Abstract
The histone-like nucleoid structuring (H-NS) protein and its analogues bind large stretches of horizontally acquired AT-rich DNA in a broad range of bacterial species. Binding by H-NS silences the promoters within such DNA that would otherwise deplete the cellular pool of RNA polymerase. Selective de-repression can occur when sequence-specific DNA-binding proteins locally disrupt H-NS function; this mechanism is important for the regulation of many virulence genes. In this issue of Molecular Microbiology, Rangarajan and Schnetz show that when transcription from a neighbouring region invades an H-NS-bound locus, it can disrupt local H-NS repression. Moreover, they show that de-repression occurs in a dose-dependent manner, and they demonstrate a natural example of this in Escherichia coli. This finding has important implications for H-NS function and its impact on genome evolution.
ABBREVIATED SUMMARY
The histone-like nucleoid structuring (H-NS) protein and its analogues silence transcription across large stretches of horizontally acquired AT-rich DNA in a broad range of bacterial species. In this issue of Molecular Microbiology, Rangarajan and Schnetz show that when transcription from a neighbouring region invades an H-NS-bound locus, it can disrupt local H-NS repression. This finding has important implications for H-NS function and its impact on genome evolution.
H-NS and transcription: a complex relationship
The Histone-like Nucleoid Structuring (H-NS) protein is an abundant DNA-binding protein found in Escherichia coli and closely related species. H-NS preferentially coats large AT-rich regionsof the genome (Grainger et al., 2006; Lucchini et al., 2006; Navarre et al., 2006; Oshima et al., 2006; Kahramanoglou et al., 2011). Although H-NS is not widely distributed among prokaryotes, functionally analogous proteins have been identified in a broad range of bacteria (Singh et al., 2016); analogues include MvaT/U in pseudomonads (Castang et al., 2008), Lsr2 in mycobacteria (Gordon et al., 2010), and Rok in bacilli (Smits and Grossman, 2010). The primary function of H-NS and its analogues is believed to be transcriptional repression (Dorman, 2007; Landick et al., 2015). Since H-NS binds to AT-rich DNA, this repression is associated with horizontally acquired genes, which often have higher AT-content than the genome as a whole (Lucchini et al., 2006; Navarre et al., 2006; Oshima et al., 2006). Failure to repress horizontally acquired genes can substantially reduce fitness (Ali et al., 2014). In addition to silencing transcription of the genes themselves, H-NS silences transcription of numerous promoters within these genes. Indeed, the majority of H-NS-repressed promoters are inside genes and/or far from gene starts (Singh et al., 2014). Consequently, loss of H-NS is highly toxic due to redistribution of active RNA polymerases (RNAPs) from housekeeping genes to intragenic promoters that would ordinarily be silenced (Lamberte et al., 2017).
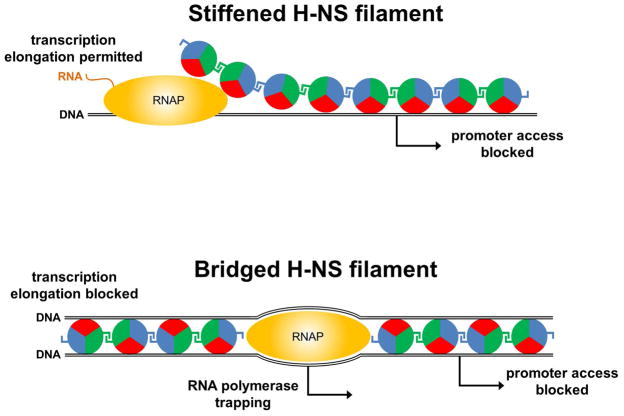
H-NS binds the DNA minor groove using an arginine side chain that resides within a hook-like motif (Gordon et al., 2011) (red sectors in Figure 1). Furthermore, H-NS can interact with itself via two distinct protein surfaces, allowing “daisy chaining” of individual protomers (blue and green sectors in Figure 1) (Arold et al., 2010). These properties of H-NS account for its ability to coat large stretches of DNA (Figure 1). Two distinct modes of DNA binding have been described. First, H-NS binding can unfurl the DNA into extended linear filaments (Figure 1, top). This is commonly referred to as the “stiffening” mode of binding (Amit et al., 2003; Liu et al., 2010). Second, H-NS can simultaneously bind two distant sections of DNA by creating nucleoprotein “bridges” (Figure 1, bottom) (Dame et al., 2006). In vitro, H-NS can switch between the two modes of binding based on the concentration of divalent cations (Liu et al., 2010; van der Valk et al., 2017). Osmolarity and H-NS-interacting proteins have also been proposed to modulate the mode of H-NS binding (van der Valk et al., 2017). Importantly, it is unknown which of the two confirmations is adopted at different loci in vivo.
Figure 1. Models for repression of transcription initiation and elongation by H-NS.
The top panel shows stiffened, linear H-NS filaments that are unable to block transcription elongation but can block access of initiating RNAP to promoters. It is unclear is the passage of RNAP (travelling from left to right) through such filaments transiently remodels the H-NS-DNA complex or if H-NS is completely displaced. The bottom panel shows H-NS complexed with DNA in a bridged conformation. Such nucleoprotein does not allow passage of transcribing RNA polymerase, and may trap RNA polymerase at promoters, and/or occlude access. The DNA is shown as a black line, and promoters as arrows. H-NS is shown by circles coloured red, green and blue. The red sector of the circle indicates the H-NS DNA-binding determinant, whilst the green and blue sectors represent the two distinct regions that H-NS uses for self-association.
H-NS represses transcription in several ways, and these appear to be linked to the stiffening and bridging modes of DNA binding. The primary mode of H-NS repression is at the level of transcription initiation; promoters that are bound by H-NS are often transcriptionally silenced. This can be due either to occlusion of RNAP (potentially associated with bridging or stiffening), or RNAP trapping (associated with bridging) (Figure 1). H-NS has also been shown to prevent transcription elongation in vitro by strongly increasing pausing of RNAP (Kotlajich et al., 2015). Importantly, this function of H-NS requires that it bind in the bridging mode (Figure 1, bottom). Although H-NS inhibition of transcription elongation has not been directly observed in vivo, H-NS binding is associated with sites of Rho-dependent transcription termination in vivo (Peters et al., 2012), suggesting that H-NS effects on transcription elongation occur in cells.
Anti-silencing and potential roles of overlapping transcription
There are numerous described examples of DNA-binding proteins that selectively de-repress transcription of H-NS-silenced genes by binding to promoter regions (Stoebel et al., 2008). These examples are often associated with the expression of virulence factors encoded by AT-rich DNA. In some instances, de-repression has been proposed to occur by large-scale displacement of H-NS (Turner and Dorman, 2007), whereas in other examples, de-repression is believed to require only local remodeling of H-NS, with the extended H-NS oligomer remaining bound to DNA (Will et al., 2014; Newman et al., 2018). These latter studies are most consistent with H-NS binding in the stiffening mode, and it has been proposed that H-NS-DNA complexes are more resistant to de-repression in the bridged form than the stiffened form (Walthers et al., 2011). This hypothesis is consistent with the observed differences between the effect of bridged and stiffened H-NS-DNA complexes on transcription elongation in vitro (Kotlajich et al., 2015). Nonetheless, since no studies have addressed the mode of H-NS binding in vivo, mechanisms of de-repression are incompletely described.
In this issue of Molecular Microbiology, Rangarajan and Schnetz describe a novel mechanism of H-NS de-repression. They demonstrate that transcription elongation that extends across H-NS-silenced promoters can lead to de-repression in vivo in E. coli (Figure 2). De-repression by such invading transcription occurs in a dose-dependent manner, such that increased levels of invading transcription lead to greater de-repression. Rangarajan and Schnetz show that this form of de-repression occurs not only in plasmid constructs, but also at the native bgl promoter that is de-repressed by transcription invading from the upstream pst-phoU transcript. These observations fundamentally change our view of how H-NS-silenced promoters are regulated, and raise many important questions about the mechanism of H-NS repression/de-repression, and the impact of H-NS on genome evolution.
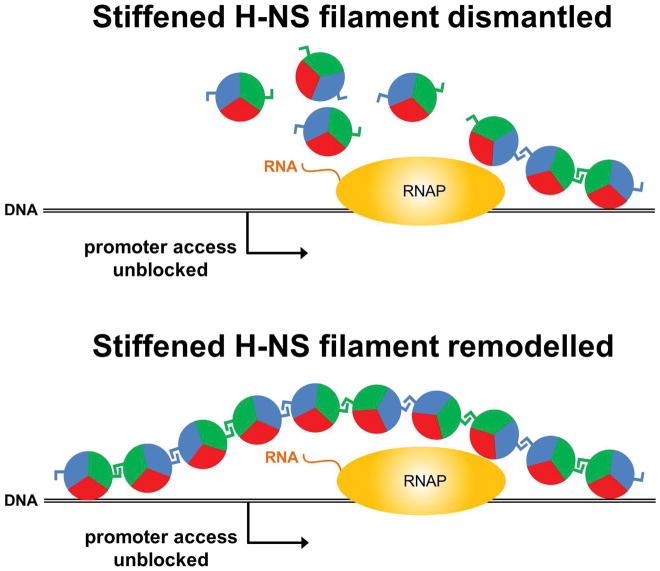
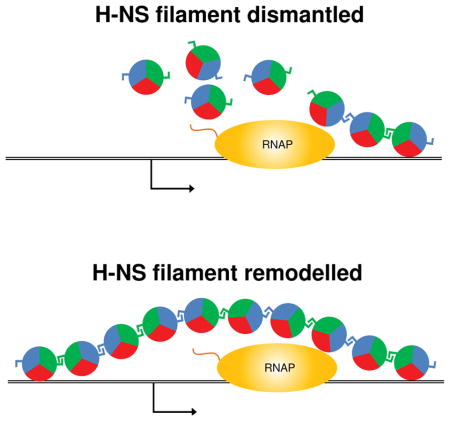
Figure 2. Models for de-repression of H-NS bound promoters by invading transcription.
An elongating RNAP (travelling from left to right) de-represses promoters (arrows) within DNA loci (black lines) bound by H-NS (circles). It is not clear if de-repression involves complete dismantlement of H-NS filaments (top) or if H-NS filaments are instead transiently remodeled (bottom). In the latter scenario, interactions between H-NS molecules would be maintained (blue and green sectors) whilst the interaction between H-NS and the DNA is transiently broken (red sectors).
Although it is unknown whether H-NS binds DNA in vivo in the bridged or stiffened mode, the observation of overlapping transcription disrupting H-NS-mediated repression strongly suggests that these promoters are bound by stiffened filaments, because bridged complexes inhibit transcription elongation in vitro (Kotlajich et al., 2015). Assuming H-NS is bound at these loci in stiffened filaments, it will be interesting to determine whether elongating RNAPs completely displace H-NS from the DNA as they transcribe this region (Figure 2, top) or whether H-NS is locally remodeled (Figure 2, bottom), as has been suggested for some cases where DNA-binding proteins that counteract H-NS-mediated silencing (Will et al., 2014; Newman et al., 2018). If the examined promoters are indeed bound by stiffened H-NS filaments, there may be other promoters that are bound by bridged complexes, and hence would likely be resistant to de-repression by overlapping transcription. Moreover, since H-NS can switch between binding modes depending upon the conditions (Liu et al., 2010; van der Valk et al., 2017), H-NS-silenced promoters may be differentially susceptible to de-repression by invading transcription depending on the growth conditions of the cells. If growth-dependent differences in de-repression are observed, this may be a useful way to infer the mode of H-NS binding at specific loci in vivo.
While the mode of H-NS binding to silenced promoters may impact their susceptibility to overlapping transcription, the mode of overlapping transcription may also be important. For example, the dose-dependence of overlapping transcription on de-repression may simply reflect repeated displacement of H-NS from the DNA. Indeed, high levels of transcription may prevent reassociation of H-NS with the DNA. An alternative explanation is that dose-dependence results from the increased processivity of RNAP in highly transcribed regions (Epshtein and Nudler, 2003). This increased processivity may facilitate elongation of RNAP through H-NS-bound regions, and the accompanying remodeling or displacement of H-NS. Coupling of translation and transcription is also known to increase RNAP processivity (Proshkin et al., 2010). Therefore, whether or not the overlapping transcript is translated may impact the level of de-repression. It is noteworthy that not all H-NS-bound promoters are de-repressed by overlapping transcription. For instance, upstream of the ehxCABD operon in E. coli O157, there are multiple promoters that are all silenced by H-NS. However, transcription from one promoter does not de-repress those adjacent; their proximity sterically hinders the binding of RNAP. In this scenario, H-NS can enhance transcription by optimally positioning RNAP at the desirable promoter (Singh and Grainger, 2013).
Implications for genome evolution
The impact of overlapping transcription on H-NS-mediated silencing is likely to affect the genomic locations at which horizontally acquired genes can stably integrate. For example, if a horizontally acquired gene was subject to invading transcription from an adjacent gene/operon, H-NS-mediated repression could be relieved at a cost to cell fitness. Alternatively, the impact of overlapping transcription on H-NS-mediated silencing may promote genome evolution by facilitating regulatory coupling between adjacent genes/operons. For example, a horizontally acquired gene may be selectively de-repressed when the upstream gene/operon is transcribed. This would allow for immediate integration of horizontally acquired genes into regulatory networks, without the need to acquire binding sites for transcription factors in the promoter region.
H-NS plays an important role in genome evolution by silencing the transcription of horizontally acquired genes and the promoters that are often found within them (Lamberte et al. 2017). If a horizontally acquired gene is subject to de-repression as a result of overlapping transcription, this also has implications for transcription from intragenic promoters. AT-rich genes typically contain many intragenic promoters that are ordinarily silenced by H-NS (Singh et al., 2014; Lamberte et al., 2017). If a horizontally acquired gene is de-repressed, it is likely that all the internal promoters will also be de-repressed due to the overlapping transcription of the mRNA. This would likely impose a fitness cost, since de-repression of H-NS-silenced intragenic promoters diverts RNAP away from housekeeping genes (Lamberte et al., 2017). It may also impact the expression of neighbouring genes, since H-NS-silenced promoters within genes can potentially transcribe mRNAs for downstream genes (Chintakayala et al., 2013; Lamberte et al., 2017).
Concluding remarks
In conclusion, the work of Rangarajan and Schnetz reveals a novel mechanism by which H-NS-mediated silencing can be reversed. Many details of this process are left to be discovered, and future studies will likely shed light on the mechanisms of DNA binding and repression by H-NS. Lastly, the observation that overlapping transcription can counteract H-NS-mediated repression suggests that other complexes that translocate along DNA, such as the DNA replication machinery, may have similar effects.
Acknowledgments
JTW was supported by National Institutes of Health grant 5R01GM114812. DCG was supported by BBSRC grants BB/N014200/1 and BB/N005961/1.
References
- Ali SS, Soo J, Rao C, Leung AS, Ngai DHM, Ensminger AW, Navarre WW. Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog. 2014;10:e1004500. doi: 10.1371/journal.ppat.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci U S A. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc Nat Acad Sci USA. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintakayala K, Singh SS, Rossiter AE, Shahapure R, Dame RT, Grainger DC. E. coli Fis protein insulates the cbpA gene from uncontrolled transcription. PLoS Genet. 2013;9:e1003152. doi: 10.1371/journal.pgen.1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Noom MC, Wuite GJL. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- Gordon BRG, Li Y, Cote A, Weirauch MT, Ding P, Hughes TR, et al. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc Natl Acad Sci U S A. 2011;108:10690–10695. doi: 10.1073/pnas.1102544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BRG, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, et al. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Goldberg MD, Busby SJW. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, et al. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlajich MV, Hron DR, Boudreau BA, Sun Z, Lyubchenko YL, Landick R. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. eLife. 2015:4. doi: 10.7554/eLife.04970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberte LE, Baniulyte G, Singh SS, Stringer AM, Bonocora RP, Stracy M, et al. Horizontally acquired AT-rich genes in Escherichia coli cause toxicity by sequestering RNA polymerase. Nat Microbiol. 2017;2:16249. doi: 10.1038/nmicrobiol.2016.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R, Wade JT, Grainger DC. H-NS and RNA polymerase: a love-hate relationship? Curr Opin Microbiol. 2015;24:53–59. doi: 10.1016/j.mib.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS Mediates the Silencing of Laterally Acquired Genes in Bacteria. PLoS Pathog. 2006;18:2. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Newman SL, Will WR, Libby SJ, Fang FC. The curli regulator CsgD mediates stationary phase counter-silencing of csgBA in Salmonella Typhimurium. Mol Microbiol. 2018 doi: 10.1111/mmi.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Milstein JN, Navarre WW. Xenogeneic Silencing and Its Impact on Bacterial Genomes. Annu Rev Microbiol. 2016;70:199–213. doi: 10.1146/annurev-micro-102215-095301. [DOI] [PubMed] [Google Scholar]
- Singh SS, Grainger DC. H-NS can facilitate specific DNA-binding by RNA polymerase in AT-rich gene regulatory regions. PLoS Genet. 2013;9:e1003589. doi: 10.1371/journal.pgen.1003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014 doi: 10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Grossman AD. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 2010;6:e1001207. doi: 10.1371/journal.pgen.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- Turner EC, Dorman CJ. H-NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. J Bacteriol. 2007;189:3403–3413. doi: 10.1128/JB.01813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk RA, van der Vreede J, Qin L, Moolenaar GF, Hofmann A, Goosen N, Dame RT. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. eLife. 2017:6. doi: 10.7554/eLife.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walthers D, Li Y, Liu Y, Anand G, Yan J, Kenney LJ. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J Biol Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will WR, Bale DH, Reid PJ, Libby SJ, Fang FC. Evolutionary expansion of a regulatory network by counter-silencing. Nat Commun. 2014;5:5270. doi: 10.1038/ncomms6270. [DOI] [PMC free article] [PubMed] [Google Scholar]