Abstract
Transposable elements (TEs) are no longer considered to be ‘junk’ DNA. Here, we review how TEs can impact gene regulation systematically. TEs encode various regulatory elements that enables them to regulate gene expression. RJ Britten and EH Davidson hypothesised that TEs can integrate the function of various transcriptional regulators into gene regulatory networks. Uniquely TEs can deposit regulatory sites across the genome when they transpose, and thereby bring multiple genes under control of the same regulatory logic. Several studies together have robustly established that TEs participate in embryonic development, and oncogenesis. We discuss the regulatory characteristics of TEs in context of evolution to understand the extent of their impact on gene networks. Understanding these features of TEs is central to future investigations of TEs in cellular processes and phenotypic presentations, which are applicable to development and disease studies. We re-visit the Britten-Davidson ‘gene-battery’ model and understand the genetic and transcriptional impact of TEs in innovating gene regulatory networks.
Keywords: gene regulation, transposable elements
1. Introduction
Transposable elements (TEs) are repetitive sequences (sometimes of viral origin) that have resided in eukaryotic genomes for millions of years.[1,2] By definition, most TEs (e.g.: DNA transposons, Long-terminal repeat (LTR) retrotransposons, and Long Interspersed Elements (LINE)) contain its own promoter and regulatory sequence that transcribes its genes encoding machinery for their movement in the host genome. Some TEs (e.g.: Short Interspersed Elements; SINEs) that lack their own transposition machinery are mobilised non-autonomously using another TE’s transposition machinery.[3–5] The initial discovery of TEs was made by Barbara McClintock’s seminal maize experiments, in which she found that TEs altered the colour of maize kernels by its transposition.[6,7] Most TEs identified in the human genome lack the ability to actively transpose; rare cases of insertional mutagenesis caused by actively mobilising TEs have been observed in the human genome.[8–11] TEs constitute 50% of the human genome sequence - most of which are present due to drift, however it is still unclear whether TEs play an active role in the genome, and to what extent they play such a role. Considerable efforts to study and investigate the role of TEs in the genome are ongoing, and will ultimately highlight the functional role of TEs in cellular processes, in both development and disease.[12,13]
TEs are often referred to as the double-edged sword, owing to their ability to have harmful effects on gene expression and yet be the source of novel sequence in the genome.[14,15] When TEs transpose, the cell is at risk of potentially lethal insertional mutagenesis, where a TE inserts into an essential gene and disrupts its expression.[7,16] TEs that are not lethal to the host could alternatively be neutral, or beneficial. For instance, TE sequences have often contributed promoters, and exons to the host genome, which have innovated existing gene regulatory programmes.[17,18] In human, TEs are found in 4% of protein-coding genes and 25% promoters.[19,20] Understanding the functional role of immobile TEs in the genome is central to determining why TEs are widespread in the genome, and what role they have in the host genome today?
Knowing what evolutionary pressures act on TEs is important for understanding what roles TEs might have in the genome.[21,22] The ‘junk’ DNA term, often colloquially used on TEs, referred to the lack of purifying selection pressures on TEs.[23] This implied that TEs are not under evolutionary selection, like protein-coding genes. However this does not rule-out that TEs were possibly once under purifying selection, which explains how TEs are so widespread in the genome.[24,25] Although TEs are not being selected for, it is important to acknowledge that TEs have not been eradicated from the genome. An important aspect of TEs that is essential for its mobilisation, and subsequently large representation in the genome is its ability to self-replicate by using the host machinery to encode its transposition machinery (i.e., ‘Selfish DNA’ hypothesis).[26–28] The substantial representation of TEs in genomes today is a testament to their ability to replicate more than the host that they reside in.[29,30] It is noteworthy here that that the contribution of TEs to the host genome sequence might be more than current estimates owing to limitations in identifying TE sequences. TEs need to resemble the ‘original’ TE adequately to be technically-identified – therefore TEs that have accumulated too many mutations that make them less like their ‘original’ TE, are more difficult to identify.[31] Yet, there are various articles have described the role of TEs as facilitators in genome evolution.[28,32,33]
Britten and Davidson proposed the ‘gene-battery’ model in which they elucidated a theoretical framework for the regulation of gene expression in eukaryotes.[34] This seminal work laid the foundation for understanding how TEs might impact gene expression regulation, by integrating cellular signals into gene regulatory networks.[1,34,35] In this Review, we characterise the ability of TEs to regulate gene expression, and its observed regulatory potential. We aim to understand the extent to which, and estimate the contexts in which TEs might function in the ‘gene-battery’ model and impact gene expression regulation.
2. Britten-Davidson’s ‘gene-battery’ model – the foundation for TE’s role in gene expression regulation
The theoretical model of gene expression regulation postulated by RJ Britten and EH Davison in 1969, has been one of the guiding models for defining gene expression regulation in multicellular eukaryotes. After Barbara McClintock’s initial discovery of TEs impacting the colour of the maize kernel, the ‘gene-battery’ model supported the ability of TEs to participate in cellular processes.[36–38] The ‘gene-battery’ model was elucidated at a time when TEs were still largely considered to be ‘junk DNA’ (1972) or genomic parasites (i.e., ‘Selfish’ DNA; 1980s), and largely disregarded in genetic studies.[23,26,27] Britten and Davidson hypothesized a role for repetitive sequences in mediating gene expression regulation in multicellular eukaryotes owing to their repetitiveness, ability to integrate cellular signals, and mobilise effectively.[35] Multicellular eukaryotes were integral to the model, since these organisms have large genome sequences, and often a substantial proportion of repetitive sequence.[39,40] When the model was postulated, a few studies had demonstrated that repetitive sequences were transcribed in differentiated cell types, and in cell-type specific patterns.[35]
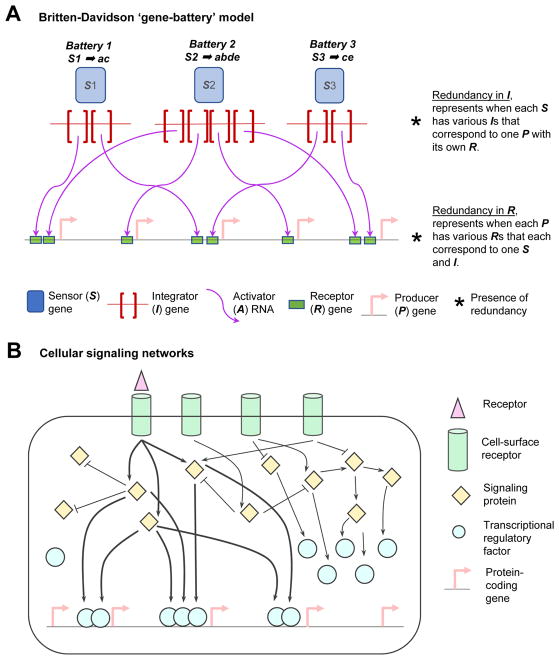
The ‘gene-battery’ model described a framework for understanding the regulation of groups of genes (i.e., batteries of genes) that specifies a particular cell type. In the ‘gene-battery’ model Britten and Davidson defined a ‘gene’ as a certain part of the genome which has a specific function, which need not be limited to the commonly-known function of defining a protein’s primary structure. The model is composed of five components or ‘genes’: sensor genes (e.g. hormones, and molecules involved in inter- or intra- cellular control), integrator genes (synthesizes activator RNAs in response to activity in sensor genes and coordinates the activity of downstream genes; e.g. regulatory factors), activator RNAs (binds at receptor genes; e.g. biochemical form of regulatory factors), receptor genes (linked to producer genes and regulates its transcriptional activity; e.g. promoters, and possibly enhancers), and producer genes (transcribed genes; e.g. haemoglobin subunit and tRNA molecules; Figure 1A). Together the set of producer genes, which is activated based on the activity of a specific sensor gene and its associated downstream regulators (i.e., the integrator gene and activator RNA), is defined as ‘battery’ of ‘genes’ (Table 1).
Figure 1.
Cellular signalling pathways and gene expression regulation. A: In 1969, RJ Britten and EH Davidson hypothesized a theoretical model consisting of five ‘genes’ – sensor (S), integrator (I), activator (A) RNA, receptor (R), and producer (P). A ‘battery’ comprises of various P and its regulators that are activated by a single S in response to a cellular cue. For example, Battery 1 targets producer genes (P) ‘a’ and ‘b’, based on the signal provided by sensor gene (S) ‘1’. The five gene-components work co-ordinately to activate various batteries of genes involved in specific biological processes (refer to Table 1). The five ‘gene’ types are listed in the figure key. The goal of these components is to effect gene expression upon stimulation from certain environmental cues. Integration of cellular signals and regulatory sites can be performed at the level of the Integrator (I), or Receptor (R). B: Cellular signalling networks as found in eukaryotes. The complexity of this system enables effective transmission of signals from the environment to the gene-level to deploy various gene expression programs in the cell. Here, the binding of a receptor at the surface of the cell, triggers the activation of a cascade of signalling proteins, which in turn activate various transcriptional regulatory factors. The active transcriptional regulatory factors bind at specific genes and regulating the gene’s expression, in response to the external stimulus.
Table 1.
Defining ‘gene-batteries’, based on the Britten-Davidson model. Here, each ‘battery’ is defined as a set of producer genes (P) that are activated by a single sensor gene (S). Therefore, there is one ‘battery’ for every sensor gene (S). Ticks (✔) represent producer genes (P) that are activated by a particular sensor gene. This table corresponds to the sensor-producer (S-P) associations depicted in Figure 1A.
| Sensor (S) gene | Producer gene ‘a’ | Producer gene ‘b’ | Producer gene ‘c’ | Producer gene ‘d’ | Producer gene ‘e’ | |
|---|---|---|---|---|---|---|
| Battery 1 | 1 | ✔ | ✔ | |||
| Battery 2 | 2 | ✔ | ✔ | ✔ | ✔ | |
| Battery 3 | 3 | ✔ | ✔ |
A collection of such gene-batteries was hypothesized to define a particular cellular state, by the integration of the functionality of multiple batteries (Table 2). Cellular differentiation is affected by external cues and signals (e.g. hormones), which in turn co-ordinately activate hundreds of genes in different parts of the genome (Figure 1B). In their model, Britten and Davidson describe two modes of integration of signals that vary based on where the redundancy of information lies.[34] First, there could be redundancy in receptor genes, where a producer gene is included in the battery if it has the receptor gene associated with the sensor gene regulating the battery. Second, there could be redundancy in integrator genes, where a producer gene is included in a battery when the sensor gene regulating the battery has an integrator gene corresponding to the producer gene.[1,35] The first mode of functioning permits coordinated regulation of producer genes. However, the second mode of functioning is possibly the more powerful mode of functioning as it integrates several diverse producer genes into one battery.
Table 2.
Interpreting ‘gene-batteries’ in the context of the biological process that it regulates. In their model, Britten and Davidson suggested that the coordinated activation of several ‘batteries’ of genes results in specific biological processes. Here, each Biological Process (row) is a result of the activation of one or more ‘batteries’. In other words, each biological process is in turn a combination of different sensor and producer genes.
| Battery 1 | Battery 2 | Battery 3 | Sensors (S) | Producers (P) | |
|---|---|---|---|---|---|
| Biological Process A | ✔ | ✔ | 1, 3 | a, c, e | |
| Biological Process B | ✔ | 2 | a, b, d, e | ||
| … | … | … | … | … | … |
| Biological Process Z | ✔ | ✔ | ✔ | 1, 3 | a, b, c, d, e |
Here, an important aspect of the ‘gene-battery’ model is the role of repetitive sequences in gene regulatory networks. Other than the role of TEs in contributing new material for the evolution of gene-regulatory sequences (producer genes), this model hypothesizes a role for TEs in the integration of cellular signals, at the levels of receptor genes and integrator genes. As models of gene regulatory network evolution are still debated, the ‘gene-battery’ model and the role of TEs in this model becomes important to understand.[41]
3. Widespread enrichment of transcription factor (TF) binding sites in TEs
The first genome-wide identification of TF binding sites occurring in TEs was observed for - p53, a tumour-suppressor gene. Up to 30% of p53 in vitro binding sites are encoded in primate-specific endogenous retrovirus (ERV) long-terminal repeats (LTRs).[42] At a time, when most sequence-alignment algorithms discarded reads mapping to multiple genomic loci, this finding showed that TEs can be analysed based on uniquely-mapping sequencing reads too. Among many hundred thousand of human LTRs, 0.5% of the LTRs contribute a p53 binding motif, which represents 30% of the in vitro binding sites for p53 -- this demonstrates the power of TEs in innovating existing transcriptional networks.
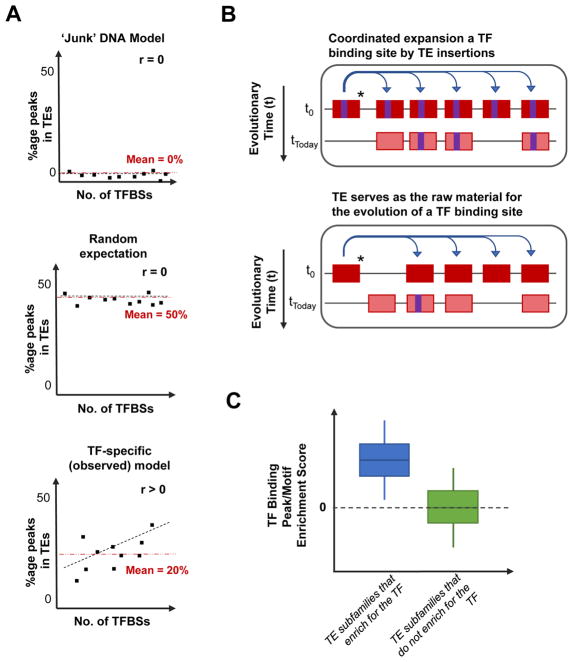
Subsequently, several other studies also demonstrated that TEs contribute binding sites for other TFs, including pluripotency factors (OCT4, NANOG), and genome-organisation and insulator-protein CTCF.[43–45] The finding of TEs with binding sites for TFs such as OCT4, NANOG and CTCF can be explained by the fact that TEs are epigenetically de-repressed in the germline and embryonic stages, and these TFs are active thereby promoting the spread of TEs.[46] There could also be unknown germline functions for several TFs that might have binding sites in TEs. These studies further encouraged the possibility that TEs encoded binding sites for various TFs, but the extent to which TEs contained TF binding sites was still unknown. We investigated ChIP-seq binding profiles in vitro of 26 TFs in two cell types in human and mouse, and quantified the contribution of TF binding sites by TEs.[47,48] On average, 20% (range: 2–40%) of a TF’s in vitro binding sites is likely to occur in TEs.[49] To understand if TEs have systematically shaped transcriptional networks by depositing TF binding sites for various TFs, it is critical to set an expectation (Figure 2A). Based on the ‘junk’ DNA hypothesis, it can be expected that TEs have no contribution to TF binding sites. Alternatively, assuming that TEs constitute ~50% of the human genome sequence, by chance 50% of a TF’s binding sites can be expected to occur in TEs.
Figure 2.
Model for understanding the evolution of TF binding sites in TEs. A: Setting the expectation for how many TF binding sites exist in TEs. Looking at various TFs (each represented by a dot in the panels), under the model of TEs being ‘junk’ DNA (upper panel), the expectation is that all TEs will have no TF binding sites because TEs are non-functional sequences. Alternatively, another common expectation is that TEs constitute almost half of the human genome sequence, therefore by chance, 50% of all TF binding sites will occur in TEs (centre panel; labelled “Random Expectation”). However, the observation is quite contrary to the previous two models (bottom panel; labelled “TF-specific (observed)). We have observed that the percentage of TF binding sites occurring in TEs ranges from 2% to 40% (average: 20%; red dotted-line in the panel) in both human and mouse cell types. It is noteworthy that the percentage of TF binding sites in TEs positively correlates with the number of TF binding sites in the genome. B: Co-ordinated expansion of TE subfamilies causing an increase in the number of TE-derived TF binding sites in the genome (upper panel;), versus TEs evolving TF binding sites by neutral evolution (lower panel). In the two models, a TE (red rectangle) capable of transposing (*) is shown to distribute across the genome. A TE with a TF binding sites (purple rectangle) will spread the TF binding site along with the TE as it transposes (upper panel). Alternatively, a TE without a binding site could serve as raw material for the evolution of new TF binding sites, based on mutations acquired (lower panel). C: Schematic representation of the enrichment scores of TEs belonging to a subfamily that enriches for a TF’s binding site (ChIP-seq peaks of sequence motifs), or TEs belonging to a subfamily that does not enrich for TF binding sites.
We observe that the fraction of TF binding sites derived from TEs is TF-specific. Enrichment studies demonstrate the ability of TEs to co-ordinately impact transcriptional networks, thereby potentially ‘rewiring’ existing gene regulatory networks. From previous studies, enrichment analyses have defined certain TF-TE associations in which a specific TE subfamily enriches for ChIP-seq binding sites and binding motifs for specific TFs. In this model, TF binding motifs in transposing-TEs are deposited across the genome, as the TE transposes (labelled “co-ordinated expansion of a TE”; Figure 2B). The mobility in TE subfamilies containing TF binding motifs serves as an evolutionary vehicle for the TF to gain new target binding sites, and consequently potential target genes to control.
Alternatively, the evolution of new TF binding sites could occur by evolutionary mutational processes that can occur in any genomic sequence that is not under purifying selection. TEs are widely thought to be raw material for the evolution of new TF binding sites, based on mutational processes under neutral evolution (labelled “TE serves as the raw material for the evolution of a TF binding site”; Figure 2B).[50] However, the model of neutral evolution of TF binding sites cannot facilitate the impact on multiple genes in a gene regulatory network, as found in the former model of rewiring facilitated by a TE subfamily. The former model serves as an efficient mechanism for the host cell to utilise the existing regulatory information in TEs, and also rewire of existing gene regulatory networks. The difference in these two models is observed in the differences in enrichment scores of TF binding in TEs belonging to subfamilies where multiple elements contain the binding site, compared to individual TEs that encode TF binding sites (Figure 2C).[49] In 2007, Wang et. al. speculated that TEs might be the reason for p53 being a master regulator, based on the gained access to controlling genes provided by TE-derived TF binding sites.[42] We also observed that the more number of binding sites that a TF has, there is also a higher fraction of the sites found in TEs (lowest panel of Figure 2A), suggesting that the TFs might have gained ‘master’ regulator status through TE-mediated expansion of their binding sites.[49]
Just as importantly, TEs have also been observed to lose binding sites for certain proteins, in what is well-known as the evolutionary arms race.[51] The evolutionary arms race is a widely-studied evolutionary biology concept that describes a constant competition between two entities that are co-evolving to outcompete each other. Jacobs et al., demonstrated that certain primate KRAB zinc-finger protein genes (known suppressors of TE activity), undergo a rapid evolution to outcompete the spread of two TE families – SINE-VNTR-Alu (SVA) and LINE (L1) elements. While one KZNF protein evolved structurally to suppress SVA elements, another KZNF protein lost its ability to suppress L1 elements, when the L1 family lost the binding site for the protein. A similar observation has been made in the evolution of the immune system (major histocompatibility complex – MHC) and TEs.[52,53] Together, these examples represent our understanding of the role TEs can have in genome evolution, and the constant contention between TEs and the host genome.
4. TEs can deposit modules of TF binding sites to integrate the function of multiple related-TFs in specific biological pathways
Britten and Davidson hypothesized the theoretical role of TEs in impacting gene regulatory networks.[34] However, the lack of genome-wide data for TF binding made it difficult to garner evidence for the role of TEs in this model. Through years of comparative genomics, and with the completion of various genome sequences, we learnt that TEs represent a sizeable fraction of regulatory sequences, and thousands of TEs undergo purifying selection.[54,55] Subsequently, as mentioned in the previous section, we also learned that TEs contain binding sites for TFs and have epigenetic signatures of transcriptional enhancers. In some cases, the biochemical activity of these TEs were validated using classical luciferase reporter assays.[42,56] More recently, with the advent of high-throughput experimental assays the regulatory activity of more TEs can be dissected.[57]
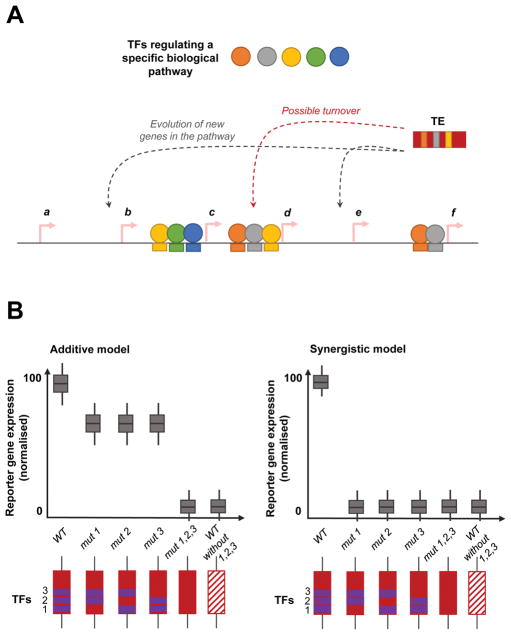
Cis-regulatory modules are widely known for their ability to integrate the effects of multiple TFs and coordinate the expression of sets of protein-coding genes for a given biological process. As described by Britten and Davidson, repetitive sequences like TEs are uniquely able to co-ordinate gene expression regulation across the genome (Figure 3A). TEs carrying ‘ready-to-use’ modules of TF binding sites in a particular pathway, provides an efficient evolutionary mechanism for the cell to evolve new target genes.[50] Recently we identified a cis-regulatory module of pluripotency TF binding sites encoded in a class of mouse-specific ERV sequences.[58] Binding sites for Oct4 and Nanog have been identified in TEs in the mouse genome, previously.[44] We discovered certain mouse-specific LTRs (missing in rats) that contained in vitro binding sites for three of the five pluripotency TFs studied – Esrrb, Klf4, Nanog, Oct4 and Sox2.[59]
Figure 3.
Transposable elements mediated evolution of cis-regulatory modules. A: Gene regulatory networks are intricately coordinated processes that involve the participation of various regulatory transcription proteins and factors (TFs; filled circles) that collectively act upon a set of genes (arrows labelled a–f). Multiple regulatory factors regulate the expression of individual genes by binding at specific sequence motifs in the vicinity of the gene. Transposable elements (TEs) carrying a module of TF binding sites offer a unique evolutionary mechanism to rewire the existing networks by incorporating new genes under the regulatory control of the factors. Additionally, TEs could also be used at sites with an existing module of binding sites by repurposing it, provided it does not negatively impact the gene and its function. B: Demonstrating the synergism among TF binding sites in a module, compared to an additive model. In the additive model, the effect of the various TF binding sites on gene expression is the sum of its effects. Whereas in the synergistic model, the effect of the various TF binding sites on gene expression is greater than the sum of its individual effects. Here, we display a schematic of reporter gene expression (to measure the effect of an enhancer on gene expression; y-axis) where a TE (red rectangle) contains binding sites for 3 TFs (numbered 1–3; represented by purple rectangles). The effect of mutations on TF binding motifs that disrupt the motif (labelled ‘mut’; depicted by absent purple rectangles) are shown in comparison to the wild-type sequence (labelled ‘WT’; positive control); and WT sequence that has naturally (over evolutionary time) lost the TF binding sites (negative control).
Using CRE-seq, a massively parallel reporter assay, we were able to dissect the regulatory interactions between the binding motifs and establish a cis-regulatory module of Esrrb, Klf4, and Sox2 binding sites encoded in these TEs.[57] Studies dissecting the cis-regulatory logic and grammar of pluripotency TF binding sites in mouse ESCs identified cooperative interactions between Klf4 with Esrrb, and Sox2.[60] We observed synergism between the three TFs, such that the presence of all three motifs were required for the regulatory potential of the TE (Figure 3B left panel). Alternatively, in an additive model, the loss of any one binding motif would not eliminate the activity of the TE (Figure 3B right panel). The synergy between the TF binding motifs demonstrate that TEs are also capable of ‘modular’ regulatory behaviour, where a TE integrates the effects of multiple TFs in a transcriptional network. This is one example of the ‘gene-battery’ model.
5. Ancestral TEs contain binding sites for TFs and the potential to regulate gene expression
Understanding the evolution of TEs and their associated regulatory sites is important to understand the impact of TEs on the host genome. Although it is well-known that TEs have widely contributed TF binding sites to the regulatory landscape of the genome, it is poorly understood how TEs have evolved these regulatory sites and the associated regulatory potential. Simply put, there are two models that could explain the presence of TF binding sites in TEs – one in which the TEs enter the genome with the regulatory site and spread their regulatory sequence as it transposes, and one in which the TE gains the binding site, via mutational processes.[50] The evolutionary model of TEs and their associated regulatory sites could be a combination of these two models, or somewhere in the spectrum between these two models.
In an approximation of the ancestral state of the TE, we and others have used the RepBase-consensus sequence as a proxy.[58,61] Since it is not feasible to identify the true ancestral state of the TE from many millions of years ago, for now we will have to resolve to using in silico estimations of the ancestral state of the TE.[62] The RepBase-consensus sequence is manually generated from existing genome sequences with identifiable repetitive sequences, this approximation assumes a specific state of the ancestral TE.[63] For a first-pass, this is a reasonable assumption, but more stringent and rigorous reconstruction of the ancestral sequence, or its most-likely states will be better for analysing the ancestral TE.
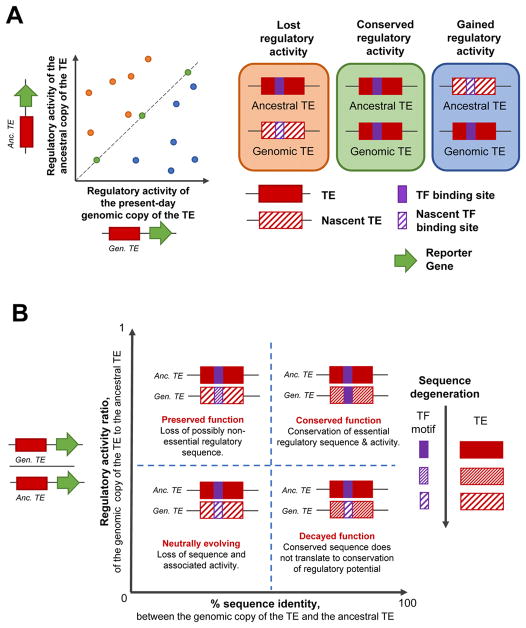
The approximated ancestral state of TEs are also capable of regulating gene expression in a reporter gene assay setting. Using sequence information built from a consensus-like method, motif analyses have revealed in a couple of cases that the ‘ancestral’ TE also contained binding motifs for the TFs that has binding motifs in the present-day genomic copies.[49,58,61,64] To test the regulatory potential of ancestral sequences in vitro, gene synthesis was combined with reporter-gene assays and revealed that the ‘ancestral’ TEs are also capable of regulating gene expression in vivo. This suggests that the observed regulatory potential in present-day copies of the TE likely originates from the ‘ancestral’ TE’s regulatory potential (Figure 4A). Comparing the regulatory potential of a few present day genomic copies with their corresponding ‘ancestral’ sequence showed that majority of the genomic copies had lower regulatory potential (labelled “Lost regulatory activity” in Figure 4A). In this first comparison of its type, there were only four TE subfamilies and ten genomic copies that were that were analysed, and a more comprehensive analysis will provide a better understanding of the evolution of the regulatory potential in TEs. The comparison between present-day genomic copies of the TE, and their ancestral states need not only be made at the regulatory potential level alone, but also at the sequence-level. TEs can have various levels of sequence identity, although their corresponding regulatory potential varies (Figure 4B). When the regulatory activity is similar between the ancestral and genomic copies of the TE, it is most likely that sequence changes in the TE sequence is mostly altering non-essential regulatory sequences (labelled “Preserved function” and “Conserved function” in Figure 4B), while alternatively varied regulatory activity could be due to sequence changes affecting both regulatory and non-regulatory sequence in the TE (labelled “Neutrally evolving” and “Decayed function” in Figure 4B).
Figure 4.
Evolution of gene regulatory activity of TEs, compared to the ‘ancestral’ TE. A: Comparing the normalised regulatory activity (as measured by reporter assays) of the ancestral state (y-axis) and the present-day genomic copy (x-axis) of the TE. Data-points falling on the diagonal represent genomic copies of the TE that have the same regulatory activity as their ancestral state, which likely has most of the sequence also conserved (green dots; panel labelled “Conserved regulatory activity”). Data-points that fall above the diagonal represent TE copies that have lost the ancestral regulatory activity, possibly by corresponding loss of regulatory sequences in the TE (orange dots; panel labelled “Lost regulatory activity”). Lastly, data-points falling below the diagonal represent TE copies that have gained regulatory activity, possibly be acquiring sequence mutations that enhance a nascent TF binding site and thereby provide the TE with increased regulatory activity. B: Understanding the evolution of the TE at a sequence- and regulatory-activity levels. Comparison of the sequence identity (%; x-axis) and ratio of regulatory activities (y-axis) between present-day genomic copies of the TE and the ancestral TE. We categorised the data based on the four quadrants and indicate the biological implication of data-points falling in each quadrant. The legend on the side depicts the sequence degeneration between the ancestral and genomic copies of the TE (filled boxes representing least/no degeneration, and patterned boxes representing degenerating sequence).
6. Impacting phenotypes - gene expression and biological processes
The first known phenotypic impact attributed to TEs was in altering the colour of the maize kernel, as discovered by Barbara McClintock.[6,7] This phenotypic impact is attributed to the mobility of TEs in the genome. Today, most eukaryotic TEs are immobile remnants of previous TE insertions, and are possible sources of regulatory innovation. Non-coding genetic (cis-regulatory) elements are difficult to ascribe a phenotypic role to, owing to the current difficulty in associating these regulatory elements to a target gene, and a cellular or organismal phenotype. This issue is only exacerbated in the cases of TEs, which are only now beginning to be analysed in genome-wide assays. However, the potential of TEs to impact phenotypes is exciting since TEs are capable of evolving ‘gene-batteries’ and expanding TF binding sites, and thereby altering gene expression in the genome.
Genome-editing tools such as CRISPR-Cas9 enable the testing of hypothesis by biological validation of the effect that the editing (i.e., deletion or mutation) of a candidate TE has on gene expression, and potentially cellular phenotypes. Recently, a significant functional role has been attributed to TEs in immune-related processes.[61] The MER-subfamily (MER41) of ERVs in mammalian genomes contain binding sites for STAT1, which is induced by interferon to regulate their target genes. CRISPR-Cas9-mediated deletion of these MER41 elements significantly reduced the expression of their target genes.
TEs differ in their frequency and form in different species, and therefore could have diverse evolutionary impact in various organisms (Figure 5A). We identified a mouse-specific LTRs encoding modules of pluripotency TF binding sites that was associated with a gene (akap12) specifically expressed mouse embryonic stem cells (ESC), and not human ESCs or other mouse cell types. Deletion of the TE using CRISPR-Cas9, demonstrated a significant decrease in the expression level. Chuong et al., also observed different “MER41-like” elements in different species, suggesting a species-specific impact of TEs on gene expression patterns. More comprehensive comparative transcriptome analyses will be able to better-define the role of TEs in gene expression differences between species, and eventually understand the impact of these differences on phenotypic differences between the species. Since 99% of TE-derived TF binding sites are species-specific, cross-species multi-omic analyses would enable a better understanding of the species-specific phenotypic effects of TEs.
Figure 5.
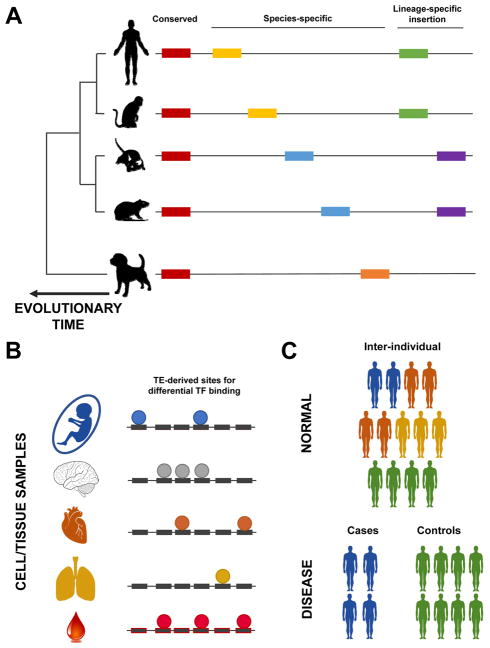
Elucidating the effect of TEs on phenotypes, considering (A) evolution, (B) cell-types and cell-fate decisions, and (C) at the population-level. A: TEs (represented by coloured rectangles) are largely present in eukaryotic genomes, but vary in their specificity. On the one hand, some TE insertions are conserved across evolution in terms of their presence in a species, and their orthology in the genome sequence (labelled “Conserved” in the figure). On the other hand, there are other TE instances that are species-specific, in terms of the presence of a subfamily in a species (orange rectangle), and insertions (yellow rectangles in human and mouse, and blue rectangles in mouse and rat). There are also lineage-specific TEs, for example whose subfamily was active in primate (green rectangles) and rodents (purple rectangles). B: Cell/tissue-type specificity in TE’s biochemical activity. Different cell types have the same genome, but have differences in the activity of TFs (coloured circles). Here, we depict the differences in TF binding on TEs in different cell types. C: Lastly, a less explored area of TE-biology is the differences in TE biochemical activity and effects on gene expression regulation in normal populations (upper panel), and patients with diseases (lower control).
TE co-option is central to their ability to impact phenotypes. Evidence suggests that ancient mammalian TEs have been co-opted into a hormone-responsive transcriptional networks associated with pregnancy in placental mammals. In 2010, Lynch et. al., characterised ESCs from different species at various stages of evolution, and observed placental mammal-specific expression patterns (upregulated and downregulated gene expression) that was associated with endometrial expression patterns of genes near MER20 insertions.[64] More recently, Lynch and colleagues analysed uterine tissues for genes showing endometrial expression and identified that many cis-regulatory elements that regulate cell-fate processes in preparation for pregnancy, are derived from ancient mammalian TEs.[12] In other cases, TEs have been found to require mutations before being adopted by the host for host functions (i.e., epistatic capture).[65] Additionally, ERV sequences encoding envelope genes have been co-opted and involved in placenta development.[66,67] Other than mammals there is also evidence of TEs being co-opted and impacting gene regulatory networks in fruit fly.[68]
TEs, especially ERVs are known to be involved in pluripotency and early development (Figure 5B). Epigenetic modifications, including DNA methylation, is considered to be a key regulator of TE-activity, especially during early mammalian development. However, ERVs are among the initial genomic regions to be transcribed in mouse embryos,[69] when TEs are assumed to be under strong epigenetic suppression. During pre-implantation development, different classes of primate-specific ERVs are expressed, which is later ceased during differentiation.[15,70,71] Additionally, ERV-derived long non-coding RNAs (lncRNA) are involved in pluripotency in human ESCs.[72–74] More than two-thirds of lncRNAs in mouse, human and zebrafish include exonic TE sequences, suggesting an extensive role for TEs in cellular transcriptomes.[17] TEs are normally kept in check in differentiated tissues by epigenetic mechanisms of suppression of TE activity,[32] however, tissue-specific epigenetic derepression has been observed in TEs in some cell types (Figure 5B).[56]
Other than in development, TEs have also a potential role in diseases; 65 known human diseases have been attributed to de novo TE insertions.[75,76] Epigenetic depression (global reduction in DNA methylation) during aging and tumorigenesis is a conducive environment for the activity of otherwise-silenced TEs, which can be as genomic instability (i.e., DNA breaks and translocations) and transcriptional activity (LINE elements in various cancers).[77] Along with TE activity in cancer, TEs have also been identified to encode transcripts upon epigenetic inhibition by cancer treatments (i.e., DNA methyltransferase inhibitors and histone deacetylase inhibitors).[13,78,79] Additionally, TEs need to be studied in other cases of epigenetic derepression, including aging.
A lesser known function of TEs, is at the population-level, including in diseases (Figure 5C). On the one-hand, actively transposing TEs could generate polymorphic TE-insertions within the population. Although most TEs in humans are not actively transposing, it is still unknown what effects and functions polymorphic TE insertions have within a population. One of the most well-known examples, is in the case of haemophilia in humans.[16] On the other hand, non-transposing TEs could also have inter-individual differences in terms of their epigenetic status, TF binding, and therefore potential to impact transcription and cellular processes. This aspect of TEs is still not well-characterised, and could open an avenue for studying TEs at the population level.
7. Discussion
Barbara McClintock’s seminal work in maize first identified TEs in the 1940s. She referred to TEs as ‘controlling elements’ - “normal components of the chromosome responsible for controlling, differentially, the time and type of activity of individual genes”.[7,15] With results from several studies, it is becoming more apparent that immobile TEs (as observed in most genomes today) also have the ability to control the expression of genes in various cellular contexts, and in different species. The cellular and organismal impact of TEs, and their differences - in composition, abundance, and genomic distribution - remains to be investigated. Britten and Davidson provided the first theoretical model for how TEs might influence gene regulatory networks via the regulatory sequences that they encode, and their mobility.
Gene regulation differences underlies gene expression variability in organisms.[80] Over many millions of years of evolution, there has been vast phenotypic diversity in observed in organisms. Evidence of closely related organisms having large differences in their genome size clearly obscures the association of genome size increase and proportional organismal organisation in eukaryotes. It is now well-known that genome size and organism size do not scale proportionally, and this is commonly referred to as the C-value paradox, or C-value enigma.[39,81,82] Various explanations have been provided for the increase in DNA-content in various organisms, one of which is for diversifying gene regulation as opposed to increasing the number of protein-coding genes.[34] Differences in regulatory relationships and interactions between species could play a pivotal role in diversifying organismal phenotypes. Although the exact contribution of gene expression changes to speciation and adaptation is still being uncovered, it is widely-accepted that differences in gene expression patterns underlie the evolution of morphological phenotypes and other complex traits in organisms.[83] Studies from closely related Drosophila species reveal that many cis-regulatory changes, rather than widespread trans-regulatory changes drive gene expression differences.[80,84–87] The ability of TEs to ‘rewire’ gene regulatory networks systematically (based on the ‘gene-battery’ model), and their differences between species make TEs a good candidate for investigation to understand the evolution and divergence of species.
We have discussed in detail in this review the ability of TEs to encode transcriptional enhancers. It might only be a coincidence that the first eukaryotic enhancers were discovered in animal viruses. The enhancer in the SV40 virus (Simian vacuolating virus 40) was used by the virus to harness the host’s transcriptional machinery, upon infection for its replication.[88] The SV40 enhancer demonstrates a key feature of enhancers in its synergism of TF binding sites, which integrates various cellular signalling processes in the host cell to increase the virus’ replication. Therefore, it is not surprising when TEs are also found having the same potential and features to regulate gene expression.[58,64] The same features of enhancers where they can integrate various cellular signalling processes are also found in TEs. The ‘gene-battery’ model hypothesized a role for TEs in systematically integrating signalling pathways.
Central to this advance in our understanding of TEs is the advent of newer technologies and methods that can assay the genome.[89] Initially, sequence-alignment pipelines would commonly discard sequencing reads mapping to TEs (i.e., repetitive sequences, that reads would map non-uniquely to), and therefore TEs were masked. Several alignment pipelines were developed to harness reads mapping to multiple genomic locations by estimating the most-likely mapping location on the genome.[90–92] However, with improved sequencing-read lengths, and paired-end sequencing, more information (i.e., sequencing reads) on TEs can be captured. Currently, most TE-studies analyse only uniquely-mapping sequencing reads. Uniquely mapping sequencing reads in fact contain adequate information, as most TEs have sufficient unique sequence to map reads to specific TEs without any ambiguity. Vast effort has already been made by various public efforts to assay specific aspects of the genome - transcripts (RNA-seq and CAGE-seq), protein-DNA interaction, chromatin/epigenetic state, and chromatin organisation – and this provides the means for TEs to be interrogated in numerous ways.[13,17,42–45,49,56,58,93]
The challenge lies in taking ahead computational findings and predictions of TEs (in the context of the ‘gene-battery’ model and otherwise) from multi-omic datasets to the cell. CRISPR-Cas9 technologies are gaining wide popularity. In the future, targeted validation of the ‘gene-battery’ model will be done by deleting elements of candidate TE subfamilies and estimating the impact of these deletions on cellular phenotypes. With this ability, similar analyses can be done between species. The impact of TEs in the differences of the wiring of gene-regulatory networks between species will provide a better understanding of the role of TEs in species divergence and evolution.
From 1969, the field has made vast progress in determining the role that TEs have in gene expression regulation. From several studies, we have learnt that TEs have biochemical activity in the cell and TEs are not always epigenetically repressed. Although TEs are exogenous, they are distinctive in their inherent regulatory functions and the potential to be biochemically active, and this feature is utilised by the cell to integrate and coordinate various other biochemical processes in the cell. With evidence of the ‘gene-battery’ model, we can now probe systematic influences of TEs in the genome and gene regulatory networks. The future lies in identifying the regulatory interactions that TEs have in different cellular contexts (in normal development and disease), and its impact on organismal phenotypes.
8. Conclusions and Outlook
With the advent and progress in sequencing technologies, it is relatively easier to analyse repetitive sequences and transposable elements (TEs), also in the context of the ‘gene-battery’ model. Although we are still deciphering the various role of TEs in cellular biology, we have indicative evidence of it providing the cell with regulatory sequence that is at the disposal of the cell. These sequences could be utilised in gene expression regulation, gene sequence, biochemical activity in the cell, chromatin state and chromatin organisation, in both normal development and disease. It is fair to say that from most genome-wide studies, a compendium of TEs role in various cellular contexts and developmental stages can be generated to determine the extent to which TEs impact cellular processes.
The ‘gene-battery’ model provides the basis for understanding how TEs might be involved in evolution – at the genetic and genomic level, and possibly the organismal level. Whereas at the genetic level TEs can modulate gene-specific expression levels, at the genomic level TEs can impact networks of genes (i.e., ‘gene-batteries’). In this model TE subfamilies impact many genes in a synchronized manner, and identifying this could impact our understanding of cellular processes. The potential avenues of exploration include TEs role in transcription regulation (as we have extensively discussed here), but also in transcript generation, chromatin state, and chromatin organisation. Single-cell analyses in the future, could shed light on the role of TEs generating cell-to-cell variation in transcriptional output, and cellular functions. Importantly, the ‘gene-battery’ model makes a compelling case to study this in a genome-wide manner, and not only in a locus-specific manner.
Other than at the cellular level, it still remains to be understood what role TEs have in organismal phenotypes. Since there are differences in TE composition between species, it is likely that TEs are involved in facilitating organismal differences between species. TE subfamilies not only differ in their presence in species, but also their genomic positions and sequence composition. These differences will have varied effects on gene expression regulation and could be connected with species’ evolution.
In conclusion, since the discovery of TEs, the field has moved far away from the notion of TEs being ‘junk’ DNA. We now understand some of the roles that TEs have in biology, and have a lot more to unravel. The future of big-data genomics holds a huge opportunity for detailed deciphering of newer and varied biological roles of TEs, potentially impacting cellular and organismal phenotypes. The ‘gene-battery’ model set the foundation for understanding the impact of TEs on gene regulatory networks, and now with multi-omic, genome-wide datasets we can effectively examine the model.
Acknowledgments
V.S. was supported by a fellowship from the EMBL Interdisciplinary Postdoc (EIPOD) programme under Marie Skłodowska-Curie actions COFUND (grant number 664726). T.W. was supported by NIH grants 5R01HG007354, 5R01HG007175, 5R01ES024992, 2U01CA200060, U24ES026699, U01 HG009391 and American Cancer Society Research Scholar grant RSG-14-049-01-DMC.
Abbreviations
- TE
transposable element
- TF
transcription factor
- ESC
embryonic stem cell
- LINE
long interspersed element
- LTR
long-terminal repeat
- SINE
short interspersed element
- CRISPR
Clustered Regularly Interspacsed Short Palindromic Repeats
Footnotes
We have no conflicts of interest to declare.
References
- 1.Britten RJ, Kohne DE. Science (80- ) 1968;161:529. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- 2.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. Nat Rev Genet. 2007;8:973. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 3.Slotkin RK, Martienssen R. Nat Rev Genet. 2007;8:272. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 4.Wessler SR. Proc Natl Acad Sci USA. 2006;103:17600. doi: 10.1073/pnas.0607612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feschotte C, Pritham EJ. Annu Rev Genet. 2007;41:331. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClintock B. Proc Natl Acad Sci. 1950;36:344. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClintock B. Cold Spring Harb Symp Quant Biol. 1956;21:197. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Deininger PL, Moran JV, Batzer MA, Kazazian HH. Curr Opin Genet Dev. 2003;13:651. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 9a.Ewing D, Kazazian HH. Genome Res. 2010 doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson SR, Morell S, Faulkner GJ. Annu Rev Genet. 2014:1. doi: 10.1146/annurev-genet-120213-092412. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford a, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan a, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian a, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson a, Deadman R, Deloukas P, Dunham a, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt a, Jones M, Lloyd C, McMurray a, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall a, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra Ma, Mardis ER, Fulton La, Chinwalla aT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty a, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen a, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs Ra, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama a, Hattori M, Yada T, Toyoda a, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal a, Platzer M, Nyakatura G, Taudien S, Rump a, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan a, Qin S, Davis RW, Federspiel Na, Abola aP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans Ga, Athanasiou M, Schultz R, Roe Ba, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey Ja, Bateman a, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones Ta, Kasif S, Kaspryzk a, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght a, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit aF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams a, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld a, Wetterstrand Ka, Patrinos a, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J. Nature. 2001;409:860. [Google Scholar]
- 12.Lynch VJ, Nnamani MC, Kapusta A, Brayer K, Plaza SL, Mazur EC, Emera D, Sheikh SZ, Grutzner F, Bauersachs S, Graf A, Young SL, Lieb JD, DeMayo FJ, Feschotte C, Wagner GP. Cell Rep. 2015;10:551. doi: 10.1016/j.celrep.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brocks D, Schmidt CR, Daskalakis M, Sik Jang H, Shah NM, Li D, Li J, Zhang B, Hou Y, Laudato S, Lipka DB, Schott J, Bierhoff H, Assenov Y, Helf M, Ressnerova A, Saiful Islam M, Lindroth AM, Haas S, Essers M, Imbusch CD, Brors B, Oehme I, Witt O, Mallm J-P, Rippe K, Will R, Weichenhan D, Stoecklin G, Gerh C, Oakes CC, Wang T, Plass C. 2017 doi: 10.1038/ng.3889. [DOI] [Google Scholar]
- 14.Wolff F, Leisch M, Greil R, Risch A, Pleyer L. Cell Commun Signal. 2017;15:13. doi: 10.1186/s12964-017-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuong EB, Elde NC, Feschotte C. Nat Rev Genet. 2016;18:71. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE, Antonarakis Stylianos E. Nature. 1988;332:164. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 17.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. PLoS Genet. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. Nature. 2006;441:87. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 19.Nekrutenko A, Li WH. Trends Genet. 2001;17:619. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 20.Van De Lagemaat LN, Landry JR, Mager DL, Medstrand P. Trends Genet. 2003;19:530. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Le Rouzic A, Boutin TS, Capy P. 2007;104:19375. doi: 10.1073/pnas.0705238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Rouzic A, Capy P. Elements. 2006 doi: 10.1007/7050. [DOI] [Google Scholar]
- 23.Ohno S. Evol Genet Syst Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- 24.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Genome Res. 2005;15:1034. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genome R, Project S. Nature. 2004;428:493. [Google Scholar]
- 26.Orgel LE, Crick FH. Nature. 1980;284 doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 27.Doolittle WF, Sapienza C. Nature. 1980;284:601. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 28.Hurst GD, Werren JH. Nat Rev Genet. 2001;2:597. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 29.McLaughlin RN, Malik HS. J Exp Biol. 2017;220:6. doi: 10.1242/jeb.148148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biemont C. Genetics. 2010;1093:1085. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Koning aPJ, Gu W, Castoe Ta, Batzer Ma, Pollock DD. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver KR, Greene WK. Bio essays. 2009:703. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- 33.Kazazian HH. Science. 2004;303:1626. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 34.Britten RJ, Davidson EH. Science. 1969;165:349. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 35.Davidson EH, Britten RJ. Science (80- ) 1979;204:1052. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- 36.Raveendran S. PNAS. 2012;109:20198. [Google Scholar]
- 37.Fedoroff NV. 1989;56:181. doi: 10.1016/0092-8674(89)90891-x. [DOI] [PubMed] [Google Scholar]
- 38.Fedoroff N, Wessler S, Shure M. 1983:235. doi: 10.1016/0092-8674(83)90226-x. [DOI] [PubMed] [Google Scholar]
- 39.Fedoroff NV. Science (80- ) 2012;338 doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 40.Canapa A, Barucca M, Biscotti MA, Forconi M. 2016:217. doi: 10.1159/000444429. [DOI] [PubMed] [Google Scholar]
- 41.Lynch VJ. Science (80- ) 2016;351:1029. doi: 10.1126/science.aaf2977. [DOI] [PubMed] [Google Scholar]
- 42.Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. Proc Natl Acad Sci U S A. 2007;104:18613. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JLL, Ruan Y, Wei CLL, Ng HH, Liu ET, Yen LL, Srinivasan KG, Chew JLL, Ruan Y, Wei CLL, Huck HN, Liu ET. Genome Res. 2008;18:1752. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunarso G, Chia NYY, Jeyakani J, Hwang C, Lu X, Chan YSS, Ng HHH, Bourque G. Nat Genet. 2010;42:631. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. Cell. 2012;148:335. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zamudio N, Bourc’his D. Heredity (Edinb) 2010;105:92. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 47.Yue F, Cheng Y, Al E, et al. Nature. 2014;515:355. [Google Scholar]
- 48.Cheng Y, Ma Z, Kim B-HB-H, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, Euskirchen G, Lin S, Lin Y, Visel A, Kawli T, Yang X, Patacsil D, Keller Ca, Giardine B, Kundaje A, Wang T, La Pennacchio LA, Weng Z, Hardison RC, Snyder MP. Nature. 2014;515:371. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MPP, Wang T. Genome Res. 2014;24:1963. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feschotte C. Nat Rev Genet. 2008;9:397. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. Nature. 2014 doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doxiadis GGM, De Groot N, Bontrop RE. 2008;82:6667. doi: 10.1128/JVI.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Oosterhout C. 2009;103:190. doi: 10.1038/hdy.2009.46. [DOI] [PubMed] [Google Scholar]
- 54.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Trends Genet. 2003;19:68. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 55.Lowe CB, Bejerano G, Haussler D. Proc Natl Acad Sci U S A. 2007;104:8005. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, Zhou X, Lee HJ, Maire CL, Ligon KL, Gascard P, Sigaroudinia M, Tlsty TD, Kadlecek T, Weiss A, O’Geen H, Farnham PJ, Madden PaF, Mungall AJ, Tam A, Kamoh B, Cho S, Moore R, Hirst M, Marra Ma, Costello JF, Wang T. Nat Genet. 2013:1. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. PNAS. 2012;109:19498. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundaram V, Choudhary MNK, Pehrsson E, Xing X, Fiore C, Pandey M, Maricque B, Udawatta M, Ngo D, Chen Y, Paguntalan A, Ray T, Hughes A, Cohen BA, Wang T. Nat Commun. 2017;8:14550. doi: 10.1038/ncomms14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Cell. 2008;133:1106. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 60.Fiore C, Cohen BA. Genome Res. 2016;26:778. doi: 10.1101/gr.200733.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chuong EB, Elde NC, Feschotte C. Science (80- ) 2016;351:1083. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jurka J, Kapitonov VV, Pavlicek a, Klonowski P, Kohany O, Walichiewicz J. Cytogenet Genome Res. 2005;110:462. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 63.Jurka J. Curr Opin Struct Biol. 1998;8:333. doi: 10.1016/s0959-440x(98)80067-5. [DOI] [PubMed] [Google Scholar]
- 64.Lynch VJ, Leclerc RD, May G, Wagner GP. Nat Genet. 2011;43:1154. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- 65.Emera D, Wagner GP. Brief Funct Genomics. 2012;11:267. doi: 10.1093/bfgp/els013. [DOI] [PubMed] [Google Scholar]
- 66.Blaise S, de Parseval N, Benit L, Heidmann T. Proc Natl Acad Sci. 2003;100:13013. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. Proc Natl Acad Sci. 2004;101:1731. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ellison C, Bachtrog D. Science (80- ) 2013;342 doi: 10.1126/science.1239552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Nature. 2012;487:57. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Göke J, Jung M, Behrens S, Chavez L, O’Keeffe S, Timmermann B, Lehrach H, Adjaye J, Vingron M. PLoS Comput Biol. 2011;7:e1002304. doi: 10.1371/journal.pcbi.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, Reijo Pera RA, Wysocka J. Nature. 2015;522:221. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Xie G, Singh M, Ghanbarian AT, Raskó T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, Schumann GG, Chen W, Lorincz MC, Ivics Z, Hurst LD, Izsvák Z. Nature. 2014 doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 73.Santoni FA, Guerra J, Luban J. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M, Tokunaga Y, Nakamura M, Watanabe A, Yamanaka S, Takahashi K. Proc Natl Acad Sci. 2014;111:12426. doi: 10.1073/pnas.1413299111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ostertag EM, HHK Annu Rev Genet. 2001;35:501. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 76.Cordaux R, Batzer MA. Nat Rev Genet. 2009;10:691. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burns KH. Nat Rev Cancer. 2017;17:415. doi: 10.1038/nrc.2017.35. [DOI] [PubMed] [Google Scholar]
- 78.Mager DL, Lorincz MC. Nat Genet. 2017;49:974. doi: 10.1038/ng.3902. [DOI] [PubMed] [Google Scholar]
- 79.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Buhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Mergoub T, Chan TA, Baylin SB, Strick R. Cell. 2015;162:974. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wittkopp PJ, Haerum BK, Clark AG. Nature. 2004;430:85. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 81.Swift H. Proc Natl Acad Sci U S A. 1950;36:643. doi: 10.1073/pnas.36.11.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gregory TR. Evolution (N Y) 2002;56:121. doi: 10.1111/j.0014-3820.2002.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 83.Villar D, Flicek P, Odom DT. Nat Publ Gr. 2014;15:221. doi: 10.1038/nrg3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wittkopp PJ, Haerum BK, Clark AG. Genetics. 2008;178:1831. doi: 10.1534/genetics.107.082032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wittkopp PJ, Kalay G. Nat Rev Genet. 2012;13:59. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 86.Wittkopp PJ, Haerum BK, Clark AG. Nat Genet. 2008;40:346. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- 87.Gompel N, Prud B, Wittkopp PJ, Kassner VA, Carroll SB. 2005:481. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 88.Levine M. Curr Biol. 2010;20:R754. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goodwin S, McPherson JD, McCombie WR. Nat Rev Genet. 2016;17:333. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. 2014:1. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuan Y, Norris C, Xu Y, Tsui K, Ji Y, Liang H. 2012;13 doi: 10.1186/1471-2164-13-S8-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ekblom R, Wolf JBW. Evol Appl. 2014 doi: 10.1111/eva.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neems DS, Garza-Gongora AG, Smith ED, Kosak ST. Proc Natl Acad Sci. 2016:201521826. doi: 10.1073/pnas.1521826113. [DOI] [PMC free article] [PubMed] [Google Scholar]