Abstract
Background
The past two decades have seen an increased incidence of squamous cell carcinoma of the head and neck (HNSCC) in a non-traditional, low-risk patient population (i.e. ≤ 45 years of age, no substance use history), owing to a combination of human papillomavirus (HPV) infection and individual genetic variation.
Methods
Articles positing genetic variants as contributing factors in HNSCC incidence in low-risk, non-traditional patients were identified using a PubMed search, reviewed in detail, and concisely summarized herein.
Results
Recent data suggest that common polymorphisms in DNA repair enzymes, cell-cycle control proteins, apoptotic pathway members, and Fanconi anemia-associated genes likely modulate susceptibility to HNSCC development in low-risk, non-traditional patients.
Conclusions
At present, there is a lack of robust, comprehensive data on genetic drivers of oncogenesis in low-risk patients and a clear need for further research on genetic alterations underlying the rising incidence of HNSCC in low-risk, non-traditional patients.
Keywords: Genetics, Germline, Hereditary, Personalized Medicine, HNSCC
Introduction
Each year, approximately 600,000 incident cases of head and neck squamous cell carcinoma (HNSCC) are diagnosed worldwide, leading to more than 350,000 deaths.1 HNSCC encompasses malignancies of the upper aerodigestive tract (i.e. oral cavity, oropharynx, hypopharynx, and larynx) and classically afflicts older, Caucasian males with heavy tobacco and/or alcohol use history.2–4 In this population, chronic exposure to carcinogens results in accumulation of somatic alterations in an assortment of oncogenes (e.g. PIK3CA), tumor suppressors (e.g. TP53, CDKN2A, NOTCH1), and cell-cycle regulators (e.g. CCND1) that increase risk of HNSCC over time.5 Despite the high prevalence of tobacco and alcohol use in the general population, only a small proportion of individuals will ultimately develop HNSCC, indicating that genetic variants may modulate susceptibility in individual patients.
Over the last two decades there has been an increasing prevalence of HNSCC in a non-traditional, low-risk patient population (i.e. ≤ 45 years of age, no substance use history). A rapid rise in high-risk human papillomavirus (hrHPV)-associated HNSCC accounts for the majority of these cases,6,7 though there remains an hrHPV-independent subset of low-risk patients who lack identifiable etiological factors.8,9 The latter population is seen with particular frequency in cancers of the oral cavity (OCC).10 While differences in genetic drivers of oncogenesis between these younger, low-risk patients and their older, traditional counterparts require further elucidation, current evidence suggests that clinical outcomes (i.e. disease-specific survival) are similar and comparably poor in these two patient groups.8,11
Recently, the International Head and Neck Cancer Epidemiology (INHANCE) consortium aimed to compare the role of family history of HNSCC, tobacco, and alcohol as oncologic risk factors among a large cohort of young (≤ 45 years) and old (≥ 45 years) patients.12 Importantly, a higher proportion of oral tongue and oral cavity cancers was seen in young HNSCC patients. Compared to older patients, these cancers were found to be less attributable to tobacco and alcohol use and had a higher association with a family history of early-onset HNSCC. As such, differences in HNSCC etiology among low-risk, non-traditional patients and their older counterparts are evident. The association of HNSCC with family history in the setting of a weaker (or absent) contribution from tobacco and alcohol suggests a hereditary, germline component present in young patients that is absent in the traditional HNSCC cohort.
Herein, we present an overview of the current landscape of genetic determinants of HNSCC in low-risk patients, a population defined by young patient age and status as non-smokers/drinkers. We discuss genetic polymorphisms in DNA repair enzymes and apoptotic pathway members, as well as known heritable forms of HNSCC, specifically familial HNSCC and Fanconi anemia (FA)-associated HNSCC. Additionally, we address genetic susceptibility to HNSCC in relation to hrHPV serology, relevant to the soaring national prevalence of viral infection and epidemiologic rise in hrHPV-associated HNSCC. Finally, we conclude with an acknowledgment of current barriers to the understanding of genetic determinants of HNSCC in low-risk patients and implications for future research in this area.
Genetic Variants in DNA Repair Enzymes in Low-Risk HNSCC Patients
Pathways for DNA damage repair fall into three broad categories determined by the nature and extent of genomic damage: base excision repair (BER), nucleotide excision repair (NER), and double-strand break repair (DSBR). Perturbations in cellular DNA repair pathways lead to genomic instability, aberrant cell function, and unchecked replication, all of which can promote tumor development.13 Inherited diseases compromising DNA repair pathways, such as Fanconi anemia (DSBR), xeroderma pigmentosum (NER), and BRCA1/2-associated cancer syndromes (DSBR) drastically increase an individual’s lifetime risk of developing multiple cancers of different sites.14 Functioning DNA repair machinery is essential to combat the accumulation of tumor-promoting mutations induced by chronic exposure to carcinogens in tobacco smoke, as in the case of HNSCC.15 However, even in non-tobacco users, polymorphisms in DNA repair genes may underlie oncogenesis by compromising genomic integrity, as clearly illustrated in the aforementioned hereditary syndromes. Current evidence on the association of polymorphisms in genes of DNA repair pathways with risk of HNSCC in low-risk patients is conflicting.
A systematic review and meta-analysis by Flores-Obando et al15 examined associations in polymorphisms in DNA damage response genes with risk of HNSCC development in 30 case-control studies comprising roughly 8,000 patients with oral, pharyngeal, or laryngeal cancers and 12,000 matched controls. Focusing their analysis on DNA repair enzymes in the NER (XPA, XPD, XPC, XPF, ERCC1), BER (XRCC1), and DSBR (XRCC3) pathways, the authors reported an increased risk of HNSCC associated with mutations in XPD codon 312 and XRCC1 codon 399 in Caucasian and Asian populations and mutations in XRCC1 codon 194 solely in Asian patients, though notably each of these associations was only marginally statistically significant. An updated meta-analysis with extensive subgroup stratification by smoking status, HNSCC subsite, and ethnicity by Lou et al17 differed from Flores-Obando et al in that the XRCC1 codon 399 polymorphism had no significant impact on HNSCC susceptibility among 7,000 HNSCC patients (compared against 10,000 controls), alone or in an interactive fashion with smoking. However, the authors did note that XRCC1 R194W homozygosity and tobacco use interacted to produce a statistically significant increase in HNSCC risk in the patients studied. While these results failed to support a clear susceptibility to HNSCC attributable to polymorphisms in genes of DNA repair machinery, they do suggest that subgroup stratification by smoking status may uncover correlations within low-risk populations. A second response to Flores-Obando et al found no link between XPD codon 312 polymorphism and HNSCC susceptibility in a meta-analysis of nine case-control studies totaling 2,700 HNSCC patients and 4,500 controls.19 However, stratification by age, smoking, or drinking status was not shown in this report, thus limiting the ability to interpret the data in the context of these critical variables.
A recent genome-wide association study (GWAS) of European HNSCC patients within the INHANCE consortium identified a novel polymorphism at 4q21 (rs1494961) significantly associated with susceptibility to HNSCC in the roughly 2,000 patients analyzed.19 This site is positioned within the HEL308 gene, a DNA-dependent ATPase and helicase important for DNA intra-strand cross-linking repair.20 Sequencing of rs1494961 in an additional independent cohort of 5,600 lung cancer cases and 9,300 controls showed that this polymorphism was associated with risk of lung cancer, suggesting that this variant may play an important role in tobacco-associated cancers in general. On subgroup stratification, the association of the rs1494961 variant with HNSCC risk was maintained in patients less than 50 years of age, though was lost in never-users of tobacco or alcohol, precluding any definitive conclusions regarding the role of rs1494961 in young, low-risk HNSCC patients. A second GWAS by Liang et al21 identified a positive, multiplicative interaction between HEL308 polymorphisms and cigarette use of greater than 70 pack-years in 600 HNSCC cases, signifying that this genetic variant is less relevant to a low-risk, non-traditional patient population. Finally, a follow-up GWAS of a relatively small Chinese HNSCC population revealed no association between rs1494961 polymorphism and HNSCC susceptibility (Table 1).22
TABLE 1.
Summary of DNA Repair Pathway Gene Polymorphisms Investigated in Low-Risk, Non-Traditional HNSCC Patients
| Gene Investigated | Study Design | Case/Control Size | Strength of HNSCC Association OR (95 % CI) |
Subgroup Stratification |
|---|---|---|---|---|
| XRCC1 R194W (Homozygous) | Meta-analysis of 30 Studies (Flores-Obando et al16) | 7,291/12,052 | 1.78 (1.13–2.82) | Asians |
| Meta-analysis of 29 Studies (Lou et al17) | 6,719/9,627 | 0.91 (0.77–1.08) | N/A (Total Population) | |
| 2.53 (1.16–5.53) | Smokers | |||
| XRCC1 R399Q (Homozygous & Heterozygous) | Meta-analysis of 30 Studies (Flores-Obando et al16) | 7,291/12,052 | 1.14 (1.01–1.27) | Caucasians |
| Meta-analysis of 29 Studies (Lou et al17) | 6,719/9,627 | 0.99 (0.90–1.09) | N/A (Total Population) | |
| 0.7 (0.43–1.15) | Smokers | |||
| XPD D312N (Heterozygous) | Meta-analysis of 30 Studies (Flores-Obando et al16) | 7,291/12,052 | 1.14 (1.01–1.29) | N/A (Total Population) |
| Meta-analysis of 9 Studies (Hu et al18) | 2,670/4,452 | 1.11 (0.99–1.24) | N/A (Total Population) | |
| HEL308 (Homozygous & Heterozygous) | GWAS of European Studies in INHANCE Consortium (McKay et al19) | 8,605/16,226 | 1.13 (1.08–1.17) | N/A (Total Population) |
| 1.19 (1.08–1.31) | Age < 50 Years | |||
| 1.03 (0.93–1.14) | Never smokers | |||
| 1.04 (0.93–1.18) | Never drinkers | |||
| HEL308 (Homozygous) | Case-Control Study of Patients in the Boston, MA Area (Liang et al21) | 575/676 | 0.78 (0.57–1.06) | N/A (Total Population) |
| Case-Control Study of Chinese Nationals (Yuan et al22) | 397/900 | 0.98 (0.82–1.18) | N/A (Total Population) |
Clearly, future confirmative studies and mechanistic investigations are needed to clarify inconsistencies in these posited associations between DNA repair polymorphisms (e.g. HEL308) and HNSCC in low-risk, non-traditional patients. Such studies should stratify included HNSCC cases and controls by relevant clinical factors such as age, tobacco use, and HNSCC subsite that may help uncover complicated gene-by-gene or gene-by-environment effects and generate robust, generalizable associations between polymorphisms in DNA repair machinery and HNSCC in low-risk patients.
Genetic Variants in Apoptotic Pathway Members in Low-Risk HNSCC Patients
Programmed cell death by apoptosis serves as a critical barrier to cancer development and progression.23 Apoptotic signals are cued by various physiological stressors (e.g. DNA damage, growth-factor deprivation) and trigger cellular suicide by caspase cascade activation. Dysfunction in this native defense mechanism promotes tumorigenesis and confers metastatic potential and treatment resistance in cancer cells.24,25 One of the key regulators of apoptosis is p53 (encoded by the TP53 gene), a tumor suppressor that induces cell-cycle arrest and apoptosis in response to DNA damage, oncogene activation, hypoxia, and numerous other stimuli.26 Loss-of-function mutation in TP53 is one of the most common acquired genetic events across all human cancers, including HNSCC, in which whole-exome sequencing has identified somatic silencing mutations in TP53 in over 70 % of tumors.5,27
In addition to acquired, tumor-promoting mutations in TP53, two germline polymorphisms at codon 72 of TP53 generating an arginine (p53Arg) or proline (p53Pro) residue at this position are common in the human population.28 The p53Arg and p53Pro variants harbor minimal conformational differences in protein structure, yet several reports have posited that these polymorphisms may be important determinants of response to chemotherapies and differential oncologic outcomes.29–31 Intriguingly, HNSCC cases have shown bias towards p53Arg homozygosity30 and patients with this alteration respond less favorably to platinum-based chemo-radiotherapy, perhaps due to negative influence of p53Arg on p73-dependent initiation of apoptosis.31 At present however, potential influences of p53 polymorphisms on HNSCC susceptibility alone, particularly in low-risk patients, have yet to be investigated.
The p53 upregulated modular of apoptosis (PUMA) plays a similar crucial role in promoting apoptosis through interaction with the anti-apoptotic Bcl-2. PUMA expression is upregulated by p53, though likely has both p53-dependent and p53-independent roles in HNSCC as the expression of PUMA is sufficient to prevent HNSCC growth in vitro regardless of p53 mutational status.32 At present, no somatic mutations within coding regions of PUMA have been identified in HNSCC, though two polymorphisms (rs3810294 and rs2032809) in the gene’s promoter region have been proposed as potential risk factors for incident HNSCC in Caucasian populations.33 These polymorphisms may result in differential binding affinities of transcription factors to the PUMA promoter, though the consequential effect on PUMA expression is still unclear. In a cohort study of 380 HNSCC cases and 335 controls, increased HNSCC risk was seen in patients with HPV16 seropositivity and a variant PUMA gene, an effect modification that was particularly pronounced in never-smokers, never-drinkers, young patients (< 58 years), and in oropharyngeal subsites.33 Mechanistically, the HPV16 E6 oncoprotein subverts apoptosis in infected cells by interfering with the p53/PUMA/Bax cascade, though functional evidence of any direct interaction between PUMA and E6 has yet to be elucidated.34
Lastly, polymorphisms in the promoter of baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5, also known as survivin) gene, have been implicated as risk factors for the development of HNSCC in Asian populations.35 In HNSCC in vitro models and primary tumors, somatic mutations and overexpression of BIRC5 correlates with resistance to conventional chemoradiotherapy,36,37 though how the promotor polymorphisms (at present, 5 identified in OCC,38 1 in nasopharyngeal carcinoma39) relate to BIRC5 expression has not yet been evaluated. Importantly, of currently identified BIRC5 promoter polymorphisms, there is conflicting evidence on their association with HNSCC in low-risk patients. One investigation found that these polymorphisms confer elevated HNSCC risk in non-smoking, non-drinking populations,39 while another found that BIRC5 variants confer susceptibility to HNSCC only in an interactive fashion with betel quid chewing or tobacco use.38 These opposing conclusions highlight the need for further studies examining the association of genetic variants in apoptotic pathways with HNSCC in low-risk patients from multiple independent cohorts.
Familial/Hereditary HNSCC
The vast majority of HNSCC are sporadic cancers, attributable to known etiologic factors such as tobacco, alcohol, and HPV. At present, there is a paucity of data on the possible role of family history and importance of inheritance patterns in HNSCC risk, particularly in low-risk patients. Previously mentioned herein, a 2015 INHANCE consortium study analyzed risk factors for incident HNSCC among 25 case-control studies and identified a convincing association between family history of early-onset cancer and incident HNSCC in ever-smokers less than 45 years of age (OR: 2.27, 95 % CI: 1.26 – 4.10).12 This association was not present among older (≥ 45 years) HNSCC patients (OR: 1.10, 95 % CI: 0.91 – 1.31). Interestingly, an earlier INHANCE case-control study examining 9,000 HNSCC patients and 14,000 healthy controls similarly found that a family history of HNSCC in a first-degree relative conferred an increased risk of incident HNSCC in the patients examined (OR: 2.2, 95 % CI: 1.6 – 3.1 for siblings, OR: 1.5, 95 % CI: 1.1 – 1.8 for parents).40 Importantly, this relationship was markedly stronger among HNSCC patients with positive family history and tobacco and alcohol use (OR: 7.2, 95 % CI: 5.5 – 9.5). These studies suggest that a familial/hereditary component to HNSCC may be attributable to inherited sensitivity towards tobacco- and alcohol-related carcinogens, thus this relationship is less relevant to a low-risk, never-smoker/never-drinker HNSCC population.
Inherited mutations in the CDKN2A gene, encoding the important tumor suppressor and cell-cycle regulating protein p16/INK4A, have been implicated in familial syndromes conferring a significantly increased lifetime risk of melanoma, pancreatic, lung, and breast cancer as well as of HNSCC.41, 42 Though the literature is limited, two recent case reports identified germline CDKN2A mutations in probands with HNSCC and melanoma amongst an extensive family history.43,44 In the first, a novel single-nucleotide deletion (c.106delG) resulted in a premature stop codon (p.Ala36ArgfsX17).43 In the second, a missense mutation (G302T) at exon 2 of CDKN2A with loss of heterozygosity was implicated.44 A third CDKN2A germline variant termed p16-Leiden results in a 19-nucleotide deletion (c.225_243del19) and significantly heightens lifetime risk of HNSCC development (RR: 18.8, 95 % CI: 6.05 – 58.2).45 Inherited CDKN2A loss-of-function mutations clearly elevate HNSCC risk in low-risk, never-smokers. Again however, risk of HNSCC in CDKN2A mutation carriers is amplified even more in ever-smokers, suggesting that such mutations may impair carcinogen metabolism and is thus less relevant to a low-risk HNSCC population.46
Nasopharyngeal carcinoma (NPC) is a poorly-differentiated squamous cell carcinoma of the posterior nasopharynx with a well-documented propensity for familial clustering.47 A review of candidate-gene approaches and GWAS to identify genomic loci conferring increased NPC risk by Bei et al48 identified ITGA9 at chromosome 3p, HLA-B/C and MICA at chromosome 6p, and CKDN2A/B at chromosome 9q. A second study by Xiong et al49 supported a link between familial variants in tumor suppressor gene clusters at chromosome 3p21 and NPC risk. Though further studies validating these findings have yet to be performed, it stands to reason that such germline variants likely contribute to NPC development in younger patients who lack identifiable risk factors.
Fanconi Anemia (FA) Association with Low-Risk HNSCCC Patients
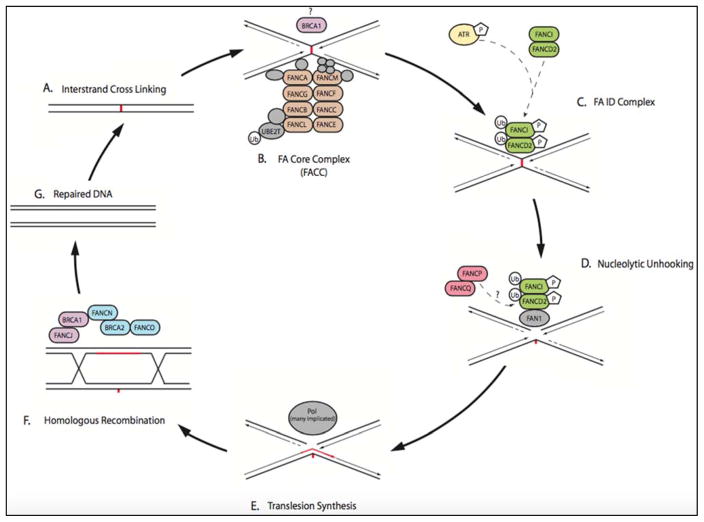
Fanconi anemia (FA) is an inherited bone marrow failure syndrome caused by biallelic inactivation of one of 17 genes (FANCA to FANCQ) inherited in an autosomal recessive or x-linked recessive pattern.50 The FANC genes normally encode proteins that maintain genomic stability by repairing interstrand crosslinks (ICLs) in DNA (Figure 1). With an incidence of roughly 1 in 100,000–250,000 births, FA leads to a spectrum of phenotypic abnormalities that include congenital malformations (e.g. short stature, microcephaly) and cytopenias.53,54 FA patients are also particularly susceptible to development of malignancies, including leukemia, esophageal cancer and HNSCC. The risk of HNSCC in FA, particularly of OCCs, is elevated roughly 500–700 times over the general population.55 The median age of onset of HNSCC in FA patients is a young 33 years of age and the vast majority (roughly 84%) of these patients are never-smokers/never-drinkers, suggesting minimal environmental contribution to HNSCC risk.56
FIGURE 1. Interstrand Crosslinking (ICL) Repair Pathway Defective in Fanconi Anemia Patients (Figure based on models put forth by Wang50, Ceccaldi51, and Garner52).
A. Interstrand crosslinking (ICL) occurs.
B. FA nuclear core complex (FACC) is a large, multi-subunit, E3 ubiquitin ligase (enzymatic activity provided by FANCL). FACC is recruited to the site of ICL detected by stalled replication fork. FANCM activates ATR via phosphorylation. ATR functions downstream to phosphorylate FANCI in the ID complex. Many models propose BRCA1 associates with the initial FACC to assist with CMG helicase removal, allowing the replication fork to approach the ICL site more closely.
C. FACC recruits the phosphorylated FA ID complex (FANCI and FANCD2), and the FACC monoubiquitinates the FA ID complex.
D. Nucleolytic “unhooking” of the ICL occurs through recruitment of downstream nucleases FAN1 (no mutations currently identified in FA patients), and multi-subunit FANCP(SLX4)/FANCQ(XPF). FAN1 binds mono-ubiquitinated FANCD2. FNACP(SLX4) contains ubiquitin binding sites and potentially interacts with FANCD2 as well. The role behind association of multiple nucleases, and their individual significance in nucleolytic processing, is unclear.51
E. Translesion synthesis (TLS) is performed by TLS-specific polymerases that are recruited to the nuclear foci. Multiple TLS polymerases have been implicated, including REV1 and POLζ.51 No mutations in TLS-specfic polymerases have been identified in FA patients to our knowledge (1).
F. Homologous recombination repair occurs through the recruitment of many repair factors. These include the FANCN(PALB2)/FANCD1(BRCA2)/FANCO(RAD51C) complex and FANCS(BRCA1)/FANCJ(BRIP1) which functions as a helicase and inhibitor of non-homologous end joining.
G. DNA has been successfully repaired by components in the FA pathway.
Investigation into the association between FA and HNSCC begets questioning of whether inherited or sporadic FANC mutations may drive HNSCC development in low-risk, non-traditional patients without FA.57 Trembly et al confirmed lower expression of FANCA and FANCG in OCC xenografts from young patients (< 40 years) HNSCC patients without FA as compared to an older patient (> 60 years) cohort and healthy controls.58 The authors posited that attenuated expression of FANCA and FANCG may explain mechanistic differences in tumorigenesis between young, low-risk patients and their traditional HNSCC counterparts through defective carcinogen metabolism or DNA repair capabilities. Recently Chandrasekharappa et al screened 417 patients for germline FA mutations in head and neck cancer patients under 50 years of age. They identified 44% of patients had FA gene variants with 26% having variants predicted to be damaging. Additionally, they identified an increased mutational burden amongst three FA genes (FANCD2, FANCE and FANCL) relative to the normal population; all of which are associated with other malignancies.[PMID: 286978401] Additionally, a recent investigation59 utilizing 17 HNSCC cell lines found that FANC inactivation promotes genomic instability and tumorigenesis in only a small minority of non-FA HNSCC, prompting other authors to posit epigenetic modifications of FANC genes as potential drivers of sporadic HNSCC in non-FA patients.60,61
Transcription of FANC genes appears to be regulated in synchrony with the cell cycle, as E2F, Rb, and ATR modulate expression of FA pathway components in human cancer cell lines.62 Additionally, FANC-encoded proteins form complexes with the products of BRCA1/2 genes (implicated in hereditary breast and ovarian cancer) to exert their ICL-repair functions.63,64 In future studies, mutational status of upstream and downstream components of the FA pathway must be considered when clarifying associations between FANC aberrations and HNSCC risk in non-traditional patients.
Analysis of a 528-patient cohort of HNSCC cases from The Cancer Genome Atlas (TCGA) dataset using cBioPortal failed to demonstrate a consistent correlation between alterations in FANC genes and HNSCC (Figure 1, Table 2).65,66 However, only 42 of these 528 (8.0 %) HNSCC patients were less than 45 years of age, so drawing definitive conclusions regarding genetic profiling in low-risk, non-traditional HNSCC patients is challenging. Despite the rising prevalence of a young, low-risk HNSCC population, the lack of genomic data from these patients precludes identification of causative genetic associations, prediction of clinical outcomes, and trials of personalized therapeutics.
TABLE 2.
Alteration Frequency of FA Pathway and FA Pathway-Associated Genes Within the 528-Patient TCGA HNSCC Provisional Dataset Retrieved Using cBioPortal.65,66 (Color-coding of genes in the table corresponds to color-coding of gene products in Figure 1 depicting the FA-pathway)
| Gene (Alias) | Number of Cases Altered (504 Total) | Percent of Cases Altered |
|---|---|---|
|
| ||
| FA Pathway | ||
| FANCA | 11 | 2.20% |
| FANCB | 18 | 4% |
| FANCC | 13 | 2.60% |
| FANCD1 (BRCA2) | 27 | 5% |
| FANCD2 | 13 | 2.60% |
| FANCE | 3 | 0.60% |
| FANCF | 5 | 1% |
| FANCG | 26 | 5% |
| FANCI | 9 | 1.80% |
| FANCJ (BRIP1) | 15 | 3% |
| FANCL | 6 | 1.20% |
| FANCM | 14 | 2.80% |
| FANCN (PALB2) | 8 | 1.60% |
| FANCO (RAD51C) | 3 | 0.60% |
| FANCP (SLX4) | 10 | 2% |
| FANCQ (ERCC4) | 5 | 1% |
| FANCS (BRCA1) | 13 | 2.60% |
|
| ||
| FA Pathway-Associated | ||
| BRCA2 | 27 | 5% |
| BRCA1 | 13 | 2.60% |
| RB | 28 | 6% |
| ATR | 76 | 15% |
The association between HNSCC and FA patients, as well as the association between altered FANC genes and non-FA, low-risk HNSCC patients, is in need of further characterization. To our knowledge, there has been limited investigation into the association of FANC genes with HNSCC in non-FA patients, with even fewer of these analyses considering low-risk patients specifically. In future mechanistic investigations of the FA pathway in HNSCC, it will be important to differentiate between somatic and germline mutations in FANC within non-FA, HNSCC patients.
E2F Polymorphisms in Low-Risk HNSCC Patients
The E2F family of transcription factors play a central role in modulation of cell-cycle progression and DNA synthesis and repair.67 In quiescent cellular states, the Rb tumor suppressor protein (Rb) binds to E2F, preventing it from activing transcription machinery.68 In the absence of Rb, or when Rb is sequestered by the HPV oncogenic protein E7, E2F becomes free to facilitate transcription of target genes that modulate the G1/S transition.69 Dysregulation of the Rb-E2F pathway is seen almost universally across all human cancer types.
A recent investigation into the association of common polymorphisms in E2F1 and E2F2, respectively, in 1,100 HNSCC cases and 1,090 healthy controls sought to correlate these genotypes with susceptibility to incident HNSCC.70 The authors identified a statistically significant dose-response relationship conferring increased risk of incident HNSCC in patients harboring a greater number of these unique genetic variants (OR: 1.62, 95 % CI: 1.14 – 2.30 for individuals with 9–10 polymorphisms vs. individuals with 0–4, p = 0.045). Importantly, the joint effect of multiple E2F polymorphisms was particularly pronounced among adults less than 57 years of ages (OR: 1.74, 95 % CI: 1.07 – 2.85), never-smokers (OR: 1.85, 95 % CI: 1.07 – 3.17), never-drinkers (OR: 2.19, 95 % CI: 1.22 – 3.95), and those with a family history of cancer in a first-degree relative (OR: 1.64, 95 % CI: 1.05 – 2.57). These results suggest a convincing, additive effect of genetic variation within E2F genes on HNSCC risk, with enhancement of susceptibility in low-risk patients.
The authors of a recent investigation hypothesized that a common polymorphism (rs3213180) in the 3′UTR miRNA binding site of E2F1 would be associated with risk of OCC and oropharynx cancer (OPSCC) and HPV status of OPSCC, given the aforementioned role of HPV E7 in the Rb-E2F pathway.71 Patients with E2F1 rs3213180 polymorphism had an increased risk of developing OCC and OPSCC (OR: 3.3, 95 % CI: 2.4 – 4.6) regardless of HPV status. In individuals with HPV seropositivity and this E2F1 polymorphism, never-smokers/drinkers had a particularly pronounced risk of OPSCC compared to ever-smokers/drinkers (7.5-fold increased risk in never- vs. ever-smokers, 4.9-fold increased risk in never- vs. ever-drinkers). In contrast, HPV seronegative individuals with E2F1 polymorphisms had similar risk of OCC and OPSCC when comparing subsets of never-smokers/drinkers to ever-smokers/drinkers (0.97-fold decreased risk in never- vs. ever-smokers, 0.8-fold decreased risk in never- vs. ever-drinkers). These complex interactions suggest that HNSCC susceptibility in HPV-positive, never-smokers/drinkers is due at least in part to specific interactions between variant E2F1-encoded proteins and the HPV E7 oncogene.
hrHPV and Low-Risk HNSCC Patients
The incidence of hrHPV-associated HNSCC, particularly of oropharyngeal subsites, has soared in recent years and is most often seen in patients who lack traditional risk factors, namely tobacco and alcohol use.6,72,73 Progression from oropharyngeal HPV infection to malignancy takes in excess of a decade and occurs in a small but significant minority of patients, indicating that genetic variation among individuals may play a pivotal role in differential risk of malignant transformation.74,75
Transforming growth factor-beta (TGFβ) is an anti-inflammatory cytokine with important roles in inflammation and immune responses that ultimately lead to HPV clearance or immune evasion and viral persistence.76 Three distinct polymorphisms in the TGFβ gene (C509T, T869C, and G915C) have been shown to increase plasma levels of circulating TGFβ in human subjects.77 As TGFβ suppresses pro-inflammatory responses mediated by Th1 and Th2 lymphocytes, elevated TGFβ levels may compromise immune surveillance and control of HPV-infected cells.78 This immunosuppressive effect may be augmented by HPV-induced transcription of immunosuppressive cytokines, including TGFβ, as has been shown in cervical cancer models.79 As such, Guan et al 80 hypothesized that TGFβ polymorphisms may contribute to differential genetic susceptibility to hrHPV-associated HNSCC. The authors genotyped TGFβ polymorphisms and confirmed HPV16 status in 200 primary tumors of the oropharynx. Patients with variant TGFβ genotypes were more than twice as likely to have an HPV16-positive tumor (OR: 2.28, 95 % CI: 1.16 – 4.50) compared with patients with wild-type TGFβ. Furthermore, a stratified analysis showed enhancement of this association among patients < 54 years of age (OR: 4.07, 95 % CI: 1.52 – 10.9), never-smokers (OR: 3.76, 95 % CI: 1.15 – 12.3), and never-drinkers (OR: 5.01, 95 % CI: 1.03 – 24.3). Similarly, four common polymorphisms (rs1800629, rs1799724, rs1800630, rs1799964) in the gene promoter for tumor necrosis factor-alpha (TNFα), a versatile pro-inflammatory cytokine involved in viral defense, have been proposed as susceptibility biomarkers for HPV16-associated oropharyngeal cancers in young, never-smokers/drinkers.81 Finally, intriguing preliminary reports suggest that functional genetic polymorphisms of microRNAs (miRNAs) likely contribute to variations in miRNA-mediated immune function and inflammation crucial for antiviral defense.82,83 Taken together, the aforementioned studies provide evidence that genetic variation in immune-related genes augments the risk of hrHPV-mediated malignant transformation, particularly in young, low-risk HNSCC populations.
The critically-important tumor suppressor p53 has been called the “guardian of the genome” for its role in activation of DNA repair pathways and initiation of growth arrest and apoptosis in response to genomic instability.84 Inactivating TP53 gene aberrations are oncogenic drivers in the vast majority of HNSCC and portend worse oncologic outcomes.5,27 The HPV E6 oncoprotein complexes with human E6-associated protein (E6AP) to ubiquitinate p53, leading to its proteosomal degradation, a necessary cellular process for HPV-mediated carcinogenesis. As discussed above, a common polymorphism is observed at codon 72 of p53, replacing an arginine residue with proline; this amino acid change may influence the protein’s susceptibility to E6-mediated degradation.85 Variant genotypes at this p53 locus enhance susceptibility to oropharyngeal cancers in an interactive and multiplicative fashion with HPV16 seropositivity and never-smoking status (OR: 22.5, 95 % CI: 4.8 – 106.2).85 Two linked polymorphisms (G4C14-to-A4T14) in exon 2 of p73, a close family member of p53 that activates the promoter of several p53-responsive genes, confers similarly enhanced susceptibility to oropharyngeal cancers in HPV16-positive, never-smokers/drinkers.86 Young (i.e. < 50 years), HPV16-positive individuals who harbor combined variants in both p53 and p73 and are never-smokers/drinkers have an even greater risk of incident oropharyngeal cancers.87–89 Finally, MDM2 and MDM4 promoter polymorphisms synergize with HPV seropositivity to increase oropharyngeal cancer risk in young, never-smokers/drinkers.90,91 MDM2 and MDM4 are p53-pathway members that inhibit p53-mediated transcriptional activity and target p53 for proteosomal degradation, thus inhibiting DNA damage repair, cellular growth arrest and apoptosis.91
Components of the intrinsic apoptotic pathway may also be important contributors to differential risk of oropharyngeal cancer in young, HPV16-positive, never-smokers/drinkers. Overexpression of Mcl-1 and deficiency of NOXA, anti- and pro-apoptotic members of the Bcl-2 family respectively, offer p53-dependent protection from apoptosis in multiple myeloma models, though confirmatory studies of this relationship in HNSCC are lacking.92,93 Genetic variants in Mcl1 and NOXA promoter regions may render cells more susceptible to HPV E6-mediated interference with the p53-NOXA-Mcl1 axis. To test this hypothesis, Zhou et al94 analyzed four functional polymorphisms in the NOXA (rs9957673, rs45589496) and Mcl-1 (rs9803935, rs3738485) promoters in 372 cases of oropharyngeal cancer and 315 healthy controls. Their results suggested a joint effect on oropharyngeal cancer risk in young, never-smoking/drinking patients with multiple NOXA/Mcl-1 polymorphisms and HPV-16 seropositivity.
Conclusions
The rise in the number of low-risk, non-traditional HNSCC patients in recent years should be met with a spirited effort to characterize novel germline and somatic genomic events that drive oncogenesis in this population. The current literature provides preliminary evidence of risk variants that may contribute to HNSCC development in low-risk patients, though conclusions are limited by conflicting findings, lack of consistent subgroup stratification and small sample sizes.
Moving forward, we postulate that germline alterations in genes such as BRCA1, p53, and E2F, which are implicated as oncologic drivers following somatic inactivation across human cancers, are likely to be ubiquitous and strongly conserved among young, low-risk HNSCC cases (Table 3). Dedicated investigation of genetic aberrations in large populations of low-risk, non-traditional HNSCC patients is imperative to advance our understanding of mechanisms of carcinogenesis, risk-stratify patients, and improve oncologic outcomes in this population. Alternatively, HNSCC development in low-risk patients may reflect exposure to unknown environmental factors, unclassified carcinogens, or a more complex interplay between multiple environmental influences and genetic variation.
TABLE 3.
Alteration Frequency of Genes Examined Within This Review, as Reported in the TCGA Cohort and in Morris et al, a Precision Oncology Sequencing Study of Recurrent and Metastatic HNSCC from Memorial Sloan Kettering Cancer Center.
| Gene (Alias) | TCGA HNSCC Cohort5 (504 Cases) | Morris et al95 (132 Cases) | ||
|---|---|---|---|---|
|
| ||||
| Number of Cases Altered | Percent of Cases Altered | Number of Cases Altered | Percent of Cases Altered | |
| DNA Repair Genes | ||||
| XRCC1 | 5 | 1% | 0 | 0 % |
| XPD (ERCC2) | 7 | 1.4% | 2 | 1.5 % |
| HEL308 (HELQ) | 10 | 2% | 0 | 0 % |
|
| ||||
| Apoptosis Related | ||||
| TP53 | 363 | 72% | 62 | 47 % |
| PUMA (BCC3) | 1 | 0.2% | 0 | 0 % |
| BIRC5 | 0 | 0% | 0 | 0 % |
|
| ||||
| Familial HNSCC Related | ||||
| CDKN2A | 270 | 54% | 32 | 24 % |
| CDKN2B | 143 | 28% | 10 | 8 % |
| ITGA9 | 6 | 1.2% | 0 | 0 % |
| HLA-B | 24 | 5% | 0 | 0 % |
| HLA-C | 5 | 1% | 0 | 0 % |
| MICA | 6 | 1.2% | 0 | 0 % |
|
| ||||
| Cell-Cycle Related | ||||
| E2F1 | 17 | 3% | 0 | 0 % |
| E2F2 | 4 | 0.8% | 0 | 0 % |
|
| ||||
| HPV Associated | ||||
| MDM2 | 24 | 5% | 3 | 2.3 % |
| MDM4 | 5 | 1% | 0 | 0 % |
| MCL1 | 13 | 2.6% | 9 | 7 % |
| NOXA (PAIMP1) | 22 | 4% | 1 | 0.8 % |
| PUMA (BBC3) | 1 | 0.2% | 0 | 0 % |
| TFGB1 | 4 | 0.8% | 0 | 0 % |
| TP53 | 363 | 72% | 62 | 47 % |
| TP73 | 8 | 1.6% | 0 | 0 % |
Many of these genes had high alteration rates (e.g. TP53, CDKN2A), though those postulated to enhance HNSCC susceptibility in low-risk patients were less commonly altered in the overall TCGA cohort. The HNSCC TCGA provisional cohort had very few younger, low-risk patients (42 individuals ≤ 45 years of age at diagnoses).
Identifying pathogenic alterations in the low-risk HNSCC population will improve efforts to combat cancer progression. These studies may allow identification of at-risk patients from an early age, leading to subsequent modification of environmental and lifestyle factors, and may precede the development of novel, targeted therapies. Further research on low-risk, non-traditional HNSCC patients is necessary and will ideally reveal a combination of actionable genetic drivers and modifiable lifestyle or environmental risk factors, thereby providing a means to reduce the prevalence, morbidity, and mortality of this disease.
Acknowledgments
Grant Support: Dr. J. Chad Brenner received funding from NIH Grants T32-DC005356, U01-DE025184, P30-CA046592 and R01-CA194536. Joshua Smith received funding from NIH grant T32-DC 5356-15. M. Ludwig received funding from NIH grant F31 CA206341-01A1.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Allen C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3(4):524–48. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blot WJ, McLaughlin JK, Winn DM, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48(11):3282–7. [PubMed] [Google Scholar]
- 3.Hashibe M, Brennan P, Benhamou S, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99(10):777–89. doi: 10.1093/jnci/djk179. [DOI] [PubMed] [Google Scholar]
- 4.Brennan JA, Mao L, Hruban RH, et al. Molecular assessment of histopathological staging in squamous-cell carcinoma of the head and neck. N Engl J Med. 1995;332:429–35. doi: 10.1056/NEJM199502163320704. [DOI] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol. 2015;33(29):3235–42. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mourad M, Jetmore T, Jategaonkar AA, Moubayed S, Moshier E, Urken ML. Epidemiological trends of head and neck cancer in the United States: a SEER population study [published online May 22 2017] J Oral Maxillofac Surg. doi: 10.1016/j.joms.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel SC, Carpenter WR, Tyree S, et al. Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J Clin Oncol. 2011;29(11):1488–94. doi: 10.1200/JCO.2010.31.7883. [DOI] [PubMed] [Google Scholar]
- 9.Harris SL, Kimple RJ, Hayes DN, Couch ME, Rosenman JG. Never-smokers, never-drinkers: unique clinical subgroup of young patients with head and neck squamous cell cancers. Head Neck. 32(4):499–503. doi: 10.1002/hed.21220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zafereo ME, Xu L, Dahlstrom KR, et al. Squamous cell carcinoma of the oral cavity often overexpresses p16 but is rarely driven by human papillomavirus. Oral Oncol. 2016;56:47–53. doi: 10.1016/j.oraloncology.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verschuur HP, Irish JC, O’Sullivan B, Goh C, Gullane PJ, Pintilie M. A matched control study of treatment outcome in young patients with squamous cell carcinoma of the head and neck. Laryngoscope. 1999;109(2):249–58. doi: 10.1097/00005537-199902000-00015. [DOI] [PubMed] [Google Scholar]
- 12.Toporcov TN, Znaor A, Zhang ZF, et al. Risk factors for head and neck cancer in young adults: a pooled analysis in the INHANCE consortium. Int J Epidemiol. 2015;44(1):169–85. doi: 10.1093/ije/dyu255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 14.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 15.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 16.Flores-Obando RE, Gollin SM, Ragin CC. Polymorphisms in DNA damage response genes and head and neck cancer risk. Biomarkers. 2010;15(5):379–99. doi: 10.3109/13547501003797664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lou Y, Peng W, Cao DS, Xie J, Li HH, Jiang ZX. DNA repair gene XRCC1 polymorphisms and head and neck cancer risk: An updated meta-analysis including 16344 subjects. PLoS One. 2013;8(9):e74059. doi: 10.1371/journal.pone.0074059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu YY, Yuan H, Jiang GB, et al. Associations between XPD Asp312Asn polymorphism and risk of head and neck cancer: A meta-analysis based on 7,122 subjects. PLoS One. 2012;7(4):e35220. doi: 10.1371/journal.pone.0035220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKay JD, Truong T, Gaborieau V, et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011;7(3):e.1001333. doi: 10.1371/journal.pgen.1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tafel AA, Wu L, McHugh PJ. Human HEL308 localizes to damaged replication forks and unwinds lagging strand structures. J Biol Chem. 2011;286(18):15832–40. doi: 10.1074/jbc.M111.228189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang C, Marsit CJ, Houseman EA, et al. Gene-environment interactions of novel variants associated with head and neck cancer. Head Neck. 2012;34(8):1111–118. doi: 10.1002/hed.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan H, Ma H, Lu F, et al. Genetic variants at 4q23 and 12q24 are associated with head and neck cancer risk in China. Mol Carcinog. 2013;52:E2–9. doi: 10.1002/mc.21929. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat Rev Cancer. 2009;9(7):501–7. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 25.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26(9):1324–37. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Simpson ER, Brown KA. p53: Protection against tumor growth beyond effects on cell cycle and apoptosis. Cancer Res. 2015;75(23):5001–7. doi: 10.1158/0008-5472.CAN-15-0563. [DOI] [PubMed] [Google Scholar]
- 27.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marin MC, Jost CA, Brooks LA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25(1):47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan A, Syed N, Gasco M, et al. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004;23:3328–337. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- 30.Perrone F, Mariani L, Pastore E, et al. p53 codon 73 polymorphisms in human papillomavirus-negative and human papillomavirus-positive squamous cell carcinomas of the oropharynx. Cancer. 2007;109(12):2461–5. doi: 10.1002/cncr.22702. [DOI] [PubMed] [Google Scholar]
- 31.Bergamaschi D, Gasco M, Hiller L, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 32.Hoque MO, Begum S, Sommer M, et al. PUMA in head and neck cancer. Cancer Lett. 2003;199(1):75–81. doi: 10.1016/s0304-3835(03)00344-6. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Z, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Genetic variants of a BH3-only pro-apoptotic gene, PUMA, and risk of HPV16-associated squamous cell carcinoma of the head and neck. Mol Carcinog. 2012;51:E54–64. doi: 10.1002/mc.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt M, Butz K, Dymalla S, Semzow J, Hoppe-Seyler F. Inhibition of Bax activity is crucial for the antiapoptotic function of the human papillomavirus E6 oncoprotein. Oncogene. 2006;25(29):4009–15. doi: 10.1038/sj.onc.1209429. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Huang L, Xu Y, et al. Association between surviving-31G>C promotor polymorphism and cancer risk: a meta-analysis. Eur J Hum Genet. 2012;20(7):790–95. doi: 10.1038/ejhg.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knauer SK, Unruhe B, Karczewski S, et al. Functional characterization of novel mutations affecting surviving (BIRC5)-mediated therapy resistance in head and neck cancer patients. Hum Mutat. 2013;34(2):395–404. doi: 10.1002/humu.22249. [DOI] [PubMed] [Google Scholar]
- 37.Konopka K, Spain C, Yen A, Overlid N, Gebremedhin S, Duzgunes N. Correlation between the levels of survivin and surviving promotor-driven gene expression in cancer and non-cancer cells. Cell Mol Biol Lett. 2009;14(1):70–89. doi: 10.2478/s11658-008-0034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng CJ, Hsieh YH, Chen MK, Tsai CM, Lin CW, Yang SF. Survivin SNP-carcinogen interactions in oral cancer. J Dent Res. 2012;91(4):358–63. doi: 10.1177/0022034512438402. [DOI] [PubMed] [Google Scholar]
- 39.Ma F, Zhang H, Zhai Y, et al. Functional polymorphism -31C/G in the promoter of BIRC5 gene and risk of nasopharyngeal carcinoma among chinese. PloS One. 2011;6(2):e16748. doi: 10.1371/journal.pone.0016748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Negri E, Boffetta P, Berthiller J, et al. Family history of cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology consortium. Int J Cancer. 2009;124:394–401. doi: 10.1002/ijc.23848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potrony M, Puig-Butille JA, Aguilera P, et al. Increased prevalence of lung, breast and pancreatic cancers in addition to melanoma risk in families bearing the CDKN2A mutation: Implications for genetic counseling. J Am Acad Dermatol. 2014;71(5):888–95. doi: 10.1016/j.jaad.2014.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soura E, Eliades PJ, Shannon K, Stratigos AJ, Tsao H. Hereditary melanoma: Update on syndromes and management: Emerging melanoma cancer complexes and genetic counseling. J Am Acad Dermatol. 2016;74(3):411–20. doi: 10.1016/j.jaad.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabanillas R, Astudillo A, Valle M, et al. Novel germline CDKN2A mutation associated with head and neck squamous cell carcinomas and melanomas. Head Neck. 2013;35(3):80–4. doi: 10.1002/hed.21911. [DOI] [PubMed] [Google Scholar]
- 44.Vinarsky V, Fine RL, Assaad A, et al. Head and neck squamous cell carcinoma in FAMMM Syndrome. Head Neck. 2009;31:1524–7. doi: 10.1002/hed.21050. [DOI] [PubMed] [Google Scholar]
- 45.Potjer TP, Kranenburg HE, Bergman W, et al. Prospective risk of cancer and the influence of tobacco use in carriers of the p16-Leiden germline variant. Eur J Hum Genet. 2015;23(5):711–4. doi: 10.1038/ejhg.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helgadottir H, Hoiom V, Jonsson G, et al. High risk of tobacco-related cancers in CDKN2A mutation-positive melanoma families. J Med Genet. 2014;51(8):545–52. doi: 10.1136/jmedgenet-2014-102320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gajwani BW, Devereaux JM, Beg JA. Familial clustering of nasopharyngeal carcinoma. Cancer. 1980;46(10):2325–7. doi: 10.1002/1097-0142(19801115)46:10<2325::aid-cncr2820461035>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 48.Bei J, Jia W, Zeng Y. Familial and large-scale case-control studies identify genes associated with nasopharyngeal carcinoma. Semin Cancer Biol. 2012;22(2):96–106. doi: 10.1016/j.semcancer.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 49.Xiong W, Zeng ZY, Xia JH, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64(6):1972–4. doi: 10.1158/0008-5472.can-03-3253. [DOI] [PubMed] [Google Scholar]
- 50.Wang AT, Smogorzewska A. Snapshot: Fanconi anemia and associated proteins. Cell. 2015;160:354. doi: 10.1016/j.cell.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 51.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17(6):337–49. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 52.Garner E, Smogorzewska A. Ubiquitylation and the Fanconi anemia pathway. FEBS Lett. 2011;585(18):2853–60. doi: 10.1016/j.febslet.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mamrak NE, Shimamura A, Howell NG. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 2017;31:93–99. doi: 10.1016/j.blre.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kutler DI, Singh B, Satagopan J, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101(4):1249–56. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 55.Velleuer E, Dietrich R. Fanconi anemia: young patients at high risk for squamous cell carcinoma. Mol Cell Pediatr. 2014;1:9. doi: 10.1186/s40348-014-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kutler DI, Auerbach AD, Satagopan J, et al. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg. 2003;129(1):106–12. doi: 10.1001/archotol.129.1.106. [DOI] [PubMed] [Google Scholar]
- 57.Romick-Rosendale LE, Lui VW, Grandis JR, Wells SI. The Fanconi anemia pathway: repairing the link between DNA damage and squamous cell carcinoma. Mutat Res. 2013;743:78–88. doi: 10.1016/j.mrfmmm.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tremblay S, Pintor Dos Reis P, Bradley G, et al. Young patients with oral squamous cell carcinoma: study of the involvement of GSTP1 and deregulation of the Fanconi anemia genes. Arch Otolaryngol Head Neck Surg. 2006;132(9):958–66. doi: 10.1001/archotol.132.9.958. [DOI] [PubMed] [Google Scholar]
- 59.Stoepker C, Ameziane N, van der Lelij, et al. Defects in the Fanconi Anemia pathway and chromatid cohesion in head and neck cancer. Cancer Res. 2015;75(17):3543–53. doi: 10.1158/0008-5472.CAN-15-0528. [DOI] [PubMed] [Google Scholar]
- 60.Smith IM, Mithani SK, Mydlarz WK, Chang SS, Califano JA. Inactivation of the tumor suppressor genes causing the hereditary syndromes predisposing to head and neck cancer via promotor hypermethylation in sporadic head and neck cancers. ORL J Otorhinolaryngol Relat Spec. 2010;72(1):44–50. doi: 10.1159/000292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23(4):1000–4. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- 62.Hoskins EE, Gunawardena RW, Habash KB, et al. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene. 2008;27(35):4798–808. doi: 10.1038/onc.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cel. 2001;7(2):249–62. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 64.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 65.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polager S, Ginsberg D. E2F – at the crossroads of life and death. Trends Cell Biol. 2008;18(11):528–35. doi: 10.1016/j.tcb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Polager S, Ginsberg D. p53 and E2F: partners in life and death. Nat Rev Cancer. 2009;9:738–48. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- 69.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9(2):95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 70.Meixia L, Zhensheng L, Hongping Y, et al. Combined effects of E2F1 and E2F2 polymorphisms on risk and early onset of squamous cell carcinoma of the head and neck. Mol Carcinog. 2012;51:E132–41. doi: 10.1002/mc.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan Y, Sturgis EM, Zhu L, et al. A functional variant at the miRNA binding site in E2F1 gene is associated with risk and tumor HPV16 status of oropharynx squamous cell carcinoma. Mol Carcinog. 2017;56:1100–106. doi: 10.1002/mc.22576. [DOI] [PubMed] [Google Scholar]
- 72.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Souza G, Kreimer AR, Viscidi R, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356(19):1944–956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 74.Kreimer AR, Johansson M, Waterboer T, et al. Evaluation of human papillomavirus antibodies and risk of subsequent head and neck cancer. J Clin Oncol. 2013;31(21):2708–715. doi: 10.1200/JCO.2012.47.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson KS, Dahlstrom KR, Cheng JN, et al. HPV16 antibodies as risk factors for oropharyngeal cancer and their association with tumor HPV and smoking status. Oral Oncol. 2015;51(7):662–7. doi: 10.1016/j.oraloncology.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.White RA, Malkoski SP, Wang XJ. TGFβ signaling in head and neck squamous cell carcinoma. Oncogene. 2010;29(40):5437–446. doi: 10.1038/onc.2010.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaklamani VG, Baddi L, Liu J, et al. Combined genetic assessment of transforming growth factor-β signaling pathway variants may predict breast cancer risk. Cancer Res. 2005;65:3454–61. doi: 10.1158/0008-5472.CAN-04-2961. [DOI] [PubMed] [Google Scholar]
- 78.Mills KH, McGuirk P. Antigen-specific regulatory T cells – their induction and role in infection. Semin Immunol. 2004;16:107–17. doi: 10.1016/j.smim.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 79.Alcocer-Gonzalez JM, Berumen J, Tamez-Guerra R, et al. In vivo expression of immunosuppressive cytokines in human papillomavirus-transformed cervical cancer cells. Viral Immunol. 2006;19:481–91. doi: 10.1089/vim.2006.19.481. [DOI] [PubMed] [Google Scholar]
- 80.Guan X, Sturgis EM, Lei D, et al. Association of TGF-β1 genetic variants with HPV16-positive oropharyngeal cancer. Clin Cancer Res. 2010;16(5):1416–422. doi: 10.1158/1078-0432.CCR-09-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jin L, Sturgis EM, Zhang Y, et al. Association of tumor necrosis factor-alpha promotor variants with risk of HPV-associated oral squamous cell carcinoma. Mol Cancer. 2013;12:80. doi: 10.1186/1476-4598-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song X, Sturgis EM, Liu J, et al. MicroRNA variants increase the risk of HPV-associated squamous cell carcinomas of the oropharynx in never smokers. PLoS One. 2013;8(2):356622. doi: 10.1371/journal.pone.0056622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Sturgis EM, Sun Y, et al. A functional variant at miRNA-122 binding site in IL-1α 3′ UTR predicts risk and HPV-positive tumours of oropharyngeal cancer. Eur J Cancer. 2015;51(11):1415–23. doi: 10.1016/j.ejca.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 85.Ji X, Neumann AS, Sturgis EM, et al. p53 codon 72 polymorphism associated with risk of human papillomavirus-associated squamous cell carcinoma of the oropharynx in never-smokers. Carcinogenesis. 2008;29(4):875–9. doi: 10.1093/carcin/bgn039. [DOI] [PubMed] [Google Scholar]
- 86.Chen X, Sturgis EM, Etzel CJ, Wei Q, Li G. p73 G4C14-to-A4T14 polymorphism and risk of human papillomavirus associated squamous cell carcinoma of the oropharynx in never smokers and never drinkers. Cancer. 2008;113(12):3307–314. doi: 10.1002/cncr.23976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen X, Sturgis EM, El-Naggar AK, Wei Q, Li G. Combined effects of the p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms on the risk of HPV16-associated oral cancer in never-smokers. Carcinogenesis. 2008;29(11):2120–5. doi: 10.1093/carcin/bgn191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z, Sturgis EM, Zhang Y, et al. Combined p53-related genetic variants together with HPV infection increases oral cancer risk. Int J Cancer. 2012;131(3):E251–8. doi: 10.1002/ijc.27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Z, Sturgis EM, Guo W, et al. Association of combined p73 and p53 genetic variants with tumor HPV16-positive oropharyngeal cancer. PLoS One. 2012;7(4):e35522. doi: 10.1371/journal.pone.0035522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu H, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Modifying effect of MDM4 variants on risk of HPV16-associated squamous cell carcinoma of the oropharynx. Cancer. 2012;118(6):1684–692. doi: 10.1002/cncr.26423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen X, Sturgis EM, Lei D, Dahlstrom K, Wei Q, Li G. HPV seropositivity synergizes with MDM2 variants to increase the risk of oral squamous cell carcinoma. Cancer Res. 2010;70(18):7199–208. doi: 10.1158/0008-5472.CAN-09-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gomez-Bougie P, Wuilleme-Toumi S, Menoret E, et al. Noxa Up-regulation and Mcl-1 cleavage are associated to apoptosis induction by bortezomib in multiple myeloma. Cancer Res. 2007;67(11):5418–424. doi: 10.1158/0008-5472.CAN-06-4322. [DOI] [PubMed] [Google Scholar]
- 93.Skoda C, Erovic BM, Wachek V, et al. Down-regulation of Mcl-1 with antisense technology alters the effect of various cytotoxic agents used in treatment of squamous cell carcinoma of the head and neck. Oncol Rep. 2008;19(6):1499–503. [PubMed] [Google Scholar]
- 94.Zhou Z, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Genetic variants of NOXA and MCL1 modify the risk of HPV16-associated squamous cell carcinoma of the head and neck. BMC Cancer. 2012;12:159. doi: 10.1186/1471-2407-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morris L, Chandramohan R, West L, et al. The molecular landscape of recurrent and metastatic head and neck cancers: Insights from a precision oncology sequencing platform. JAMA Oncol. 2017;3(2):244–55. doi: 10.1001/jamaoncol.2016.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]