Abstract
Objectives
Oesophageal squamous cell carcinoma (OSCC) is an aggressive malignancy and the major histological subtype of oesophageal cancer. Although recent large-scale genomic analysis has improved the description of the genetic abnormalities of OSCC, few targetable genomic lesions have been identified, and no molecular therapy is available. This study aims to identify druggable candidates in this tumour.
Design
High-throughput small-molecule inhibitor screening was performed to identify potent anti-OSCC compounds. Whole-transcriptome sequencing (RNA-Seq) and chromatin immunoprecipitation sequencing (ChIP-Seq) were conducted to decipher the mechanisms of action of CDK7 inhibition in OSCC. A variety of in vitro and in vivo cellular assays were performed to determine the effects of candidate genes on OSCC malignant phenotypes.
Results
The unbiased high-throughput small-molecule inhibitor screening led us to discover a highly potent anti-OSCC compound, THZ1, a specific CDK7 inhibitor. RNA-Seq revealed that low-dose THZ1 treatment caused selective inhibition of a number of oncogenic transcripts. Notably, further characterisation of the genomic features of these THZ1-sensitive transcripts demonstrated that they were frequently associated with super-enhancer (SE). Moreover, SE analysis alone uncovered many OSCC lineage-specific master regulators. Finally, integrative analysis of both THZ1-sensitive and SE-associated transcripts identified a number of novel OSCC oncogenes, including PAK4, RUNX1, DNAJB1, SREBF2 and YAP1, with PAK4 being a potential druggable kinase.
Conclusions
Our integrative approaches led to a catalogue of SE-associated master regulators and oncogenic transcripts, which may significantly promote both the understanding of OSCC biology and the development of more innovative therapies.
INTRODUCTION
Oesophageal squamous cell carcinoma (OSCC) is one of the most common and aggressive GI malignancies. 1,2 Due to a lack of understanding of the molecular basis and limited treatment options, the prognosis for patients with OSCC has not improved for decades.3 Recently, researchers, including ourselves, have determined the genomic landscape of OSCC and identified a number of driver events; however, genetic alterations of drug targets are infrequent in patients with OSCC, except those affecting PIK3CA and FGFR1.4–8 Clearly, alternative molecular approaches are needed to further elucidate the pathogenesis of OSCC for developing more innovative and effective regimens.
Transcription factors and cofactors interact with enhancers to control specific gene expression programmes, which are fundamental to cell biology. Recently, a special group of enhancers, termed super-enhancers (SEs),9,10 have been identified in many cell types. SEs recruit an exceptionally large number of transcription factors/cofactors, and they differ from typical enhancers (TEs) in size, transcription factor density and ability to induce transcription.11,12 SEs are frequently associated with key lineage-specific genes that control cell state and differentiation in normal somatic cells.13 Interestingly, SEs are also found to drive the expression of a few critical oncogenes in several types of tumour cells, such as STAT3,10 MYCN,14 and TAL1.15 However, little is known regarding whether and how SEs drive the pathogenesis of OSCC.
Interestingly, expression of a few SE-associated oncogenes was reported to be particularly vulnerable to transcriptional inhibition, offering a potential cancer targeting approach.12,16 One of the possible mechanisms is that these SE-associated oncogenes are addicted to continuous active transcription, allowing for selective effects before a global blockade of transcription is achieved.
In this study, we performed high-throughput small-molecule inhibitor screening, and identified THZ1, a newly developed covalent CDK7 inhibitor,16 as a potent anti-OSCC compound. Interestingly, we observed that low-dose THZ1 treatment elicited selective effects against genes significantly enriched in processes important in cancer biology. We further characterised the SE landscape in OSCC cells and found that THZ1-sensitive transcripts were significantly more frequently associated with SEs. Finally, through integrative analysis of the gene transcription signature and SE features, we established a functional genomic approach to discover novel oncogenes in OSCC.
MATERIALS AND METHODS
Human cell lines
KYSE cell line series were provided by Dr Y Shimada (Kyoto University, Japan), and TE-5 and TE-7 cells were provided by Dr Koji Kono (Cancer Science Institute of Singapore, Singapore). KYSE series cell lines were cultured in RPMI-1640 medium; TE-5, TE7 and HEK293T were maintained in Dulbecco’s modified Eagle medium. All media were supplemented with 10% fetal bovine serum (Invitrogen, San Diego, California, USA), penicillin (100 U/mL) and streptomycin (100 mg/mL).
Xenograft assays in NOD-SCID gamma mice
Twelve 6-week old, female NOD-SCID gamma (NSG) mice were subcutaneously (s.c.) injected with 1×106 KYSE510 cells on their dorsal flanks, with each mouse carrying two explants. After 10–15 days, mice were randomly separated into two groups and treated with either vehicle or the inhibitor. KPT-9274 was administered orally (150 mg/kg, twice daily, 5 days/week), and THZ1 was administered intraperitoneally (i.p.) (10 mg/kg, twice daily). The size of the tumours was measured every 4 days for a total of 4 weeks after treatment. At the end of experiments, mice were euthanised and examined for s.c. xenograft tumour growth. Immunohistochemical (IHC) analysis was performed on 5 mm sections of paraffin-embedded s.c. tumours.
Tumour metastasis assay
Twelve 6-week-old female NSG mice were injected with 1×106 KYSE510 cells through the tail vein. Once cancer cells metastasised into the lungs (around 45 days after initial injection), they were randomly separated into two groups and treated with either vehicle or THZ1 (twice daily, 10 mg/kg). At the end of metastasis assays, mice were euthanised and examined. Metastasis nodules in lung tissues were fixed in Bouin’s solution, embedded in paraffin, cut into 5 mm sections and stained with H&E. All of the animal experiments in this study were approved by the Institutional Animal Care and Use Committee (IACUC), National University of Singapore.
High-throughput small-molecule inhibitor screening
Cell lines were screened for sensitivity against a panel of 104 small-molecule inhibitors as previously described.17 Briefly, cells were exposed to graded concentrations of each drug in 384 well plates with a seeding density of 250 cells per well in 50 μL final volume per well. Each drug was tested across an eight-point, threefold interval dose–response curve (including the no-drug control). After 3 days, relative abundance of viable cells was quantified in each well using a tetrazolium-based MTS assay. All absorbance values from the MTS assay were normalised to the average of 48 wells per plate containing no drug, and these normalised values were used to fit a third-order polynomial curve to each drug dose–response. IC50 values were interpreted from this curve fit to assess the relative sensitivity of each cell line to each drug.
Chromatin immunoprecipitation sequencing data analysis
Chromatin immunoprecipitation sequencing (ChIP-Seq) reads were aligned to human reference genome (build GRCh37/hg19) using Bowtie Aligner. ChIP-Seq peaks were identified using MACS (Model-Based Analysis of ChIP-seq) by considering reads mapped only once at a given locus. Wiggle files were generated using read pileups for every 50 base pair bins. These wiggle files were normalised in terms of reads per million (rpm) by dividing tag counts in each bin by the total number of reads (in millions, duplicates removed). Wiggle files were converted into bigwig files using wigToBigWig tool (http://hgdownload.cse.ucsc.edu/admin/exe/) and visualised in Integrative Genomics Viewer (http://www.broadinstitute.org/igv/home). SEs were identified using ROSE (https://bitbucket.org/youngcomputation/rose). Closely spaced peaks (except those within 2 kb of TSS) within a range of 12.5 kb were merged together, followed by the measurement of input and H3K27Ac signals. These merged peaks were ranked by H3K27Ac signal and then classified into SEs or TEs. Both SEs and TEs were assigned to the nearest Ensemble genes. The ChIP sequencing files have been deposited into Gene Expression Omnibus (GSE76861).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using GSEA standalone desktop programme. An expression matrix was created containing expression values at zero and 6 h (upon 50 nM THZ1 treatment). All SE-associated genes were used as a ‘gene set database’. GSEA was run with parameter ‘Metric for ranking genes’ set to ‘log2_Ratio_of_classes’ to calculate enrichment score for SE-associated genes.
RESULTS
High-throughput small-molecule inhibitor screening identified small-molecules with potent anti-OSCC properties
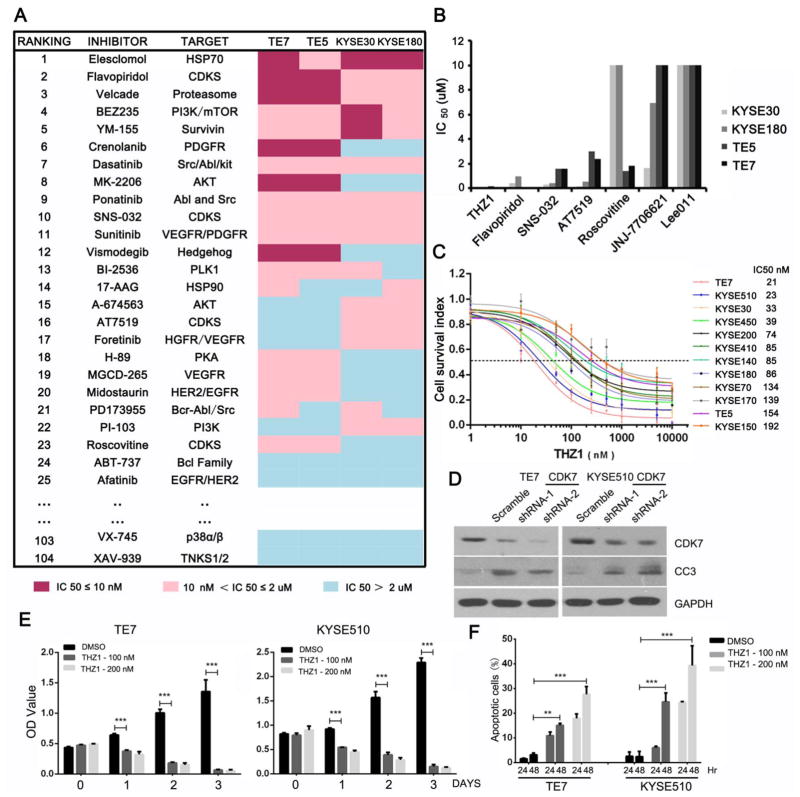
To identify small-molecule inhibitors with antineoplastic effects against OSCC cells in an unbiased manner, we first assembled a focused collection of 104 compounds that had broad targeting coverage of the kinome.17,18 Four OSCC cell lines were subjected to the high-throughput screen, and IC50 values were measured. As a result, 23 inhibitors from different families showed significant anticancer effect in at least two cell lines (figure 1A, see online supplementary table S1). Many of the kinase inhibitors previously shown to have anti-OSCC properties in vitro, including those targeting PI3K/AKT/MTOR pathway,19,20 HSP family21,22 or RTK,23,24 also demonstrated potent activities here, validating the methodology of our high-throughput approach (see online supplementary figure S1). As CDK inhibitors have shown promising therapeutic merit in several other types of tumours but are less well-studied in OSCC, we focused on this category of chemicals. To determine whether specific or pan CDK inhibitors were effective, we tested a total of seven compounds targeting different CDKs. Notably, all four cell lines were highly sensitive to THZ1, a new covalent CDK7 inhibitor. In addition, OSCC cells were also sensitive to the treatment with flavopiridol and SNS-032, both of which suppressed CDK7 activity among other targets (figure 1B, see online supplementary figure S2). Dose– response experiments using a panel of 12 OSCC cells showed that they were highly sensitive to THZ1 treatment, with IC50 values ranging from 21 to 192 nM (figure 1C). We further depleted CDK7 expression by shRNA-mediated knockdown in TE7 and KYSE510 and confirmed that CDK7 is essential for both the survival and proliferation of OSCC cells (figure 1D, see online supplementary figure S3). As TE7 and KYSE510 cells were among the most sensitive ones to THZ1 treatment, we focused our analysis on these two cell lines in the following studies.
Figure 1.
High-throughput small-molecule screen of four oesophageal squamous cell carcinoma (OSCC) cell lines identified THZ1 with potent anti-OSCC property. (A) Heatmap showed the IC50 values of 104 small-molecule inhibitors in TE7, TE5, KYSE30 and KYSE180 cells. Due to space limitations, only part of the results is shown, and the rest of the results are provided in online supplementary table S1. (B) IC50 values of various CDK inhibitors against OSCC cell lines. (C) Dose–response curves of 12 OSCC cell lines to THZ1 treatment (3 days). Data were represented as mean ±SD of three replicates. (D) TE7 and KYSE510 cells were infected with lentivirus encoding either shRNA against CDK7 or control shRNA (Scramble), and subjected to immunoblotting analysis with indicated antibodies. CC3 denotes Cleaved Caspase-3. (E) Proliferation assay (MTT) showed the effects of THZ1 treatment in OSCC cell lines at indicated time points. Bars represent mean±SD of three experiments performed in triplicate wells (***p<0.001). (F) OSCC cells were treated with either THZ1 or DMSO, and apoptosis was determined by Annexin V–FITC/PI staining. Bars represented mean±SD of three experiments (**p<0.01, ***p<0.001).
THZ1 showed powerful antineoplastic properties against OSCC cells
Recent studies showed that THZ1 potently suppressed cell proliferation and tumour growth in four different types of malignancies through inactivation of RNA polymerase II (RNAPII)-mediated transcription initiation and elongation, including small cell lung cancer,12 neuroblastoma,14 T cell acute lymphoblastic leukaemia16 and triple negative breast cancer.25 To evaluate the antineoplastic properties of THZ1 against OSCC cells, we first performed cell cycle analysis and observed G2/M phase arrest upon THZ1 treatment (see online supplementary figure S4). THZ1 treatment also resulted in profound inhibition of cell proliferation and induction of massive apoptosis (figure 1E, F).
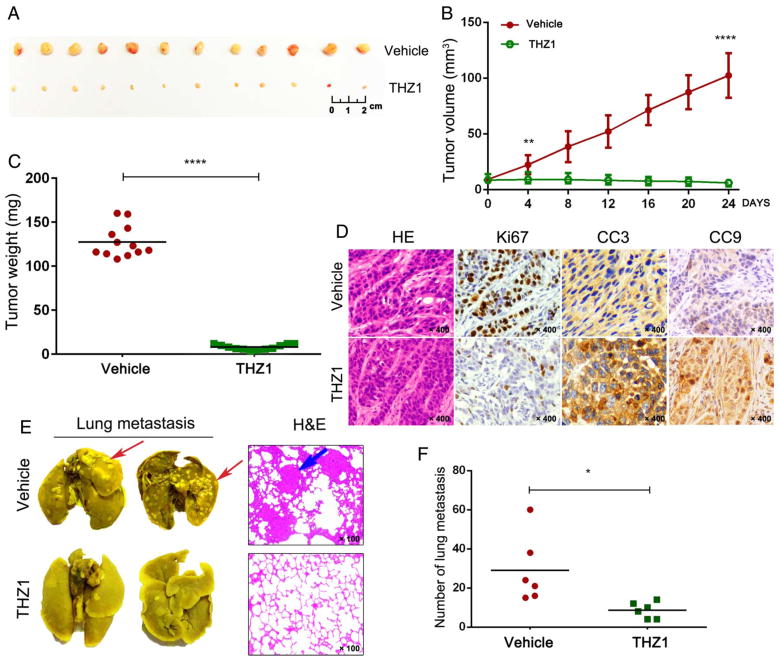
We subsequently tested the antitumour effects of THZ1 in NSG murine models, where each mouse carried two explants formed by KYSE510 cells. The mice received either vehicle or THZ1 twice daily (10 mg/kg). Strikingly, THZ1 completely suppressed OSCC tumour growth in vivo (figure 2A–C). Importantly, no significant loss of body weight (see online supplementary figure S5) or other common toxic effects (e.g., diarrhoea, rash, etc.; data not shown) were observed. IHC analysis confirmed the dramatic decrease of cell proliferation and increase of apoptosis in the xenografts upon THZ1 administration (figure 2D).
Figure 2.
THZ1 suppressed the growth of KYSE510 xenografts and inhibited lung metastasis in NOD-SCID gamma (NSG) murine model. (A) Photographs of tumours from both vehicle and THZ1 (twice daily, 10 mg/kg, 24 days) treatment groups. (B) Tumour growth curves of mice treated with either vehicle or THZ1. Data represent mean±SD (****p<0.0001). (C) Weights of tumours from each mice at the endpoint (****p<0.0001). (D) H&E and immunohistochemical (IHC) staining of Ki67, CC3 and Cleaved Caspase-9 (CC9) in tumour tissue sections. Original magnification, ×400. (E) Representative images of lungs after Bouin’s fixation (left) and H&E staining of tissue sections (right). Original magnification, ×100. Representative tumour lesions are indicated by arrows. (F) Visible lung metastases were counted and graphed (*p<0.05).
We performed additional in vivo experiments to investigate further the effect of THZ1 on distal metastasis of OSCC cells. In control group, most mice developed visually observable lung metastatic nodules in 69 days. In contrast, the THZ1-treated mice had no or few tumour nodules in their lungs (figure 2E, F). These results demonstrated that THZ1 possessed very strong antineoplastic activities against OSCC cells both in vitro and in vivo.
Global and selective transcription repression by CDK7 inhibition in OSCC cells
We next aimed to understand the mechanisms underlying the cytotoxic effects of THZ1 on OSCC cells. CDK7 regulates transcriptional processing through recognising and phosphorylating the initiation-associated serine 5 (S5) and serine 7 (S7) and elongation-associated serine 2 (S2) of the RNAPII C-terminal domain (RNAPII CTD).26,27 Indeed, we observed decreased phosphorylation S5, S7 and S2 of RNAPII in both TE7 and KYSE510 cells in a dose- and time-dependent manner after THZ1 treatment (figure 3A). We next examined the effect of CDK7 inhibition on gene expression profile by performing whole-transcriptome sequencing (RNA-Seq) (figure 3B–E, see online supplementary tables S2 and S3). As expected, high-dose THZ1 (200 nM, i.e., complete inhibition of RNAPII) resulted in global downregulation of steady-state mRNA levels at 6 h (figure 3D, E). Interestingly, we found that low-dose THZ1 (50 nM, i.e., partial inhibition of RNAPII) elicited downregulation of a group of transcripts in a gene-selective fashion at early time points (figure 3B, C). We termed this group (transcripts which decreased over twofold with low-dose treatment at 6 h) ‘THZ1-sensitive transcripts’.
Figure 3.
THZ1 treatment inhibited RNA polymerase II (RNAPII)-mediated transcription in oesophageal squamous cell carcinoma (OSCC). (A) Immunoblotting analysis of RNAPII C-terminal domain (CTD) phosphorylation in TE7 and KYSE510 cells which were treated with either THZ1 or DMSO at indicated time points (left), with indicated concentrations (right). (B) Heatmap showing expression changes (log2 fold) of all active transcripts upon either DMSO or THZ1 (50 nM) treatment at indicated time points. (C) Box plots of log2 fold changes in global gene expression in TE7 and KYSE510 cells treated with either DMSO or THZ1 (50 nM) at indicated time points. (D) Heatmap and (E) box plots comparing the effect of high-dose (200 nM) and low-dose (50 nM) THZ1 on global mRNA abundance. (F) Enriched gene ontology (GO) functional categories of THZ1-sensitive transcripts.
Previous studies with other types of THZ1-sensitive cancers found that low-dose THZ1 treatment led to selective inactivation of lineage-specific and oncogenic genes. Based on these finding, we hypothesised that in the context of OSCC, THZ1-sensitive transcripts might not be random but have important biological relevance. Specifically, we speculated that THZ1-sensitive transcripts might consist of a set of oncogenic transcripts or pathways conferring the sensitivity of OSCC cells to low-dose THZ1 treatment. To address this and to analyse comprehensively which sets of genes were particularly sensitive to CDK inhibition, we performed gene ontology (GO) analysis of THZ1-sensitive transcripts. Notably, genes involved in transcription regulation, DNA repair and apoptosis regulation were among the most sensitive to CDK7 inhibition in both OSCC cell lines (figure 3F), highlighting their crucial roles in OSCC biology.
We next asked whether these THZ1-sensitive transcripts were associated with any genomic features. It had been reported that master transcription factors and cofactors which played key roles in cell identity and malignant phenotypes were frequently associated with SEs. Interestingly, in other cancer types, SE-associated transcripts were found to be particularly sensitive to transcriptional perturbation.9–11,16 Thus, we hypothesised that these THZ1-sensitive transcripts might be driven by SEs and conferred the sensitivity to THZ1 treatment in OSCC. However, SE-associated transcriptional events and their biological relevance in the context of pathogenesis of OSCC remain unknown.
Characterisation of SE landscape in OSCC cells
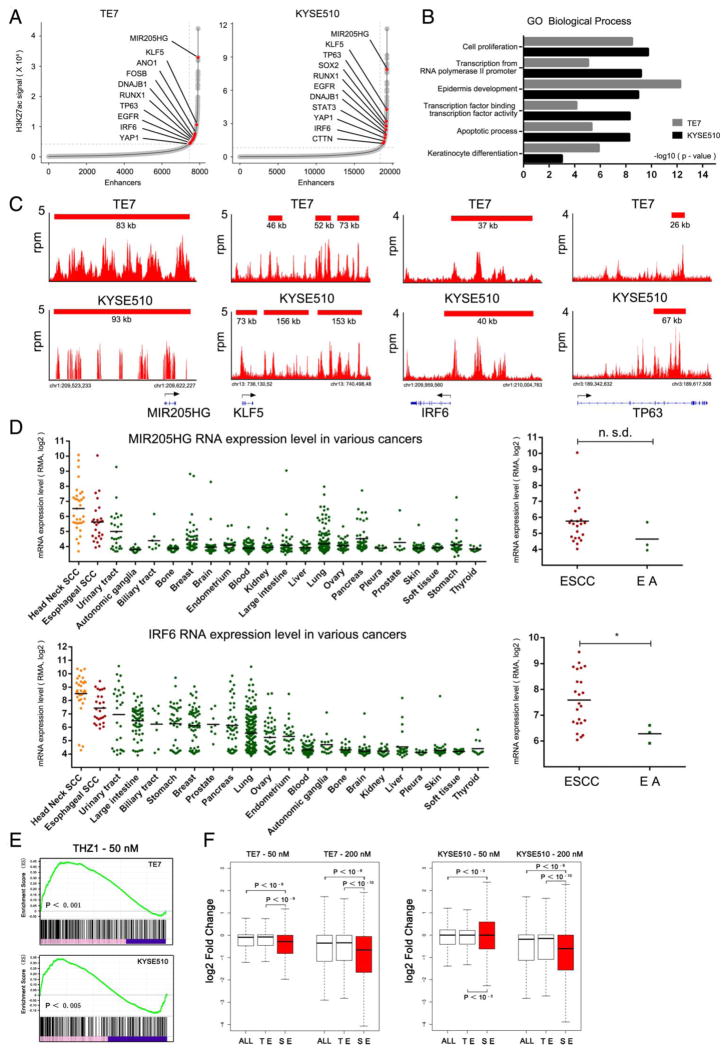
To characterise SE-associated transcripts in OSCC, we performed ChIP-Seq using the antibody recognising H3K27ac modifications in OSCC cells which were also profiled by RNA-Seq. As a result, we annotated 444 and 855 SE-associated transcripts in TE7 and KYSE510 cell lines, respectively (figure 4A, see online supplementary table S4). Notably, we readily observed that many of these transcripts were lineage-specific transcription factors acting as master regulators of keratinocyte cell differentiation, such as KLF5,28 TP6329 and IRF630 (figure 4A,C). This was concordant with the fact that both of these cell lines are squamous cell type. Furthermore, a number of well-defined OSCC oncogenes were also associated with SEs, such as TP63,31 EGFR,32 ANO1,33 SOX2,34 FSCN135 and CTTN36 (figure 4A, C, see online supplementary figure S6). To determine this enrichment more comprehensively, we performed GO analysis. Importantly, squamous-cell unique biological processes, including epidermis development and keratinocyte differentiation, were highly enriched in both cell lines (figure 4B). In addition, genes involved in cancer-related functions such as cell proliferation and apoptosis were also significantly overrepresented in SE-associated transcripts. Moreover, we identified many novel SE-associated transcripts, including both coding and non-coding RNAs, whose functions have not been reported in the setting of OSCC biology.37 For example, non-coding RNA MIR205HG was associated with one of the biggest SEs in both cells (figure 4C). As SEs were reported to drive lineage-specific expression of key transaction factors in somatic cells, we next analysed and found that a number of SE-associated transcripts, such as MIR205HG, IRF6, TP63 and SOX2, showed a lineage-specific expression pattern, with highest expression levels in squamous cell cancers, including OSCC and head/neck squamous carcinoma (figure 4D, see online supplementary figure S7A, B). To confirm further this expression pattern, we compared OSCC with oesophageal adenocarcinoma (OA) and found that many SE-associated transcripts identified in our OSCC cells were more highly expressed in the squamous cell type (representative results shown in figure 4D, see online supplementary figure S7). Together, these results suggest that in OSCC cells, SE-associated transcripts contain lineage-specific master regulators and oncogenic transcripts; many of which displayed tissues-specific expression patterns.
Figure 4.
Characterisation of super-enhancer (SE)-associated genes in TE7 and KYSE510 cell lines. (A) Hockey stick plots of oesophageal squamous cell carcinoma (OSCC) cells showing input normalised, rank ordered H3K27ac signals for SE-associated genes. (B) Gene ontology (GO) analysis of enriched biological processes for SE-associated genes in OSCC cells. (C) H3K27ac chromatin immunoprecipitation sequencing (ChIP-Seq) binding profiles of representative SE-associated genes in both cell lines. (D) Left, mRNA expression level of representative transcripts (MIR205HG and IRF6) across various types of human cancer cells; Right, MIR205HG and IRF6 expression in OSCC compared with OA cell lines. Data were retrieved from CCLE project from Broad Institute. (E) Gene set enrichment analysis of THZ1-sensitive and SE-associated transcripts. (F) Box plots showing log2 fold changes for transcripts associated with total pool of all enhancers (ALL), typical enhancers (TEs) and SEs upon THZ1 treatment at 6 h.
Identification of novel SE-associated oncogenes in OSCC
We next asked whether SE-associated transcripts are disproportionately sensitive to CDK7 inhibition. GSEA for all active genes upon low-dose THZ1 treatment showed that the majority of the genes in the leading edge were SE-associated transcripts, and they were most sensitive to THZ1 treatment (figure 4E). Moreover, the abundance of SE-associated transcripts were downregulated to a significantly higher degree upon THZ1 treatment, compared with those associated with TEs (figure 4F, see online supplementary table S5). These results suggested that SE-associated transcripts were particularly sensitive to partial inhibition of transcription, and SE-associated oncogenes might be responsible for the sensitivity of OSCC cells to low-dose THZ1 treatment.
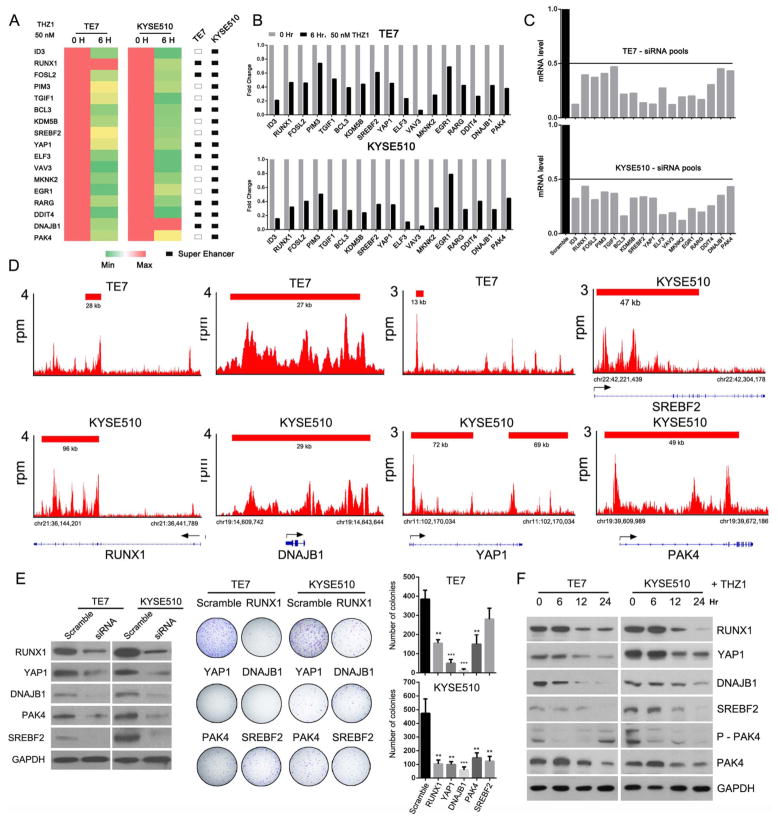
We hypothesised that further in-depth analysis of the expression dynamics of SE-associated transcripts during transcription inhibition might identify novel SE-associated oncogenes in OSCC, as it is reasonable to assume that SE-associated oncogenes would be strongly expressed in OSCC tumours and highly dependent on continuous transcription. Thus, we required that the candidate novel oncogenes be: (i) associated with SEs, (ii) ranked among the top 15% of all actively expressed transcripts in RNA-Seq results and (iii) highly sensitive to low-dose THZ1 treatment at 6 h. As shown in figure 5A, 17 candidate oncogenes were selected. Quantitative PCR results validated their hypersensitivity to transcription inhibition (figure 5B). In sharp contrast, TE-associated genes were either only modestly decreased or remained unaltered upon exposure to THZ1 (see online supplementary figure S8).
Figure 5.
Integrative analysis of chromatin immunoprecipitation sequencing (ChIP-Seq) with whole-transcriptome sequencing (RNA-Seq) results to screen for novel super-enhancer (SE)-associated oncogenes. (A) Heatmap and (B) qRT-PCR validation of RNA-Seq results of candidate SE-associated genes. (C) qRT-PCR showed that the mRNA levels of candidate genes were reduced significantly by their corresponding siRNAs (RNA from cells was harvested after 48 h of siRNA exposure). (D) H3K27ac ChIP-seq profiles of five candidate oncogenes (RUNX1, DNAJB1, YAP1, SREBF2 and PAK4) in TE7 and KYSE510 cells. (E) Left panel, western blot showing efficacy of siRNAs. Middle panel, representative results of colony formation assays of oesophageal squamous cell carcinoma (OSCC) cells transfected with indicated siRNAs. Right panel, graphic display of colony formation results. Means±SD of three independent experiments are shown (**p<0.01, ***p<0.001). (F) Immunoblotting analysis shows that the protein levels of RUNX1, YAP1, DNAJB1, SREBF2 and PAK4 were decreased by THZ1 treatment.
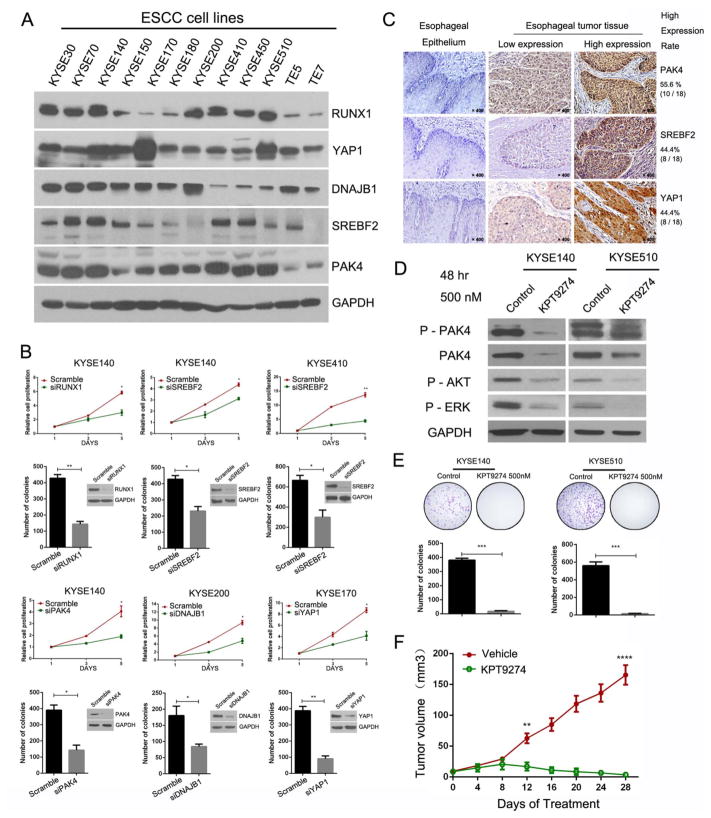
To test their biological functions in OSCC cells, we silenced each one of these transcripts by siRNA-mediated knockdown and performed cell proliferation assay in these two representative cell lines (figure 5C, see online supplementary table S6). As a result, we identified four candidate genes, namely, RUNX1, YAP1, DNAJB1 and PAK4, which contributed to the proliferation of the two OSCC cells (figure 5D, E, see online supplementary figure S9). We also confirmed that these four proteins were significantly decreased in both cell lines by THZ1 treatment (figure 5F). Worthwhile to note, SREBF2 was associated with SEs in KYSE510 but not TE7 cells; and protein and mRNA levels of this gene in KYSE510 cells were consistently highly expressed compared with TE7 cells (figure 6A). Importantly, SREBF2 only promoted the growth of KYSE510 but not TE7 cells (figure 5E, see online supplementary figure S9), underscoring the ability of our approaches to delineate the connections between expression pattern, SE feature and biological relevance.
Figure 6.
Functional relevance of candidate super-enhancer (SE)-associated genes in oesophageal squamous cell carcinoma (OSCC) cell lines. (A) Immunoblotting analysis shows protein expression level of RUNX1, YAP1, DNAJB1, SREBF2 and PAK4 in 12 OSCC cell lines. (B) MTT (upper rows) and colony formation (lower rows) assays of indicated cells transfected with indicated siRNAs. (C) Representative immunohistochemical (IHC) results of YAP1, SREBF2 and PAK4 expression in OSCC samples and normal oesophageal epithelium. (D) OSCC cells were treated with KPT-9274 (500 nM, 48 h) and subjected to immunoblotting analysis with indicated antibodies. (E) Antiproliferation effect of KPT-9274 (500 nM) against KYSE140 and KYSE510 cells as assayed by colony formation. Representative colonies (top row) and quantification of colonies (bottom row). Data were represented as mean±SD (n=3 experiments done in triplicate, ***p<0.001). (F) OSCC (KYSE510) tumour growth curve of mice treated with either vehicle or KPT-9274. Data represent as means±SD (**p<0.01, ****p<0.0001).
To characterise further the biological functions of these five novel candidate oncogenes in OSCC, we measured their protein levels in a panel of 12 OSCC cell lines and identified additional cell lines with high expression of each gene (figure 6A). We next chose additional representative cell lines which expressed candidate genes at high levels for functional assays. Importantly, MTT and colony formation analysis showed that each of these five genes was required for cell proliferation in at least three different OSCC cell lines (figure 6B).
We next determined whether these five SE-associated oncogenes are up-regulated in OSCC specimens compared with non-malignant oesophageal epithelium. Importantly, IHC staining showed that PAK4, SREBF2 and YAP1 proteins were highly expressed in OSCC tissues but were markedly lower in adjacent normal oesophageal epithelium (figure 6C). We also examined the expression of RUNX1 and DNAJB1 proteins; however, their antibodies failed to generate specific signals in the experiments (data not shown).
Identification of PAK4 as an SE-associated candidate drug target in OSCC
The p21-activated kinases (PAKs) belong to Ser/Thr protein kinase family, which contains six members in humans (PAK1–6). Overexpression of PAK4 has been implicated in cancer progression by activating oncogenic signalling pathways, such as RAF/MEK/ERK and PI3K/AKT.38,39 We were particularly interested in this candidate SE-associated kinase as its small-molecule inhibitors were shown to have antineoplastic activity in colon, lung, breast and gastric cancers.40,41 However, its roles and therapeutic value in OSCC cells are yet to be explored. As mentioned earlier, depletion of the expression of PAK4 by siRNA inhibited OSCC cell proliferation (figures 5E and 6B, see online supplementary figure S9). To examine the therapeutic value of targeting PAK4, we investigated its novel small-molecule, orally available inhibitor named KPT-9274.42 Dose–response studies using OSCC cells expressing high PAK4 levels found most of them sensitive to the compound, with IC50 values ranging from 180 to 612 nM (see online supplementary figure S10A). After confirming its on-target effect (figure 6D), we showed that KPT-9274 robustly inhibited cell proliferation and clonogenic growth of OSCC cells (figure 6E, see online supplementary figure S10B). To explore the anti-OSCC effect of KPT-9274 in vivo, NSG mice with KYSE510 xenografts were treated orally with either vehicle or KPT-9274 (150 mg/kg, twice daily, 5 days/week). Strikingly, after 28 days, KPT-9274 almost completely suppressed the tumour growth in these mice and induced massive apoptosis in the xenografts (figure 6F, see online supplementary figures S10C–E). Importantly, no systemic toxicity was observed during continuous administration for 4 weeks, and the body weight showed no significant change (see online supplementary figure S10F).
DISCUSSION
Developing novel molecular-based targeted therapies is one of the most important strategies to treat highly aggressive cancers such as OSCC. Sadly, we and others have shown that the genomic alterations in OSCC often cause inactivation of tumour suppressor genes rather than activation of druggable oncogenes. 4,6,7,33 In this study, we demonstrated that targeting transcriptional regulation, as opposed to specific genomic lesions, might provide therapeutic value against this malignancy.
Based on unbiased high-throughput screening with small-molecule inhibitors, we identified that OSCC cells displayed exceptional sensitivity to CDK7 inhibition, which was extensively validated by in vitro and in vivo assays. We were initially intrigued by the observations that inhibition of general gene transcription by low-dose THZ1 produced much less cytotoxicity in healthy tissues than in OSCC xenograft (see online supplementary figure S5). Recent investigations found that partial transcription inhibition resulted in selective rather than global suppression of those transcripts which were highly dependent on continuous transcription in several types of THZ1-sensitive tumours, but not unresponsive cancers nor non-malignant cells.12,14,16,25 We reasoned that the exceptional sensitivity of several OSCC cells to low-dose THZ1 treatment might be conferred by a set of oncogenes which were extremely addicted to transcriptional activation and were thus particularly vulnerable to CDK7 inhibition. Indeed, RNA-seq profiles showed that low-dose THZ1 treatment led to selective inhibition of a number of well-known OSCC oncogenes, many of which are transcriptional factors, such as TP63,31 CTTN,36 SOX2,43 PLK1,44 STAT345 and ID146 (see online supplementary tables S2 and S3). Interestingly, we observed that many non-coding RNAs with high-level expression were also particularly vulnerable to THZ1 treatment, such as MIR205HG, TP53TG1 and MIR210HG, whose functions are still unknown. Therefore, our approach may help nominate novel oncogenic non-coding RNAs in OSCC cells.
Although a set of oncogenes with extreme addiction to RNA Pol-II mediated transcription might explain the sensitivity of OSCC cells to CDK7 inhibition, we still wondered what caused the continuous transcriptional activation of these oncogenes in OSCC cells. To probe the mechanism, we first performed GO analysis and found that THZ1-sensitive transcripts were enriched in processes that regulate gene transcription, DNA repair and cell apoptosis, all of which are critical for cancer biology. As oncogenes involved in these cellular functions have been shown to be associated with SEs in other types of tumours, we hypothesised that SEs might be responsible for the robust transcriptional activation of THZ1-sensitive transcripts in OSCC cells.
Hnisz et al10 have created a catalogue of SEs for different types of human cells and tissues, OSCC was not investigated. Here, we characterised the SE landscape of OSCC, and identified that SE-associated transcripts are enriched in processes which are exclusive to SCC biology, such as epidermis development and keratinocyte differentiation, as well as cell proliferation and apoptosis. Interestingly, we also discovered some novel SE-associated non-coding mRNAs. For example, MIR205HG was associated with one of the largest SEs in OSCC cells. Expression profiling also supported the uniqueness of this non-coding RNA in SCC.
We reasoned that as both THZ1-sensitive, SE-associated cohort of transcripts contain oncogenic factors important for OSCC malignant phenotype, an integrative interrogation of these two datasets might identify novel candidate oncogenes. Through a series of functional experiments, we showed that 5 out 17 candidate SE-associated genes, namely, RUNX1, YAP1, DNAJB1, PAK4 and SREBF2, were important for the proliferation of OSCC cells. In addition, we confirmed the overexpression of some of these proteins in OSCC compared with non-malignant oesophageal epithelium. Since we only tested the phenotypes of cell proliferation, candidates implicated in other cellular functions such as migration, invasion and epithelial– mesenchymal transition might be overlooked and need further investigations to characterise.
Transcription factor RUNX1 is a well-studied differentiation regulator and tumour suppressor in haematopoietic cells. However, its functions in normal and malignant epithelial cells remain obscure and inconclusive. For example, it was reported to promote the proliferation of skin squamous cancer cells47,48 but inhibit breast cancer cells.49 Interestingly, RUNX1 gene is frequently deleted in EA and RUNX1 suppressed the proliferation of EA cells.50,51 In sharp contrast, here we show that RUNX1 is an SE-associated oncogene and promotes cell proliferation in OSCC. These results again underscore the ability of our integrative approaches to discern cell type-specific gene functions.
Similarly, DNAJB1 is poorly studied in human cancers and appears to have seemingly opposite roles. Specifically, as a protein implicated in stimulating the ATPase activity of Hsp70s, investigators showed that DNAJB1 inhibited p53-mediated apoptosis by destabilising PDCD5 in lung cancer.52 In contrast, Qi et al53 found that it could decrease cell proliferation in a p53-dependent manner in breast cancers. Our data revealed that as an SE-associated oncogene, DNAJB1 was highly expressed in OSCC compared with other human cancers (see online supplementary figure S11), and it significantly promoted the growth and proliferation of OSCC cells.
Last, our systematic approach identified a druggable SE-associated oncogene, PAK4. Both in vitro and in vivo experiments confirmed that its small-molecule inhibitor, KPT-9274, dramatically suppressed OSCC cell viability and induced massive apoptosis. These data suggested the potential therapeutic value of targeting PAK4 for clinical management of patients with OSCC.
In aggregate, the current study addressed both basic and translational questions, which are all highly novel and unexplored in the context of OSCC biology. Specifically, our results provide an important molecular foundation to understand the transcriptional landscape of OSCC and a catalogue of novel oncogenic transcripts, both of which are valuable for the OSCC research community. Moreover, our work may help establish the therapeutic merit of targeting SE-associated oncogenic transcription programme for OSCC treatment.
Supplementary Material
Significance of this study.
What is already known on this subject?
The genomic landscape of oesophageal squamous cell carcinoma (OSCC) has been established; however, genetic alterations of actionable targets are infrequent in this malignancy.
Super-enhancers (SEs) recruit an exceptionally large number of transcription factors/cofactors, and they differ from typical enhancers in size, transcription factor density and ability to induce transcription.
SEs are found to be associated with key lineage-specific master regulators in normal somatic cells as well as with a few critical oncogenes in several types of tumour cells.
What are the new findings?
The SE landscape is established in OSCC cells, and many SE-associated, squamous-specific master regulators and novel oncogenic transcripts are identified.
Targeting SE-associated transcription activation by a small-molecule CDK7 inhibitor, THZ1, shows powerful antineoplastic properties against OSCC cells.
PAK4 is an SE-associated candidate drug target in OSCC.
How might it impact on clinical practice in the foreseeable future?
Through characterising the transcriptional abnormalities in OSCC, our study suggests that targeting transcriptional activation programme rather than specific genomic lesions might provide better efficacy in the clinical management of solid tumours with highly complex genomes, such as OSCC.
This work may help establish the potential therapeutic merit of targeting SE-associated oncogenic transcription programme through a small-molecule CDK7 inhibitor, THZ1, for the treatment of patients with OSCC.
Acknowledgments
The authors thank Anand D Jeyasekharan (a member of Singapore Gastric Cancer Consortium) for his helpful advice. The authors also thank Hazimah Binte Mohd Nordin for help with mouse work.
Funding This work was funded by the Singapore Ministry of Health’s National Medical Research Council (NMRC) under its Singapore Translational Research (STaR) Investigator Award to HPK and by NMRC Individual Research Grant (NMRC/1311/2011) and National Natural Science Foundation of China (81520108023, 81330052) to M-RW. D-CL was supported by Donna and Jesse Garber Awards for Cancer Research and National Center for Advancing Translational Sciences UCLA CTSI Grant UL1TR000124. This study was partially supported by a generous donation from the Melamed family.
Footnotes
Contributors D-CL, Y-YJ and HPK designed the study, analysed and discussed the data and wrote the manuscript. Y-YJ, MH, LX, L-WD performed experiments. LS, XX and M-RW coordinated sample collection and processing. WS and EB provided KPT-9274 and discussed its related results. AM and HY performed bioinformatical analysis. JWT helped conduct high-throughput small-molecule inhibitor screening. YC, J-FX, DK and W-WC discussed the data.
Competing interests WS and EB are employees of Karyopharm Therapeutics Inc.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.He YT, Hou J, Chen ZF, et al. Study on the esophageal cancer incidence and mortality rate from 1974–2002 in Cixian, China. Zhonghua liu xing bing xue za zhi. 2006;27:127–31. [PubMed] [Google Scholar]
- 2.Zhao P, Dai M, Chen W, et al. Cancer trends in China. Jpn J Clin Oncol. 2010;40:281–5. doi: 10.1093/jjco/hyp187. [DOI] [PubMed] [Google Scholar]
- 3.Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet. 2013;381:400–12. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 4.Lin DC, Hao JJ, Nagata Y, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467–73. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin DC, Wang MR, Koeffler HP. Targeting genetic lesions in esophageal cancer. Cell Cycle. 2014;13:2013–14. doi: 10.4161/cc.29458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 7.Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–5. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal N, Jiao Y, Bettegowda C, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovén J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapuy B, McKeown MR, Lin CY, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–90. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen CL, Kwiatkowski N, Abraham BJ, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer cell. 2014;26:909–22. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amaral PP, Bannister AJ. Re-place your BETs: the dynamics of super enhancers. Mol Cell. 2014;56:187–9. doi: 10.1016/j.molcel.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Chipumuro E, Marco E, Christensen CL, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–39. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansour MR, Abraham BJ, Anders L, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwiatkowski N, Zhang T, Rahl PB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–20. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyner JW, Yang WF, Bankhead A, III, et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 2013;73:285–96. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bicocca VT, Chang BH, Masouleh BK, et al. Crosstalk between ROR1 and the Pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell. 2012;22:656–67. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Xi Q, Chen Y, et al. A dual mTORC1 and mTORC2 inhibitor shows antitumor activity in esophageal squamous cell carcinoma cells and sensitizes them to cisplatin. Anticancer Drugs. 2013;24:889–98. doi: 10.1097/CAD.0b013e328363c64e. [DOI] [PubMed] [Google Scholar]
- 20.Liu SG, Wang BS, Jiang YY, et al. Atypical protein kinase Ciota (PKCiota) promotes metastasis of esophageal squamous cell carcinoma by enhancing resistance to Anoikis via PKCiota-SKP2-AKT pathway. Mol Cancer Res. 2011;9:390–402. doi: 10.1158/1541-7786.MCR-10-0359. [DOI] [PubMed] [Google Scholar]
- 21.Wang XT, Bao CH, Jia YB, et al. BIIB021, a novel Hsp90 inhibitor, sensitizes esophageal squamous cell carcinoma to radiation. Biochem Biophys Res Commun. 2014;452:945–50. doi: 10.1016/j.bbrc.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Ui T, Morishima K, Saito S, et al. The HSP90 inhibitor 17-N-allylamino-17- demethoxy geldanamycin (17-AAG) synergizes with cisplatin and induces apoptosis in cisplatin-resistant esophageal squamous cell carcinoma cell lines via the Akt/XIAP pathway. Oncol Rep. 2014;31:619–24. doi: 10.3892/or.2013.2899. [DOI] [PubMed] [Google Scholar]
- 23.Sato F, Kubota Y, Natsuizaka M, et al. EGFR inhibitors prevent induction of cancer stem-like cells in esophageal squamous cell carcinoma by suppressing epithelial-mesenchymal transition. Cancer Biol Ther. 2015;16:933–40. doi: 10.1080/15384047.2015.1040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Lan T, Zhang W, et al. Dasatinib enhances cisplatin sensitivity in human esophageal squamous cell carcinoma (OSCC) cells via suppression of PI3K/AKT and Stat3 pathways. Arch Biochem Biophys. 2015;575:38–45. doi: 10.1016/j.abb.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang T, Kwiatkowski N, et al. CDK7-Dependent Transcriptional Addiction in Triple-Negative Breast Cancer. Cell. 2015;163:174–86. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larochelle S, Amat R, Glover-Cutter K, et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol. 2012;19:1108–15. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–43. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oishi Y, Manabe I, Tobe K, et al. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Truong AB, Kretz M, Ridky TW, et al. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–97. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson RJ, Dixon J, Malhotra S, et al. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38:1329–34. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe H, Ma Q, Peng S, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest. 2014;124:1636–45. doi: 10.1172/JCI71545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Gu J, Jiang P, et al. DNMT1-microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res. 2015;21:854–63. doi: 10.1158/1078-0432.CCR-14-1740. [DOI] [PubMed] [Google Scholar]
- 33.Shi ZZ, Shang L, Jiang YY, et al. Consistent and differential genetic aberrations between esophageal dysplasia and squamous cell carcinoma detected by array comparative genomic hybridization. Clin Cancer Res. 2013;19:5867–78. doi: 10.1158/1078-0432.CCR-12-3753. [DOI] [PubMed] [Google Scholar]
- 34.Gen Y, Yasui K, Nishikawa T, et al. SOX2 promotes tumor growth of esophageal squamous cell carcinoma through the AKT/mammalian target of rapamycin complex 1 signaling pathway. Cancer Sci. 2013;104:810–16. doi: 10.1111/cas.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie JJ, Xu LY, Zhang HH, et al. Role of fascin in the proliferation and invasiveness of esophageal carcinoma cells. Biochem Biophys Res Commun. 2005;337:355–62. doi: 10.1016/j.bbrc.2005.09.055. [DOI] [PubMed] [Google Scholar]
- 36.Luo ML, Shen XM, Zhang Y, et al. Amplification and overexpression of CTTN (EMS1) contribute to the metastasis of esophageal squamous cell carcinoma by promoting cell migration and anoikis resistance. Cancer Res. 2006;66:11690–9. doi: 10.1158/0008-5472.CAN-06-1484. [DOI] [PubMed] [Google Scholar]
- 37.Song JH, Meltzer SJ. Long non-coding RNA SS-001 regulates proliferation, cell cycle, and migration in esophageal adenocarcinoma cells. Cancer Res. 2013;73(8 Suppl):1839. [Google Scholar]
- 38.Siu MK, Chan HY, Kong DS, et al. p21-activated kinase 4 regulates ovarian cancer cell proliferation, migration, and invasion and contributes to poor prognosis in patients. Proc Natl Acad Sci USA. 2010;107:18622–7. doi: 10.1073/pnas.0907481107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabusa H, Brooks T, Massey AJ. Knockdown of PAK4 or PAK1 inhibits the proliferation of mutant KRAS colon cancer cells independently of RAF/MEK/ERK and PI3K/AKT signaling. Mol Cancer Res. 2013;11:109–21. doi: 10.1158/1541-7786.MCR-12-0466. [DOI] [PubMed] [Google Scholar]
- 40.Murray BW, Guo C, Piraino J, et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci USA. 2010;107:9446–51. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Wang J, Guo Q, et al. LCH-7749944, a novel and potent p21-activated kinase 4 inhibitor, suppresses proliferation and invasion in human gastric cancer cells. Cancer Lett. 2012;317:24–32. doi: 10.1016/j.canlet.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Senapedis W, Landesman Y, Schenone M, et al. Identification of Novel Small Molecules as Selective PAK4 Allosteric Modulators (PAMs) by Stable Isotope Labeling of Amino acids in Cells (SILAC) Eur J Cancer. 2014;50:156. [Google Scholar]
- 43.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Du XL, Wang CJ, et al. Reciprocal activation between PLK1 and Stat3 contributes to survival and proliferation of esophageal cancer cells. Gastroenterology. 2012;142:521–30. e3. doi: 10.1053/j.gastro.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 45.Du XL, Yang H, Liu SG, et al. Calreticulin promotes cell motility and enhances resistance to anoikis through STAT3-CTTN-Akt pathway in esophageal squamous cell carcinoma. Oncogene. 2009;28:3714–22. doi: 10.1038/onc.2009.237. [DOI] [PubMed] [Google Scholar]
- 46.Li B, Tsao SW, Li YY, et al. Id-1 promotes tumorigenicity and metastasis of human esophageal cancer cells through activation of PI3K/AKT signaling pathway. Int J Cancer. 2009;125:2576–85. doi: 10.1002/ijc.24675. [DOI] [PubMed] [Google Scholar]
- 47.Scheitz CJ, Lee TS, McDermitt DJ, et al. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31:4124–39. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoi CS, Lee SE, Lu SY, et al. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol Cell Biol. 2010;30:2518–36. doi: 10.1128/MCB.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Brugge JS, Janes KA. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc Natl Acad Sci USA. 2011;108:E803–12. doi: 10.1073/pnas.1103423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu J, Ajani JA, Hawk ET, et al. Genome-wide catalogue of chromosomal aberrations in barrett’s esophagus and esophageal adenocarcinoma: a high-density single nucleotide polymorphism array analysis. Cancer Prev Res (Phila) 2010;3:1176–86. doi: 10.1158/1940-6207.CAPR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dulak AM, Schumacher SE, van Lieshout J, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–93. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui X, Choi HK, Choi YS, et al. DNAJB1 destabilizes PDCD5 to suppress p53-mediated apoptosis. Cancer Lett. 2015;357:307–15. doi: 10.1016/j.canlet.2014.11.041. [DOI] [PubMed] [Google Scholar]
- 53.Qi M, Zhang J, Zeng W, et al. DNAJB1 stabilizes MDM2 and contributes to cancer cell proliferation in a p53-dependent manner. Biochim Biophys Acta. 2014;1839:62–9. doi: 10.1016/j.bbagrm.2013.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.