences contained a lysine at this position that is the generic amino acid for this position in the majority (74%) of the nonsubtype C strains. During the subsequent decade, the frequency of the residue histidine 29 reduced further in subtype C from 34% to 13% with a concomitant increase in the proportion of the generic amino acid lysine from 18% to 30%. The presence of histidine was seen in only one of the 53 subjects of the southern Indian clinical cohort in the recent years, suggesting continued selection against histidine at this location. A negative selection is also evident against the presence of a proline at position 60 in subtype C, which is being replaced by the generic amino acid glutamine. A similar trend of signature amino acid replacement at serine 57 with the generic amino acid arginine appears to have begun in subtype C. Thus, at three different locations in subtype C Tat, it appears that the subtype-specific marks are being lost and subtype-generic marks are being expressed.
At present, it is unclear why a specific amino acid residue selectively emerges as a signature residue in the first place and why such a residue can subsequently be subjected to negative selection. The replacement of the SAR serine at position 57 with the generic arginine residue can perhaps be explained. The position 57 constitutes the last amino acid residue of the BD that governs many biological functions of Tat, including the nuclear translocation, TAR binding, extracellular Tat uptake, and protein dimerization.2 The BD, as the name suggests, is rich in basic amino acid residues, especially arginine. In all the HIV-1 genetic subtypes, position 57 is occupied by an arginine residue except subtype C where the arginine is substituted by a serine. The impact of such an unusual substitution on the biological functions of Tat is not known. Presently, we note that in subtype C, the frequency of serine at position 57 appears to be gradually decreasing from 88% to 83% to 81% during 1986–2004, 2005–2014, and 2011–2013, respectively, with a concomitant rise in the frequency of the generic amino acid arginine from 8% to 12% for the same time periods. An arginine at position 57 is probably expected to be associated with stronger nuclear localization compared to the presence of a serine. The functional significance of the presence of serine at this position needs a methodical evaluation.
A detailed evaluation of serine 31 demonstrated that a key consequence of the cysteine-to-serine substitution at this position is the loss of chemotactic capability of subtype C Tat.25 This study provides a proof-of-concept that a single amino acid residue can impart drastically differential biological function on Tat. Likewise, Dey et al. demonstrated that positions 35 and 39 in Tat are functionally associated and coevolved in the viral subtypes.35 Although a vast majority of nonsubtype C viral strains contain a glutamine residue at position 35 that appears to be critical for efficient recruitment of P-TEFb to the transcription complex, in subtype C alone, this position is replaced by a leucine that is not efficient in recruiting P-TEFb. Likewise, at position 39, most of the nonsubtype C viral strains contain either an isoleucine, leucine, or threonine residue, unlike subtype C, which contains a glutamine residue at this location. The glutamine residue at this location appears to be inefficient in driving the phosphorylation of the C-terminal domain of RNAPII.35 Although the presence of the two residues at the two positions in subtype C Tat appears to dampen the transactivation function of Tat individually, when present together, these two inefficient residues appear to compensate for each other's deficiencies.35 It is, however, not known why the presence of the amino acids at these two locations has been reversed in subtype C compared with other HIV-1 subtypes.
In a previous report, examining the tat sequences derived from India, we suggested the changing profile of the SAR at two locations in Tat, positions 29 and 60 in the Indian context.6 This work confirms these changes at the global level using Tat sequences downloaded from the databases and by determining the Tat sequences from a different southern Indian clinical cohort. In addition, this work extends the changing profile to two more positions in subtype C Tat, positions 57 and 63. The changing SAR profile does not seem to be unique to subtype C Tat, but is also applicable to Tat from the other subtypes. A signature analysis of the subtype B tat sequences downloaded from the databases shows a similar changing profile of SAR at select positions between 1986–2004 and 2005–2014 time frames (data not presented). While the frequencies of the aspartic acid and valine residues at positions 61 and 67 have reduced from 70% and 60% to 61% and 44%, respectively, the frequency of the asparagine at position 24 appears to have increased marginally. A cross-sectional analysis of Tat exon 1 sequences derived from 120 HIV-1-seropositive subjects of a northern Indian clinical cohort identified K24T and H29Y Tat variations and minor variants constituted by F38L, Q39M, K41N, and G42A.39 However, none of these Tat variations was found in our southern Indian clinical cohort. In a different publication, the same group reported the presence of several Tat variations in a cross-sectional cohort of 21 subjects of a northern Indian clinical cohort, including N24T, K29Y, L35P, G44S, S46F, P68L, and F38L.40 Of the variations, only S46F was observed in only 4 of the 53 subjects in our cohort.
Unlike the downloaded sequences, the 74 tat sequences derived from the Indian cohort here are associated with necessary clinical information regarding several confounding factors, including the stage (asymptomatic) of the viral infection, the absence of opportunistic infections, the availability of CD4 cell count, and the plasma viral load for each of the subjects. Importantly, longitudinal samples from at least 16 subjects have been drawn at two or three points at a 6-month interval. The sequences available from the extant database are much more diverse with respect to these clinical parameters; hence examining the selection pressure using such sequences could have been of limited value. In addition, a recent publication performed the selection pressure analysis for the positively selected residues using the global tat sequences downloaded from the LANL database.41,42 Furthermore, Rossenkhan et al. examined a cohort of 20 subjects of Botswana, consisting of eight acutely infected and 12 randomly selected seroconverted individuals, for the selection pressures on Tat exon 1. They report that residues 3, 4, 21, 24, 29, 39, and 68 in tat exon 1 have been under positive selection pressure. In addition, different profiles of SAR could be identified between the primary and chronic phases of subtype C infection.43
In summary, using subtype C Tat as a model viral factor, we demonstrated that the profile of the SAR has been undergoing a progressive transformation at selective sites without apparent changes in the frequencies of the other signature residues in the same protein. How the changing signature amino acid profile is going to impact the functions of a viral protein and as a consequence the replication competence of the viral strains, needs further evaluation. It will also be of interest to understand the mechanisms that operate at the molecular level, imposing the selective pressure for or against a specific amino acid residue at a given position. The regional genetic divergence of HIV-1 and associated evolutionary pressures could allude to the presence of specific host factors in certain populations that may exert a selective pressure on viral evolution in one direction or another.
Sequence Data
The 53 tat sequences amplified from individual subjects and used in this study are available from GenBank under the accession Nos. KM496583-KM496600, KM496602-KM496605, and KM496607-KM496658.
Acknowledgments
S.S. is a recipient of a research fellowship from The Department of Biotechnology and M.M. from The Council of Scientific and Industrial Research, The Government of India. This work was supported by intramural funds from JNCASR and a grant to U.R. from The Department of Science and Technology, The Government of India (DST/INT/SAFR/MEGA-P(5)/2011). We thank Arthur Ruiz for critical reading of the article.
Authors' Contributions
M.M. performed the signature amino acid and dn/ds analysis and prepared Figure 1 and Table 2; S.P.G.A. and S.S. performed molecular phylogenetic analyses and prepared Figures 2 and 3 and Table 1; V.R.P., K.G.M., S.S., S.S., and U.R. contributed to reagents/materials/analysis tools and conceived, designed the experiments, and wrote the article. All authors read and approved the final article.
Author Disclosure Statement
The authors declare no competing financial interests.
References
- 1.Hemelaar J, Gouws E, Ghys PD, Osmanov S: Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011;25:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, et al. : Impact of Tat genetic variation on HIV-1 disease. Adv Virol 2012;2012:123605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu D J, Buve A, Baggs J, van der Groen G, Dondero TJ: What role does HIV-1 subtype play in transmission and pathogenesis? An epidemiological perspective. AIDS 1999;13:873–881 [DOI] [PubMed] [Google Scholar]
- 4.Korber B, Myers G: Signature pattern analysis: A method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses 1992;8:1549–1560 [DOI] [PubMed] [Google Scholar]
- 5.Flys TS, et al. : Quantitative analysis of HIV-1 variants with the K103N resistance mutation after single-dose nevirapine in women with HIV-1 subtypes A, C, and D. J Acquir Immune Defic Syndr 2006;42:610–613 [DOI] [PubMed] [Google Scholar]
- 6.McCormick-Davis C, Dalton SB, Singh DK, Stephens EB: Comparison of Vpu sequences from diverse geographical isolates of HIV type 1 identifies the presence of highly variable domains, additional invariant amino acids, and a signature sequence motif common to subtype C isolates. AIDS Res Hum Retroviruses 2000;16:1089–1095 [DOI] [PubMed] [Google Scholar]
- 7.Neogi U, et al. : Genetic characterization of HIV-1 Tat exon 1 from a southern Indian clinical cohort: Identification of unique epidemiological signature residues. AIDS Res Hum Retroviruses 2012;28:1152–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Neill E, et al. : Dynamic evolution of the human immunodeficiency virus type 1 pathogenic factor, Nef. J Virol 2006;80:1311–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shankarappa R, et al. : Human immunodeficiency virus type 1 env sequences from Calcutta in eastern India: Identification of features that distinguish subtype C sequences in India from other subtype C sequences. J Virol 2001;75:10479–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibellini D, Vitone F, Schiavone P, Re MC: HIV-1 tat protein and cell proliferation and survival: A brief review. New Microbiol 2005;28:95–109 [PubMed] [Google Scholar]
- 11.Kuppuswamy M, et al. : Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res 1989;17:3551–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel AD, Bredt DS, Pabo CO: Tat protein from human immunodeficiency virus forms a metal-linked dimer. Science 1988;240:70–73 [DOI] [PubMed] [Google Scholar]
- 13.Frankel AD, et al. : Dimerization of the tat protein from human immunodeficiency virus: A cysteine-rich peptide mimics the normal metal-linked dimer interface. Proc Natl Acad Sci U S A 1988;85:6297–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalantari P, et al. : Thioredoxin reductase-1 negatively regulates HIV-1 transactivating protein Tat-dependent transcription in human macrophages. J Biol Chem 2008;283:33183–33190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, et al. : HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J 2002;21:6801–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rana TM, Jeang KT: Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch Biochem Biophys 1999;365:175–185 [DOI] [PubMed] [Google Scholar]
- 17.Marzio G, et al. : HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A 1998;95:13519–13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng L, et al. : Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology 2000;277:278–295 [DOI] [PubMed] [Google Scholar]
- 19.Wong K, et al. : HIV-1 Tat interactions with p300 and PCAF transcriptional coactivators inhibit histone acetylation and neurotrophin signaling through CREB. J Biol Chem 2005;280:9390–9399 [DOI] [PubMed] [Google Scholar]
- 20.Mitola S, et al. : Identification of specific molecular structures of human immunodeficiency virus type 1 Tat relevant for its biological effects on vascular endothelial cells. J Virol 2000;74:344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barillari G, et al. : The Tat protein of human immunodeficiency virus type 1, a growth factor for AIDS Kaposi sarcoma and cytokine-activated vascular cells, induces adhesion of the same cell types by using integrin receptors recognizing the RGD amino acid sequence. Proc Natl Acad Sci U S A 1993;90:7941–7945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubartelli A, et al. : HIV-I Tat: A polypeptide for all seasons. Immunol Today 1998;19:543–545 [DOI] [PubMed] [Google Scholar]
- 23.Jeang KT, Xiao H, Rich EA: Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem 1999;274:28837–28840 [DOI] [PubMed] [Google Scholar]
- 24.Gao F, et al. : A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol 1998;72:5680–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranga U, et al. : Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol 2004;78:2586–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satishchandra P, et al. : Profile of neurologic disorders associated with HIV/AIDS from Bangalore, south India (1989–96). Indian J Med Res 2000;111:14–23 [PubMed] [Google Scholar]
- 27.Wadia RS, et al. : Neurological manifestations of HIV disease. J Assoc Physicians India 2001;49:343–348 [PubMed] [Google Scholar]
- 28.Campbell GR, Watkins JD, Singh KK, Loret EP, Spector SA: The human immunodeficiency virus type 1 subtype C Tat fails to induce intracellular calcium flux and induces reduced tumor necrosis factor production from monocytes. J Virol 2007;81:5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandhi N, et al. : Differential effects of HIV type 1 clade B and clade C Tat protein on expression of proinflammatory and antiinflammatory cytokines by primary monocytes. AIDS Res Hum Retroviruses 2009;25:691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong JK, Campbell GR, Spector SA: Differential induction of interleukin-10 in monocytes by HIV-1 clade B and clade C Tat proteins. J Biol Chem 2010;285:18319–18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao VR, et al. : HIV-1 clade-specific differences in the induction of neuropathogenesis. J Neurosci 2008;28:10010–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra M, Vetrivel S, Siddappa NB, Ranga U, Seth P: Clade-specific differences in neurotoxicity of human immunodeficiency virus-1 B and C Tat of human neurons: Significance of dicysteine C30C31 motif. Ann Neurol 2007;63:366–376 [DOI] [PubMed] [Google Scholar]
- 33.Albini A, et al. : HIV-1 Tat protein mimicry of chemokines. Proc Natl Acad Sci U S A 1998;95:13153–13158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, et al. : NMDA receptor activation by HIV-Tat protein is clade dependent. J Neurosci 2008;28:12190–12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dey SS, et al. : Mutual information analysis reveals coevolving residues in Tat that compensate for two distinct functions in HIV-1 gene expression. J Biol Chem 2012;287:7945–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siepel A, C, et al. : A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses 1995;11:1413–1416 [DOI] [PubMed] [Google Scholar]
- 37.Korber B: HIV Signature and sequence variation analysis. In: Computational Analysis of HIV Molecular Sequences, Ch. 4 (Rodrigo AG, Learn GH, eds.) Kluwer Academic, 2000, pp. 55–72 [Google Scholar]
- 38.Rao VR, et al. : Clade C HIV-1 isolates circulating in Southern Africa exhibit a greater frequency of dicysteine motif-containing Tat variants than those in Southeast Asia and cause increased neurovirulence. Retrovirology 2013;10:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronsard L, et al. : Molecular and genetic characterization of natural HIV-1 Tat Exon-1 variants from North India and their functional implications. PLoS One 2014;9:e85452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lata S, et al. : Effect on HIV-1 gene expression, Tat-Vpr interaction and cell apoptosis by natural variants of HIV-1 Tat exon 1 and Vpr from Northern India. PLoS One 2013;8:e82128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy CN, et al. : Molecular characterization of full-length Tat in HIV-1 subtypes B and C. Bioinformation 2015;11:151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy CN, Khandaker I, Oshitani H: Intersubtype genetic variation of HIV-1 Tat exon 1. AIDS Res Hum Retroviruses 2015;31:641–648 [DOI] [PubMed] [Google Scholar]
- 43.Rossenkhan R, et al. : Tat exon 1 exhibits functional diversity during HIV-1 subtype C primary infection. J Virol 2013;87:5732–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]