Abstract
Purpose
To describe in detail a dataset consisting of serial four‐dimensional computed tomography (4DCT) and 4D cone beam CT (4DCBCT) images acquired during chemoradiotherapy of 20 locally advanced, nonsmall cell lung cancer patients we have collected at our institution and shared publicly with the research community.
Acquisition and validation methods
As part of an NCI‐sponsored research study 82 4DCT and 507 4DCBCT images were acquired in a population of 20 locally advanced nonsmall cell lung cancer patients undergoing radiation therapy. All subjects underwent concurrent radiochemotherapy to a total dose of 59.4–70.2 Gy using daily 1.8 or 2 Gy fractions. Audio‐visual biofeedback was used to minimize breathing irregularity during all fractions, including acquisition of all 4DCT and 4DCBCT acquisitions in all subjects. Target, organs at risk, and implanted fiducial markers were delineated by a physician in the 4DCT images. Image coordinate system origins between 4DCT and 4DCBCT were manipulated in such a way that the images can be used to simulate initial patient setup in the treatment position. 4DCT images were acquired on a 16‐slice helical CT simulator with 10 breathing phases and 3 mm slice thickness during simulation. In 13 of the 20 subjects, 4DCTs were also acquired on the same scanner weekly during therapy. Every day, 4DCBCT images were acquired on a commercial onboard CBCT scanner. An optically tracked external surrogate was synchronized with CBCT acquisition so that each CBCT projection was time stamped with the surrogate respiratory signal through in‐house software and hardware tools. Approximately 2500 projections were acquired over a period of 8–10 minutes in half‐fan mode with the half bow‐tie filter. Using the external surrogate, the CBCT projections were sorted into 10 breathing phases and reconstructed with an in‐house FDK reconstruction algorithm. Errors in respiration sorting, reconstruction, and acquisition were carefully identified and corrected.
Data format and usage notes
4DCT and 4DCBCT images are available in DICOM format and structures through DICOM‐RT RTSTRUCT format. All data are stored in the Cancer Imaging Archive (TCIA, http://www.cancerimagingarchive.net/) as collection 4D‐Lung and are publicly available.
Discussion
Due to high temporal frequency sampling, redundant (4DCT and 4DCBCT) data at similar timepoints, oversampled 4DCBCT, and fiducial markers, this dataset can support studies in image‐guided and image‐guided adaptive radiotherapy, assessment of 4D voxel trajectory variability, and development and validation of new tools for image registration and motion management.
Keywords: 4D imaging, computed tomography, cone beam computed tomography lung
1. Introduction
The prevalence of imaging in modern radiation therapy has enabled an improved understanding of geometric variation of the patient anatomy during the treatment course. The resulting widespread development of innovative methods to manage this variation during the treatment course is broadly known as image‐guided radiation therapy (IGRT). In‐room and onboard CT imaging supports characterization of patient anatomy in the treatment position prior to each fraction, enabling improved radiation targeting accuracy,1, 2 evaluation of the validity of the initial treatment plan,3 and assessing response to therapy.4, 5 Offline IGRT, such as resimulation of the patient, can be used to modify the treatment plan directly6 or integrate mid‐treatment multimodality imaging such as positron emission tomography and magnetic resonance imaging into this replanning process.7, 8 However, a variety of treatment site‐specific issues require careful design of the IGRT process to efficiently and effectively manage geometric variation. For example, breathing motion during imaging and delivery must be managed for thoracic and upper abdominal sites. Furthermore, anatomical variation is highly variable across patients, so a strategy that works for a single patient or clinic may not be appropriate for another. Testing of new IGRT strategies and tools can therefore be challenging, as it is often unknown what the best frequency and approach might be for the patient population without prospective evaluation in the clinic.
Fortunately, IGRT strategies and tools can often be thoroughly tested retrospectively using a database of patient images through a strategy such as a virtual clinical trial.6, 9 IGRT interventions, such as replans, couch shifts, etc. that don’t actively impact the patient geometry can be tested through such means. However, although there are numerous such published works, there is little publicly available data for conducting IGRT virtual clinical trials. Data that are available tends to be of low temporal frequency sampling (e.g., once or twice during the treatment course) or at inconsistent intervals from patient to patient.
The issue of lack of data is particularly critical in studies of IGRT in lung cancer. Because of the issues of breathing‐induced tissue motion, four dimensional (4D) imaging such as 4D fan beam CT (4DCT) and 4D cone beam CT (4DCBCT) are often required to appropriately characterize motion. These datasets, being larger in size and requiring typically higher imaging doses, are less frequently available.
The purpose of this work was to describe in detail a high‐frequency 4DCT and 4D cone beam CT (4D CBCT) dataset of 20 lung cancer patients we have collected at our institution and shared publicly with the research community. This dataset has previously been used by us for a variety of IGRT‐related studies, including virtual clinical trials of adaptive radiotherapy,6 testing of deformable image registration,10 a variety of studies describing inter‐ and intrafraction variation of targets and normal tissue,11, 12, 13, 14 and testing of 4DCBCT‐based lung ventilation imaging.15
2. Acquisition and validation methods
2.A. Overview of dataset
Throughout this work, we use the following terminology. An image refers to the complete reconstructed 4D scan, whereas a phase refers to a single 3D frame in this 4D image. A phase is therefore a 3D CT image, and a 4D image is thus composed of several (usually ten for our dataset) individual phases. In DICOM terminology (see Section 3), an image corresponds to a study and a phase to a series based on the way these data are stored in this dataset.
The dataset consists of serial 4DCT and 4DCBCT images of 20 locally advanced nonsmall cell lung cancer patients undergoing radiochemotherapy at the VCU Massey Cancer Center in the Department of Radiation Oncology, from 2008 through 2012. All patients provided informed consent and were enrolled on an IRB‐approved, NCI‐sponsored prospective imaging study. The subjects were imaged at or near the time of simulation with 4DCT. Onboard 4DCBCT images were acquired on all subjects for most treated fractions (average 25, range 2 to 35 per patient). Thirteen of the 20 subjects also underwent repeat 4DCT imaging once or more during treatment (average 6, range 3 to 8 per patient). Figure 1 shows example 4DCT and 4DCBCT phase images for two subjects. Targets and risk structures were delineated by a physician on 4DCT images. See Table 1 for detailed image counts for each subject.
Figure 1.
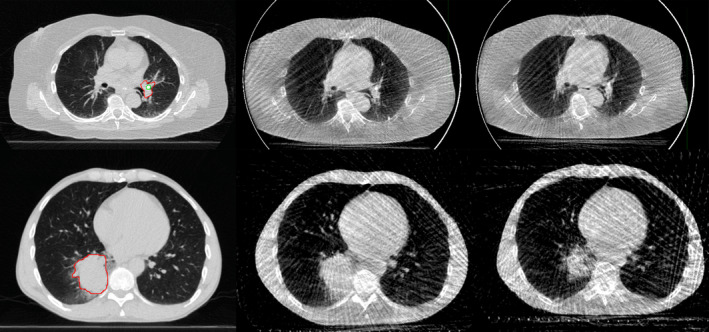
Example images from the dataset. Top row, left to right: 4DCT, 4DCBCT at first fraction, 4DCBCT at end of treatment for P104. Bottom row, corresponding images for P114. All images from the end of inhalation breathing phase. The gross tumor volume outline is shown on 4DCT for both patients. For P104, an implanted fiducial marker is also outlined.
Table 1.
Number and type of 4D images available in the VCU 4D Lung dataset. Each of the 4DCT and 4DCBCT images is composed of 10 phase images
| Subject | Number of 4DCT images (delineated) | Number of 4DCBCT images | Total images |
|---|---|---|---|
| 100 | 1 (1) | 33 | 34 |
| 101 | 1 (1) | 10 | 11 |
| 102 | 1 (1) | 16 | 17 |
| 103 | 3 (1)a | 35 | 38 |
| 104 | 1 (1) | 31 | 32 |
| 105 | 1 (1) | 33 | 34 |
| 106 | 1 (1) | 31 | 32 |
| 107 | 5 (5) | 19 | 24 |
| 108 | 5 (5) | 23 | 28 |
| 109 | 5 (5) | 28 | 33 |
| 110 | 5 (5) | 29 | 34 |
| 111 | 4 (4) | 30 | 34 |
| 112 | 6 (6) | 34 | 40 |
| 113 | 5 (5) | 2 | 7 |
| 114 | 5 (5) | 28 | 33 |
| 115 | 6 (6) | 28 | 34 |
| 116 | 5 (5) | 28 | 33 |
| 117 | 8 (8) | 26 | 34 |
| 118 | 6 (6) | 25 | 31 |
| 119 | 8 (8) | 18 | 26 |
| Total | 82 (80) | 507 | 589 |
Subject 103 had an initial 4DCT for planning prior to marker implantation, and a resimulation mid‐treatment. These scans are included in the database but were not delineated.
2.B. Acquisition
2.B.1. Clinical parameters
Table 2 lists clinical treatment information for all subjects, including prescription and fractionation, stage, tumor location and size. All patients underwent curative intent radiochemotherapy to a total dose of 59.4–70.2 Gy using daily 1.8 or 2 Gy fractions. Because the imaging dose was significantly higher than typical for a clinical course (approximately 4–5% of the prescription dose), imaging dose was estimated for each subject during planning and included in the treatment plan for physician assessment and approval.
Table 2.
Clinical and treatment information for each subject. (RUL = right upper lung, RLL = right lower lung, LUL = left upper lung, LLL = left lower lobe)
| Subject | T | N | M | Overall stage | Location | Total dose (Gy) | Number of fractions | Dose per fraction (Gy) | Tumor volume (cc) |
|---|---|---|---|---|---|---|---|---|---|
| 100 | 4 | 3 | 0 | IIIB | RUL | 62.6 | 32 | 2.0 (1.8 for 7) | 75 |
| 101 | 4 | 3 | 0 | IIIB | RUL | 66.6 | 37 | 1.8 | 27 |
| 102 | 4 | 2 | 0 | IIIB | LUL | 66 | 33 | 2 | 171 |
| 103 | 4 | 2 | 0 | IIIB | RUL | 66.6 | 37 | 1.8 | 58 |
| 104 | 2 | 3 | 0 | IIIB | LLL | 70 | 35 | 2 | 47 |
| 105 | 3 | 1 | 0 | IIIA | LLL | 70 | 35 | 2 | 33 |
| 106 | 0 | 2 | 0 | IIIA | mediastinum | 66 | 33 | 2 | 143 |
| 107 | 2 | 2 | 0 | IIIA | LUL | 63 | 35 | 1.8 | 18 |
| 108 | 2 | 2 | 0 | IIIA | RUL | 70.2 | 39 | 1.8 | 12 |
| 109 | 4 | 2 | 0 | IIIB | RLL | 59.4 | 33 | 1.8 | 392 |
| 110 | 2 | 3 | 0 | IIIB | RLL | 62 | 31 | 2 | 55 |
| 111 | 3 | 1 | 0 | IIIA | RLL | 64 | 32 | 2 | 75 |
| 112 | 3 | 2 | 0 | IIIA | RUL | 63 | 35 | 1.8 | 31 |
| 113 | 1 | 1 | 0 | IIA | LLL | 66 | 33 | 2 | 78 |
| 114 | 2 | 2 | 0 | IIIA | RLL | 66 | 33 | 2 | 179 |
| 115 | 1 | 3 | 0 | IIIB | RUL | 66.6 | 37 | 1.8 | 7 |
| 116 | 3 | 2 | 0 | IIIA | LUL | 70 | 35 | 2 | 33 |
| 117 | 3 | 2 | 0 | IIIA | RUL | 66 | 33 | 2 | 10 |
| 118 | 3 | 2 | 0 | IIIA | RUL | 66 | 33 | 2 | 13 |
| 119 | 4 | 3 | 0 | IIIB | RUL | 66 | 33 | 2 | 142 |
2.B.2. 4DCT
4DCT images were acquired on a 16‐slice helical CT simulator (Brilliance Big Bore, Philips Medical Systems, Andover, MA, USA) as respiration‐correlated CTs. An external respiration signal (Real‐time Position Management (RPM), Varian Medical Systems, Inc., Palo Alto, CA, USA) was used to acquire the respiratory signal and to sort the raw data into 10 breathing phases (0 to 90%) using a phase sorting approach. The 0% phase corresponds to end of inhalation. The reconstructed slice thickness was 3 mm for all images and in‐plane spacing was 0.98 to 1.17 mm. The technique was 120 kVp for all scans, 50 to 114 mA, and 3.53 to 5.83 ms.
Audio‐visual biofeedback was performed during all 4DCT acquisitions in all subjects using a prototype system.16 A training RPM waveform was acquired during the first 4DCT acquisition, and used as the reference waveform for guidance during all subsequent imaging sessions.
The 4DCT images are stored with numerical identifiers (series description, see Section 3 for details) in the form SXXX, where XXX is a number starting at 300. Note that this identifier does not necessarily correspond with the study (acquisition) date and therefore images should not be ordered by series identifier to simulate treatment order. Instead, the study date should be used to order images. The study dates have been changed from the actual acquisition dates to protect patient confidentiality by adding a fixed offset to the acquisition date of each scan. Modification was done in this manner to preserve the relative time between acquisitions.
All subjects had a 4DCT acquired either as the clinical planning image, or near the time of simulation. Four subjects were planned using free‐breathing CT prior to the study 4DCT being acquired. For these four subjects, a study 4DCT was selected as a reference image, which we term the planning 4DCT. For these four subjects, Table 3 notes the time from simulation until acquisition of this study planning 4DCT. Either way, the planning image does not necessarily have a numerical identifier of S300, due to either scan acquisition issues or other factors. Table 3 lists the numerical identifier for the image considered as the planning CT for study purposes and provides the time from image acquisition until treatment start.
Table 3.
Planning 4DCT information for each subject
| Subject | Planning 4DCT study identifier | Is planning CT? | Acquisition time to treatment start time (days) | Comments |
|---|---|---|---|---|
| 100 | S300 | Y | 14 | |
| 101 | S300 | Y | 9 | |
| 102 | S300 | Y | 18 | |
| 103 | S301 | Y | 13 | |
| 104 | S300 | Y | 15 | |
| 105 | S300 | Y | 13 | |
| 106 | S301 | Y | 18 | |
| 107 | S300 | N | −2 | 15 days after simulation |
| 108 | S304 | N | 0 | 12 days after simulation |
| 109 | S302 | Y | 12 | |
| 110 | S300 | Y | 14 | |
| 111 | S301 | Y | 12 | |
| 112 | S301 | Y | 26 | |
| 113 | S300 | N | −16 | 30 days after simulation |
| 114 | S300 | N | 0 | 12 days after simulation |
| 115 | S300 | Y | 13 | |
| 116 | S301 | Y | 14 | |
| 117 | S300 | Y | 10 | |
| 118 | S306 | Y | 12 | |
| 119 | S300 | Y | 14 |
‘Is planning CT?’ denotes if this image was used as the clinical planning CT. Acquisition time to treatment start time gives the number of days from acquisition of this image to the start of treatment (first fraction). See also supplemental material for timing information.
For subjects with repeat 4DCT during treatment, these 4DCT images were registered to the planning 4DCT based on bony anatomy in the treatment planning system (Philips Pinnacle v9.0). See (Supplemental material) includes a table which lists the cumulative dose delivered through the acquisition date for each of these repeat during‐treatment 4DCT images.
As all 4DCT images were acquired on the same scanner, a Hounsfield unit to electron density calibration curve reproduced from the clinical treatment planning system is listed in Table 4 to facilitate treatment planning on these images.
Table 4.
Hounsfield unit to relative electron density calibration curve for the 4DCT images in this study
| CT intensity (HU) | Electron density relative to water |
|---|---|
| −1000 | 0.00 |
| −705 | 0.30 |
| −569 | 0.41 |
| −95 | 0.92 |
| −42 | 0.98 |
| 0 | 1.00 |
| 40 | 1.05 |
| 86 | 1.11 |
| 225 | 1.14 |
| 227 | 1.16 |
| 473 | 1.34 |
| 845 | 1.56 |
| 1283 | 1.82 |
| 5812 | 5.76 |
| 9035 | 8.00 |
| 9754 | 8.50 |
2.B.3. 4DCBCT
4D‐CBCT images were acquired on a commercial CBCT scanner (On‐Board Imager v1.3; Varian Medical Systems, Inc.) after in‐house modification. The same external breathing surrogate used for 4DCT was integrated into the CBCT acquisition system to stamp each CBCT projection with the surrogate respiratory signal through in‐house software and hardware tools. Approximately 2000–2500 projections were acquired over a period of 8–10 minutes in half‐fan mode with half bow‐tie filter. The technique was 125 kVp, 20 mA, and 20 ms in a single 360 slow gantry arc. The rotational gantry speed was varied using the technique described by Lu et al. 17 so that the angular sampling frequency was similar for each subject and independent of breathing period. Using the external surrogate, the CBCT projections were sorted into 10 breathing phases (0 to 90%, phase‐based binning). As with 4DCT, the 0% phase corresponded with end of inhalation. 4DCBCT was reconstructed using an in‐house Feldkamp‐Davis‐Kress (FDK) reconstruction algorithm, with minimal preprocessing (median filtering for noise reduction). Similar to 4DCT acquisition, audio‐visual biofeedback was performed during all 4DCBCT acquisitions. Figure 2 shows several phase images from two 4DCBCT images acquired during the first and last fractions for a single subject.
Figure 2.
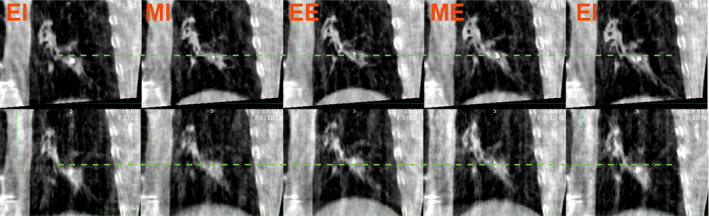
Example 4DCBCT phase images from the dataset, subject P105. Top row: First fraction 4DCBCT. Bottom row: 4DCBCT during 32nd fraction. The dotted line shows the superior tumor border at end of inhalation. The amplitude of motion is higher in the later 4DCBCT, as evidenced by a more superior position and end of exhalation. Left to right, approximate breathing phase: EI ‐ End inhalation phase, MI ‐ Mid inhalation phase, EE ‐ End exhalation phase, ME ‐ Mid exhalation phase, EI. These correspond to the 0%, 20%, 40%, 60%, and 80% phases, respectively.
The 4DCBCT image headers were generated so that aligning the planning 4DCT to the 4DCBCT by the origin in the image header will simulate the actual patient setup prior to CBCT imaging.
Analogous to the 4DCT images, the numerical identifier in the series description is formatted SXXX; however, XXX for 4DCBCT images starts at 100. For similar reasons as for the 4DCT images, study date rather than numerical identifier should be used to order the images by acquisition time. Supplemental material17 includes a table which lists the delivered cumulative dose through the date of acquisition for each 4DCBCT image. For example, for the image acquired at the second fraction for a 2 Gy dose per fraction, this table would list 4 Gy as the delivered cumulative dose for this image. The table is organized by study identifier and subject number. Note that several subjects have multiple 4DCBCTs acquired on the same day, which are pre‐ and post‐treatment images. The order of these two images can be found through the DICOM acquisition time tag.
2.B.4. Fiducial marker description and implantation procedure
Seven of the patients had two to four 0.35 mm diameter (either 10 or 20 mm length) gold fiducial markers (Visicoil, IBA Dosimetry, Bartlett, TN, USA) implanted in or near the tumor or involved lymph nodes. The markers were implanted prior to 4DCT imaging by a pulmonologist using either endobronchial ultrasound‐guided bronchoscopy or electromagnetic navigational bronchoscopy (Covidien superDimension, Minneapolis, MN, USA), with the patient under conscious sedation. 4DCT images were then acquired on the same day. Details about marker location, stability, and analysis of marker to target variation can be found in Roman et al. 11
2.B.5. Physician‐delineated planning structures
A single experienced radiation oncologist (EW) supervised delineation of all target and organ at risk (OAR) structures in the 4DCT datasets. The gross tumor volume and any visible involved lymph nodes were delineated on all phases of all 4DCT images. However, because of the very large size of the dataset, organs at risk were contoured on only a subset of images. Table 5 lists the number of delineated phases on each 4DCT, for each image and structure. The delineated targets and organs at risk are named as listed in Table 5 with ‘_cXX’ appended, where ‘XX’ is the 4DCT phase from 00 to 90. For example, the tumor on the 40% phase is named ‘Tumor_c40’.
Table 5.
Count of target, organ at risk, and other structures delineated on the 4DCT images. A count of 10 means all 10 phases of the 4DCT are delineated. A count of 1 means only the 0% phase has been delineated. (LN = lymph node, Esoph = esophagus, LL = left lung, RL = right lung, Trach = trachea, Vert = vertebral body)
| Subject | Image | Targets | Markers | Organs at risk | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor | LN | LN2 | A | B | C | D | Carina | Cord | Esoph | Heart | LL | RL | Trach | Vert | |||
| 100 | S300 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 1 | 121 | ||
| 101 | S300 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 1 | 121 | ||
| 102 | S300 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 1 | 121 | ||
| 103 | S301 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 1 | 101 | ||||
| 104 | S300 | 10 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 1 | 46 | |||||
| 105 | S300 | 10 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 55 | |||||
| 106 | S301 | 10 | 10 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 65 | ||||
| 107 | S300 | 10 | 10 | 10 | 1 | 10 | 1 | 10 | 10 | 10 | 72 | ||||||
| 107 | S301 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 70 | ||||||||
| 107 | S302 | 10 | 10 | 10 | 10 | 1 | 1 | 10 | 52 | ||||||||
| 107 | S303 | 10 | 10 | 10 | 10 | 1 | 1 | 10 | 52 | ||||||||
| 107 | S304 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 70 | ||||||||
| 108 | S301 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 108 | S302 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 108 | S303 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 108 | S304 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 45 | ||||||
| 108 | S305 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 109 | S300 | 10 | 10 | 10 | 30 | ||||||||||||
| 109 | S301 | 10 | 10 | 10 | 30 | ||||||||||||
| 109 | S302 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 35 | |||||||
| 109 | S304 | 10 | 10 | 10 | 30 | ||||||||||||
| 109 | S305 | 10 | 10 | 10 | 30 | ||||||||||||
| 110 | S300 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 45 | ||||||
| 110 | S301 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 110 | S303 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 110 | S305 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 110 | S306 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 111 | S301 | 10 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 55 | |||||
| 111 | S302 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 111 | S303 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 111 | S304 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 112 | S301 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 45 | ||||||
| 112 | S302 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 112 | S303 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 112 | S304 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 112 | S305 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 112 | S306 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 113 | S300 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 113 | S301 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 113 | S302 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 113 | S303 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 113 | S304 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 114 | S300 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 45 | ||||||
| 114 | S301 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 114 | S302 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 114 | S303 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 114 | S304 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 115 | S300 | 10 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 10 | 54 | ||||||
| 115 | S301 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 115 | S302 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 115 | S303 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 115 | S304 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 115 | S305 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 116 | S300 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 116 | S301 | 10 | 10 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 10 | 55 | |||||
| 116 | S302 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 116 | S303 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 116 | S304 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 117 | S300 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 117 | S301 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 117 | S302 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 117 | S303 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 117 | S304 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 117 | S305 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 117 | S306 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 117 | S308 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 118 | S302 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 118 | S303 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 118 | S304 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 118 | S306 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 118 | S307 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 118 | S308 | 10 | 10 | 10 | 10 | 40 | |||||||||||
| 119 | S300 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 119 | S301 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 119 | S302 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 119 | S303 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 119 | S304 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 119 | S305 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 119 | S306 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| 119 | S307 | 10 | 10 | 10 | 10 | 10 | 50 | ||||||||||
| Grand Total | 800 | 720 | 220 | 70 | 70 | 40 | 10 | 800 | 51 | 101 | 52 | 83 | 83 | 20 | 755 | 3875 | |
For the seven patients with implanted fiducial markers, these are labeled as markers A–D, and also are listed in Table 5. Markers, where visible, were delineated on 4DCT.
2.C. Validation
2.C.1. Processing and quality assurance
After acquisition, 4DCT images were immediately reviewed for phase sorting artifact. If present, the end inhalation tags placed by the Philips CT simulator were manually reviewed and modified, if necessary. The image was again reconstructed and reviewed. Due to the high volume of imaging, if this process did not correct artifact we did not reacquire 4DCT, but instead accepted this image.
4DCBCT data in the form of raw projections and external surrogate signal were reviewed after acquisition by physics personnel (SB, GH) for fidelity. Several issues were identified and corrected when possible. The RPM system automatically identifies breathing phase. In 52 acquisitions (10 different patients) out of 507, the system incorrectly identified the phase in at least a portion of the respiratory trace. These errors were corrected manually, and the 4DCBCT images were reconstructed using the corrected phase tags. Projection angle errors were identified and corrected in 5 scans by plotting the projection angle vs. projection number and locating projections that had substantial errors relative to adjacent projections. Incomplete projections were identified in four scans which could not be corrected. These images were removed from the dataset. After all error correction, each reconstructed 4DCBCT image was reviewed visually.
Subject P113 had metal hardware implanted in the spine which generated artifact on 4DCBCT that was determined to limit use of these images. Because of this issue, 4DCBCT imaging was discontinued in this subject after two fractions at start of treatment. However, the subject remained on study for the 4DCT portion.
All 4DCT and 4DCBCT data were then transferred to a research database. 4DCT were initially reconstructed on the clinical CT simulator, deidentified and anonymized, and then moved in DICOM format to the research database. For 4DCBCT data, raw projection and RPM data were deidentified and anonymized and then moved to the research database and reconstructed in DICOM format. The reconstructed 4DCBCT were then moved to the research database. A Pinnacle database was then constructed for each patient to enable contouring.
A data acquisition and processing log was kept by study personnel for each subject so that data integrity could be verified in the Pinnacle database against the log of acquired data. A changelog of all data (unprocessed and processed) was kept for both images and structures. Out of 590 acquired images, one 4DCT image was unable to be transferred into the database, leaving 589 images for processing and analysis.
The shifts applied to the 4DCT image origin (see Section 2.B.3) were verified by registering each 4DCBCT image to the planning 4DCT image and comparing the resulting measured shift to the clinically applied shift stored in the clinical record and verify system.
Delineated structures were delineated by several physicians, but reviewed by a single radiation oncologist (EW). To reduce intra‐observer variation, each structure was initially delineated manually on a single phase image but then copied to other phases through rigid registration for subsequent phases in the same image. The structure was then adjusted manually to conform to the appropriate phase image.
2.C.2. Known issues and limitations
There are several known issues and limitations of this dataset, based on the large size and required high frequency of imaging.
First, sorting artifact is prevalent in the 4DCT images. While this makes some analyses challenging, it does reproduce the clinical situation where such artifact is widespread.18 Due to the already‐high imaging dose from the study, we chose not to reacquire 4DCT images for study purposes even if artifact was identified.
Second, 4DCBCT, not being commercially available on our treatment units, was acquired in a noncommercial mode. Thus, 4DCBCT image quality may not be of clinical quality due to the use of in‐house reconstruction with only minimal processing. Furthermore, despite the high angular sampling rate, reconstruction of 10 phases resulted in approximately 250 projections per phase. Thus, the 4DCBCT phase images all have noticeable view‐aliasing streak artifacts due to angular undersampling which limits signal to noise ratio and low contrast detectability. Due to this limited image quality in 4DCBCT images, normal tissue structures were not delineated in these images. Tumor and markers were delineated in 4DCBCT,11 but are not included in the current dataset. We are planning to process these for eventual inclusion in the TCIA dataset.
3. Data format and usage notes
The Cancer Imaging Archive (TCIA) at the National Institutes of Health was selected as the repository for long term storage of this dataset.19 All data were exported from Pinnacle in DICOM (images) and DICOM‐RT (structures) format. Although the dataset was initially deidentified and anonymized at VCU, subsequently TCIA processed the dataset further to ensure all potentially confidential data were removed.20
Imaging data are stored with an anonymized patient name and identifier. Subjects are labeled as PXXX, where XXX is the subject from 100 to 119. The PatientID DICOM tag is similarly labeled. Images can be identified by the DICOM study date tag, which as mentioned was offset but preserves relative time between images. Also, the DICOM series description contains the patient ID, study ID, and 4D phase. This tag is structured, for example, as P4 P100 S102 I0, Gated, 40.0%. P4 identifies an internal project identifier at VCU. P100 identifies the subject as 100. S102 identifies the study as study 102. Because the study identifier is in the 100s, it is a 4DCBCT image (300 and up are reserved for 4DCT, see Section 2.B.2). I0 is an internal identifier that can be ignored. Finally, the phase is identified as a percentage [0, 90]. DICOM tags related to acquisition such as the technique, image orientation, and spacing were preserved during deidentification and anonymization.
When downloaded, at the time of publication, the TCIA software stores the images on the user's system in a directory hierarchy first by subject then by series UID. This directory structure is dependent on the TCIA software, and may change in the future. To reorganize the dataset on disk, it is best to use software that can parse the entire directory structure and select DICOM files by the series description and other identifiers described in the previous paragraph. Many commercial image analysis software packages have such functionality. A free alternative is DicomBrowser (http://nrg.wustl.edu/software/dicom-browser).
The dataset can be found as collection 4D‐Lung at the TCIA website http://www.cancerimagingarchive.net/. Alternatively, the dataset can be accessed by digital object identifier (DOI) http://doi.org/10.7937/K9/TCIA.2016.ELN8YGLE.
The total size of the dataset is 183 GB, with 6690 individual DICOM files consisting of 5890 image files and 800 RTSTRUCT files. DICOM and DICOM‐RT compatibility have been tested with Philips Pinnacle and MIM Maestro (MIM Software, Cleveland, OH, USA).
4. Discussion
In spite of the limitations, this dataset has several advantages over similar datasets. First, 4DCBCT is available in a large number of treated fractions in many of the subjects. The 4DCBCT images themselves are acquired with very fine angular sampling, consisting of roughly 2000–2500 projections and being acquired over 8–10 minutes compared to approximately 700 projections and 2 minutes for a typical clinical scan. The 4DCBCT therefore have less view‐aliasing (streaking) artifact than comparable clinical 4DCBCT scans. These factors would allow the dataset to be of value for testing motion management strategies, particularly the effect of interfraction changes on motion management.
Second, in many patients, there are 4DCT images acquired on or near the same day as 4DCBCT images. This allows the high‐quality 4DCT data to be used to validate strategies and tools applied on the 4DCBCT images. For example, tools to improve CBCT to CT registration and localization21 could be tested in 4DCBCT but validated in 4DCT. Stability of image features and consistency of these features between CT and CBCT could be compared to assess performance of radiomics strategies.
Third, fiducial markers were implanted in seven subjects, were delineated in the 4DCT images for these patients, and are visible in the 4DCBCT images. Such markers can be used to validate a variety of studies, including image registration and tumor tracking, as well as evaluation of the fidelity of the markers as fiducials themselves.11 In separate work, we segmented the markers in some of the CBCT projections, and reconstructed the 3D marker positions during CBCT acquisition.22 These data can be used to measure instantaneous translation and rotation of lung tumors,14 for example. Although not included in the TCIA dataset, these data (both projections and marker traces) can be obtained by contacting the authors.
Finally, due to the high frequency of imaging, this dataset may be most useful for testing of IGRT strategies such as online or offline correction protocols and for simulating adaptive radiotherapy and related tools such as deformable image registration and dose mapping.
5. Conclusions
In summary, we have constructed a dataset consisting of serial 4DCT and 4DCBCT images acquired during chemoradiotherapy of 20 locally advanced, nonsmall cell lung cancer patients and corresponding delineated targets and organs at risk on a subset of these images. The dataset has been archived in a standardized format in the publicly available TCIA. This dataset was used to test a variety of image‐guided, adaptive, and motion management strategies for radiotherapy and should be of use to the research community for related studies.
Conflicts of interest
Virginia Commonwealth University has a research agreement with Philips Medical Systems, and a licensing agreement with Varian Medical Systems. EW receives royalties from UpToDate. JFW is Editor‐in‐Chief of Medical Physics and is supported by a contract from the American Association of Physicists in Medicine.
Supporting information
Data S1: Fractiondoses.
Acknowledgments
We first thank the patients who participated in this study. We also thank the VCU research staff and radiation therapists who organized and performed the imaging, and we thank Ken Clark, John Freymann, Justin Kirby, and Kirk Smith at the TCIA and Jim Deye at the NCI for their help in designing and hosting the public access to this dataset. This work was supported in part by research grants from the National Cancer Institute of the National Institutes of Health under award numbers P01CA116602 and R01CA166119. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Purdie TG, Bissonnette JP, Franks K, et al. Cone‐beam computed tomography for on‐line image guidance of lung stereotactic radiotherapy: localization, verification, and intrafraction tumor position. Int J Radiat Oncol Biol Phys. 2007;68:243–252. [DOI] [PubMed] [Google Scholar]
- 2. Grills IS, Hugo G, Kestin LL, et al. Image‐guided radiotherapy via daily online cone‐beam CT sub‐ stantially reduces margin requirements for stereotactic lung radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70:1045–1056. [DOI] [PubMed] [Google Scholar]
- 3. Kwint M, Conijn S, Schaake E, et al. Intra thoracic anatomical changes in lung cancer patients during the course of radiotherapy. Radiother Oncol. 2014;113:392–397. [DOI] [PubMed] [Google Scholar]
- 4. Bral S, De Ridder M, Duchateau M, et al. Daily megavoltage computed tomography in lung cancer radiotherapy: correlation between volumetric changes and local outcome. Int J Radiat Oncol Biol Phys. 2011;80:1338–1342. [DOI] [PubMed] [Google Scholar]
- 5. Brink C, Bernchou U, Bertelsen A, Hansen O, Schytte T, Bentzen SM. Locoregional control of non‐small cell lung cancer in relation to auto‐ mated early assessment of tumor regression on cone beam computed tomography. Int J Radiat Oncol Biol Phys. 2014;89:916–923. [DOI] [PubMed] [Google Scholar]
- 6. Weiss E, Fatyga M, Wu Y, et al. Dose escalation for locally advanced lung cancer using adaptive radiation therapy with simultaneous integrated volume‐adapted boost. Int J Radiat Oncol Biol Phys. 2013;86:414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grootjans W, de Geus‐Oei L‐F, Troost EGC, Visser EP, Oyen WJG, Bussink J. PET in the management of locally advanced and metastatic NSCLC. Nat Rev Clin Oncol. 2015;12:395–407. [DOI] [PubMed] [Google Scholar]
- 8. Hermans BCM, Persoon LCGG, Podesta M, Hoebers FJP, Verhaegen F, Troost EGC. Weekly kilovoltage cone‐beam computed tomography for detection of dose discrepancies during (chemo)radiotherapy for head and neck cancer. Acta Oncol. 2015;54:1483–1489. [DOI] [PubMed] [Google Scholar]
- 9. Guckenberger M, Wilbert J, Richter A, Baier K. Potential of adaptive radiotherapy to escalate the radiation dose in combined radiochemotherapy for locally advanced non‐small cell lung cancer. Int J Radiat Oncol Biol Phys. 2011;79:901–908. [DOI] [PubMed] [Google Scholar]
- 10. Balik S, Weiss E, Jan N, et al. Eval‐ uation of 4‐dimensional computed tomography to 4‐dimensional cone‐beam computed tomography deformable image registration for lung cancer adaptive radiation therapy. Int J Radiat Oncol Biol Phys. 2013;86:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roman NO, Shepherd W, Mukhopadhyay N, Hugo GD, Weiss E. Interfractional positional variability of fiducial markers and primary tumors in lo‐ cally advanced non‐small‐cell lung cancer during audiovisual biofeedback radiotherapy. Int J Radiat Oncol Biol Phys. 2012;83:1566–1572. [DOI] [PubMed] [Google Scholar]
- 12. Jan N, Balik S, Hugo GD, Mukhopadhyay N, Weiss E. Interfraction displacement of primary tumor and involved lymph nodes relative to anatomic landmarks in image guided radiation therapy of locally advanced lung cancer. Int J Radiat Oncol Biol Phys. 2014;88:210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jan N, Hugo GD, Mukhopadhyay N, Weiss E. Respiratory motion variability of primary tumors and lymph nodes during radiotherapy of locally advanced non‐small‐cell lung cancers. Radiat Oncol. 2015;10:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang C‐Y, Tehrani JN, Ng JA, Booth J, Keall P. Six degrees‐of‐freedom prostate and lung tumor motion measurements using kilovoltage intrafraction monitoring. Int J Radiat Oncol Biol Phys. 2015;91:368–375. [DOI] [PubMed] [Google Scholar]
- 15. Kipritidis J, Hugo G, Weiss E, Williamson J, Keall PJ. Measuring interfraction and intrafraction lung function changes during radiation therapy using four‐dimensional cone beam CT ventilation imaging. Med Phys. 2015;42:1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Venkat RB, Sawant A, Suh Y, George R, Keall PJ. Development and preliminary evaluation of a prototype audiovisual biofeedback device incorporating a patient‐ specific guiding waveform. Phys Med Biol. 2008;53:N197–N208. [DOI] [PubMed] [Google Scholar]
- 17. Lu J, Guerrero TM, Munro P, et al. Four‐dimensional cone beam CT with adaptive gantry rotation and adaptive data sampling. Med Phys. 2007;34:3520–3529. [DOI] [PubMed] [Google Scholar]
- 18. Yamamoto T, Langner U, Loo BW, Shen J, Keall PJ, Loo BW Jr. Retrospective analysis of artifacts in four‐dimensional CT images of 50 abdominal and thoracic radiotherapy patients. Int J Radiat Oncol Biol Phys. 2008;72:1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freymann J, Kirby J, Perry JH, Clunie DA, Jaffe CC. Image data sharing for biomedical research–meeting HIPAA requirements for De‐identification. J Digit Imaging. 2012;25:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robertson SP, Weiss E, Hugo GD. A block matching‐based registration algorithm for localization of locally advanced lung tumors. Med Phys. 2014;41:041704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poulsen PR, Fledelius W, Keall PJ, et al. A method for robust segmentation of arbitrarily shaped radiopaque structures in cone‐beam CT projections. Med Phys. 2011;38:2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Fractiondoses.


