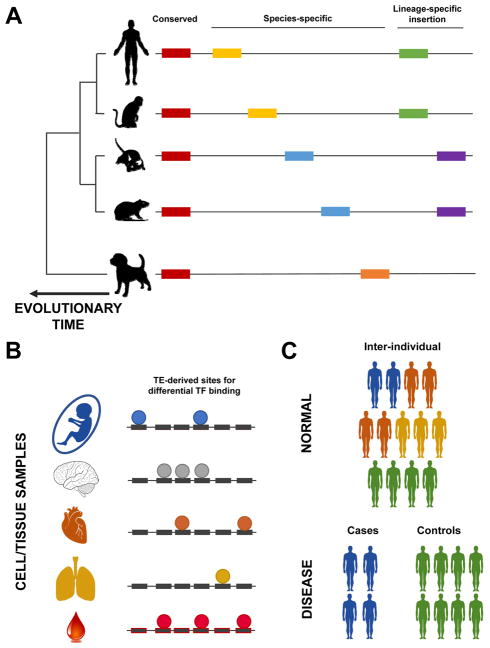
Figure 5.
Elucidating the effect of TEs on phenotypes, considering (A) evolution, (B) cell-types and cell-fate decisions, and (C) at the population-level. A: TEs (represented by coloured rectangles) are largely present in eukaryotic genomes, but vary in their specificity. On the one hand, some TE insertions are conserved across evolution in terms of their presence in a species, and their orthology in the genome sequence (labelled “Conserved” in the figure). On the other hand, there are other TE instances that are species-specific, in terms of the presence of a subfamily in a species (orange rectangle), and insertions (yellow rectangles in human and mouse, and blue rectangles in mouse and rat). There are also lineage-specific TEs, for example whose subfamily was active in primate (green rectangles) and rodents (purple rectangles). B: Cell/tissue-type specificity in TE’s biochemical activity. Different cell types have the same genome, but have differences in the activity of TFs (coloured circles). Here, we depict the differences in TF binding on TEs in different cell types. C: Lastly, a less explored area of TE-biology is the differences in TE biochemical activity and effects on gene expression regulation in normal populations (upper panel), and patients with diseases (lower control).