Abstract
Volatile metabolites are currently under investigation as potential biomarkers for the detection and identification of pathogenic microorganisms, including bacteria, fungi, and viruses. Unlike bacteria and fungi, which produce distinct volatile metabolic signatures associated with innate differences in both primary and secondary metabolic processes, viruses are wholly reliant on the metabolic machinery of infected cells for replication and propagation. In the present study, the ability of volatile metabolites to discriminate between respiratory cells infected and uninfected with virus, in vitro, was investigated. Two important respiratory viruses, namely respiratory syncytial virus (RSV) and influenza A virus (IAV), were evaluated. Data were analyzed using three different machine learning algorithms (random forest (RF), linear support vector machines (linear SVM), and partial least squares-discriminant analysis (PLS-DA)), with volatile metabolites identified from a training set used to predict sample classifications in a validation set. The discriminatory performances of RF, linear SVM, and PLS-DA were comparable for the comparison of IAV-infected versus uninfected cells, with area under the receiver operating characteristic curves (AUROCs) between 0.78 and 0.82, while RF and linear SVM demonstrated superior performance in the classification of RSV-infected versus uninfected cells (AUROCs between 0.80 and 0.84) relative to PLS-DA (0.61). A subset of discriminatory features were assigned putative compound identifications, with an overabundance of hydrocarbons observed in both RSV- and IAV-infected cell cultures relative to uninfected controls. This finding is consistent with increased oxidative stress, a process associated with viral infection of respiratory cells.
Keywords: virus, VOCs, metabolomics, comprehensive two-dimensional gas chromatography, GC×GC, mass spectrometry
1. Introduction
Infections of the lower respiratory tract, including both influenza and pneumonia, are among the top 10 leading causes of death in the United States [1], and pneumonia remains one of the world’s leading causes of death for children under the age of five [2]. According to the Centers for Disease Control and Prevention (CDC), approximately 30% of acute respiratory infections of viral etiology in the United States (roughly 47 million cases annually) are inappropriately treated with antimicrobial therapies that are not effective against viral pathogens [3–5]. Furthermore, it is estimated that a causative pathogen is identified in only approximately 40% of pneumonia cases overall, and a subset of these cases for which a pathogen could not be identified are likely of viral etiology [6]. A diagnostic capable of rapidly distinguishing between infections of viral, bacterial, or fungal etiology could inform the clinical management of individuals with respiratory infections, potentially reducing the inappropriate use of antibiotics for viral infections [7, 8].
Limitations of currently-available diagnostic tools for the detection of lower respiratory infections are mainly related to the difficulty of obtaining an adequate sputum sample (e.g., sputum is not produced by most children) and in differentiating between infection and colonization in the setting of a positive result [9]. Specifically, one must be careful when interpreting the results obtained from tests that specifically target organisms such as Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae, or certain fungi (i.e., Candida), as up to 20% of healthy individuals can be asymptomatically colonized [10]. Several rapid, multiplex diagnostic tests for organism detection are commercially available [8, 11, 12], but their role at present is limited, since, in addition to the previously-mentioned shortcomings, they lack proper evaluation of their selectivity and specificity, mainly due to the absence of an indisputable gold standard techniques for the identification of many pathogens [8, 10–13].
To-date, most assays for the detection of respiratory viruses have focused on the identification of either virally-derived nucleic acids (e.g., multiplex PCR, such as PneumoVir®) or antigens (e.g., rapid influenza immunoassays, such as DirectigenTM EZ Flu A + B). Recently, however, volatile metabolites in exhaled breath have been investigated as potential alternative biomarkers for pathogen detection and identification. For example, volatile metabolites in breath are widely used in the diagnosis of Helicobacter pylori gastritis [14], and are under investigation for the diagnosis of both acute and chronic respiratory infections [15]. In the murine model, it has been shown that volatile metabolites can discriminate between respiratory infections caused by common bacterial pathogens, including H. influenzae, Klebsiella pneumoniae, Legionella pneumophila, Moraxella catarrhalis, Pseudomonas aeruginosa, S. aureus, and S. pneumoniae [16–18]. However, unlike bacteria, which produce distinct volatile metabolic signatures derived from fundamental differences in components of both core and secondary metabolism [19], viruses are entirely reliant on the metabolic machinery of infected cells. Several transcriptomics studies have demonstrated that different infectious agents (both viruses and bacteria) trigger specific pattern-recognition receptors expressed on host immune cells, activating different transcriptional factors that activate specific metabolic programs [20–30]. For instance, the cytokine profile induced by influenza A virus (IAV) infection in infants is distinct from the profile induced by respiratory syncytial virus (RSV) [30]. In light of these findings, we hypothesized that volatile metabolic signatures could differentiate between virally-infected and uninfected cells. In addition to assessing the diagnostic utility of such an approach, the study of volatile metabolites produced during infection has the potential to generate insight into viral pathogenesis.
To-date, few studies have focused on the identification of volatile metabolites produced by cell cultures infected with virus (i.e., influenza, RSV, human rhinovirus, adenovirus, and herpes simplex) [31–35]. These studies involved basic characterization of the headspace of infected cell culture versus uninfected cell culture, but did not evaluate the discrimination capability of the volatile metabolites produced during infection. The aim of this study is therefore to generate volatile fingerprints of cell cultures infected with virus (both RSV and IAV) and to evaluate their discrimination capability. Volatile metabolites were extracted from the headspace using solid-phase microextraction (SPME) and then separated and identified by comprehensive two-dimensional gas chromatography (GC×GC) hyphenated with a ToF mass spectrometer (MS). The present study represents a novel application of this technique, which is particularly well-suited for the analysis of complex mixtures and is amongst the most powerful analytical tools available today for the analysis of volatile metabolites [36]. Using different machine learning algorithms, we were able to identify volatile metabolic patterns that could discriminate between cells infected with virus and those that were uninfected.
2. Materials and methods
2.1. Viral infection of human cell lines
2.1.1. RSV infection
Six-well microtiter plates were seeded with HEp-2 cells (a human laryngeal cancer cell line) from the American Type Culture Collection (ATCC®, CL-23™) (4 × 105 cells/well) to be 70%–80% confluent in 24 h. Human RSV (ATCC® VR-1540™) was diluted to a multiplicity of infection (MOI) of 0.3 in phosphate-buffered saline. HEp-2 cells were maintained in a growth media consisting of Minimum Essential Medium (MEM) (Corning CellGro 15-010) containing penicillin (100 units ml−1) and streptomycin (100 μg ml−1) (Hyclone, Pittsburgh, PA, USA), and 10% fetal bovine serum (FBS). For viral infection, the culture supernatant was removed, and cells were inoculated with 0.5 ml of the viral suspension. Plates were incubated at 37 °C with a 5% CO2 atmosphere, with gentle shaking/rocking every 30 min for 1.5 h. After this initial incubation, the supernatant was aspirated and each single well was overlaid with 3.0 ml of MEM containing penicillin-streptomycin and 2% FBS (Corning CellGro 15-010). At 5, 24, 48, and 72 h after the initial inoculation, a microtiter plate was sampled by collecting 2.5 ml of media from each well in a 10 ml air-tight glass vial sealed with a PTFE/silicone cap (both from Sigma-Aldrich) and frozen at −30 °C. At each sampling time, six replicates each of RSV-infected and uninfected cells were collected.
2.1.2. IAV infection
Aliquots of 500 000 MLE-Kd cells (a mouse lung epithelial cell line) maintained in 100 μl of 1X Dulbecco’s Modified Eagle Medium (DMEM) (containing glucose, L-glutamine, and sodium pyruvate; Mediatech) were infected on ice with 10 μl of a stock of A/PR8/34 H1N1 influenza virus, titrated at ∼1 ×108 TCID50 (tissue culture infective dose 50%) units per ml for 20 min, corresponding to an MOI of 1. The suspensions were pipetted into 6-well polystyrene tissue culture plates containing 3 ml per well of pre-warmed complete media (1X DMEM, 10% FBS, 200 U each of penicillin and streptomycin, and 2 mM extra L-glutamine) (Hyclone). Plates were swirled to mix and incubated at 37 °C with a 5% CO2 atmosphere. At 24, 49, 72, and 122 h, 2.5 ml supernatant for each well were collected into a 10 ml air-tight glass vial sealed with a PTFE/silicone cap (Sigma-Aldrich) and frozen at −30 °C. Controls consisting of uninfected cells in media were incubated and collected in parallel. At each sampling time, six replicates each of IAV-infected and uninfected cells were collected.
2.2. Sample preparation
All samples were analyzed within one month of collection. Volatile metabolites were extracted using a divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) df 50/30 μm, 2 cm length fiber from Supelco (Bellefonte, PA, USA). The fiber was conditioned before use. Samples (agitated at 250 rpm) were incubated for 15 min at 37 °C before fiber exposure for 30 min at the same temperature. The fiber was introduced into the GC injector for thermal desorption for 1 min at 250 °C in splitless mode.
2.3. Analytical instrumentation
A Pegasus 4D (LECO Corporation, St. Joseph, MI) GC ×GC time-of-flight (TOF) MS instrument with an Agilent 6890N GC, and an MPS autosampler (Gerstel, Linthicum Heights, MD, USA) equipped with a cooled sampler tray (4 °C), was used. The primary column was an Rxi-624Sil (60 m ×250 μm ×1.4 μm) connected in series with a Stabilwax secondary column (1 m ×250 μm ×1.4 μm) from Restek (Bellefonte, PA, USA). The carrier gas was helium, at a flow rate of 2 ml min−1. The primary oven temperature program was 35 °C (hold 1 min) ramped to 230 °C at a rate of 5 °C min−1. The secondary oven and the thermal modulator were offset from the primary oven by +5 °C and +25 °C, respectively. A modulation period of 2.5 s (alternating 0.75 s hot and 0.5 s cold) was used. The transfer line temperature was set at 250 °C. A mass range of m/z 30 to 500 was collected at a rate of 200 spectra/s following a 3 min acquisition delay. The ion source was maintained at 200 °C. Data acquisition and analysis was performed using ChromaTOF software, version 4.50 (LECO Corp.).
2.4. Processing and analysis of chromatographic data
Chromatographic data were processed and aligned using ChromaTOF. For peak identification, a signal-to-noise cutoff was set at 50:1 in at least one chromatogram and a minimum of 20:1 S/N ratio in all others. The resulting peaks were identified by a forward search of the NIST 2011 library. For putative peak identification, a forward match score of ≥800 (of 1000) was required. For the alignment of peaks across chromatograms, maximum first and second-dimension retention time deviations were set at 6 s and 0.2 s, respectively, and the inter-chromatogram spectral match threshold was set at 600. Compounds eluting prior to 4 min and artifacts (e.g., siloxane, phthalates, etc) were removed prior to statistical analysis.
A mixture of normal alkanes (C6–C20), and the Grob mixture (containing Methyl decanoate (CAS#: 110-42-9), Methyl undecanoate (CAS#: 1731-86-8), Methyl dodecanoate (CAS#: 111-82-0), Decane (CAS#: 124-18-5), Undecane (CAS#: 1120-21-4), 2,6-Dimethylaniline (CAS#: 87-62-7), 2,6-Dimethylphenol (CAS#: 576-26-1), 2-Ethylhexanoic acid (CAS#: 149-57-5), Nonanal (CAS#: 124-19-6), and 1-Octanol (CAS#: 111-87-5)) (Supelco, Bellefonte, PA, USA) were analyzed every 20 runs to calculate the linear retention index (LRI) [37] and evaluate the instrument and SPME performance, respectively. The same SPME and GC methods were used, except for the SPME exposition time which was shorter (5 min) to avoid excessive overload of the fiber.
Discriminatory features were tentatively identified based on mass spectral similarities to the NIST 2011 mass spectral library, with a match score ≥800 (of 1000) required for putative identifications. In addition, at least one of the following two criteria were required: (I) a probability ≥5000 out of 10 000, and/or (II) an experimentally-determined LRI in agreement (i.e., in the ±10 range), with data reported using the same stationary phase. For the latter information, three main sources were used, namely [38], an application note (http://blog.restek.com/wp-content/uploads/2013/04/624silms.pdf), and the Pro EZGC® Chromatogram Modeler (http://restek.com/proezgc) (the latter two both from Restek). Most hydrocarbons were generally assigned as ‘alkylated hydrocarbons’, as it is almost impossible to assign them a specific name based only on the mass spectra similarity, due to the intense fragmentation of this class of compounds into the MS ion source. However, the chemical class of these compounds can be assigned by considering both their location in the two-dimensional chromatogram and their mass spectral fragmentation pattern.
2.5. Statistical analysis
All statistical analyses were performed using R v3.3.2 (R Foundation for Statistical Computing, Vienna, Austria). Prior to statistical analyses, the relative abundance of compounds across chromatograms was normalized using Probabilistic Quotient Normalization [39]. Data was randomly subdivided into discovery (training) and validation (test) sets 100 times, with 2/3 of samples included in the discovery set, and the remaining 1/3 in the validation set. Three machine learning algorithms were used to identify the most highly discriminatory volatile metabolites and predict the class (i.e., cells infected with virus versus uninfected cells) to which samples in the validation set belonged, namely: random forest (RF) [40], support vector machines with a linear kernel (linear SVM) [41], and partial least-squares discriminant analysis (PLS-DA) [42]. Mean decrease in accuracy, feature weights, and variable importance in projection were used as the measures of variable importance for RF, linear SVM, and PLS-DA, respectively [43]. For each of the 100 discovery/validation splits, volatile compounds were ranked according to their discriminatory ability, and different feature inclusion thresholds were compared (e.g., top 10%, 20% and 30%, etc) in terms of predictive ability. A compromise between the number of features included and model accuracy was obtained via the inclusion of the top 20% of features. The class probabilities were used to generate receiver operating characteristic (ROC) curves, and from these ROC curves, sensitivities, specificities, and area under the ROC curve (AUROC) were calculated. The optimal thresholds for class probabilities were calculated using Youden’s J statistic [44], rather than the 0.5 cutoff that is traditionally applied to two-class classification problems. K-means clustering was used to identify groups of volatile metabolites that exhibited similar changes in relative concentration as a function of time, with the relative concentration defined as the difference in the chromatographic area (calculated based on the unique mass, A) between cells infected with virus and uninfected cells (Ainfected–Auninfected). The elbow method was used to estimate the optimal number of clusters for k-means clustering [45].
3. Results and discussion
Prior to the statistical analysis of headspace volatiles, the stability of the HS-SPME GC×GC-ToF MS system was assessed using the Grob mixture, both in term of retention time shift and area repeatability. A coefficient of variation (CV %) below 0.2% and 2% were obtained for first and second dimension times, respectively, for all peaks except for 1-octanol, which presented a higher shift in the second dimension (about 20%, standard deviation of 0.2 s). This shift was taken into account in setting the alignment matching parameters. A variation of the area ≤15% was obtained for all standards considered.
3.1. RSV: discrimination between infected and uninfected cells
To identify volatile metabolic fingerprints that were discriminatory between cells infected with RSV and uninfected HEp-2 cells, the chromatographic data were first pre-processed to remove artifacts, reducing the total number of peak features from 358 to 216. These features were used for further data analysis. RF, linear SVM, and PLS-DA, were used to identify the most highly discriminatory volatile metabolites in the discovery set, and predict the class to which samples in the validation set belonged. This process was repeated 100 times using unique discovery/validation splits for each iteration, and the most highly discriminatory volatile metabolites (top 20%, corresponding to 43 features per iteration) were retained and used to predict the class (i.e., virally-infected cells versus uninfected cells, pooling together the different time points) to which samples in the validation set belonged.
The performance of these models was visualized by generating a ROC curve using the validation set class probabilities for each sample, and from these, the AUROC, as well as optimal sensitivities and specificities, were calculated (figure 1(A)).
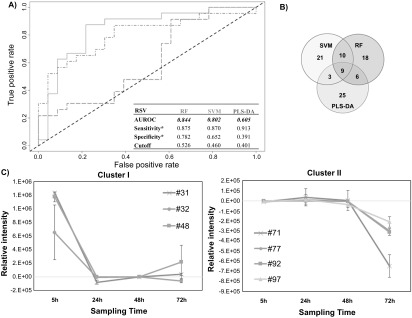
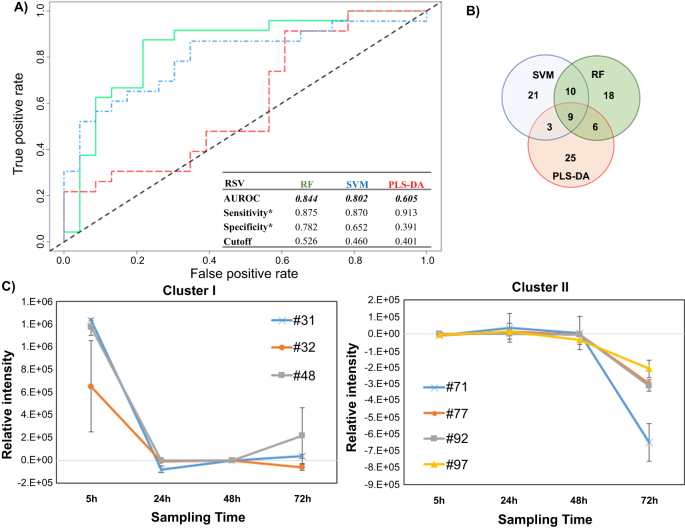
Figure 1.
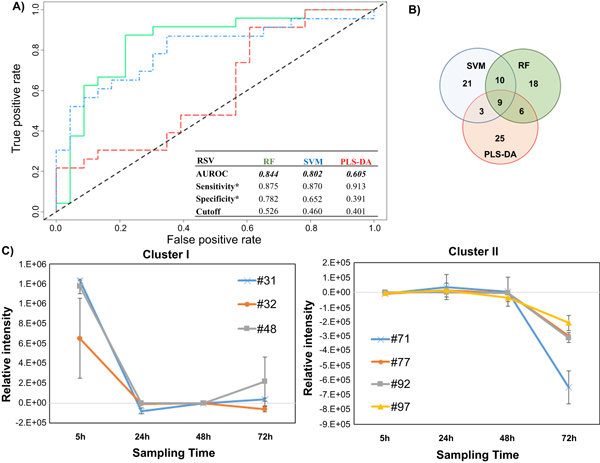
Differential production of volatile metabolites by RSV-infected and uninfected HEp-2 cells. (A) ROC curve for the discrimination between infected and uninfected cells using random forest (RF, green), support vector machines with a linear kernel (linear SVM, blue), and partial least-squares discriminant analysis (PLS-DA, red). *Optimal sensitivity and specificity of each statistical model are calculated at the given class prediction cutoff (ranging from 0 to 1). (B) Venn Diagram of the top 20% of features selected as the most discriminatory between RSV-infected and uninfected cells using RF, SVM, and PLS-DA algorithms. (C) Differences in the composition of headspace volatile molecules as a function of sampling time (5, 24, 48, and 72 h), for two sample clusters identified via k-means clustering. The remaining cluster reported in supplementary figure 2. # features codification as in table 1.
The AUROCs were generated using the class probabilities for validation set samples and were similar for RF and linear SVM (0.844 and 0.802, respectively), while PLS-DA performed relatively poorly (0.605). The optimal thresholds for class probabilities ranged from 0.401 for PLS-DA to 0.526 for RF. At these optimal thresholds, RF achieved the highest specificity (0.782) relative to either linear SVM or PLS-DA (0.652 and 0.391, respectively), while PLS-DA achieved the highest sensitivity (0.913) relative to either RF or linear SVM (0.875 and 0.870), albeit with poor overall model performance.
To assess the contribution of incubation time to the model performance, we considered the average prediction accuracies for samples at each of the four-time points evaluated independently (supplementary figure S1 is available online at stacks.iop.org/JBR/12/026015/mmedia). RF yielded the highest mean sample classification accuracy at three of four sampling times (5, 48, and 72 h), while SVM yielded the highest accuracy at 24 h. PLS-DA yielded the lowest classification accuracy at all sampling points. Of note, classification accuracy was most highly variable at 72 h, probably related to the confounding effect of natural senescence (and possibly cell death) of the in vitro cell culture, irrespective of the infection process.
The top discriminatory features obtained from the three models were compared to evaluate possible overlap. The number of features selected from discovery set samples to predict the classification of validation set samples was held constant across all three machine learning algorithms (n = 43, corresponding to the top 20% of discriminatory features). In total, 92 distinct volatile metabolites were included in the selected features for one or more algorithm, of which nine (10%) were in common across all three algorithms, 10 (11%) between SVM and RF only, six (7%) between RF and PLS-DA only, and three (3%) between SVM and PLS-DA only. The remaining 64 (70%) were unique to a single algorithm (figure 1(B)). The ranks of these discriminatory features varied considerably between algorithms. For example, the most discriminatory feature from RF and PLS-DA was identified as hexadecane, which ranked 7th for SVM, while pentadecane, which ranked 1st for SVM, had lower ranks for both RF (2nd) and PLS-DA (4th). A comprehensive listing of all discriminatory volatile metabolites with their feature importance ranks across all three machine learning algorithms is presented in table 1.
Table 1.
List of the discriminatory volatile metabolites putatively identified, along with their importance ranks from each machine learning algorithms (RF, SVM, and PLS-DA).
| RSV Rank | IAV Rank | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | RF | PLS | SVM | RF | PLS | SVM | Compound | Class | Formula | CAS | Forward similarity | Reverse similarity | Probability | LRI exp | LRI lit | 1tR (min:s) | 2tR (s) | Reference |
| 1 | 34 | Unknown | 467 | 04:02 | 0.8 | |||||||||||||
| 2 | 9 | Unknown | 467 | 04:03 | 1.3 | |||||||||||||
| 3 | 18 | 8 | Unknown | 468 | 04:04 | 0.8 | ||||||||||||
| 4 | 16 | 16 | 6 | Unknown | 469 | 04:06 | 0.7 | |||||||||||
| 5 | 20 | Unknown | 473 | 04:14 | 0.7 | |||||||||||||
| 6 | 42 | 22 | Unknown | 474 | 04:16 | 0.7 | ||||||||||||
| 7 | 35 | 5 | 13 | 10 | Unknown | 475 | 04:18 | 1.2 | ||||||||||
| 8 | 20 | 14 | Acetaldehyde | Ald | C2H4O | 75-07-0 | 863 | 905 | 8534 | 476 | 04:20 | 0.8 | [46, 47, 54] | |||||
| 9 | 12 | 34 | 11 | Unknown | 476 | 04:21 | 2.1 | |||||||||||
| 10 | 17 | Unknown | 476 | 04:22 | 0.6 | |||||||||||||
| 11 | 25 | 19 | Unknown | 477 | 04:22 | 0.9 | ||||||||||||
| 12 | 34 | 25 | Unknown | 477 | 04:23 | 0.7 | ||||||||||||
| 13 | 17 | Unknown | 483 | 04:36 | 0.7 | |||||||||||||
| 14 | 33 | Unknown | 487 | 04:43 | 0.8 | |||||||||||||
| 15 | 8 | 29 | 23 | Alkylated hydrocarbon | Hyd | 469 | 04:52 | 0.6 | ||||||||||
| 16 | 19 | Unknown | 478 | 05:07 | 0.7 | |||||||||||||
| 17 | 33 | 24 | Unknown | 487 | 05:22 | 0.8 | ||||||||||||
| 18 | 31 | 11 | 23 | 30 | Ethanol | Alc | C2H6O | 64-17-5 | 820 | 836 | 5124 | 500 | 506 | 05:45 | 1.3 | [46, 51, 56] | ||
| 19 | 21 | Unknown | 509 | 06:00 | 0.7 | |||||||||||||
| 20 | 18 | Furan | Het-Cyc | C4H4O | 110-00-9 | 801 | 905 | 9424 | 512 | 511 | 06:05 | 0.9 | [47] | |||||
| 21 | 4 | 2 | 2-Propenal | Ald | C3H4O | 107-02-8 | 808 | 860 | 9010 | 519 | 523 | 06:17 | 1.0 | [47] | ||||
| 22 | 29 | Propanal | Ald | C3H6O | 123-38-6 | 846 | 851 | 7605 | 522 | 526 | 06:23 | 0.8 | [38, 46, 47, 52] | |||||
| 23 | 15 | 32 | Acetone | Ket | C3H6O | 67-64-1 | 965 | 967 | 9537 | 526 | 530 | 06:30 | 0.9 | [33, 46, 47] | ||||
| 24 | 32 | 4 | Unknown | 529 | 06:35 | 0.9 | ||||||||||||
| 25 | 6 | 31 | 20 | Unknown | 532 | 06:40 | 0.7 | |||||||||||
| 26 | 34 | Unknown | 539 | 06:51 | 0.8 | |||||||||||||
| 27 | 14 | Unknown | 545 | 07:01 | 0.8 | |||||||||||||
| 28 | 35 | 4 | 2-Propanol | Alc | C3H8O | 67-63-0 | 872 | 910 | 7258 | 552 | 542 | 07:14 | 1.1 | [31, 52] | ||||
| 29 | 39 | 8 | 13 | 1-Propanol | Alc | C3H8O | 71-23-8 | 867 | 912 | 7823 | 553 | 07:16 | 1.1 | [50, 51] | ||||
| 30 | 31 | Unknown | 556 | 07:21 | 0.9 | |||||||||||||
| 31 | 9 | 2 | Unknown | 556 | 07:21 | 1.1 | ||||||||||||
| 32 | 41 | 11 | 2-methyl-Pentane | Hyd | C6H14 | 107-83-5 | 923 | 932 | 7352 | 557 | 564 | 07:23 | 0.7 | [46, 50–52] | ||||
| 33 | 38 | Alkylated hydrocarbon | Hyd | 561 | 07:28 | 0.7 | ||||||||||||
| 34 | 1 | 3-Hydroxy-3-methyl-2-butanone | Ket | C5H10O2 | 115-22-0 | 815 | 837 | 6977 | 561 | 07:29 | 1.0 | |||||||
| 35 | 10 | Unknown | 575 | 07:53 | 1.1 | |||||||||||||
| 36 | 4 | 5 | 3-methyl-Pentane | Hyd | C6H14 | 96-14-0 | 868 | 878 | 3671 | 576 | 581 | 07:55 | 0.7 | [46, 51] | ||||
| 37 | 23 | 24 | 26 | 2,3-dihydro-Furan | Het-Cyc | C4H6O | 1191-99-7 | 844 | 913 | 5941 | 591 | 08:20 | 0.9 | [19] | ||||
| 38 | 7 | Unknown | 599 | 08:34 | 0.7 | |||||||||||||
| 39 | 17 | 13 | 2-Butenal | Ald | C4H6O | 4170-30-3 | 819 | 872 | 5104 | 605 | 08:46 | 0.8 | [46] | |||||
| 40 | 11 | 29 | 9 | Alkylated hydrocarbons | Hyd | 606 | 08:48 | 0.7 | ||||||||||
| 41 | 1 | 17 | 32 | n-Hexane | hyd | C6H14 | 110-54-3 | 911 | 921 | 8003 | 606 | 600* | 08:48 | 0.7 | [47, 50, 52] | |||
| 42 | 40 | Alkylated hydrocarbons | Hyd | 622 | 09:20 | 0.7 | ||||||||||||
| 43 | 36 | Alkylated hydrocarbons | Hyd | 622 | 09:21 | 0.7 | ||||||||||||
| 44 | 23 | 10 | Alkylated hydrocarbons | Hyd | 96-37-7 | 628 | 09:32 | 0.7 | ||||||||||
| 45 | 27 | Disulfide, bis[1-(methylthio)ethyl] | S-Com | C6H14S4 | 69078-77-9 | 802 | 851 | 5684 | 630 | 09:37 | 1.2 | [57] | ||||||
| 46 | 25 | 3 | 2-Butanone | Ket | C4H8O | 78-93-3 | 890 | 904 | 7723 | 633 | 09:42 | 0.9 | [47, 50, 55] | |||||
| 47 | 15 | 28 | Methyl-cyclopentane | Hyd | C6H12 | 96-37-7 | 891 | 905 | 4872 | 639 | 638 | 09:55 | 0.7 | [50, 51] | ||||
| 48 | 6 | 34 | 36 | Methyl sulfone | S-Com | C2H6O2S | 67-71-0 | 823 | 832 | 6612 | 639 | 09:56 | 2.1 | [49] | ||||
| 49 | 38 | Formic acid, propyl ester | Est | C4H8O2 | 110-74-7 | 809 | 829 | 7358 | 647 | 10:10 | 0.9 | |||||||
| 50 | 5 | 21 | Tetrahydrofuran | Het-Cyc | C4H8O | 845 | 864 | 4102 | 652 | 655 | 10:18 | 0.8 | [47] | |||||
| 51 | 11 | Unknown | 654 | 10:26 | 2.0 | |||||||||||||
| 52 | 23 | Alkylated hydrocarbons | Hyd | 662 | 10:43 | 0.7 | ||||||||||||
| 53 | 12 | Alkylated hydrocarbon | Hyd | 667 | 10:52 | 0.7 | ||||||||||||
| 54 | 29 | 9 | 2-methyl-hexane | Hyd | C7H16 | 856 | 864 | 4872 | 673 | 674 | 11:05 | 0.7 | [51] | |||||
| 55 | 18 | Cyclohexane | Hyd | C6H12 | 110-82-7 | 849 | 867 | 1590 | 676 | 673 | 11:10 | 0.7 | [51] | |||||
| 56 | 10 | 26 | 26 | 9 | Benzene | Aro | C6H6 | 71-43-2 | 883 | 911 | 7847 | 684 | 684 | 11:28 | 0.9 | [46, 47, 51] | ||
| 57 | 28 | Unknown | 691 | 11:42 | 0.9 | |||||||||||||
| 58 | 28 | 3-methyl-butanal | Ald | C5H10O | 96-17-3 | 882 | 884 | 5914 | 701 | 694 | 12:02 | 0.8 | [47, 51] | |||||
| 59 | 14 | 27 | Alkylated hydrocarbons | Hyd | 731 | 13:13 | 0.7 | |||||||||||
| 60 | 27 | Alkylated hydrocarbons | Hyd | 731 | 13:13 | 0.7 | ||||||||||||
| 61 | 43 | 2,5-dimethyl hexane | Hyd | C8H18 | 592-13-2 | 802 | 823 | 3156 | 734 | 737 | 13:20 | 0.7 | ||||||
| 62 | 22 | Alkylated hydrocarbons | Hyd | 755 | 14:10 | 0.7 | ||||||||||||
| 63 | 24 | Alkylated hydrocarbons | Hyd | 757 | 14:15 | 0.7 | ||||||||||||
| 64 | 10 | Alkylated hydrocarbons | Hyd | 762 | 14:28 | 0.7 | ||||||||||||
| 65 | 17 | 2-methyl heptane | Hyd | C8H18 | 592-27-8 | 833 | 854 | 3824 | 766 | 767 | 14:36 | 0.7 | [46] | |||||
| 66 | 16 | 3-methyl heptane | Hyd | C8H18 | 589-81-1 | 826 | 845 | 3421 | 774 | 774 | 14:55 | 0.7 | [46, 51] | |||||
| 67 | 19 | 27 | Toluene | Aro | C7H8 | 108-88-3 | 841 | 877 | 4083 | 794 | 795 | 15:42 | 0.9 | [47, 52] | ||||
| 68 | 21 | Alkylated hydrocarbon | Hyd | 806 | 16:12 | 0.7 | ||||||||||||
| 69 | 24 | 19 | 13 | Unknown | 818 | 16:40 | 0.7 | |||||||||||
| 70 | 31 | Alkylated hydrocarbons | Hyd | 821 | 16:47 | 0.7 | ||||||||||||
| 71 | 26 | 37 | Alkylated hydrocarbons | Hyd | 823 | 16:52 | 0.7 | |||||||||||
| 72 | 7 | Alkylated hydrocarbons | Hyd | 830 | 17:07 | 0.7 | ||||||||||||
| 73 | 40 | Unknown | 837 | 17:25 | 1.0 | |||||||||||||
| 74 | 30 | 16 | 15 | Hexanal | Ald | C6H12O | 66-25-1 | 841 | 868 | 5463 | 840 | 540 | 17:32 | 0.9 | [46, 47, 51] | |||
| 75 | 18 | 2,4-dimethyl-Heptane | Hyd | C9H20 | 2213-23-2 | 852 | 892 | 3592 | 844 | 844 | 17:40 | 0.7 | [47] | |||||
| 76 | 35 | 43 | 2,4-Dimethyl-1-heptene | Hyd | C9H20 | 19549-87-2 | 834 | 861 | 3120 | 847 | 847 | 17:47 | 0.7 | [46, 48] | ||||
| 77 | 20 | 22 | 2-methyl-Octane | Hyd | C9H20 | 3221-61-2 | 819 | 893 | 4734 | 865 | 873 | 18:30 | 0.7 | [47] | ||||
| 78 | 13 | Alkylated hydrocarbons | Hyd | 875 | 18:54 | 0.7 | ||||||||||||
| 79 | 32 | Unknown | 885 | 19:18 | 0.8 | |||||||||||||
| 80 | 8 | 7 | 15 | Ethylbenzene | Aro | C8H10 | 100-41-4 | 840 | 886 | 4996 | 890 | 889 | 19:28 | 0.9 | [38, 51] | |||
| 81 | 3 | 31 | 3 | p-Xylene | Aro | C8H10 | 106-42-3 | 812 | 832 | 2691 | 898 | 897 | 19:48 | 0.9 | [47, 51] | |||
| 82 | 28 | Alkylated hydrocarbons | Hyd | 918 | 20:33 | 0.7 | ||||||||||||
| 83 | 25 | o-Xylene | Aro | C8H10 | 95-47-6 | 815 | 847 | 2264 | 926 | 924 | 20:51 | 1.0 | [31, 47, 51] | |||||
| 84 | 33 | 39 | Styrene | Aro | C8H8 | 100-42-5 | 909 | 922 | 4797 | 928 | 926 | 20:55 | 1.1 | [47, 51] | ||||
| 85 | 15 | Alkylated hydrocarbons | Hyd | 930 | 21:00 | 0.7 | ||||||||||||
| 86 | 27 | Unknown | Est | C5H10O2 | 934 | 21:08 | 0.9 | |||||||||||
| 87 | 33 | Alkylated hydrocarbons | Hyd | 936 | 21:12 | 0.7 | ||||||||||||
| 88 | 21 | Benzene, (1-methylethyl)- | Aro | C9H12 | 98-82-8 | 818 | 861 | 4654 | 953 | 954 | 21:51 | 0.9 | ||||||
| 89 | 25 | Alkylated hydrocarbons | Hyd | 965 | 22:18 | 0.7 | ||||||||||||
| 90 | 27 | 30 | Alkylated hydrocarbons | Hyd | 978 | 22:45 | 0.7 | |||||||||||
| 91 | 2 | 33 | 2 | Decane | Hyd | C10H22 | 124-18-5 | 821 | 853 | 768 | 1000 | 1000* | 23:35 | 0.7 | [47, 51] | |||
| 92 | 30 | Alkylated hydrocarbons | Hyd | 1014 | 24:05 | 0.7 | ||||||||||||
| 93 | 26 | 17 | Alkylated hydrocarbons | Hyd | 1020 | 24:17 | 0.7 | |||||||||||
| 94 | 18 | 14 | Alkylated hydrocarbons | Hyd | 1024 | 24:25 | 0.7 | |||||||||||
| 95 | 14 | Benzaldehyde | Ald | C7H6O | 100-52-7 | 802 | 900 | 5753 | 1030 | 24:38 | 1.5 | [31, 34, 47, 52] | ||||||
| 96 | 43 | Alkylated hydrocarbons | Hyd | 1049 | 25:17 | 0.7 | ||||||||||||
| 97 | 42 | 35 | 10 | Alkylated hydrocarbons | Hyd | 1059 | 25:37 | 0.7 | ||||||||||
| 98 | 31 | Alkylated hydrocarbons | Hyd | 1065 | 25:50 | 0.7 | ||||||||||||
| 99 | 39 | Benzonitrile | Aro | C7H5N | 100-47-0 | 807 | 837 | 5265 | 1068 | 1071 | 25:56 | 1.6 | [34] | |||||
| 100 | 38 | 2-ethyl-1-hexanol | Alc | C8H18O | 104-76-7 | 871 | 883 | 6374 | 1079 | 26:18 | 1.1 | [48] | ||||||
| 101 | 5 | Ketone | Ket | 1094 | 26:50 | 0.9 | ||||||||||||
| 102 | 22 | 7 | Undecane | Hyd | C11H24 | 1120-21-4 | 841 | 868 | 1477 | 1100 | 1100* | 27:03 | 0.7 | |||||
| 103 | 18 | 21 | Alkylated adehyde | Ald | C5H10O | 1101 | 27:05 | 1.5 | ||||||||||
| 104 | 12 | Alkylated hydrocarbons | Hyd | 1102 | 27:06 | 0.7 | ||||||||||||
| 105 | 9 | Alkylated hydrocarbons | Hyd | 1102 | 27:07 | 0.7 | ||||||||||||
| 106 | 28 | Alkylated hydrocarbons | Hyd | 1109 | 27:19 | 0.7 | ||||||||||||
| 107 | 34 | Alkylated hydrocarbons | Hyd | 1126 | 27:52 | 1.0 | ||||||||||||
| 108 | 19 | 16 | Alkylated hydrocarbon | Hyd | 1126 | 27:53 | 0.6 | |||||||||||
| 109 | 8 | Unknown | 1128 | 27:57 | 0.9 | |||||||||||||
| 110 | 5 | 6 | Unknown | 1142 | 28:25 | 1.4 | ||||||||||||
| 111 | 4 | Nonanal | Ald | C9H18O | 124-19-6 | 846 | 839 | 5616 | 1150 | 1147* | 28:40 | 0.9 | [47, 49, 52, 53] | |||||
| 112 | 7 | 12 | 16 | 19 | 3 | Dodecane | Hyd | C12H26 | 112-40-3 | 836 | 859 | 788 | 1200 | 1200* | 30:17 | 0.7 | [38] | |
| 113 | 30 | Unknown | 1219 | 30:52 | 0.6 | |||||||||||||
| 114 | 12 | Alkylated hydrocarbons | Hyd | 1214 | 30:43 | 0.7 | ||||||||||||
| 115 | 22 | Alkylated hydrocarbons | Hyd | 1246 | 31:39 | 0.7 | ||||||||||||
| 116 | 32 | Alkylated hydrocarbons | Hyd | 1251 | 31:50 | 0.7 | ||||||||||||
| 117 | 12 | Unknown | 1256 | 31:59 | 0.7 | |||||||||||||
| 118 | 6 | Unknown | 1261 | 32:07 | 0.8 | |||||||||||||
| 119 | 11 | 41 | 25 | Alkylated hydrocarbons | Hyd | 1265 | 32:15 | 0.7 | ||||||||||
| 120 | 42 | Unknown | 1275 | 32:32 | 0.7 | |||||||||||||
| 121 | 13 | Alkylated benzene | Hyd | C14H22 | 1280 | 32:42 | 0.8 | [51] | ||||||||||
| 122 | 37 | 37 | 35 | Alkylated hydrocarbons | Hyd | 1281 | 32:44 | 0.7 | ||||||||||
| 123 | 15 | Alkylated hydrocarbons | Hyd | 1309 | 33:33 | 0.7 | ||||||||||||
| 124 | 20 | 40 | Alkylated hydrocarbons | Hyd | 1327 | 34:04 | 0.7 | |||||||||||
| 125 | 1 | 20 | 26 | Alkylated hydrocarbons | Hyd | 1327 | 34:04 | 0.7 | ||||||||||
| 126 | 2 | 4 | Tetradecane | Hyd | C14H30 | 629-59-4 | 840 | 864 | 3120 | 1400 | 1400* | 36:09 | 0.7 | |||||
| 127 | 3 | 28 | 1 | Alkylated hydrocarbons | Hyd | 1401 | 36:09 | 0.7 | ||||||||||
| 128 | 35 | 14 | Alkylated hydrocarbons | Hyd | 1411 | 36:25 | 0.7 | |||||||||||
| 129 | 24 | Unknown | 1454 | 37:34 | 0.9 | |||||||||||||
| 130 | 3 | 5 | 2 | Pentadecane | Hyd | C15H32 | 629-62-9 | 844 | 859 | 1010 | 1500 | 1500* | 38:48 | 0.7 | ||||
| 131 | 23 | Unknown | 1552 | 40:09 | 0.9 | |||||||||||||
| 132 | 41 | 29 | Alkylated hydrocarbons | Hyd | 1554 | 40:12 | 0.7 | |||||||||||
| 133 | 21 | 36 | 6 | Unknown | 1591 | 41:08 | 1.6 | |||||||||||
| 134 | 1 | 1 | 7 | Hexadecane | Hyd | C16H34 | 544-76-3 | 819 | 828 | 1520 | 1601 | 1600* | 41:24 | 0.8 | ||||
| 135 | 22 | Unknown | 1636 | 42:15 | 0.7 | |||||||||||||
| 136 | 30 | 8 | 33 | Alkylated hydrocarbons | Hyd | 1642 | 42:23 | 0.7 | ||||||||||
| 137 | 32 | Unknown | 1701 | 43:50 | 0.9 | |||||||||||||
| 138 | 24 | 29 | Unknown | 1715 | 44:10 | 0.9 | ||||||||||||
The relative concentration (Ainfected–Auninfected) of all 92 discriminatory metabolites (putatively identified through mass spectral matching) was calculated at each time point individually. K-means clustering was used to identify metabolites with similar behavior as a function of time. Three main clusters were identified. Cluster I included three metabolites (#31: molecule not identified, #32: 2-methyl-pentane, #48: methyl sulfone) which were in highest abundance at the beginning of the infection process (5 h), and subsequently decreased and remained relatively constant between 24 and 72 h (figure 1(C)). Cluster II included four (#71: 2,4-dimethyl-heptane, #77: 4-methyl-octane, #92: alkylated hydrocarbon, #97: alkylated hydrocarbon), which remained relatively constant between 5 and 48 h, and then substantially decreased at 72 h (figure 1(C)). Of note, for features in cluster II, increased expression was observed in the uninfected cells (rather than decreased expression in RSV-infected cells) at 72 h. Finally, cluster III encompassed the remaining 84, which exhibited no clear temporal trend (supplementary figure S2).
3.2. Influenza A: discrimination between infected and uninfected cells
The chromatographic data obtained for the comparison of cells infected with IAV versus uninfected MLE-Kd were pre-processed to remove artifacts, reducing the total number of peak features from 278 to 177. The performance of the models were visualized by generating ROC curves using the validation set class probabilities for each sample, and from these, the AUROCs, as well as optimal sensitivities and specificities, were calculated (figure 2(A)). The AUROCs were similar across the three algorithms employed, with SVM yielding the best overall performance (0.825), followed by RF (0.806), and PLS-DA (0.783). At the optimal classification probability thresholds, sensitivities and specificities were 0.792 and 0.792 for RF (optimal cut-off of 0.499), 0.708 and 0.875 for linear SVM (optimal cut-off of 0.530), and 0.708 and 0.708 for PLS-DA (optimal cut-off of 0.514).
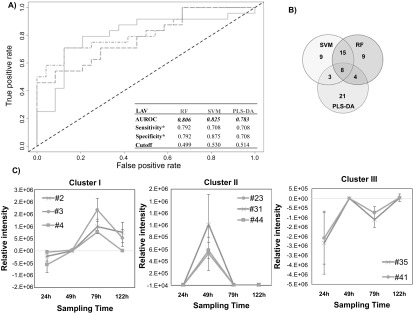
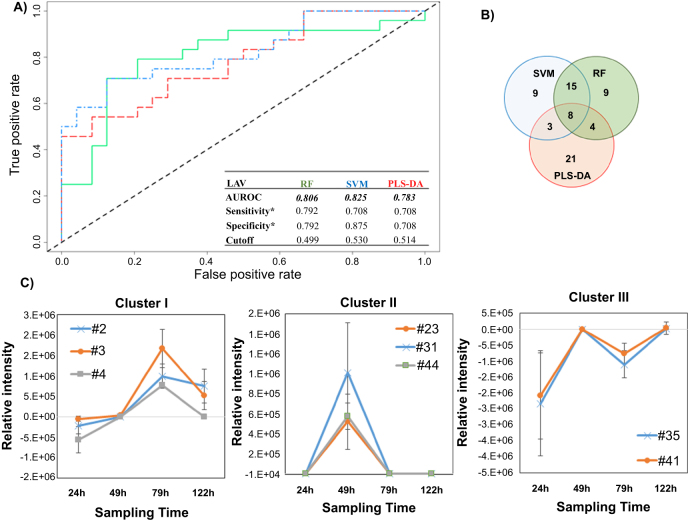
Figure 2.
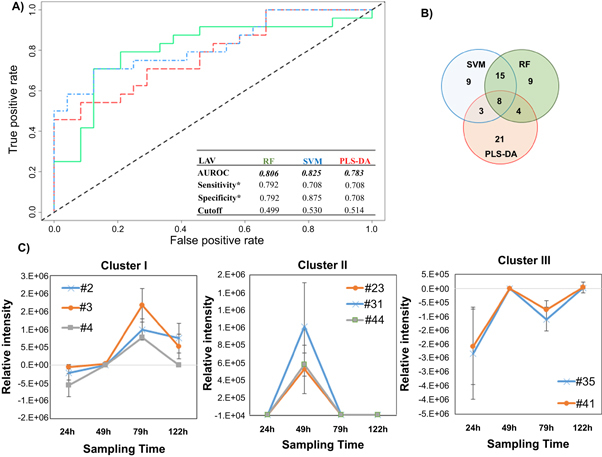
Differential production of volatile metabolites by IAV-infected and uninfected MLE-Kd cells. (A) ROC curve for the discrimination between infected and uninfected cells using random forest (RF, green line), support vector machines with a linear kernel (linear SVM, blue line), and partial least-squares discriminant analysis (PLS-DA, red line). *Optimal sensitivity and specificity of each statistical model are calculated at the given class prediction cutoff (ranging from 0 to 1). (B) Venn Diagram of the top 20% features selected as the most discriminatory between IVA-infected cell line and uninfected control using RF. SVM, and PLS-DA algorithm. (C) Most significant clusters obtained by k-Means Clustering analysis of the difference between infected and uninfected cell cultures. Remaining cluster is reported in supplementary figure 4. # features codification as in table 1.
The most highly discriminatory volatile metabolites (top 20%, corresponding to 35 features) were retained and used to predict to which class samples in the validation set belonged. In total, 67 distinct volatile metabolites were included across RF, linear SVM, and PLS-DA, of which eight (12%) were common between all three algorithms, 15 (22%) between SVM and RF only, four (6%) between RF and PLS-DA only, and three (4%) between SVM and PLS-DA only. The remaining 39 (58%) were unique to a single algorithm (figure 2(B)). Of note, while the most discriminatory features identified from RF and SVM are similar in feature importance rank, (e.g., features #127 and #91, which ranked 1st and 2nd using linear SVM, and 3rd and 2nd using RF, respectively), the top five features obtained using PLS-DA are not included in the top 20% for either RF or SVM, with the exception of #83, which was ranked 31st using RF.
The contribution of incubation time to model performance was evaluated by considering the average prediction accuracies for samples at each time points (24, 49, 79 and 122 h) independently (supplementary figure S3). A general descending trend over time can be observed, with a median approximating 0.5 for all three models at 122 h. PLS-DA yielded the highest mean sample classification accuracy at 49 h with very low variability, while RF yielded optimal classification accuracy at 24 and 79 h. SVM showed large variability at all time points but represented the optimal classification model at 122 h. As with the cell cultures infected with RSV, the variability of prediction increased for the last time point (122 h) for all algorithms, probably due to changes in metabolite production linked to cellular senescence and death.
The relative concentrations (Ainfected–Auninfected) of the 67 selected discriminatory metabolites (putatively identified through mass spectral matching) as a function of time were again evaluated using k-means clustering algorithm, and four main clusters were extrapolated. In the first cluster, three volatile metabolites (#2, #3, #4, all molecules not identified) were included, whose relative abundance increased between 24 and 72 h, before a decrease by 122 h (figure 2(C)). The second cluster included three (#23: acetone, #31: molecule not identified, #44: alkylated hydrocarbon) that were detected at 49 h only, and not detected at the remaining time points. The third cluster included two (#35: not identified, #41: n-hexane) that increased between 24 and 49 h then decreased at 79 h only to increase again by 122 h. The relative concentrations of these latter features were negative across all time points, indicating that they were more highly abundant in uninfected controls. We therefore hypothesize that they were related to cell line aging rather than infection. Further studies are necessary to explain this behavior. The fourth cluster included the remaining 59 metabolites which demonstrated no clear trend as a function of time (supplementary figure S4).
3.3. Putative identifications of discriminatory volatile metabolites
Combining all the features selected from the different models used for discriminating between cells infected with virus (both RSV and IAV) versus uninfected cells, a list of 138 metabolites (20 in common between the two virally-infected cell lines) were generated and tentatively identified according to the criteria reported in the Materials and Methods. Sixty-five (47%) were classified as hydrocarbons, nine (7%) as aldehydes, eight (6%) as aromatic compounds, four (3%) as alcohols, four (3%) as ketones, three (2%) as heterocyclic compounds, two (1%) as sulfur-containing compounds, two (1%) as esters, and finally 41 (30%) as unknowns. It is interesting to note that hydrocarbons comprised a greater proportion of discriminatory metabolites in the comparison of RSV-infected versus non-infected HEp-2 cells relative to the comparison of IAV-infected versus non-infected MLE-Kd cells (56 of 95 compounds (59%) for RSV, versus 18 of 67 (27%) in the IAV experiment). All other chemical classes were similarly represented in the two set of experiments.
Five compounds (i.e., acetone, 2-propanol, o-xylene, benzaldehyde, and benzonitrile) have previously been reported in the headspace of cell cultures infected with viruses (three of which in cells infected with IAV, namely 2-propanol, o-xylene, and benzaldehyde) [31, 33, 34], while forty have been reported in the headspace of cell cultures more generally (mostly cancer cell cultures) (table 1) [33, 34, 46–58]. The relatively minimal overlap between our study and prior studies that have considered in vitro cells infected with viruses is likely related to a number of factors, such as: the low signal generated by this kind of sample, the different MOIs applied, differences in the cell lines used and viral infection performed, as well as growth conditions and media used, different SPME fiber phase composition which affects the selectivity of the extracted compounds, differences in the analytical techniques utilized, as well as the difficulty in assigning precise identifications to alkylated hydrocarbons, which are generally the most abundant chemical class.
Most of the volatile metabolites tentatively identified can be attributed to chemical classes related to the lipid oxidation pathways, namely ketones, aldeheydes, alcohols, and hydrocarbons. They have been reported to originate largely from free radical oxidative fragmentation of lipids due to oxidative stress [19, 59]. It has been shown that viral infection impairs the pro-oxidant-antioxidant balance in favor of the former by increasing the production of reactive oxygen species, in part through a NAD(P)H oxidase-dependent mechanism [60]. In particular, it has been shown that the activity of superoxide dismutase enzymes increases during viral infection, especially at a mitochondrial level [60]. This increase in reactive oxygen species is directly correlated with the formation of aliphatic hydrocarbons, which can explain the high abundance of hydrocarbons in our samples. However, these findings mainly refer to linear or iso-alkanes, while the origin of most of the alkylated hydrocarbons, which have been identified both in vitro and in vivo is still unclear [59]. An exogenous source for these compounds can also be hypothesized even if a presently undefined metabolic process cannot be excluded; further research has to be carried out to unveil this speculative idea.
Nine aldehydes were also putatively identified (i.e., 2-butenal, 2-propenal, 3-methyl-butanal, acetaldehyde, alkylated aldehyde, benzaldehyde, hexanal, nonanal, and propanal). These compounds have been related to lipid peroxidation during the inflammation process, where it is hypothesized that they serve as secondary messengers in signal transduction, gene regulation, and cellular proliferation [59, 61]. Three furan derivatives were found (i.e., furan, 2,3-dihydro-furan, and tetrahydrofuran), which were previously identified in the headspace of cell culture and bacteria [19, 59]. Two sulfur-containing compounds (i.e., methyl sulfone and bis[1-(methylthio)ethyl] disulfide) were also identified. The formation of sulfur compounds have been linked to the sulfur-containing amino acids methionine and cysteine in the transamination pathway, which is affected by an oxidative stress, causing a depletion of such sulfur-containing amino acids [62, 63].
3.4. RSV-infected HEp-2 versus IAV-infected MLE-Kd cells
A direct comparison of the volatile metabolic signatures produced by RSV and IAV infection was not possible due to differences in the composition of headspace volatiles at baseline (i.e., differences between uninfected HEp-2 and MLE-Kd cells). We attempted to identify a volatile metabolic fingerprint that could discriminate between infected cells but not discriminate between uninfected cells by using recursive feature elimination coupled to RF (RFE-RF). However, the differences at baseline were sufficiently great such that it was not possible to effectively make such a comparison. This may have resulted from numerous factors, including: (1) the use of different growth media across cell lines, (2) the comparison of a human (HEp-2) and murine (MLE-Kd) lineages, and (3) the comparison of transformed (HEp-2) versus non-transformed (MLE-Kd) lineages. RFE-RF resulted in the identification of 10 volatile metabolites that could differentiate between RSV-infected HEp-2 cells and IAV-infected MLE-Kd cells with approximately 74.9% accuracy, but which also differentiated between uninfected HEp-2 cells and uninfected MLE-Kd cells with 60.0% accuracy. Because of our inability to discriminate between uninfected HEp-2 and MLE-Kd cells, we elected to not report on those compounds that were most highly discriminatory between RSV- and IAV-infected cells, as differences in the production of these metabolites may have resulted from factors other than the type of virus used for infection.
However, we do note that 21 of the compounds reported as discriminatory overall (table 1) were discriminatory for both sets of experiments (i.e., RSV infected cells versus uninfected cells and IAV infected cells versus uninfected cells). Amongst these 21 compounds, we putatively identified seven hydrocarbons (2-methyl-pentane, dodecane, and five generic alkylated hydrocarbons), four aromatics (p-xylene, ethylbenzene, toluene, and benzene), two heterocycles (2,3-dihydrofuran and tetrahydrofuran), two alcohols (ethanol and 1-propanol), one aldehyde (acetaldehyde), and one ketone (acetone). The identities of four compounds remain unknown. Notably, ethanol, benzene, and dodecane represent the three metabolites that were identified as discriminatory by two or more machine learning algorithms in both the RSV and IAV experiments.
3.5. Study strengths and limitations
In the present study, we have evaluated the potential ability of volatile metabolites for discriminating between virally-infected and uninfected cells using three different machine learning algorithms, demonstrating the potential effectiveness of the approach.
The use of SPME coupled to GC×GC-ToF MS generated 216 and 177 features from the headspace of cells infected with RSV and IAV, respectively. The GC×GC-ToF MS system results in improvements in sensitivity and identification ability compared to conventional GC. The volatile profile obtained resulted, in part, from the specific selectivity of the SPME fiber (PDMS/Car/DVB) used, and do not necessarily mirror the real profile present in the headspace of the sample. A relatively low number of compounds herein identified have been previously reported in the literature, likely related to both biological (different MOI, growth conditions, media, and cell culture) and analytical (sample preparation and analytical determination methods) differences.
The choice of host cells was based on their permissiveness to high levels of viral replication, and under these conditions we were able to discriminate between virally-infected and uninfected cells. However, these findings do not necessarily allow for generalization to other cell types. Moreover, the use of different cell lineages for RSV and IAV infections did not allow for the comparison of infections caused by different viruses.
4. Conclusions and future perspectives
Viral infection results in the alteration of numerous biochemical pathways, a subset of which involve the production of small molecules that can cross the cell membrane and thus be detected in the headspace of an infected cell culture. Here we show that volatile compounds can be used to effectively discriminate between infected (RSV and IAV) and uninfected cells. The abundance of these discriminatory volatiles can fluctuate over time according to the infection stage, but, irrespective of the sampling time post-infection, an effective discriminatory prediction was obtained, although a decreasing accuracy was observed after 72 h or 122 h for RSV and IAV, respectively.
Future work in this area should involve investigating the utility of volatile metabolites to discriminate between infections caused by different viruses in a single cell line, as well as generate insight into viral pathogenesis. Furthermore, the use of a common cell line for culturing both viruses, specifically a non-transformed human lung epithelial cell line, will be considered. In the present experiments, different cell lines were chosen because of their ability to optimize the replication of the viruses selected, and this limited our ability to identify volatile metabolites that could differentiate between viruses. Further studies will be carried out to answer this latter question.
Supplementary Material
Acknowledgments
Financial support for this work was provided by Hitchcock Foundation and the National Institute of Health (NIH, Project#1R21AI12107601). CAR was supported by the Burroughs Wellcome Fund Institutional Program Unifying Population and Laboratory Based Sciences, awarded to Dartmouth College (Grant#1014106), and a T32 training grant (T32LM012204, PI: Christopher I Amos). P-H Stefanuto is a Marie-Curie COFUND postdoctoral fellow co-funded by the European Union and the University of Liège.
The authors gratefully acknowledge Supelco for providing the SPME fiber.
References
- 1.Price T, Schuchat A, Rothwell C J. Health, United States, 2016: With Chartbook on Long-term Trends in Health in Americans. Hyattsville, MD: National Center for Health Statistics; 2017. (DHHS Publication No 2017-1232) [PubMed] [Google Scholar]
- 2.UNICEF . Levels and Trends in Child Mortality. New York: UNICEF; 2015. pp. pp 1–30. [Google Scholar]
- 3.United States Centers for Disease Control and Prevention National Action Plan for Combating Antibiotic-Resistant Bacteria. 2015:1-63. [Google Scholar]
- 4.World Health Organization . Global Action Plan on Antimicrobial Resistance. Geneva: WHO Press; 2015. pp. pp 1–28. [Google Scholar]
- 5.CDC Antibiotic resistance threats in the United States, 2013. Current. 2013;114:1-114. [Google Scholar]
- 6.Jain S, et al. Community-acquired pneumonia requiring hospitalization among US children. New Engl. J. Med. 2015;372:835–45. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamma P D, Cosgrove S E. Addressing the appropriateness of outpatient antibiotic prescribing in the United States: an important first step. J. Am. Med. Assoc. 2016;315:1839–41. doi: 10.1001/jama.2016.4286. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization (WHO) WHO Recommendations on the Use of Rapid Testing for Influenza Diagnosis. Geneva: WHO; 2005. pp. pp 1–18. [Google Scholar]
- 9.Lode H, Schaberg T, Raffenberg M, Mauch H. Diagnostic problems in lower respiratory tract infections. J. Antimicrob. Chemother. 1993;32(Suppl A):29–37. doi: 10.1093/jac/32.suppl_A.29. [DOI] [PubMed] [Google Scholar]
- 10.Robinson J. Colonization and infection of the respiratory tract: what do we know? Paediatr. Child Health. 2004;9:21–4. doi: 10.1093/pch/9.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer K A, Perez K K, Forrest G N, Goff D A. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin. Infect. Dis. 2014;59:S134–45. doi: 10.1093/cid/ciu547. [DOI] [PubMed] [Google Scholar]
- 12.Poritz M A, et al. Filmarray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6:e26047. doi: 10.1371/annotation/468cfdcd-184c-42f7-a1d0-3b72a2f6a558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salez N, Vabret A, Leruez-Ville M, Andreoletti L, Carrat F, Renois F, de Lamballerie X. Evaluation of four commercial multiplex molecular tests for the diagnosis of acute respiratory infections. PLoS One. 2015;10:e0130378. doi: 10.1371/journal.pone.0130378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gisbert J P, Pajares J M. Review article: 13C-urea breath test in the diagnosis of Helicobacter pylori infection—a critical review. Aliment. Pharmacol. Ther. 2004;20:1001–17. doi: 10.1111/j.1365-2036.2004.02203.x. [DOI] [PubMed] [Google Scholar]
- 15.Sethi S, Nanda R, Chakraborty T. Clinical application of volatile organic compound analysis for detecting infectious diseases. Clin. Microbiol. Rev. 2013;26:462–75. doi: 10.1128/CMR.00020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Bean H D, Jimenez-Diaz J, Hill J E. Secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting of multiple bacterial lung pathogens, a mouse model study. J. Appl. Physiol. 2013;114:1544–9. doi: 10.1152/japplphysiol.00099.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Bean H, Wargo M J, Leclair L W, Hill J E. Detecting bacterial lung infections: in vivo evaluation of in vitro volatile fingerprints. J. Breath Res. 2014;7:016003. doi: 10.1088/1752-7155/7/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Jiménez-Díaz J, Bean H D, Daphtary N A, Aliyeva M I, Lundblad L K A, Hill J E. Robust detection of P. aeruginosa and S. aureus acute lung infections by secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting: from initial infection to clearance. J. Breath Res. 2013;7:037106. doi: 10.1088/1752-7155/7/3/037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz S, Dickschat J S. Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 2007;24:814–42. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- 20.Sweeney T E, Shidham A, Wong H R, Khatri P, Alto P, Alto P. A comprehensive time-course–based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci. Transl. Med. 2016;7:1–33. doi: 10.1126/scitranslmed.aaa5993. 287ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sweeney T, Purvesh K. Comprehensive validation of the FAIM3:PLAC8 ratio in time matched public gene expression data. Am. J. Respir. Crit. Care Med. 2015;192:1260–1. doi: 10.1164/rccm.201507-1321LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McHugh L, et al. A molecular host response assay to discriminate between sepsis and infection-negative systemic inflammation in critically ill patients: discovery and validation in independent cohorts. PLoS Med. 2015;12:1–35. doi: 10.1371/journal.pmed.1001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scicluna B P, et al. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am. J. Respir. Crit. Care Med. 2015;192:826–35. doi: 10.1164/rccm.201502-0355OC. [DOI] [PubMed] [Google Scholar]
- 24.Hu X, Yu J, Crosby S D, Storch G A. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc. Natl Acad. Sci. 2013;110:12792–7. doi: 10.1073/pnas.1302968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaas A K, et al. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci. Transl. Med. 2013;5:203ra126. doi: 10.1126/scitranslmed.3006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suarez N M, Bunsow E, Falsey A R, Walsh E E, Mejias A, Ramilo O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J. Infect. Dis. 2015;212:213–22. doi: 10.1093/infdis/jiv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delneste Y, Beauvillain C, Jeannin P. Innate immunity: structure and function of TLRs. Med. Sci. 2007;23:67–73. doi: 10.1051/medsci/200723167. [DOI] [PubMed] [Google Scholar]
- 28.Medzhitov R, Janeway C A J. Innate immunity: minireview the virtues of a nonclonal system of recognition. Cell. 1997;91:295–8. doi: 10.1016/S0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 29.Medzhitov R, Janeway C J. Innate immune recognition: mechanisms and pathways. Immunol. Rev. 2000;173:89–97. doi: 10.1034/j.1600-065X.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 30.Sung R Y, Hui S H, Wong C K, Lam C W, Yin J. A comparison of cytokine responses in respiratory syncytial virus and influenza A infections in infants. Eur. J. Pediatr. 2001;160:117–22. doi: 10.1007/s004310000676. [DOI] [PubMed] [Google Scholar]
- 31.Aksenov A A, Sandrock C E, Zhao W, Sankaran S, Schivo M, Harper R, Cardona C J, Xing Z, Davis C E. Cellular scent of influenza virus infection. ChemBioChem. 2014;15:1040–8. doi: 10.1002/cbic.201300695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips M, Cataneo R N, Chaturvedi A, Danaher P J, Devadiga A, Legendre D A, Nail K L, Schmitt P, Wai J. Effect of influenza vaccination on oxidative stress products in breath. J. Breath Res. 2010;4:026001. doi: 10.1088/1752-7155/4/2/026001. [DOI] [PubMed] [Google Scholar]
- 33.Schivo M, Aksenov A A, Linderholm A L, McCartney M M, Simmons J, Harper R W, Davis C E. Volatile emanations from in vitro airway cells infected with human rhinovirus. J. Breath Res. 2014;8:37110. doi: 10.1088/1752-7155/8/3/037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rochford K, Chen F, Waguespack Y, Figliozzi R W, Kharel M K, Zhang Q, Martin-Caraballo M, Hsia S V. Volatile organic compound gamma-butyrolactone released upon herpes simplex virus type -1 acute infection modulated membrane potential and repressed viral infection in human neuron-like cells. PLoS One. 2016;11:e0161119. doi: 10.1371/journal.pone.0161119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abd El Qader A, Lieberman D, Shemer Avni Y, Svobodin N, Lazarovitch T, Sagi O, Zeiri Y. Volatile organic compounds generated by cultures of bacteria and viruses associated with respiratory infections. Biomed. Chromatogr. 2015;29:1783–90. doi: 10.1002/bmc.3494. [DOI] [PubMed] [Google Scholar]
- 36.Tranchida P Q, Purcaro G, Dugo P, Mondello L. Modulators for comprehensive two-dimensional gas chromatography. TrAC—Trends Anal. Chem. 2011;30:1437–61. doi: 10.1016/j.trac.2011.06.010. [DOI] [Google Scholar]
- 37.d’Acampora Zellner B, Bicchi C, Dugo P, Rubiolo P, Dugo G, Mondello L. Linear retention indices in gas chromatographic analysis: a review. Flavour Fragrance J. 2008;23:297–314. doi: 10.1002/ffj.1887. [DOI] [Google Scholar]
- 38.Schallschmidt K, Becker R, Jung C, Bremser W, Walles T, Neudecker J, Leschber G, Frese S, Nehls I. Comparison of volatile organic compounds from lung cancer patients and healthy controls-challenges and limitations of an observational study. J. Breath Res. 2016;10:046007. doi: 10.1088/1752-7155/10/4/046007. [DOI] [PubMed] [Google Scholar]
- 39.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006;78:4281–90. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 40.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 41.Cortes C, Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–97. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- 42.Barker M, Rayens W. Partial least squares for discrimination. J. Chemom. 2003;17:166–73. doi: 10.1002/cem.785. [DOI] [Google Scholar]
- 43.Krooshof P W T, Ustun B, Postma G J, Buydens L M C. Vizualization and recovery of the (Bio)chemical interesting variables in data analysis with support vector machine classification. Anal. Chem. 2010;82:7000–7. doi: 10.1021/ac101338y. [DOI] [PubMed] [Google Scholar]
- 44.Ruopp M D, Perkins N J, Whitcomb B W, Schisterman Enrique F. Youden index and optimal cut-point estimated from observations affected by a lower limit of detection. Biometrical J. 2008;50:419–30. doi: 10.1002/bimj.200710415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alpaydın E. In: Introduction to Machine Learning. 2nd edn. Dietterich T, editor. Cambridge, MA: MIT; 2009. [Google Scholar]
- 46.Filipiak W, Mochalski P, Filipiak A, Ager C, Cumeras R, Davis C E, Agapiou A, Unterkofler K, Troppmair J. A compendium of volatile organic compounds (VOCs) released by human cell lines. Curr. Med. Chem. 2016;23:2112–31. doi: 10.2174/0929867323666160510122913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Filipiak W, Sponring A, Filipiak A, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. TD-GC-MS analysis of volatile metabolites of human lung cancer and normal cells in vitro. Cancer Epidemiol. Biomarkers Prev. 2010;19:182–95. doi: 10.1158/1055-9965.EPI-09-0162. [DOI] [PubMed] [Google Scholar]
- 48.Filipiak W, Sponring A, Mikoviny T, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. Release of volatile organic compounds (VOCs) from the lung cancer cell line CALU-1 in vitro. Cancer Cell Int. 2008;8:17. doi: 10.1186/1475-2867-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavra L, et al. Investigation of VOCs associated with different characteristics of breast cancer cells. Sci. Rep. 2015;5:13246. doi: 10.1038/srep13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwak J, et al. Volatile biomarkers from human melanoma cells. J. Chromatogr. B . 2013;931:90–6. doi: 10.1016/j.jchromb.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Sponring A, Filipiak W, Mikoviny T, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. Release of volatile organic compounds from the lung cancer cell line NCI-H2087 in vitro. Anticancer Res. 2009;29:419–26. [PubMed] [Google Scholar]
- 52.Schallschmidt K, Becker R, Jung C, Rolff J, Fichtner I, Nehls I. Investigation of cell culture volatilomes using solid phase micro extraction: options and pitfalls exemplified with adenocarcinoma cell lines. J. Chromatogr. B . 2015;1006:158–66. doi: 10.1016/j.jchromb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Mochalski P, Theurl M, Sponring A, Unterkofler K, Kirchmair R, Amann A. Analysis of volatile organic compounds liberated and metabolised by human umbilical vein endothelial cells (HUVEC) in vitro. Cell Biochem. Biophys. 2014;71:323–9. doi: 10.1007/s12013-014-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nozoe T, Goda S, Selyanchyn R, Wang T, Nakazawa K, Hirano T, Matsui H, Lee S W. In vitro detection of small molecule metabolites excreted from cancer cells using a Tenax TA thin-film microextraction device. J. Chromatogr. B . 2015;991:99–107. doi: 10.1016/j.jchromb.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 55.Sulé-Suso J, Pysanenko A, Španěl P, Smith D. Quantification of acetaldehyde and carbon dioxide in the headspace of malignant and non-malignant lung cells in vitro by SIFT-MS. Analyst. 2009;134:2419. doi: 10.1039/b916158a. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, et al. Identification of volatile biomarkers of gastric cancer cells and ultrasensitive electrochemical detection based on sensing interface of Au-Ag alloy coated MWCNTs. Theranostics. 2014;4:154–62. doi: 10.7150/thno.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies M P A, Barash O, Jeries R, Peled N, Ilouze M, Hyde R, Marcus M W, Field J K, Haick H. Unique volatolomic signatures of TP53 and KRAS in lung cells. Br. J. Cancer. 2014;111:1213–21. doi: 10.1038/bjc.2014.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wihlborg R, Pippitt D, Marsili R. Headspace sorptive extraction and GC-TOFMS for the identification of volatile fungal metabolites. J. Microbiol. Methods. 2008;75:244–50. doi: 10.1016/j.mimet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Haick H, Broza Y Y, Mochalski P, Ruzsanyi V, Amann A. Assessment, origin, and implementation of breath volatile cancer markers. Chem. Soc. Rev. 2014;43:1423–49. doi: 10.1039/C3CS60329F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hosakote Y M, Liu T, Castro S M, Garofalo R P, Casola A. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes. Am. J. Respir. Cell Mol. Biol. 2009;41:348–57. doi: 10.1165/rcmb.2008-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forman H J. Reactive oxygen species and alpha, beta-unsaturated aldehydes as second messengers in signal transduction. Ann. New York Acad. Sci. 2010;1203:35–44. doi: 10.1111/j.1749-6632.2010.05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tangerman A. Measurement and biological significance of the volatile sulfur compounds hydrogen sulfide, methanethiol and dimethyl sulfide in various biological matrices. J. Chromatogr. B . 2009;877:3366–77. doi: 10.1016/j.jchromb.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 63.Panayiotidis M I, Stabler S P, Allen R H, Pappa A, White C W. Oxidative stress-induced regulation of the methionine metabolic pathway in human lung epithelial-like (A549) cells. Mutat. Res.—Genet. Toxicol. Environ. Mutagen. 2009;674:23–30. doi: 10.1016/j.mrgentox.2008.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.